Abstract
The past decade has seen tremendous advances in both our understanding of cancer immunosuppressive microenvironments and colonic bacteria facilitated by immune checkpoint inhibitor antibodies and next generation sequencing, respectively. Because an important role of the host immune system is to communicate with and regulate the gut microbial community, it should not come as a surprise that the behavior of one is coupled to the other. In this review, we will attempt to dissect some of the studies demonstrating cancer immunotherapy modulation by specific gut microbes and discuss possible molecular mechanisms for this effect.
Keywords: gut microbiota, immune checkpoint inhibitor therapy, cancer, dendritic cells, cytotoxic T-lymphocytes
The past decade has seen tremendous advances in both our understanding of cancer immunosuppressive microenvironments and the intestinal bacteria that promote host immune responses. There is mounting evidence that the gut microbial community and the host immune system continually interact, resulting in a mutualistic shaping of both host immune responses and gut microbial taxonomic composition. We will review how patients’ gut microbes modulate the benefit of cancer immunotherapy. These exciting findings have been confirmed in animal models, and the mechanisms are being actively explored. The results may point to a noninvasive method of increasing the therapeutic index of immunotherapy with diet or probiotics.
Cancer Immune Checkpoint Inhibitor Therapy (ICT)
Until recently, cancer therapy has focused on cyto-ablative approaches, including surgery, ionizing radiation, and cytotoxic chemotherapies. The assumption was that each modality reduced host tumor burden by geometric amounts. Theoretically, if patient tumor burden can be reduced to a certain undefined level, the patient can maintain a durable remission. The hypothesis appeared to be confirmed by results with leukemia and lymphoma in mice and humans.1 However, this approach failed to explain spontaneous remissions in some cancer patients.2 Furthermore, the hypothesis fails to explain the aggressive natural history of cancer in immunosuppressed individuals.3 Further evidence for the role of the host immune system is the difference in efficacy of chemotherapy for immunocompetent versus immunocompromised rodent models.4 Finally, the difference in clinical durable remissions of allogeneic versus autologous stem cell transplants in leukemia patients points to a need for allogeneic T-lymphocytes for cure.5 The alternative theory is that host immune cells are critical in controlling cancers. For spontaneous malignancies, tumor cells evolve and alter their surface and release cytokines to create a microenvironment armed to withstand immune surveillance. Even melanoma and renal cell carcinoma (RCC) immunotherapy with interleukin (IL)-2 produces rare durable remissions.6 In most situations, therapeutic advances to reverse the immune barrier were unsuccessful.
Two groundbreaking advances have dramatically changed our detailed understanding of tumor immune resistance and led to the development of effective treatment options. Cytotoxic T-lymphocyte associated protein 4 (CTLA4) is a protein receptor that acts as an immune checkpoint and is overexpressed on regulatory T-cells (Tregs).7 Antibodies to CTLA4 partially reverse tumor immunosuppression and yield durable remissions in tumor-bearing mice and melanoma patients.8,9 Programmed death receptor-1 (PD1) is expressed on cytotoxic T-lymphocytes (CTLs) and binds PD1 ligand (PD-L1) expressed by stromal and tumor cells; this protein pair also functions as an immune checkpoint.10 Antibodies to PD1 and PD-L1 block T-lymphocyte senescence and trigger CTL activity. Using these antibodies individually or in combination, ICT produces remissions in a variety of metastatic human neoplasms (Table 1). Unfortunately, the majority of cancer patients fail to respond to ICT, and improving the response rate and response durability is the focus of our lab and others.
Table 1.
Diseases and ICT.
| Cancer Type | Immunotherapy Agents | Response Rate (%) |
|---|---|---|
| Hodgkin’s lymphoma | Nivolumab | 65 |
| Merkel cell carcinoma | Avelumab | 62 |
| Melanoma | Nivolumab + Ipilimumab | 58 |
| MSI-H/MMR Def CRC | Nivolumab + Ipilimumab | 55 |
| SC skin carcinoma | Cemiplimab | 47 |
| MSI-H/MMR Def non-CRC | Pembrolizumab | 46 |
| NSCLC High TMB or PDL1+ >50% de novo or all de novo or relapsed | Nivolumab + Iplimumab or pembrolizumab or pembrolizumab + pemetrexed/carboplatin or nivolumab | 43 or 45 or 55 or 25 |
| RCC | Nivolumab + Ipilimumab | 40 |
| HCC | Nivolumab | 20 |
| Urothelial carcinoma | Nivolumab | 20 |
| Head and neck SC carcinoma | Pembrolizumab | 16 |
| Gastric carcinoma | Pembrolizumab | 13 |
| SCLC | Nivolumab | 12 |
Abbreviations: ICT, immune checkpoint inhibitor therapy; MSI-H/MMR Def, microsatellite instability-high and mismatch repair deficient; CRC, colorectal carcinoma; SC, squamous cell; NSCLC, non–small-cell lung carcinoma; TMB, tumor mutation burden; PDL1, programmed death receptor-1 ligand; RCC, renal cell carcinoma, HCC, hepatocellular carcinoma; SCLC, small-cell lung carcinoma.
The steps mediating ICT have been partially elucidated and are associated with predictive markers. The preliminary step is acquisition of an inflammatory phenotype. Danger associated molecular patterns react with pattern recognition receptors (PRRs) on or in dendritic cells (DCs) and macrophages.11 They notify the body of the presence of either a pathogen or damage. The innate immune cells then release type I interferon (IFN) and chemokines, leading to further inflammation and infiltration of CTLs. Absence of DCs or tumor overexpression of molecules blocking DC recruitment (eg, β-catenin) is associated with ICT failure and a paucity of tumor CTLs.12 Both in vitro and in vivo studies show that the cytoplasmic DNA-cGAS-STING-IRF3-IFN pathway is critical for innate immune activity in tumors and ICT response.13 Recent efforts to combine cytolytic viro-therapy with ICT have produced encouraging improvements in response rate consistent with the important role of this step.14
A second step is presentation of tumor neo-antigens to CTLs by DCs. Patients with high tumor mutation burden and high tumor neo-antigen loads are more likely to respond to ICT.15 Interestingly, only a subset of tumor neo-antigens conveys ICT benefit, and many of these neo-antigens associated with clinical benefit match pathogen-associated peptide antigens.16 Furthermore, tumor evolution with heterogeneity of neo-antigen expression among metastases is associated with lack of ICT efficacy.17
The third step is overcoming immune checkpoints. Patients with low levels of tumor CTLA4 and high levels of tumor PD-L1 have higher response rates.18,19 Interestingly, recent studies show that anti-CTLA4 works primarily through elimination of Tregs,7 and anti-PD1 and anti-PD-L1 work via enabling CTLs to produce IFNγ, which in turn triggers DCs to release IL12, which further stimulates CTLs.10
The final effector step of immunotherapy is tumor cell execution by CTLs. Multiple mutations have been seen in tumors to escape execution, including loss of β2-microglobulin and IFN-JAK signaling via apelin receptor.20 Two recent predictive factors do not simply fit within the steps described above. High body mass index (BMI) patients have a higher ICT response rate than patients with normal or low BMI.21,22 Patients exposed to antibiotics before or during the first 60 days of ICT have much lower response rates.23-25 These unusual findings hinted at an earlier step in immunotherapy only recently discovered.
Human Intestinal Microbiota
The human colon is colonized at birth by Lactobacilli and Bifidobacteria from the mother’s vaginal wall, but within a few years acquires several hundred bacterial species dominated by members of the phyla Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, and Verrucomicrobia along with an Archaebacterium, Methanobrevibacter smithii. This microbial diversity is consistent with evolution by adaptive radiation. The commensals produce vitamin K and small carbohydrates from plant fiber polysaccharides and detoxify xenobiotics. Furthermore, they transmit vital signals to the nervous and immune systems. Each human has a slightly different population, totaling 100 trillion microbes, and a recent analysis showed a total human diversity of 4930 species.26 Most individuals show relative stability of their microbiome with transient effects of diet and longer duration effects of antibiotics.27-30
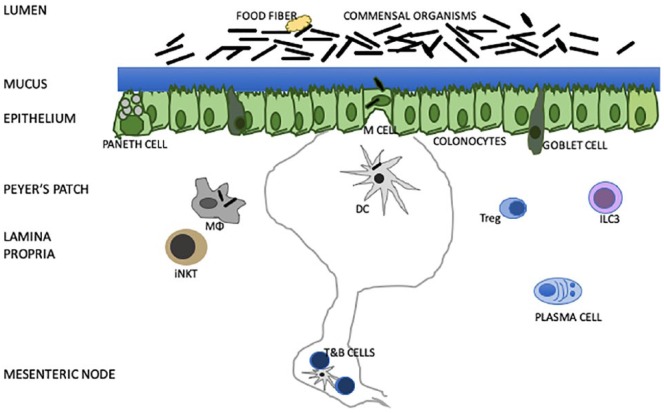
The host immune system interacts with gut bacteria at the intestinal epithelium (Figure 1). Colonocyte tight junctions and goblet cell mucin limit access of luminal organisms to the host. Paneth cell–derived defensins and RegIIIγ and submucosal plasma cell IgA destroy many invading bacteria. Special thin colonocyte M cells secrete CCL12 to attract DCs, and their smaller volume provides a window by which macrophages and DCs can sample luminal organisms and transport them to mesenteric lymph nodes.31 There, T-lymphocytes are educated via HLA-antigen-T-cell receptor signaling. The antigen-trained T-cells help B-cells return to the colon wall to yield IgA-producing plasma cells, provide appropriate Tregs, and participate in CTL control of pathogens. The genetic diversity of the multiple colonic commensals combined with the multilevel immune elements in the colon wall provide an excellent milieu for gut microbe–human immune interactions.
Figure 1.
Model of colon contents, epithelium, and submucosa with focus on immune interactions. ILC3 are group 3 innate immune cells that require RORγt, TOX, and NFIL3. Goblet cells produce a 150-µm mucin layer. Paneth cells make antimicrobial peptides: α-defensin, β-defensin, C-type lectin RegIIIα. Colonic dietary fibers contain indigestible polysaccharides. Microfold M cells are formed with RANKL and Spi-B and produce CCL20 to attract Peyers’ patch, transcytose antigen and permit egress microbes. Dendritic cells bind bacteria and bacterial antigens and transport to mesenteric lymph nodes where T- and B-cells are educated. Plasma cells in lamina propria produce IgA. Macrophages eat microbes. Tregs limit local cytotoxic T-cell responses. Not shown are colonic epithelium enteroendocrine cells. Paneth cells secrete anti-microbial peptides (AMPs) via calcium-activated potassium channel KCA3.1 or SK4 and secrete 3040 amino acid long defensins with 6 cysteine residues and 3 intramolecular disulfides. Defensins are chemokines for CCR6 positive dendritic cells and can neutralize bacterial exotoxins. Humans have 2 α-defensins, which are activated by trypsin. Lysozyme C is a glycosidase specific for peptidoglycan hydrolysis. Phospholipase A2 degrades bacterial membrane phosphatidylethanolamine and phosphatidylglycerol. Unlike defensins, RegIIIα is induced through TLR and MYD88. Bacteriocins are pore forming, induce membrane permeabilization, or degrade the peptidoglycan cell wall. Commensal bacteria produce different bacteriocins. Adapted from Belkaid and Harrison.31
Abbreviations: Tregs, regulatory T-cells;
Mouse Tumor ICT Models Showing Effects of Gut Microbiota
Several reports in the past 5 years have shown that alterations in the gut microbiota influence immunotherapy efficacy in mice (Table 2). MC38 colon tumor–bearing mice responded to anti-IL10 receptor plus CpG-oligonucleotide immunotherapy with stimulation of tumor-associated myeloid monocytes, macrophages, and DCs and release of tumor necrosis factor α (TNFα), IL1, IL12, and CXCL10. Loss of gut bacteria by using germ-free mice or treating specific pathogen–free mice with antibiotics ablated the response, the myeloid cell proliferation, and cytokine production.32 Gavage of antibiotic-treated mice with Alistipes shahii or Ruminococci reversed the inhibition. In contrast, Lactobacillus fermentum gavaged mice failed to show immunotherapy profit. No in vitro studies of these commensals with mouse myeloid cells were done.
Table 2.
Bacterial Species Associated With Enhancement ICT in Mice.
| Tumor Model | ICT | Bacterial Species | Reference |
|---|---|---|---|
| MC38 colon | Anti-IL10+CpG | Alistipes shahii, Ruminococcus | 32 |
| MCA205 sarcoma | Cyclophosphamide | Enterococcus hirae, Barnisiella intestinihominus | 33, 34 |
| B16 melanoma | Anti-PD-L1 | Bifidobacterium | 35 |
| MCA205 sarcoma | Anti-CTLA-4 | Bacteroides fragilis, Bacteroides thetaiotamicron, Burkholderia | 36 |
| MC38 colon | Anti-PD1 or anti-CTLA-4 | Ruthenibacterium lactatiformans, Eubacterium limosum, Fusobacterium ulcerans, Phascolarctobacterium succinatutens, Bacteroides uniformis, Bacteroides dorei, Paraprevotella xylaniphila, Parabacteroides johnsonii, Parabacteroides gordonii, and Alistipes senegalensis | 37 |
| BRAFV600E/PTEN−/− melanoma | Anti-PD-L1 | Responder patient FMT | 41 |
| B16 SIY melanoma | Anti-PD-L1 | Responder patient FMT | 42 |
| MCA205 sarcoma | Anti-PD1 | Responder patient FMT, Akkermansia muciniphila, Enterococcus hirae, Alistipes | 43 |
| RENCA RCC | Anti-PD1+anti-CTLA-4 | Responder patient FMT | 43 |
| RET melanoma | Anti-PD1 | Akkermansia muciniphila, Alistipes, Enterococcus hirae | 43 |
| LLC lung carcinoma | Anti-PD1 | Akkermansia muciniphila, Alistipes, Enterococcus hirae | 43 |
Abbreviations: ICT, immune checkpoint inhibitor therapy; IL, interleukin; PD-L1, programmed death receptor-1 ligand; CTLA, cytotoxic T-lymphocyte associated protein 4; PD1, programmed death receptor-1; FMT, fecal microbiota transplant; RCC, renal cell carcinoma.
MCA205 sarcoma-carrying mice treated with cyclophosphamide had IFNγ- and IL17-producing splenic and tumor T-cells associated with tumor growth arrest.33,34 The antitumor activity required MyD88 and ileal and mesenteric lymph node bacteria. Germ-free or antibiotic-treated mice lost cyclophosphamide tumor inhibition. Gavage of antibiotic-treated mice with Enterococcus hirae clone 13144 reinstated cyclophosphamide efficacy. However, other gavaged bacteria, including Parabacteroides distasonis, Lactobacillus plantarum, Lactobacillus reuteri, Lactobacillus johnsonii, other Enterococcus hirae isolates, and segmented filamentous bacteria were inactive.33,34 Potency was also seen for Enterococcus hirae clone 13144 in HPV16-E7-expressing TC1 tumor-grafted mice. Gavage with Barnesiella intestinihominis also enhanced cyclophosphamide efficacy and yielded tumor IFNγ T-cell infiltration.
B16 melanoma SQ JAX but not TAC mice treated with anti-PD-L1 antibody showed complete remissions, and the enhanced effect was transmissible by gavage with JAX feces or Bifidobacterium species.35 The gut microbial effect depended on live organisms, DC activation, and increased tumor IFNγ producing CD8+ T-cells. Interestingly, no evidence of mesenteric lymph node Bifidobacteria was observed.
Mice with established MCA205 sarcomas showed tumor shrinkage with anti-CTLA4, and this activity was lost in germ-free or antibiotic-treated animals.36 The immunotherapy responses depended on intratumoral CD11b+ DCs secreting IL12 and splenic ICOS+ Ki67+ IFNγ+ TNFα+ T-cells, and tumor infiltrating T-cells. Bacteroides fragilis and Bacteroides thetaiotamicron and Burkholderia but not Parabacteroides distasonis or Escherichia coli nor Bacteroides uniformis effectively replaced mouse gut commensals and aided immunotherapy.
Tanoue et al37 isolated human gut bacteria that increased colonic IFNγ+ T-cells. These 11 bacteria were Ruthenibacterium lactatiformans, Eubacterium limosum, Fusobacterium ulcerans, Phascolarctobacterium succinatutens, Bacteroides uniformis, Bacteroides dorei, Paraprevotella xylaniphila, Parabacteroides distasonis, Parabacteroides johnsonii, Parabacteroides gordonii, and Alistipes senegalensis. These were rare, low-abundance human microbiota components. MC38 tumors in mice responded to anti-PD1 or anti-CTLA4 antibodies, but the response was reduced with antibiotic pretreatment or use of gnotobiotic mice. Gavage with the human 11-bacterium mix (11-mix) recovered ICT efficacy and infiltration of tumors with IFNγ+ T-cells. Use of the 7 Bacteroides microbes and the 4 non-Bacteroides bacteria showed that the latter retained partial inductive effects.
These diverse studies revealed that the gut bacteria influence ICT greatly. Furthermore, multiple different bacteria stimulate DCs and T-cells in mice, and the mechanism for the immune modulation remains uncertain. There are at least 3 hypotheses for the synergy: (a) Microbial pathogen–associated molecular pattern reaction with DC PRRs leads to innate immune activation with stimulation of cross-antigen presentation and release of cytokines and chemokines; (b) molecular mimicry of bacterial antigens with tumor neo-antigens, yielding an endogenous tumor vaccine; (c) the immune-stimulatory gut bacteria may produce small molecule modulators stimulating CTL function. The preliminary preclinical studies led directly to clinical experiments.
Clinical Correlative Studies of the Gut Microbiota and ICT
Several human studies have been conducted associating gut microbial profiles and pathways with ICT (Table 3). Unfortunately, small patient numbers, unique cancer patient populations, different collection techniques, and distinctive sequencing methods limit accuracy of comparisons. Clinical assessments of autoimmune toxicities are relatively straightforward, but ICT response measurements are challenging because of short-term fluctuations and need for long-term follow-up. These factors may contribute to the diversity of findings.
Table 3.
Bacterial Species Associated With Enhancement ICT in Humans.
| Cancer Type | ICT | Bacterial Species | Reference |
|---|---|---|---|
| Melanoma | Anti-CTLA-4 | Faecalibacterium prausnitzii 12-6, Gemmiger formicilis ATCC27749, Butyrate-producing bacteria SS2-1, Ruminococcus, Lachnospiraceae, Clostridium XIVa, Blautia | 39 |
| Melanoma | Anti-PD1 + Anti-CTLA-4 | Faecalibcterium prausnitzii, Bacteroides thetaiotamicron, Holdemania filiformis, Bacteroides caccae | 40 |
| Melanoma | Anti-PD1 | Faecalibacterium prausnitzii, Ruminococcus bromiii, Porphyromonas pasteri, Clostridium hungati, Phascolarctobacterium faecium | 41 |
| Melanoma | Anti-PD1 | Enterococcus faecium, Collinsella aerofacients, Bifidobacterium adolescentis, Klebsiella pneumoniae, Veillonella parvula, Parabacteroides merde, Lactobacillus sp, Bifidobacterium longum | 42 |
| NSCLC, RCC | Anti-PD1 | Akkermansia muciniphila, Lachnospiraceae, Erisypelotrichaceae lacteria 5-2-64, Enterococus faevium, Alistipes indistinctus, Bacteroidaceae, Bacteriodes xylanisolvens, Bacteroides nordii | 43 |
Abbreviations: ICT, immune checkpoint inhibitor therapy; CTLA, cytotoxic T-lymphocyte associated protein 4; PD1, programmed death receptor-1; NSCLC, non–small-cell lung carcinoma; RCC, renal cell carcinoma.
Dubin et al38 treated 34 melanoma patients with anti-CTLA4 ICT. Stool samples were collected prior to therapy; 10/34 patients developed autoimmune colitis. Fecal gDNAs were prepared and subjected to either 16S ribosomal RNA sequencing or metagenomics shotgun sequencing (MSS). Noncolitis patients were found to have higher levels of Bacteroidetes, including Bacteroidaceae, Rikenellaceae, and Barnesiellaceae. HUMAnN genetic pathway analysis with Kyoto Encyclopedia of Gene and Genomes (KEGG) assignments revealed decreased polyamine transport and B vitamin synthesis among colitis patients.
Chaput et al39 treated 26 melanoma patients with anti-CTLA4 ICT and collected multiple fecal samples; 9/26 had long-term clinical benefit and 7/26 developed autoimmune colitis. Fecal gDNAs were 16S rRNA sequenced; peripheral blood flow cytometry was done posttherapy. Patients enriched for Faecalibacterium prausnitzii L2-6, Gemmiger formicilis ATCC27749, butyrate-producing bacteria SS2-1, Ruminococcus, Lachnospiraceae, Clostridium XIVa, and Blautia had more durable remissions and more colitis. In contrast, patients with increased Bacteroides had fewer remissions or colitis events. Peripheral blood posttherapy of responders had more ICOS+ T-cells and sCD25 and fewer Tregs. There was no clear explanation for the importance of these anti-inflammatory firmicutes either for response or autoimmune colitis.
Frankel et al40 treated 39 melanoma patients with ICT (anti-CTLA4 + anti-PD1, anti-PD1, or anti-CTLA4). There were 15/23 responses after anti-CTLA4 + anti-PD1, 7/15 responses with anti-PD1, and 1/1 response with anti-CTLA4. Pretreatment stool samples were processed for gDNAs and MSS performed on the Illumina platform. Gut bacteria associated with response included Faecalibacterium prausnitzii, Bacteroides thetaiotamicron, Holdemania filiformis, and Bacteroides caccae. KEGG analysis showed that responders had increased gut bacterial enzymes associated with fatty acid synthesis and inositol phosphate metabolism. These findings were similar to those reported by Chaput et al.39
Gopalakrishnan et al41 studied 43 melanoma patients treated with anti-PD1 antibody. There were 30 responders and 13 nonresponders. Fecal studies, including 16S rRNA sequencing and MSS and immune-phenotyping of blood and tumors, were done. Responders’ gut bacteria had increased α-diversity, and there was an abundance of Faecalibacterium prausnitzii, Ruminococcus bromii, Porphyromonas pasteri, Clostridium hungati, and Phascolarctobacterium faecium. KEGG analysis of responders’ bacteria showed increased amino acid biosynthesis. Tumor immunohistochemistry (IHC) and blood flow cytometry showed increased tumor CD8+ T-cells and CD68+HLA-Dr+CD163+ myeloid DCs and decreased blood Tregs and myeloid-derived suppressor cells. The immunomodulatory bacteria mirror those described above by Chaput et al39 and Frankel et al.40 The mechanism of how Faecalibacterium prausnitzii and other Clostridial species promote ICT action remains undefined.
Matson et al42 treated 42 melanoma patients with anti-PD1 antibody (38 patients) or anti-CTLA4 antibody (4 patients). Pretreatment fecal samples were extracted, and 16S rRNA, MSS, and quantitative polycermase chain reaction (qPCR) data obtained. Tumor samples were subjected to whole exome sequencing (WES), mRNA profiling, and IHC. QIIME and BLAST analysis established Enterococcus faecium, Collinsella aerofaciens, Bifidobacterium adolescentis, Klebsiella pneumoniae, Veillonella parvula, Parabac-teroides merdae, Lactobacillus sp, and Bifidobacterium longum as overrepresented in responders. Responder tumors had higher PD1 and PD-L1 mRNA by profiling and CD8+ T-cells by IHC. There was little correlation with previously observed stimulatory bacteria, although Veillonella and Lactobacillus are firmicutes.
Routy et al43 measured the fecal gDNA MSS with BlastN analyses on 60 non–small-cell lung carcinoma (NSCLC) and 40 RCC patients treated with anti-PD1 antibody. Responders had overabundance of Akkermansia muciniphila, Lachnospiraceae, Erisypelotricheae bacterium 5-2-64, Enterococcus faecium, Alistipes indistinctus, Bacteroides caccae, Bacteroides xylanisolvens, and Bacteroides nordii. Stool-cultured responders demonstrated increased Enterococcus hirae. Furthermore, patients exposed to antibiotics up to 60 days before or 30 days into ICT had depletion of microbes and half of the survival of patients not exposed to antibiotics.
The distinct microbial profile of NSCLC and RCC responders versus melanoma responders may indicate cancer-specific immunity gut bacteria. A single-center or cooperative group study, including multiple cancer types with a single gDNA isolation, sequencing method, and bioinformatics approach, would help resolve the question.
Translational Hybrid Studies of Patient Gut Microbiota and Murine Models
Three of the above clinical trials collected stool specimens and tested them via fecal microbiota transplantation (FMT) in ICT-treated rodent tumor models. In each case, responder FMT led to improved antitumor efficacy as well as increased tumor infiltration with CD8+ T-cells and myeloid DCs.
BRAF V600E+/PTEN−/− melanomas inoculated subcutaneously (SQ) in germ-free mice followed by gavage with re-sponder or nonresponder patient FMT and anti-PD-L1 systemic treatment was performed by Gopalakrishnan et al.41 At day 28 post–tumor inoculation, responder patient FMT treated mice had one-sixth the tumor volume of nonresponder patient FMT treated mice. Furthermore, responder FMT mice had more tumor CD8+ T-cells and tumor CD45+CD11b+Ly6G+ myeloid dendritic cells (mDCs) and fewer splenic CD11b+CD11c+ myeloid-derived suppressor cells and splenic Fox3P+CD4+ Tregs. Fecal Faecalibacterium prausnitzii was elevated in responder FMT gavaged mice by qPCR.
Germ-free mice gavaged with responder patient or nonresponder patient fecal material were inoculated with B16-SIY melanoma cells.42 Two-thirds of responder FMT mice and one-third of nonresponder FMT mice had slower tumor growth when combined with anti-PD-L1 antibody therapy.42 Splenic IFNγ+CD8+ T-cells and Batf3+ DCs were increased in responder FMT mice.
Antibiotic pretreatment followed by patient FMT and then SQ tumors and anti-PD1 or anti-PD1 + anti-CTLA4 IP yielded 50% and 40% reduced tumor growth when responder patient FMT was compared with nonresponder patient FMT for the MCA205 sarcoma and RENCA RCC models, respectively.43 When Akkermansia muciniphila, Enterococcus hirae, or Alistipes probiotics were substituted for FMT, 40% tumor growth inhibition was observed relative to ICT without probiotics for the MCA205 sarcoma, RET melanoma, and LLC Lewis lung carcinoma models. Akkermansia muciniphila gavaged mice also showed statistically significant increases in mesenteric lymph node and tumor CCR9+CD4+ T cells. Finally, anti-IL12 antibody ablated the MCA205 tumor growth inhibition and CCR9+CD4+ T-cell tumor infiltration.
These translational studies and the earlier normal human 11-mix probiotic work establish that human immunomodulatory bacteria can directly alter ICT efficacy in multiple rodent tumor models and provide preliminary evidence of a pathway by which bacteria stimulate mDCs to secrete IL12 and differentiate tumor CTLs. Subsequent clinical trials of either responder FMT or selected probiotics prior to ICT should confirm clinical benefit and immune mechanisms.
Discussion
The above studies document an association between particular gut bacteria and ICT response in mice and humans. A critical question is whether these associations are causative. The translational studies show that the clinically isolated microbes are able to enhance ICT across species. However, many questions remain.
The differences between mouse and human commensals that were connected to ICT response is not surprising because of significant differences between species for immunology, diet, and tumor biology. Only Sivan et al35 and Matson et al42 reported the same microorganisms—Bifidobacteria—in mouse and human feces of ICT responders.
The similarity of commensals associated with patient ICT response and isolates from normal Japanese individuals that trigger lamina propria IFNγ CD8+ T-cells was striking. Both studies included groups of Clostridial firmicutes and Bacteroidetes. Ruthenibacterium lactatiformans44 found in normal individuals is more than 99% identical to the keystone microbe Faecalibacterium prausnitzii that was discovered in the ICT response association studies of Chaput et al,39 Frankel et al,40 and Gopalakrishnan et al.41 Ex vivo studies suggest that Bacteroidetes digest insoluble fibers and mucins and provide acetate and other metabolites to Faecalibacterium and other firmicutes.45
Distinct immunity-promoting bacteria were found among the above studies. Matson et al42 found Bifidobacteria, whereas Gopalakrishnan et al,41 Frankel et al,40 and Chaput et al39 did not identify Bifidobacteria but instead Clostridiales and Bacteroides in ICT responders. In contrast, the work by Routy et al43 uncovered the Verrucomicrobia Akkermansia muciniphila as the sentinel organism in responders. ICT treatment of different cancers may have unique immunogenic bacteria. Alternatively, investigator-specific stool collection, gDNA extraction, sequencing, or computer analyses may lead to recognition of different “responder” bacteria.46-48
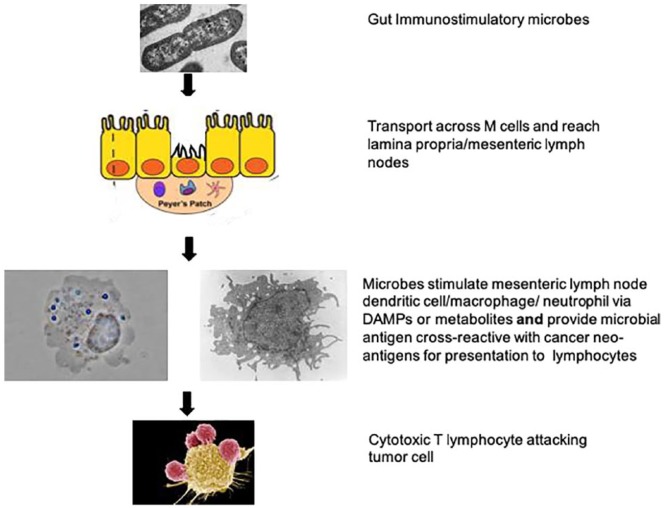
How do these bacteria accelerate antitumor immunity? One hypothesis posits that microbial products react with PRRs and activate mDCs to both release cytokines, including CXCL9/10 and IL12, and perform cross-antigen presentation to CD8+ T-lymphocytes (Figure 2). There is evidence that the cytoplasmic DNA-cGAS-STING-IRF3 pathway is critical for gut immunity and ICT function.13,49 Thus, phagocytosed microbes in lamina propria or mesenteric lymph node DCs may be digested to cytoplasmic DNA and initiate cGAS-STING signaling. Live Faecalibacterium prausnitzii activate TLR2/TLR6 ex vivo.50 There is additional evidence for TLR-MyD88-TRIFF pathways at least for cyclophosphamide immune modulation, with a dependence on gut microbe interaction in mesenteric lymph node DC via MyD88.33,34 Three of the preclinical studies showed a requirement for gut microbial signaling through mDC IL12.32,36,43 Another motivator for mDC differentiation can be gut microbial products such as the short-chain fatty acid butyrate. Butyrate producers such as Faecalibacterium prausnitzii and Ruthenibacterium lactatiformans can not only reduce inflammation but also stimulate CTL function via epigenetic targets.51,52 Future experiments should confirm the DC location whether it is the lamina propria, mesenteric lymph node, or tumor and molecular pathway—PRRs versus small-molecule epigenetic modifiers. The “tumor vaccine or molecular mimicry” hypothesis posits that the specificity of ICT-related bacteria is a result of cross-reactivity of microbial peptide antigens with tumor neo-antigens. There is mixed evidence that patients respond best to ICT if they have tumor neo-antigens that not only bind strongly to major histocompatiblity complex (MHC), but also resemble immune epitopes of microbial pathogens.53-55 Future work should correlate ICT response with presence of patient gut microbes that have peptides resembling the patient’s neo-antigen peptides.
Figure 2.
Schematic hypothesis for commensal bacteria stimulation of ICT. Live immunomodulating bacteria cross the epithelium at M cells and become internalized by mDCs. The mDCs are then activated and transported to mesenteric lymph nodes. There, they release chemokines and cytokines that recruit and stimulate CD8+ T-lymphocytes to bind the mDCs via T cell receptor (TCR) and costimulatory proteins, leading to antigen presentation via MHC class I and T-cell education. The cytotoxic T-lymphocytes then travel to tumor deposits where they attack and kill malignant cells in the presence of immune checkpoint inhibitors.
Abbreviations: ICT, immune checkpoint inhibitor therapy.
If immunity-producing bacteria are confirmed for particular cancers, how do we use that knowledge to improve ICT response? Historically, probiotics have had minimal impact on the patient’s gut microbiome.56 However, evidence of high rates of cure with FMT for Clostridium difficile colitis suggest that clinical trials are warranted of either FMT or defined bacterial mixture probiotics.57 The optimal dose, schedule, pretreatment therapy with antibiotics or bowel preps will need to be established. Two clinical trials for gut microbiota enhancement of ICT are ongoing. In one, responder donor FMT is added to ICT (NCT03353402). In the second, a Clostridium butyricum probiotic CBM588 is added to ICT (NCT03829111). A third clinical trial combines the prebiotic MegaPrebiotic containing galacto-oligosaccharides, fructo-oligosaccharides, and xylo-oligosaccharides and the probiotic MegaSpore Biotic composed of spores of Bacillus clausii, Bacillus coagulans, Bacillus subtilis, Bacillus indicis, and Bacillus licheniformis. The probiotic mixture has been shown to increase Faecalibacterium prausnitzii in ex vivo culture.58 The trial has started at the University of South Alabama. The next few years should see results of these studies in cancer patients receiving ICT. The results of these and other therapeutic clinical trials will hopefully complete Koch’s postulates for these gut microbes. Fecal sampling and qPCR can be used in some of these and in the future to confirm engraftment.59
The next decade will see further advances in cancer immunotherapy. There is a strong likelihood that one of the new combinations will include applications targeting the patient gut microbiota.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors recieved financial support from NCI grant CA204801.
ORCID iDs: Arthur E. Frankel  https://orcid.org/0000-0001-8375-0066
https://orcid.org/0000-0001-8375-0066
Amit Reddy  https://orcid.org/0000-0003-0600-5550
https://orcid.org/0000-0003-0600-5550
Brooks Rabideau  https://orcid.org/0000-0001-8253-8688
https://orcid.org/0000-0001-8253-8688
References
- 1. Skipper HE, Schabel FM, Jr, Trader MW, Laster WR, Jr, Simpson-Herren L, Lloyd HH. Basic and therapeutic trial results obtained in the spontaneous AK leukemia (lymphoma) model-end of 1971. Cancer Chemother Rep. 1974;56:273-314. [PubMed] [Google Scholar]
- 2. Ghatalia P, Morgan CJ, Sonpavde G. Meta-analysis of regression of advanced solid tumors in patients receiving placebo or no anti-cancer therapy in prospective trials. Crit Rev Oncol Hematol. 2016;98:122-136. [DOI] [PubMed] [Google Scholar]
- 3. Rossi AP, Klein CL. Posttransplant malignancy. Surg Clin North Am. 2019;99:49-64. [DOI] [PubMed] [Google Scholar]
- 4. Humeau J, Lévesque S, Kroemer G, Pol JG. Gold standard assessment of immunogenic cell death in oncological mouse models. Methods Mol Biol. 2019;1884:297-315. [DOI] [PubMed] [Google Scholar]
- 5. Suciu S, Mandelli F, de Witte T, et al. ; EORTC and GIMEMA Leukemia Groups. Allogeneic compared with autologous stem cell transplantation on the treatment of patients younger than 46 years with acute myeloid leukemia in first complete remission (CR1): an intention-to-treat analysis of the EORTC/GIMEMAAML-10 trial. Blood. 2003;102:1232-1240. [DOI] [PubMed] [Google Scholar]
- 6. Petrella T, Quirt I, Verma S, Haynes AE, Charette M, Bak K; Members of the Melanoma Disease Site Group of Cancer Care Ontario’s Program in Evidence-Based Care. Single-agent interleukin-2 in the treatment of metastatic melanoma. Curr Oncol. 2007;14:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ingram JR, Blomberg OS, Rashidian M, et al. Anti-CTLA-4-therapy requires an Fc domain for efficiency. Proc Natl Acad Sci U S A. 2018;115:3912-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734-1736. [DOI] [PubMed] [Google Scholar]
- 9. Robert C, Thomas L, Bondarenko I, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517-2526. [DOI] [PubMed] [Google Scholar]
- 10. Garris CS, Arlauckas SP, Kohler RH, et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity. 2018;49:1148-1161.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gandini S, Massi D, Mandalà M. PD-L1 expression in cancer patients receiving anti-PD-1/anti-PD-L1 antibodies: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2016;100:88-98. [DOI] [PubMed] [Google Scholar]
- 12. Luke JJ, Bao R, Sweis RF, Spranger S, Gejewski TF. Wnt/β-catenin pathway activation correlates with immune exclusion across human cancers [published online January 11, 2019]. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-18-1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H, Hu S, Chen X, et al. cGAS is essential for the antitumor effects of immune checkpoint blockade. Proc Natl Acad Sci U S A. 2017;114:1637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun L, Funchain P, Song JM, et al. Talimogene laherparepvec combined with anti-PD-1 based immunotherapy for unresectable stage III-IV melanoma. J Immunother Cancer. 2018;6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chakravarti N, Ivan D, Trinh VA, et al. High cytotoxic T lymphocyte antigen-4 and phospho-Akt expression in tumor samples predicts poor clinical outcomes in ipilimumab treated melanoma patients. Melanoma Res. 2017;27:24-31. [DOI] [PubMed] [Google Scholar]
- 19. Herbst RS, Baas P, Perez-Gracia JL, et al. Use of archival versus newly collected tumor samples for assessing PD-L1 expression and overall survival: an updated analysis of KEYNOTE-010 trial. Ann Oncol. 2019;30:281-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel SJ, Sanjana NE, Kishton RJ, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McQuade JL, Daniel CR, Hess KR, et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol. 2018;19:310-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richtig G, Hoeller C, Wolf M, et al. Body mass index may predict the response to ipilimumab in metastatic melanoma: an observational multi-centre study. PLoS One. 2018;13:e0204729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29:1437-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahmed J, Kumar A, Parikh K, et al. Use of broad-spectrum antibiotics impacts outcome in patients treated with immune checkpoint inhibitors. Oncoimmunology. 2018;7:e1507670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huemer F, Rinnerthaler G, Lang D, Hacki H, Lamprecht B, Greil R. Association between antibiotics use and outcome in patients with NSCLC treated with immunotherapeutics [published online January 24, 2019]. Ann Oncol. doi: 10.1093/annonc/mdz021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pasolli E, Asnicar F, Manara S, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography and lifestyle. Cell. 2019;176:649-662.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allaband C, McDonald D, Vásquez-Baeza Y, et al. Microbiome 101: studying, analyzing and interpreting gut microbiome data for clinicians. Clin Gastroenterol Hepatol. 2019;17:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haak BW, Lankelma JM, Hugenholtz F, Belzer C, de Vos WM, Wiersinga WJ. Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on gut microbiota in healthy humans. J Antimicrobiol Chemother. 2019;74:782-786. [DOI] [PubMed] [Google Scholar]
- 30. Palleja A, Mikkelsen KH, Forslund SK, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018;3:1255-1265. [DOI] [PubMed] [Google Scholar]
- 31. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46:562-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Daillere R, Vetizou M, Waldschmitt N, et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931-943. [DOI] [PubMed] [Google Scholar]
- 35. Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vetizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanoue T, Morita S, Plichta DR, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600-605. [DOI] [PubMed] [Google Scholar]
- 38. Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368-1379. [DOI] [PubMed] [Google Scholar]
- 40. Frankel AE, Coughlin LA, Kim J, et al. Metagenomic shotgun sequencing and unbiased metabolomics profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [DOI] [PubMed] [Google Scholar]
- 44. Shkoporov AN, Chaplin AV, Shcherbakova VA, et al. Ruthenibacterium lactatiformans gen. nov., sp. nov., an anaerobic, lactate-producing member of the family Ruminococcaceae isolated from human faeces. Int J Syst Evol Microbiol. 2016;66:3041-3049. [DOI] [PubMed] [Google Scholar]
- 45. Das P, Ji B, Kevatcheva-Datchary P, Bäckhed F, Nielsen J. In vivo co-culture of human gut bacterial species as predicted from co-occurrence network analysis. PLoS One. 2018;13:e0195161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hill CJ, Brown JR, Lynch DB, et al. Effect of room temperature vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome. 2016;4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Panek M, Paljetak HC, Barešić A, et al. Methodology challenge in studying human gut microbiota—effects of collection, storage, DNA extraction, and next generation sequencing technologies. Sci Rep. 2018;8:5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knight R, Vrbanac A, Taylor BC, et al. Best practices for analyzing microbiomes. Nat Rev Microbiol. 2018;16:410-422. [DOI] [PubMed] [Google Scholar]
- 49. Canesso MCC, Lemos L, Neves TC, et al. The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal Immunol. 2018;11:820-834. [DOI] [PubMed] [Google Scholar]
- 50. Maier E, Anderson RC, Altermann E, Roy NC. Live Faecalibacterium prausnitzii induces greater TLR2 and TLR2/6 activation then the dead bacteria in an apical anaerobic co-culture system. Cell Microbiol. 2018;20(2). doi: 10.1111/cmi.12805 [DOI] [PubMed] [Google Scholar]
- 51. Luu M, Weigand K, Ovedi F, et al. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci Rep. 2018;8:14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kespohl M, Vachharajani N, Luu M, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Front Immunol. 2017;8:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lusaka M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumor responses to checkpoint blockade immunotherapy. Nature. 2017;551:517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Balanchandran VP, Łuksza M, Zhao JN, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nathanson T, Ahuja A, Rubinsteyn A, et al. Somatic mutation and neoepitope homology in melanoma treated with CTLA-4 blockade. Cancer Immunol Res. 2017;5:84-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zmora N, Zilberman-Schapira G, Suez J, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388-1405.e21. [DOI] [PubMed] [Google Scholar]
- 57. Stripling J, Rochiguez M. Current evidence in delivery and therapeutic use of fecal microbiota transplantation in human diseases—Clostridium difficile disease and beyond. Am J Med. 2018;356:424-432. [DOI] [PubMed] [Google Scholar]
- 58. McFarlin BK, Henning AL, Bowman EM, Gary MA, Carbajal KM. Oral spore-based probiotic supplementation was associated with reduced incidence of postprandial dietary endotoxin, triglyceride and disease risk biomarkers. World J Gastrointest Pathophysiol. 2017;8:117-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koh AY. Potential for monitoring gut microbiota for diagnosing infection and graft-versus-host disease in cancer and stem cell transplant patient. Clin Chem. 2017;63:1685-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]