Potassium is an essential ion in every living cell. Even though potassium is the most abundant cation in cells, its accumulation can be toxic. Therefore, the level of potassium has to be tightly controlled. In many Gram-positive bacteria, the second messenger cyclic di-AMP plays a key role in the control of potassium homeostasis by binding to potassium transporters and regulatory proteins and RNA molecules. In the lactic acid bacterium Lactococcus lactis, none of these conserved c-di-AMP-responsive molecules are present. In this study, we demonstrate that the KupA and KupB proteins of L. lactis IL1403 are high-affinity potassium transporters and that their transport activity is inhibited by the second messenger c-di-AMP.
KEYWORDS: Lactococcus, c-di-AMP, potassium transport, second messenger
ABSTRACT
Cyclic di-AMP (c-di-AMP) is a second messenger involved in diverse metabolic processes, including osmolyte uptake, cell wall homeostasis, and antibiotic and heat resistance. In Lactococcus lactis, a lactic acid bacterium which is used in the dairy industry and as a cell factory in biotechnological processes, the only reported interaction partners of c-di-AMP are the pyruvate carboxylase and BusR, the transcription regulator of the busAB operon for glycine betaine uptake. However, recent studies uncovered a major role of c-di-AMP in the control of potassium homeostasis, and potassium is the signal that triggers c-di-AMP synthesis. In this study, we have identified KupA and KupB, which belong to the Kup/HAK/KT family, as novel c-di-AMP binding proteins. Both proteins are high-affinity potassium transporters, and their transport activities are inhibited by binding of c-di-AMP. Thus, in addition to the well-studied Ktr/Trk potassium channels, KupA and KupB represent a second class of potassium transporters that are subject to inhibition by c-di-AMP.
IMPORTANCE Potassium is an essential ion in every living cell. Even though potassium is the most abundant cation in cells, its accumulation can be toxic. Therefore, the level of potassium has to be tightly controlled. In many Gram-positive bacteria, the second messenger cyclic di-AMP plays a key role in the control of potassium homeostasis by binding to potassium transporters and regulatory proteins and RNA molecules. In the lactic acid bacterium Lactococcus lactis, none of these conserved c-di-AMP-responsive molecules are present. In this study, we demonstrate that the KupA and KupB proteins of L. lactis IL1403 are high-affinity potassium transporters and that their transport activity is inhibited by the second messenger c-di-AMP.
INTRODUCTION
Ion homeostasis is a key factor for each living cell, and potassium (K+) and glutamate are the most abundant cation and anion, respectively, in the cytosol of all cells. While glutamate can be synthesized, for potassium, the uptake is essential and tightly regulated. Bacteria have evolved different systems to accumulate potassium intracellularly to achieve stable concentrations to meet their demand for potassium in processes such as pH homeostasis, proton motive force generation, cell turgor, and protein synthesis, among many others (1). Different high- and low-affinity uptake systems are involved if the bacteria are growing in media with low and high potassium concentrations, respectively.
In Escherichia coli, there are three systems responsible for potassium uptake, namely, Trk, Kdp, and Kup. These systems have different characteristics and complement each other to allow growth under various conditions. The Trk system consists of four genes which are constitutively expressed, with trkA encoding the predominant potassium transporter at neutral pH. TrkA has low affinity for the ion and depends on ATP and proton motive force for potassium uptake (2). On the other hand, the Kdp-ATPase system includes the inducible high-affinity transporter KdpA, which is induced at low potassium concentrations and under conditions of osmotic stress (3). Finally, Kup (formerly TrkD) is also constitutive with low affinity, and its activity increases at low pH, when TrkA and Kdp activities are not sufficient (4).
In the Gram-positive model bacterium Bacillus subtilis, three main potassium transport systems can be found. KtrAB and KtrCD are high- and low-affinity transporters, respectively, and both belong to the Trk/Ktr/HTK transporter family (5). In addition, the novel high-affinity transporter KimA (formerly YdaO) has recently been characterized (6). The expression of the kimA gene as well as of the ktrAB operon is negatively regulated by a riboswitch that responds to the second messenger cyclic di-AMP (c-di-AMP) (7). An increase in the potassium concentration results in an accumulation of c-di-AMP, which in turn binds to the riboswitch and thus prevents transcription of the genes encoding the transporters beyond the riboswitch (6, 7). Interestingly, c-di-AMP also binds the regulatory subunits of KtrAB and KtrCD, upon which inhibition of the respective transporters occurs (8). As in B. subtilis, c-di-AMP regulates potassium uptake in Staphylococcus aureus by binding both to the KtrA proteins and to the KdpD sensor kinase that controls the expression of the high-affinity potassium transporter KdpABC (8, 9).
c-di-AMP was discovered as a second messenger 10 years ago (10), and its intracellular levels are tightly regulated (see references 11, to ,13 for a review). High concentrations of c-di-AMP are toxic for the cell, and the synthesizing enzymes are essential under standard growth conditions (14, 15). Deletion of the gene(s) encoding the diadenylate cyclases has been achieved only in strictly controlled minimal media in Listeria monocytogenes, B. subtilis, S. aureus, and Streptococcus agalactiae (6, 16–18). In the Gram-positive model organism B. subtilis, c-di-AMP is dispensable only at low potassium concentrations in minimal medium (6).
Lactococcus lactis is the best-characterized lactic acid bacterium (LAB), and it is intensively used in dairy fermentation (19). It is also a model microorganism used as food-grade bacterial factory and, in the last decades, it has been used to design and produce new therapeutic molecules (20, 21). In L. lactis, c-di-AMP is synthesized by the single enzyme CdaA and degraded by the phosphodiesterase GdpP (22). The cdaA gene is encoded in the widely conserved cdaAR-glmM operon, in which the gene for the regulatory protein CdaR is also present. The third gene of the operon codes for GlmM, an enzyme which converts glucosamine-6-phosphate to the peptidoglycan precursor glucosamine-1-phosphate. In addition, GlmM interacts with CdaA and modulates its activity (22). Accumulation of c-di-AMP due to the deletion of the gdpP gene encoding the c-di-AMP-degrading phosphodiesterase results in heat-resistant and salt-hypersensitive phenotypes, indicating a link between c-di-AMP and osmoadaptation in L. lactis (23). Such a strain rapidly acquires mutations resulting in osmoresistance. These mutations typically reduce the intracellular levels of c-di-AMP and affect the diadenylate cyclase CdaA, the glucosamine-6-phosphate mutase GlmM, or the expression of a c-di-AMP-exporting multidrug exporter (22, 24). A second class of suppressor mutations directly affects osmotic homeostasis. In such strains, mutations in the KupB potassium transporter result in increased potassium uptake, and the deletion of the gene encoding the BusR transcription regulator results in increased expression of the busAB operon coding for a glycine betaine transporter and in increased accumulation of this osmoprotectant (24). These observations support the idea that the control of osmotic homeostasis is a key function of c-di-AMP (25). Interestingly, the BusR protein is also a direct target of c-di-AMP. In the presence of the second messenger, it binds to the promoter region of the busAB operon to repress transcription. Thus, c-di-AMP accumulation results in reduced osmoprotectant uptake (18, 24). Moreover, c-di-AMP binds to and inhibits the pyruvate carboxylase in L. lactis (26). However, the c-di-AMP regulatory network is not yet completely understood in L. lactis, and further investigations remain crucial to elucidate the metabolic pathways which are controlled by c-di-AMP and the reason for its essentiality.
In this study, we identified KupA and KupB, two potassium transporters, as direct targets of the signaling nucleotide c-di-AMP in L. lactis IL1403. The evidence suggests that these very similar proteins have high affinity for this ion and that binding of c-di-AMP inhibits potassium uptake.
RESULTS
Analysis of the L. lactis IL1403 genome for potential c-di-AMP binding proteins.
Recently, an increasing amount of evidence has linked the essential role of c-di-AMP to the control of potassium homeostasis and osmoregulation (25). Consequently, the L. lactis IL1403 genome was analyzed for genes encoding putative potassium transporters and proteins potentially related to osmotic regulation. A search for potassium transporters encoded in the L. lactis IL1403 genome by use of the free online software UniProt (UniProtKB, UniRef, and UniParc databases) identified the proteins KupA, KupB, and YrbD. For KupB, potassium transport activity has recently been demonstrated (24). KupA and KupB belong to the KT/KUP/HAK family (27), whereas YrbD might be a voltage-gated channel. The fact, the fact that the accumulation of c-di-AMP is triggered the emergence of KupB variants with increased activity suggests that the second messenger might control KupB activity, as has been shown for other potassium transporters from S. aureus, B. subtilis, and Streptococcus pneumoniae (8, 28). Thus, we considered KupA and KupB as well as YrbD to be potential novel target proteins for c-di-AMP. In addition, we considered the putative voltage-gated potassium channel proteins YjdJ and YncB, which exhibit more than 60% identity to the potassium efflux system KefA of other Lactococcus species.
Proteins with high homology to members of the Trk/Ktr/HKT family were also evaluated. Interestingly, even though putative proteins similar to B. subtilis KtrA were found in some L. lactis strains, this family is not present in the IL1403 and MG1363 strains studied in this work. Moreover, even though the kimA (ydaO) riboswitch was already reported to be absent in Lactococcus species (29), the presence of the associated potassium transporter KimA or related proteins was considered. No KimA homologues are encoded in the L. lactis genome; however, the protein blast showed certain homology to LysP, an amino acid permease (24% identity in a 310-amino-acid region), which reflects that both proteins are members of the amino acid-polyamine-organocation (APC) superfamily.
Given the recent association of c-di-AMP to amino acid metabolism and osmoregulation (17, 30), both LysP and LysQ were also included in the list of potential c-di-AMP interaction partners. In L. monocytogenes, a mutant strain for the single c-di-AMP-synthesizing enzyme was viable in rich media due to mutations occurring in the genes oppABCDF (involved in oligopeptide import) or gbuABC (involved in glycine betaine import) (17). Therefore, the homologous OppA protein of L. lactis IL1403, which is likely to function as an oligopeptide permease, was added to the list of potential c-di-AMP targets as well.
In several Gram-positive bacteria, the PII-like signal transduction protein DarA has been identified as a c-di-AMP binding protein (8, 31, 32). However, this protein is not encoded in the L. lactis genome.
This selection resulted in a list of eight potential c-di-AMP targets that were chosen for further analysis (see Table S1 in the supplemental material).
c-di-AMP interaction with selected lactococcal proteins.
The eight selected genes were cloned using the E. coli expression vector pWH844 (Table S1). After checking the expression of all genes using strain E. coli BL21 as a host (see Fig. S1), the lysates of strains carrying the corresponding plasmids were assessed for a possible interaction with c-di-AMP using the differential radial capillary action of ligand assay (DRaCALA) (see Materials and Methods).
While the majority of the selected proteins showed no or weak interaction with c-di-AMP in the DRaCALA, the two potassium transporters KupA and KupB gave the strongest binding results and were therefore investigated further (Fig. 1A). The binding of both proteins to [32P]c-di-AMP is specific since unlabeled c-di-AMP, but not ATP, competes for binding (Fig. 1B and C). Moreover, the interaction of c-di-AMP with KupA seems to be stronger than that with KupB.
FIG 1.
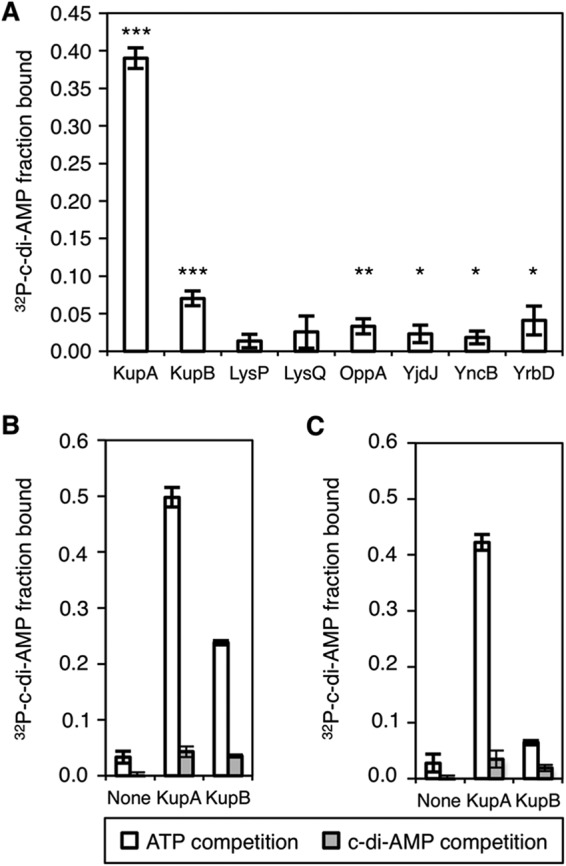
c-di-AMP interaction partners determined by DRaCALA. (A) Fraction bound of radiolabeled c-di-AMP is shown for lysates from E. coli induced overnight for the expression of the indicated gene. The significance of binding was determined using unpaired t test against buffer-only control, with P values <0.05, <0.01, and <0.001, indicated by *, **, and ***, respectively. (B and C) Specificity of KupA and KupB binding to [32P]c-di-AMP was assessed by competition assays with 100 μM ATP or 100 μM ATP and 100 μM c-di-AMP using overnight lysates (B) or from lysates obtained from freshly reinoculated cultures (C).
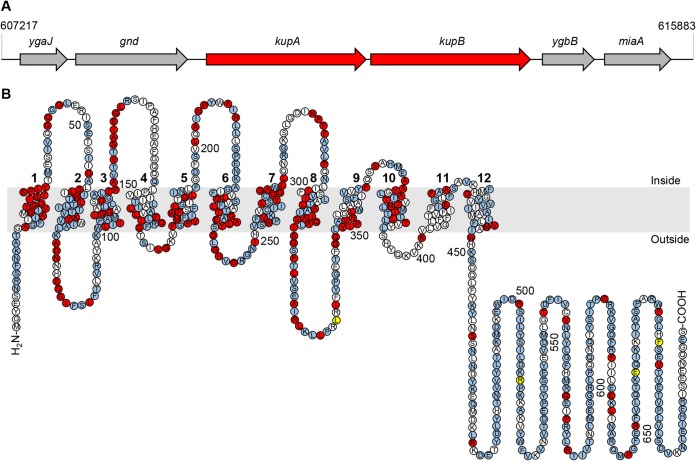
Interestingly, the genes encoding the homologous KupA and KupB proteins were found in the genome of L. lactis IL1403, where they form a putative bicistronic transcription unit separated by 223 bp (Fig. 2A). In contrast, in the related L. lactis MG1363, KupA is mutated, and only kupB encodes a functional protein (24). The amino acid sequences of KupA and KupB show 73% identity, and topology prediction using the TOPCONS software (33) suggests very similar structures with 12 transmembrane segments for these proteins, very similar to the topology of the E. coli Kup protein (34) (see Fig. 2B). Moreover, as shown in Fig. 2B, the majority of highly conserved residues are located in the N-terminal domain and particularly in the transmembrane regions.
FIG 2.
KupA and KupB in silico analysis. (A) Arrangement of the tandem kupA and kupB in the genome of L. lactis IL1403. In L. lactis MG1363, the kupA gene is inactive due to an internal stop codon. (B) Predicted membrane topology of KupA using the Protter Web server. Amino acids conserved in L. lactis KupA and KupB are highlighted in blue, and those conserved in E. coli as well are shown in red. Positions of amino acid substitutions that result in increased activity of KupB are highlighted in yellow. The cell membrane is shown in gray. The 12 transmembrane helices are numbered.
Taken together, the evidence here presented supports the fact that the L. lactis KupA and KupB K+ transporters are novel targets of c-di-AMP. Binding of c-di-AMP to several other tested proteins was weak but statistically significant, and further work with improved expression levels is needed to determine if they are true c-di-AMP receptors.
KupA and KupB of L. lactis IL1403 restore growth of an E. coli strain lacking potassium transporters.
In a first approach to address the functional properties of the KupA and KupB proteins, we performed a complementation assay with the E. coli strain LB650 (35). This strain lacks the main potassium transport systems KdpABC, Kup, TrkH, and TrkG and is unable to grow at potassium concentrations below 10 mM. The L. lactis IL1403 kupA and kupB genes were cloned into the expression vector pWH844 in a way to express tag-free proteins (see Materials and Methods). The resulting plasmids pIQ309 and pIQ310 (carrying kupA and kupB genes, respectively [see Table 2]), were introduced into E. coli LB650. The transformants were used to analyze if KupA and/or KupB expression could restore growth on complex medium in the absence of added potassium. As shown in Fig. 3, the strain harboring the empty vector pWH844 is unable to grow on LB unless it is supplemented with KCl. The introduction of either the kupA or kupB genes, however, resulted in the growth of colonies on the plates without extra addition of KCl, even in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer of the expression of the cloned genes. Apparently, the basal expression of kupA and kupB is sufficient to allow potassium uptake of the complemented mutant strains and, thus, growth. These results demonstrate that the L. lactis IL1403 KupA and KupB proteins are both active potassium transporters.
TABLE 2.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| L. lactis IL1403 | Plasmid-free strain, Trp+ | 51 |
| L. monocytogenes EGD-e | Wild type, serovar 1/2a strain | Laboratory collection |
| E. coli DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 ϕ80Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | 52 |
| E. coli BL21 | B F− ompT gal dcm lon hsdSB(rB− mB−) [malB+]K-12(λS) | 53 |
| E. coli LB650 | F− thi lacZ gal rha ΔkdpFABC5 trkD1 ΔtrkH ΔtrkG Km50 Cm30 | 35 |
| E. coli LB2003 | ΔkdpABC5 kup1 (trkD1) ΔtrkA | 37 |
| Plasmids | ||
| pBAD33 | Para Cm30 | 45 |
| pBP370 | pBAD33 cdaAlmo Cm30 | This work |
| pBP371 | pWH844 ktrC-ktrD (L. monocytogenes) Amp100a | This work |
| pBP372 | pWH844 ktrAB (B. subtilis) Cm30 | 6 |
| pBP373 | pBAD33 cdaAlmo D171N Cm30 | This work |
| pWH844 | Plac His6× Amp100 | 48 |
| pIQ309 | pWH844 kupA Amp100 | This work |
| pIQ310 | pWH844 kupB Amp100 | This work |
FIG 3.
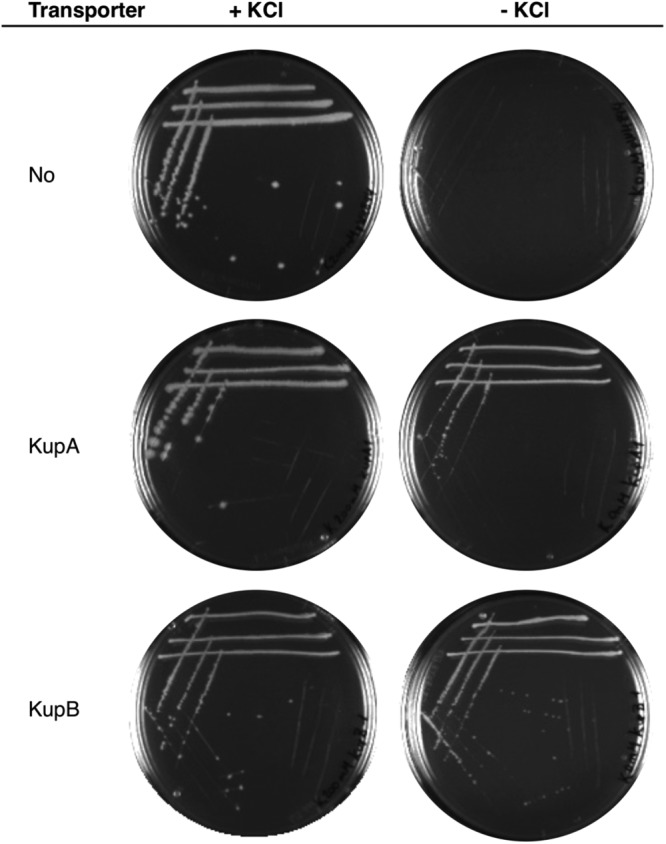
KupA and KupB restore growth of E. coli LB650 on LB plates. The strain was transformed with the empty vector pWH844 (no transporter), pIQ309 (KupA), or pIQ310 (KupB) and tested on plates with or without the supplementation of KCl (200 mM).
KupA and KupB are high-affinity potassium transporters.
In order to estimate the affinity of KupA and KupB proteins for K+, a comparison to previously characterized transporters was made. For this purpose, the high-affinity potassium transporter KtrAB from B. subtilis and putative KtrCD transporter from L. monocytogenes were employed (5, 36). The corresponding expression vectors pBP371 and pBP372, respectively, were introduced into E. coli LB650. Growth assays were then performed in modified M9 minimal salt medium, in which potassium salts were replaced by equimolar quantities of the respective sodium salt (M9mod [see Materials and Methods]). KCl was then added to the medium to final concentrations between 0.025 and 50 mM. The maximum optical density (ODmax) and μmax were then determined for E. coli LB650, harboring the empty vector pWH844, as well as the kupA (pIQ309) and kupB (pIQ310) genes. LB650 expressing KtrCD (pBP371) or KtrAB (pBP372) was included as a reference for the response of low- and high-affinity systems under the conditions studied here.
The expression of both KupA and KupB in the E. coli strain deficient in potassium transport resulted in similar phenotypes. Both transporters allowed growth even at concentrations below 0.1 mM KCl, although the growth rate and final OD600 were low (Table 1). Moreover, values close to the highest ODmax and μmax were observed at a potassium concentration of 1 mM KCl. Similar results have been obtained with the high-affinity system KtrAB of B. subtilis. On the other hand, upon expression of the low-affinity potassium transporter KtrCD, growth was detected only at concentrations of 10 mM KCl or higher, and the ODmax and μmax values were observed when at least 50 mM KCl was present in the medium. A marked difference in the growth parameters is then established with respect to high-affinity KtrA/B and Kup proteins, being more closely related to strain LB650 harboring the vector. Taken together, these results clearly demonstrate that both KupA and KupB are high-affinity potassium transporters.
TABLE 1.
ODmax and μmax parameters for E. coli LB650 derivative strains
| [K] (mM) | Value for derivative with: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pWH844 |
KupA |
KupB |
KtrAB |
KtrCD |
||||||
| ODmax | μmax (min−1) | ODmax | μmax (min−1) | ODmax | μmax (min−1) | ODmax | μmax (min−1) | ODmax | μmax (min−1) | |
| 0 | NDa | ND | 0.3 | 0.005 | 0.1 | 0.0037 | 0.4 | 0.005 | ND | ND |
| 0.025 | ND | ND | 0.4 | 0.005 | 0.2 | 0.0071 | 0.5 | 0.007 | ND | ND |
| 0.50 | ND | ND | 0.4 | 0.0089 | 0.3 | 0.01 | 0.7 | 0.008 | ND | ND |
| 0.075 | ND | ND | 0.5 | 0.011 | 0.3 | 0.011 | 0.7 | 0.01 | ND | ND |
| 0.50 | ND | ND | 0.8 | 0.013 | 0.8 | 0.014 | 0.9 | 0.01 | ND | ND |
| 1.0 | ND | ND | 1.0 | 0.016 | 1.0 | 0.013 | 0.9 | 0.007 | ND | ND |
| 10.0 | 0.8 | 0.0047 | 1.0 | 0.015 | 1.0 | 0.009 | 1.0 | 0.006 | 1.1 | 0.009 |
| 50.0 | 1.3 | 0.012 | 1.0 | 0.016 | 1.1 | 0.007 | 1.1 | 0.012 | 1.2 | 0.013 |
ND, not determined.
c-di-AMP inhibits potassium transport by the Kup proteins.
The identification of KupA and KupB as potassium transporters and their specific interaction with c-di-AMP suggested a functional role for c-di-AMP in the control of activity of the KupA and KupB proteins. Therefore, we analyzed the impact of this second messenger nucleotide on the potassium transport activity of KupA and KupB. For this purpose, a coexpression system was established in the model bacterium E. coli LB2003 (37). Similar to LB650, this strain is deficient in the three major potassium uptake systems Trk, Kup, and Kdp and is therefore not able to grow at low K+ concentrations without complementation using a gene encoding a potassium transporter. Importantly, and in contrast to strain LB650 (kanamycin MIC of 50 μg/ml [Km50], chloramphenicol MIC of 30 μg/ml [Cm30]), its mutations are clean, thus allowing the introduction of plasmids encoding kanamycin or chloramphenicol resistance determinants. Importantly, E. coli lacks c-di-AMP-synthesizing enzymes. Thus, the coexpression of cdaA (encoding the diadenylate cyclase CdaA from L. monocytogenes [CdaALmo]) and kup genes allows the analysis of the phenotypic effect of c-di-AMP on Kup proteins. An inactive diadenylate cyclase, CdaALmo*(D171N) (38), served as a negative control.
Plasmids pIQ309 (L. lactis kupA) and pIQ310 (L. lactis kupB) were used for IPTG-dependent expression of the kup genes, whereas cdaAlmo and cdaAlmo* were expressed from plasmids pBP370 and pBP373, respectively. These plasmids are derivatives of pBAD33, allowing the expression of genes under the control of an arabinose-inducible promoter. Importantly, these plasmids are compatible with the pWH844 derivatives that were used to express the potassium transporters. A system of coexpression was then established where potassium transporters and c-di-AMP cyclases could be induced by IPTG and arabinose, respectively.
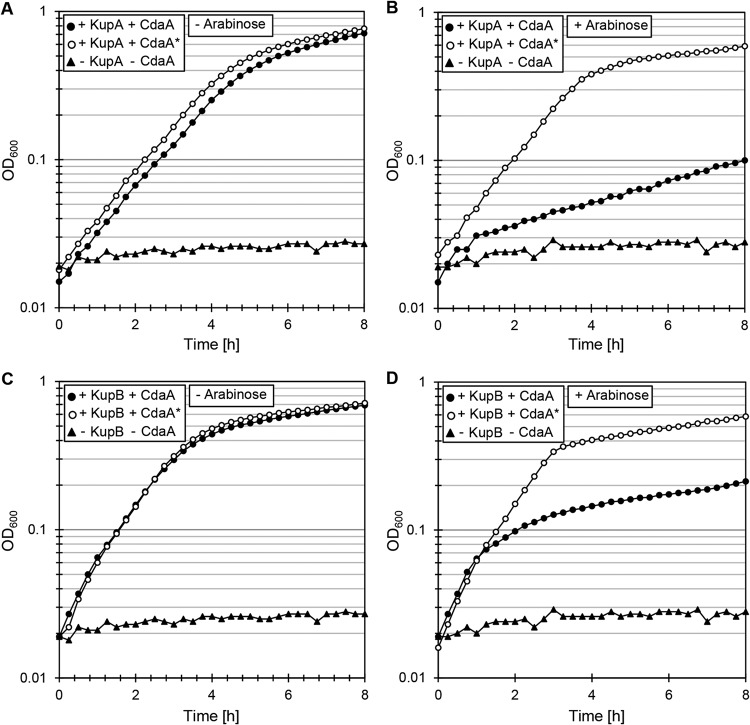
As described above, the addition of 0.1 mM KCl to M9mod medium was sufficient for growth of the bacteria if KupA or KupB was expressed. Therefore, this concentration was used to supplement the medium to assay growth of the E. coli LB2003 derivative strains. With this potassium concentration, E. coli LB2003 harboring the empty vector pWH844 and plasmid pBP370 (cdaAlmo) is not able to grow. This strain was thus used as the negative control. As shown in Fig. 4, expression of the kupA gene from pIQ309 allowed the growth of E. coli LB2003 in the presence of both the active CdaA and the inactive CdaA* proteins (Fig. 4A). However, production of c-di-AMP due to the expression of the wild-type cdaA gene (plasmid pBP370) resulted in severely impaired growth (Fig. 4B). In the absence of a functional diadenylate cyclase, and thus, of c-di-AMP (absence of inducer or expression of the inactive CdaA* variant from pBP373), growth was restored (Fig. 4B). The same experiment was performed to study the impact of c-di-AMP on KupB. In this case, the results were similar, but the growth inhibition exerted on the strain expressing KupB and the active CdaALmo (using plasmids pIQ310 and pBP370, respectively) was less pronounced (Fig. 4C and D).
FIG 4.
Inhibition of KupA and KupB transport activity by c-di-AMP. E. coli LB2003 carrying the relevant plasmids was cultivated in minimal salt M9mod medium, supplemented with 2.5 μM IPTG for kup gene induction and without arabinose (left) or with 0.005% l-arabinose (right) for induction of the cdaA alleles. (A and B) The strain carried the plasmid combinations pIQ309 (KupA)/pBP370 (CdaA), pIQ309 (KupA)/pBP373 (CdaA*), or pWH844/pBAD33 (empty vector control). (C and D) The strain carried the plasmid combinations pIQ310 (KupB)/pBP370 (CdaA), pIQ310 (KupB)/pBP373 (CdaA*), or pWH844/pBAD33 (empty vector control).
It is important to mention that the growth inhibition observed in both cases could simply result from high intracellular c-di-AMP concentrations, which have been reported to be toxic for bacteria (39). Alternatively, expression of the heterologous diadenylate cyclase CdaA might be toxic for the cell irrespective of the c-di-AMP concentration or the presence of the potassium transporters. These possibilities were tested by assaying the growth of E. coli LB2003 harboring the empty vector pWH844 and plasmid pBP370. As these bacteria were unable to grow at low potassium concentrations due to the lack of efficient transport systems, 50 mM KCl was added to the growth media. Wild-type growth was observed irrespective of the expression state of the active diadenylate cyclase CdaA (absence or presence of the inducer arabinose) (Fig. S2). As expected, the strain expressing the inactive CdaA* had a similar growth profile. Since the two strains exhibited similar growth phenotypes, as observed for the strain carrying the empty vectors pWH844 and pBAD33, the results presented here demonstrate that c-di-AMP inhibits the potassium transport activity of the KupA and KupB proteins.
DISCUSSION
In this study, we have identified two novel targets of c-di-AMP in L. lactis, the potassium transporters KupA and KupB. These proteins are the only known potassium transporters conserved in all strains of L. lactis. Interestingly, other potassium transport systems, such as the Kdp and Ktr systems, are present in other L. lactis strains. The Kup transporters are widespread in numerous species of Lactobacillus and Enterococcus, and they are present in a few species of Streptococcus and Staphylococcus as well. In contrast, no Kup potassium transporter is encoded in the genome of the more intensively studied Gram-positive bacteria, such as B. subtilis, L. monocytogenes, S. aureus, and S. pneumoniae.
In the L. lactis group, the kup genes are typically arranged in a two-copy kupA kupB operon, as in the case of L. lactis IL1403 presented here (Fig. 2A). However, some strains even possess three copies of the kup gene. Nonetheless, some strains have only one functional copy of a kup gene. This is the case in L. lactis MG1363 and its derivative NZ9000, the strain of choice for genetic engineering in L. lactis. In these bacteria, a spontaneous mutation in kupA results in a stop codon. This suggests that the presence of one single Kup protein is sufficient for potassium uptake and for satisfying the vital needs of the cell concerning cell turgor, proton motive force, and pH homeostasis.
In a recent study on the adaptation of L. lactis MG1363 to high levels of c-di-AMP, four independent mutations affecting KupB, the single Kup-type potassium transporter in this strain, were found. Each of these mutations resulted in increased activity of the KupB proteins and thus in increased potassium uptake (24). This observation fits very well with the general role of c-di-AMP in the control of potassium homeostasis. In B. subtilis, S. aureus, L. monocytogenes, and S. pneumoniae, c-di-AMP binds and thereby inhibits potassium transporters and interferes with their expression (see reference 25 for a review). Accumulation of c-di-AMP as in the case of the L. lactis gdpP mutant lacking the c-di-AMP-degrading phosphodiesterase (24) may therefore result in permanent inhibition of the potassium transporters. Two observations demonstrate that this exactly is the case, as follows: first, the selection of KupB variants with higher transport activity upon accumulation of c-di-AMP suggests that potassium uptake is a major growth-limiting factor for the gdpP mutant that is unable to degrade the c-di-AMP (24). Second, the results presented in this study clearly demonstrate that c-di-AMP indeed binds and inhibits KupB as well as the paralogous KupA protein. KupA is not active in L. lactis MG1363, which was used in the previous study (24).
In addition to potassium transporters containing an RCK_C domain and the KdpD sensor kinase (8, 9, 28), the Kup proteins are the third class of proteins involved in the control of potassium homeostasis that bind and are inhibited by c-di-AMP. Taking into account the c-di-AMP binding proteins containing a CBS domain, the PII-like proteins, and pyruvate carboxylase, six classes of proteins or domains bind this second messenger. Moreover, c-di-AMP binds the RNA molecule that serves to control the expression of high-affinity potassium transporters in B. subtilis and of a cell wall hydrolase in Streptomyces coelicolor (6, 7, 40). Thus, c-di-AMP is able to interact with a variety of biological macromolecules. The evolutionary convergence of c-di-AMP-mediated regulation of three classes of proteins and of a riboswitch in the control of potassium homeostasis is particularly striking.
This work strongly supports the idea that the control of potassium uptake is a key function of c-di-AMP in the Firmicutes. However, our picture is not yet complete. Prior to this study, only potassium transporters carrying a RCK_C domain were known to bind c-di-AMP. The discovery that the unrelated KupA and KupB proteins also bind this second messenger suggests that the additional classes of potassium transporters in the Firmicutes, i.e., KimA and KdpABC, might also be controlled by c-di-AMP. If the activity of these proteins could not be controlled in response to potassium availability for which c-di-AMP is an indicator (6, 24), the potassium uptake could not be rapidly switched off if cells encounter a sudden increase in the potassium concentration. Since high intracellular potassium concentrations are toxic for the bacteria (6, 41), control of KimA and the Kdp system at the protein activity level in addition to the established c-di-AMP-mediated regulation of their expression seems to be necessary for the rapid adaptation of the cells to changing potassium concentrations. So far, little is known about whether c-di-AMP controls the export of potassium in addition to uptake. In S. aureus, c-di-AMP binds to the CpaA potassium exporter (8), and the activity of this protein is stimulated by c-di-AMP. The binding pocket for the second messenger is strongly conserved in the corresponding proteins from other Firmicutes, including B. subtilis or Lactobacillus casei (42), suggesting that the control of potassium export by c-di-AMP is also a more general phenomenon.
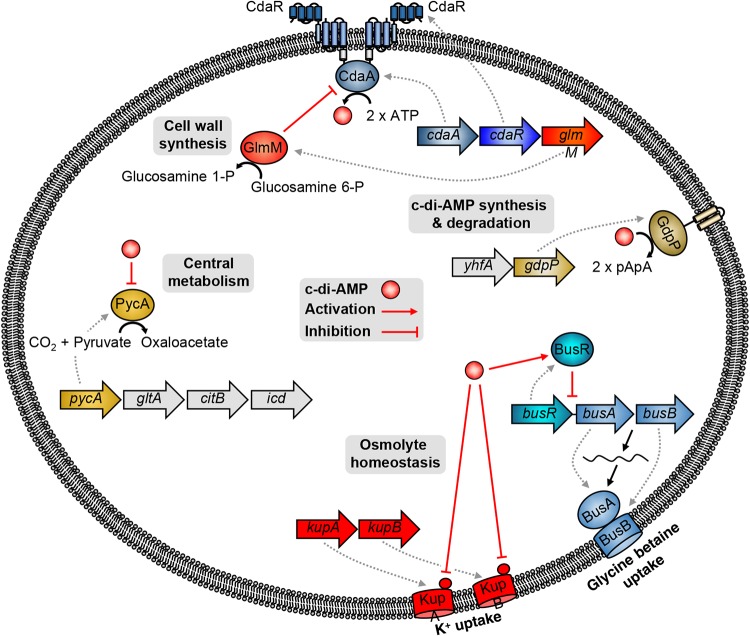
An updated view of c-di-AMP metabolism in L. lactis IL1403 is presented in Fig. 5. Even though it is well known that the synthesis and degradation of this second messenger occur through specific cyclases and phosphodiesterases (CdaA and GdpP, respectively), the potential regulation mechanism of CdaA by CdaR is still not exactly known. It has been suggested that CdaR might be a negative regulator of CdaA activity (43). The first confirmed regulator of c-di-AMP levels in L. lactis IL1403 is GlmM, which is encoded by the highly conserved cdaA-cdaR-glmM operon and acts as a negative modulator of CdaA (22). Moreover, the c-di-AMP levels in L. lactis are also controlled by the potassium concentration in a way that is not yet understood (24). In this way, c-di-AMP metabolism is connected to both potassium and cell wall homeostasis in L. lactis. Moreover, c-di-AMP was recently shown to be involved in central metabolism in L. lactis IL1403 due to the negative regulation it exerts on the pyruvate carboxylase, which is responsible for replenishing the cellular oxaloacetate pool for amino acid biosynthesis. The discovery that c-di-AMP controls the uptake of compatible solutes as well as the uptake of potassium (reference 25 and this work) puts c-di-AMP in the broader context of osmoadaptation also for L. lactis. Since other potassium transporters are present in different strains of L. lactis, it will be interesting to test whether all these systems are also subject to control by c-di-AMP, as KupA and KupB were shown to be.
FIG 5.
Schematic representation of c-di-AMP signaling in L. lactis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 2. L. lactis IL1403 was grown at 30°C in M17 medium (Oxoid) supplemented with 0.5% glucose (M17G). All E. coli strains were grown at 37°C in LB medium with continuous shaking at 200 rpm. L. monocytogenes EGD-e was grown at 37°C in brain heart infusion (BHI) broth (Sigma-Aldrich) with shaking at 200 rpm. In all cases, stocks of the strains at −80°C were propagated twice in overnight liquid cultures, which were in turn used to inoculate fresh media for the experiment of interest. Suitable antibiotics were added to the media. Experiments requiring minimal salts medium were performed in M9mod, in which M9 potassium salts were replaced with equimolar quantities of sodium salts. Thus, M9mod contains 40 mM Na2HPO4, 22 mM NaH2PO4, 18.5 mM NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 0.5 μM FeCl3, 350 μM proline, 3 μM thiamine-di-chloride, 0.66% Casamino Acids, and 0.5% glucose or 0.2% glycerol depending on the experiment (see below).
DNA manipulation.
Transformation of E. coli was performed using standard procedures (44). Plasmid DNA was extracted using the NucleoSpin extraction kit (Macherey and Nagel, Düren, Germany). Chromosomal DNA of L. monocytogenes and L. lactis was isolated using the NucleoSpin microbial DNA kit (Macherey and Nagel). Commercially available restriction enzymes, T4 DNA ligase, and DNA polymerases were used as recommended by the manufacturers. DNA fragments were purified using the PCR purification kit (Qiagen, Hilden, Germany). DNA sequences were determined by the dideoxy chain termination method (SeqLab, Göttingen, Germany).
Construction of plasmids.
For the construction of pBP370 (pBAD33 cdaAlmo), the cdaA gene of L. monocytogenes (lmo2120) was amplified from L. monocytogenes EGD-e chromosomal DNA using oligonucleotides JH51/JH52 and cloned between the XbaI/PstI sites of pBAD33 to allow l-arabinose-inducible expression (45). pBP373 (pBAD33 cdaAlmo D171N) was constructed using the same oligonucleotides for amplification, with the addition of the 5′-phosphorylated oligonucleotide JR18, for the introduction of the D171N amino acid exchange into cdaA using the combined chain reaction (46), rendering it enzymatically inactive (38). For the expression of the putative potassium transporter KtrCD from L. monocytogenes, plasmid pBP371 (pWH844-ktrCDlmo) was constructed as follows: the homologs of ktrC (lmo1023) and ktrD (lmo0993) were amplified using oligonucleotides JH59/JH60 and JH61/JH62, respectively. Oligonucleotides JH60 and JH61 share a complementary region, allowing the use of the resulting PCR products as templates in another PCR using oligonucleotides JH59/JH62. ktrC and ktrD are thereby fused together by gene splicing by overlap extension (SOE) (47) to create an artificial operon of ktrC and ktrD with their native ribosomal binding sites. The resulting construct was cloned between the EcoRI/BamHI sites of pWH844 to allow IPTG-inducible expression (48). Expression of the KupA and KupB proteins in E. coli was approached as follows: for the construction of plasmid pIQ309, the kupA gene from L. lactis IL1403 was amplified using oligonucleotides IQ682 (containing a ribosomal binding site [RBS]) and JN561. The PCR product was cloned between the EcoRI and SalI sites of plasmid pWH844, which allows elimination of the His tag. A similar protocol was used for plasmid pIQ310, harboring L. lactis IL1403 kupB gene, using primers IQ683 and JN563. Genes coding for putative c-di-AMP interaction partners were amplified from L. lactis IL1403 chromosomal DNA, using the series of primers JN554 to JN569. PCR products were then cloned into the expression vector pWH844.
Growth curves in minimal salt medium.
E. coli LB650-derived strains were propagated twice from −80°C stocks in M9mod medium supplemented with 50 mM KCl, 0.5% glucose, and the corresponding antibiotics. An overnight culture was used to inoculate fresh M9mod medium supplemented with 10 μM IPTG and 0.1 mM KCl for strains expressing the KupA and KupB proteins, 50 mM KCl for the strain expressing B. subtilis KtrAB, and 100 mM KCl for the strain expressing L. monocytogenes KtrCD and the empty vector control. When the OD600 reached 0.5, cultures were harvested and incubated for 1 h in the initial volume of fresh medium without KCl addition. After this, cells were harvested and washed three times with fresh medium with no KCl supplementation. These samples were then used to inoculate fresh M9mod medium in microplates supplemented with 1% glucose, 10 μM IPTG, and different KCl concentrations. Antibiotics were added as appropriate. Microplates were incubated at 37°C with continuous orbital shaking, and OD600 measurements were taken every 15 min.
Coexpression of Kup transporters and diadenylate cyclases.
For cultivation, 10 ml of fresh medium was inoculated at an OD600 of 0.05 from overnight cultures prepared as described above, with 0.2% glycerol as the carbon source. When strains reached an OD600 of 0.5, cultures were harvested and resuspended in same volume of M9mod medium containing 0.2% glycerol (no KCl added). Samples were incubated at 37°C for 1 h, after which two wash steps were performed. These washed samples were used for microplate inoculation supplemented with 0.1 mM KCl, 2.5 μM IPTG, and 0.005% l-arabinose when indicated. Microplates were incubated at 37°C with continuous orbital shaking, and OD600 measurements were taken every 15 min.
DRaCALA.
The selected genes were cloned into the expression vector pWH844 as described above (see Table S1). Expression of the genes upon induction using 0.1 mM IPTG was verified by analyzing the protein patterns of the expression strains by SDS-PAGE. 32P-labeled c-di-AMP synthesis was performed using purified diadenylate cyclase DisA (8). The analysis of protein-ligand interaction was performed using E. coli whole-cell lysates that were grown to an OD600 of 0.5 to 1.0 and induced for 4 h by 1 mM IPTG, as described previously (49, 50), or grown overnight in the presence of 50 μg/ml carbenicillin and 1 mM IPTG. All binding reactions were performed in 1× binding buffer (10 mM Tris [pH 8], 100 mM NaCl, 5 mM MgCl2) containing ∼10 pM [32P]c-di-AMP. Protein-ligand mixtures were spotted on nitrocellulose membrane (Amersham Hybond-ECL; GE Healthcare) and allowed to dry. The areas and intensities of the spots were quantified by exposing phosphorimager screens and scanning by a Fuji FLA-7000 phosphorimager. The competition assays were performed with 100 μM unlabeled ATP and the absence or presence of 100 μM unlabeled c-di-AMP (Axxora).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Zuzanna Grubek for help with plasmid construction. Christina Herzberg and Jan Gundlach are acknowledged for helpful discussions. We thank Agencia Nacional de Promoción Científica y Tecnológica (ANPyCT, PICT 2014-1513,), CONICET (PIP numbers 0718 and 11220150100855CO), and Sanofi-CONICET for financial support.
This work was supported by grants from the Deutsche Forschungsgemeinschaft via Priority Program SPP 1879 (to J.S. and F.M.C.) and from the National Institutes of Health NIH; grant AI133670 to V.T.L.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00028-19.
REFERENCES
- 1.Epstein W. 2003. The roles and regulation of potassium in bacteria. Prog Nucleic Acids Res Mol Biol 75:293–320. doi: 10.1016/S0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 2.Trchounian A, Kobayashi H. 2000. K+ uptake by fermenting Escherichia coli cells: pH dependent mode of the TrkA system operating. Biosci Rep 20:277–288. doi: 10.1023/A:1026493024066. [DOI] [PubMed] [Google Scholar]
- 3.Ballal A, Basu B, Apte SK. 2007. The Kdp-ATPase system and its regulation. J Biosci 32:559–568. doi: 10.1007/s12038-007-0055-7. [DOI] [PubMed] [Google Scholar]
- 4.Trchounian A, Kobayashi H. 1999. Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Lett 447:144–148. doi: 10.1016/S0014-5793(99)00288-4. [DOI] [PubMed] [Google Scholar]
- 5.Holtmann G, Bakker EP, Uozumi N, Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol 185:1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundlach J, Herzberg C, Kaever V, Gunka K, Hoffmann T, Weiß M, Gibhardt J, Thürmer A, Hertel D, Daniel R, Bremer E, Commichau FM, Stülke J. 2017. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci Signal 10:eaal3011. doi: 10.1126/scisignal.aal3011. [DOI] [PubMed] [Google Scholar]
- 7.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. 2013. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moscoso JA, Schramke H, Zhang Y, Tosi T, Dehbi A, Jung K, Gründling A. 2016. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. J Bacteriol 198:98–110. doi: 10.1128/JB.00480-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witte G, Hartung S, Büttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell 30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Corrigan RM, Gründling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 12.Pham TH, Liang ZX, Marcellin E, Turner MS. 2016. Replenishing the cyclic di-AMP pool: regulation of diadenylate cyclase activity in bacteria. Curr Genet 62:731–738. doi: 10.1007/s00294-016-0600-8. [DOI] [PubMed] [Google Scholar]
- 13.Commichau FM, Heidemann JL, Ficner R, Stülke J. 2019. Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J Bacteriol 201:e00462-18. doi: 10.1128/JB.00462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gundlach J, Mehne FM, Herzberg C, Kampf J, Valerius O, Kaever V, Stülke J. 2015. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol 197:3265–3274. doi: 10.1128/JB.00564-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh TN, Woodward JJ. 2016. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol 30:22–29. doi: 10.1016/j.mib.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteley AT, Pollock AJ, Portnoy DA. 2015. The PAMP c-di-AMP is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 17:788–798. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeden MS, Schuster CF, Bowman L, Zhong Q, Williams HD, Gründling A. 2018. Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J Biol Chem 293:3180–3200. doi: 10.1074/jbc.M117.818716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaux L, Sleiman D, Mazzuoli M-V, Gominet M, Lanotte P, Trieu-Cuot P, Kaminski P-A, Firon A. 2018. Cyclic di-AMP regulation of osmotic homeostasis is essential in group B Streptococcus. PLoS Genet 14:e1007342. doi: 10.1371/journal.pgen.1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kongo JM, Ho AJ, Malcata FX, Wiedmann M. 2007. Characterization of dominant lactic acid bacteria isolated from São Jorge cheese, using biochemical and ribotyping methods. J Appl Microbiol 103:1838–1844. doi: 10.1111/j.1365-2672.2007.03423.x. [DOI] [PubMed] [Google Scholar]
- 20.Yagnik B, Sharma D, Padh H, Desai P. 2017. Dual recombinant Lactococcus lactis for enhanced delivery of DNA vaccine reporter plasmid pPERDBY. Microbiol Immunol 61:123–129. doi: 10.1111/1348-0421.12473. [DOI] [PubMed] [Google Scholar]
- 21.Zeng Z, Yu R, Zuo F, Zhang B, Ma H, Chen S. 2017. Recombinant Lactococcus lactis expressing bioactive exendin-4 to promote insulin secretion and beta-cell proliferation in vitro. Appl Microbiol Biotechnol 101:7177–7186. doi: 10.1007/s00253-017-8410-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Pham TH, Nhiep TH, Vu NM, Marcellin E, Chakrabortti A, Wang Y, Waanders J, Lo R, Huston WM, Bansal N, Nielsen LK, Liang ZX, Turner MS. 2016. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol Microbiol 99:1015–1027. doi: 10.1111/mmi.13281. [DOI] [PubMed] [Google Scholar]
- 23.Smith WM, Pham TH, Lei L, Dou J, Soomro AH, Beatson SA, Dykes GA, Turner MS. 2012. Heat resistance and salt hypersensitivity in Lactococcus lactis due to spontaneous mutation of llmg_1816 (gdpP) induced by high-temperature growth. Appl Environ Microbiol 78:7753–7759. doi: 10.1128/AEM.02316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham HT, Nhiep NTH, Vu TNM, Huynh TAnh. N, Zhu Y, Huynh ALD, Chakrabortti A, Marcellin E, Lo R, Howard CB, Bansal N, Woodward JJ, Liang Z-X, Turner MS. 2018. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet 14:e1007574. doi: 10.1371/journal.pgen.1007574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Commichau FM, Gibhardt J, Halbedel S, Gundlach J, Stülke J. 2018. A delicate connection: c-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol 26:175–185. doi: 10.1016/j.tim.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Choi PH, Vu TMN, Pham HT, Woodward JJ, Turner MS, Tong L. 2017. Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria. Proc Natl Acad Sci U S A 114:E7226–E7235. doi: 10.1073/pnas.1704756114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabov A. 2007. Plant KT/KUP/HAK potassium transporters: single family—multiple functions. Ann Bot 99:1035–1041. doi: 10.1093/aob/mcm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. 2014. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol 196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Commichau FM, Dickmanns A, Gundlach J, Ficner R, Stülke J. 2015. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol Microbiol 97:189–204. doi: 10.1111/mmi.13026. [DOI] [PubMed] [Google Scholar]
- 30.Gundlach J, Commichau FM, Stülke J. 2018. Perspective of ions and messengers: an intricate link between potassium, glutamate, and cyclic di-AMP. Curr Genet 64:191–195. doi: 10.1007/s00294-017-0734-3. [DOI] [PubMed] [Google Scholar]
- 31.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ. 2014. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gundlach J, Dickmanns A, Schröder-Tittmann K, Neumann P, Kaesler J, Kampf J, Herzberg C, Hammer E, Schwede F, Kaever V, Tittmann K, Stülke J, Ficner R. 2015. Identification, characterization and structure analysis of the c-di-AMP binding PII-like signal transduction protein DarA. J Biol Chem 290:3069–3080. doi: 10.1074/jbc.M114.619619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato Y, Nanatani K, Hamamoto S, Shimizu M, Takahashi M, Tabuchi-Kobayashi M, Mizutani A, Schroeder JI, Souma S, Uozumi N. 2014. Defining membrane spanning domains and crucial membrane-localized acidic amino acid residues for K(+) transport of a Kup/HAK/KT-type Escherichia coli potassium transporter. J Biochem 155:315–323. doi: 10.1093/jb/mvu007. [DOI] [PubMed] [Google Scholar]
- 35.Schlösser A, Meldorf M, Stumpe S, Bakker EP, Epstein W. 1995. TrkH and its homolog, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J Bacteriol 177:1908–1910. doi: 10.1128/jb.177.7.1908-1910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 37.Stumpe S, Bakker EP. 1997. Requirement of a large K+-uptake capacity and of extracytoplasmic protease activity for protamine resistance of Escherichia coli. Arch Microbiol 167:126–136. doi: 10.1007/s002030050425. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg J, Dickmanns A, Neumann P, Gunka K, Arens J, Kaever V, Stülke J, Ficner R, Commichau FM. 2015. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J Biol Chem 290:6596–6606. doi: 10.1074/jbc.M114.630418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. 2013. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St-Onge RJ, Haiser HJ, Yousef MR, Sherwood E, Tschowri N, Al-Bassam M, Elliot MA. 2015. Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol Microbiol 96:779–795. doi: 10.1111/mmi.12971. [DOI] [PubMed] [Google Scholar]
- 41.Radchenko MV, Tanaka K, Waditee R, Oshimi S, Matsuzaki Y, Fukuhara M, Kobayashi H, Takabe T, Nakamura T. 2006. Potassium/proton antiport system of Escherichia coli. J Biol Chem 281:19822–19829. doi: 10.1074/jbc.M600333200. [DOI] [PubMed] [Google Scholar]
- 42.Chin KH, Liang JM, Yang JG, Shih MS, Tu ZL, Wang YC, Sun XH, Hu NJ, Liang ZX, Dow JM, Ryan RP, Chou SH. 2015. Structural insights into the distinct binding mode of cyclic di-AMP with SaCpaA_RCK. Biochemistry 54:4936–4951. doi: 10.1021/acs.biochem.5b00633. [DOI] [PubMed] [Google Scholar]
- 43.Rismondo J, Gibhardt J, Rosenberg J, Kaever V, Halbedel S, Commichau FM. 2016. Phenotypes associated with the essential diadenylate cyclase CdaA and its potential regulator CdaR in the human pathogen Listeria monocytogenes. J Bacteriol 198:416–426. doi: 10.1128/JB.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 45.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi W, Stambrook PJ. 1997. CCR: a rapid and simple approach for mutation detection. Nucleic Acids Res 25:2949–2951. doi: 10.1093/nar/25.14.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horton RMCZ, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 48.Schirmer F, Ehrt S, Hillen W. 1997. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol 179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roelofs KG, Wang J, Sintim HO, Lee VT. 2011. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A 108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orr MW, Lee VT. 2017. Differential radial capillary action of ligand assay (DRaCALA) for high-throughput detection of protein-metabolite interactions in bacteria. Methods Mol Biol 1535:25–41. doi: 10.1007/978-1-4939-6673-8_3. [DOI] [PubMed] [Google Scholar]
- 51.Chopin A, Chopin MC, Moillo-Batt A, Langella P. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263. doi: 10.1016/0147-619X(84)90033-7. [DOI] [PubMed] [Google Scholar]
- 52.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 53.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.