Abstract
Senescent cells (SnCs) are increasingly recognized as central effector cells in age-related pathologies. Extracellular vesicles (EVs) are potential cellular communication tools through which SnCs exert central effector functions in the local tissue environment. To test this hypothesis in a medical indication that could be validated clinically, we evaluated EV production from SnCs enriched from chondrocytes isolated from human arthritic cartilage. EV production increased in a dose-responsive manner as the concentration of SnCs increased. The EVs were capable of transferring senescence to nonsenescent chondrocytes and inhibited cartilage formation by non-SnCs. microRNA (miR) profiles of EVs isolated from human arthritic synovial fluid did not fully overlap with the senescent chondrocyte EV profiles. The effect of SnC clearance was tested in a murine model of posttraumatic osteoarthritis. miR and protein profiles changed after senolytic treatment but varied depending on age. In young animals, senolytic treatment altered expression of miR-34a, -30c, -125a, -24, -92a, -150, and -186, and this expression correlated with cartilage production. The primary changes in EV contents in aged mice after senolytic treatment, which only reduced pain and degeneration, were immune related. In sum, EV contents found in synovial fluid may serve as a diagnostic for arthritic disease and indicator for therapeutic efficacy of senolytic treatment.
Keywords: Aging, Therapeutics
Keywords: Cartilage, Osteoarthritis
Content of extracellular vesicles can serve as biomarkers for human osteoarthritis disease and predicts efficacy of senolytic therapeutics.
Introduction
Osteoarthritis (OA) is an age-related and posttraumatic degenerative joint disease that is accompanied by cartilage degradation, persistent pain, and impairment of mobility (1). Senescent cells (SnCs) are a newly implicated factor in the development of OA (2–6). Cellular senescence is characterized by a proliferation arrest, which protects against cancer, as well as other changes that can also contribute to aging phenotypes and pathologies (7, 8). SnCs accumulate with age in many tissues, including articular cartilage, where they promote pathological age-related deterioration. These and other tissue pathologies are presumably mediated by the secretion of extracellular proteases, proinflammatory cytokines, chemokines, and growth factors, termed the senescence-associated secretory phenotype (SASP), by SnCs (9, 10). The local elimination of SnCs in a murine model of posttraumatic OA (PTOA) reduced pain and increased cartilage development (4). Bridging these results to human cells, the selective removal of senescent chondrocytes improved the cartilage-forming ability of chondrocytes isolated from human arthritic tissue. Recent findings suggest that SnCs can transmit limited senescent phenotypes to nearby cells, termed secondary or paracrine senescence (11, 12). Understanding the mechanisms of this SnC transmission may inform mechanisms of OA disease causation.
Extracellular vesicles (EVs), including exosomes and microvesicles, are small membrane-limited particles (30 nm to 1 μm) that can participate in intercellular communication (13). EVs mediate local tissue development and homeostasis through the transfer of cargoes, such as proteins and microRNAs (miRs). For example, the EVs present in articular cartilage and synovial fluid can contribute to mineralization of the cartilage extracellular matrix (ECM) and formation of an inflammatory joint environment (14–16). Recently, it was reported that SnCs secrete more EVs compared with their nonsenescent counterparts (17, 18). These senescent-associated EVs may also induce senescence in neighboring cells (19). In the case of arthritis, SnCs can modulate the environment of the articular joint, increasing inflammation and ECM degradation. It is not known whether EVs secreted by SnCs in the articular joint are responsible for the progression of OA or whether they can be use as indicators of disease progression and treatment efficacy.
In this study, we found that senescent chondrocytes isolated from OA patients secrete more EVs compared with nonsenescent chondrocytes. These EVs inhibit cartilage ECM deposition by healthy chondrocytes and can induce a senescent state in nearby cells. We profiled the miR and protein content of EVs isolated from the synovial fluid of OA joints from mice with SnCs. After treatment with a molecule to remove SnCs, termed a senolytic, the composition of EV-associated miR and protein was markedly altered. The senolytic reduced OA development and enhanced chondrogenesis, and these were attributable to several specific differentially expressed miRs (miR-30c, miR-92a, miR-34a, miR-24, miR-125a, miR-150, miR-186, and miR-223) and proteins (Serpina and aggrecan). In aged animals, treatment with senolytic modulated the inflammatory response by decreasing recruitment and activation of myeloid and phagocytic cells. Collectively, these findings suggest that altered levels of synovial EV miRs and proteins are a potential mechanism by which SnCs can transfer senescence, inhibit tissue formation, and promote OA development. When isolated from synovial fluid, EVs may also be used to predict therapeutic response to senolytic therapies in the articular joint.
Results
SnCs from OA patients impair cartilage ECM production by neighboring chondrocytes through secreted factors.
To understand how senescent human chondrocytes might effect surrounding nonsenescent chondrocytes, we employed a coculture system in which cells are not in physical contact but are exposed to secreted factors. Since there are no known specific surface markers for SnCs, we sorted chondrocytes based on size (20) to isolate populations enriched in SnCs. Three relative concentrations (high, medium, low) of human senescent chondrocytes were enriched and validated by the presence of senescence-associated β-galactosidase (SA-β-Gal) activity, a marker of SnCs (21) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125019DS1).
To characterize the secretory phenotypes of nonsenescent and senescent chondrocytes isolated from arthritic human cartilages, we measured the levels of 36 secreted proteins using antibody arrays (Supplemental Figure 1B). Many proteins secreted by the high-senescent cultures were previously reported SASP factors (22, 23). These factors included growth-regulated oncogene α (GROA), IL-1A, IL-1B, and IL-6, intercellular adhesion molecule 1 (ICAM1), chemokine (C–C motif) ligand 5 (CCL5), and macrophage migration inhibitory factor (MIF). Notably, nonsenescent chondrocytes cocultured with the high-senescent chondrocyte group significantly increased expression levels of mRNAs encoding p16INK4a (also known as cyclin-dependent kinase inhibitor 2a [CDKN2A], which is a widely used SnC biomarker) (7, 8) and the SASP factor MMP3. Coculturing nonsenescent chondrocytes with SnCs also decreased chondrogenesis, as defined by Safranin-O staining for proteoglycans and type II collagen (COL2A1) mRNA levels (Supplemental Figure 1, C and D).
Similar to that in the coculture experiments, nonsenescent chondrocytes cultured in SnC-conditioned medium for 7 days also reduced proteoglycan production, as confirmed by Safranin-O and Alcian blue staining. They also produced a dose-dependent increase in SA-β-Gal activity (Supplemental Figure 1, E and F). These findings indicate that SnCs from arthritic human cartilage impair the function of, and induce bystander senescence in, nonsenescent chondrocytes, both most likely through SASP factors.
EVs secreted by senescent human chondrocytes induce bystander senescence.
Since SnCs were found to secrete EVs (17, 18), we asked whether EVs secreted by senescent human chondrocytes mediated their effects on non-SnCs. We isolated vesicles with a characteristic EV diameter of ~100 nm by differential centrifugation (Supplemental Figure 2A), as visualized by electron microscopy (Figure 1A). Over 95% of the vesicles recovered from cultures with increasing amounts of senescent chondrocytes were <100 nm in diameter. Quantitative nanoparticle tracking analysis (NTA) showed that populations enriched with senescent chondrocytes released significantly more EVs than nonsenescent chondrocyte populations (Figure 1B).
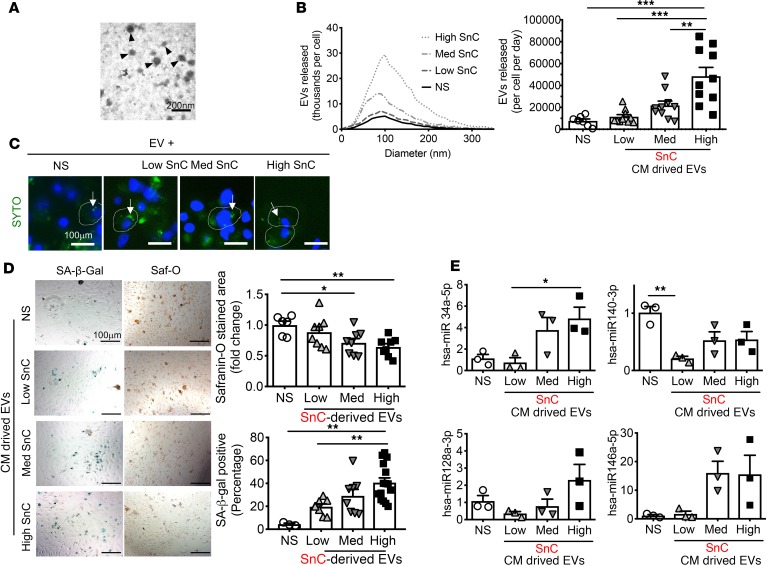
Figure 1. SnC-derived EVs inhibit cartilage ECM deposition of nonsenescent chondrocytes by transmitting cellular senescence.
(A) Representative electron micrographs of nonsenescent chondrocyte-derived EVs. Scale bar: 200 nm. (B) Size distribution and concentration of EVs from nonsenescent primary chondrocytes (NS) and low, medium, or high levels of senescent OA chondrocytes, sorted by flow cytometry, determined by nanoparticle tracking analysis. (C) Uptake by nonsenescent chondrocytes of EVs labeled with SYTO RNASelect (arrows; dye-stained EVs). Scale bar: 100 μm. (D) Representative images of SA-β-Gal and Safranin-O staining of nonsenescent chondrocytes 6 days after incubation with EVs (8 × 108) derived from conditioned medium (CM) from high, medium, or low SnC populations to measure the induction of senescence and proteoglycan levels (left). Scale bar: 100 μm. Quantification of the percentage of SA-β-Gal–positive cells (n = 3 for NS; n = 8 for low and medium; n = 15 for high) and Safranin-O–stained areas (n = 6 for NS; n = 8 for low, medium, and high) (right). (E) The expression of selected miRs (hsa-miR-140-3p, -34a-5p, -128a-3p, and -146a-5p) in EVs collected from CM of senescent and nonsenescent chondrocytes, detected by RT-qPCR (n = 3 per group). All data are expressed as mean ± SEM. Statistics in B, D, and E were performed using 1-way ANOVA and Tukey’s multiple-comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001.
We hypothesized that EVs from SnCs might be a mechanism for the transmission of senescence characteristics to non-SnCs. To test this idea, we exposed EVs isolated from senescent human chondrocytes to nonsenescent chondrocytes. The OA-derived EVs induced a senescence-like phenotype in nonsenescent chondrocytes, as determined by SA-β-Gal activity. They also reduced proteoglycan production, as measured by Safranin-O staining (Supplemental Figure 2, B and C).
To track EVs and their uptake by the normal cells, we labeled EVs isolated from low, medium, and high-senescent OA chondrocyte populations using the green RNA-selective nucleic acid stain SYTO RNASelect. We then cultured the labeled EVs for 6 hours with nonsenescent chondrocytes and confirmed EV internalization (Figure 1C). We then cultured nonsenescent chondrocytes with enriched EVs secreted by the low, medium, or high SnC populations or control EVs from nonsenescent chondrocytes for 6 days. Nonsenescent chondrocytes exposed to enriched EVs secreted by SnCs developed more senescence and reduced proteoglycan production in a dose-responsive manner (Figure 1D). These findings suggest that senescent chondrocyte EVs can induce bystander senescence, spreading senescent characteristics to neighboring cells, and, moreover, contribute to the decline in chondrocyte matrix production.
To identify EV components that may mediate the bystander effect and reduced cartilage production, we investigated extracellular miRs associated with senescence and OA. EVs from highly enriched senescent chondrocyte populations contained less human miR-140-3p (hsa-miR-140-3p) and more hsa-miR-34a-5p compared with nonsenescent chondrocyte EVs (Figure 1E). Changes in miR-140 are known to be associated with chondrocyte dysfunction and OA development (24). Low miR-140 expression impairs cartilage homeostasis, and miR-140 is regulated by SOX9, a cartilage master regulator transcription factor that promotes cartilage development and ECM production during development and repair (24, 25). miR-34a and miR-128a are associated with cellular senescence and target the CDKN1A and CDKN2A pathways, respectively (26). These data suggest that hsa-miR-34a and -140 in senescent chondrocyte-derived EVs contribute to human OA development and cartilage loss.
SnC removal decreases the secretion of EVs from human OA chondrocytes.
We previously demonstrated the ability of a senolytic small molecule (UBX0101) to clear SnCs in cultures of human OA chondrocytes. We therefore evaluated EV production after treating senescent OA chondrocytes with this molecule. Exposure to UBX0101 (43 μM) did not change EV size (Figure 2A). The mean size of EVs secreted by human OA chondrocytes 1, 2, 4, and 6 days after incubation with vehicle or UBX0101, measured by NTA, was similar (Figure 2B). However, treatment with the senolytic for 2 days significantly reduced the number of secreted EVs at all time points evaluated (Figure 2C).
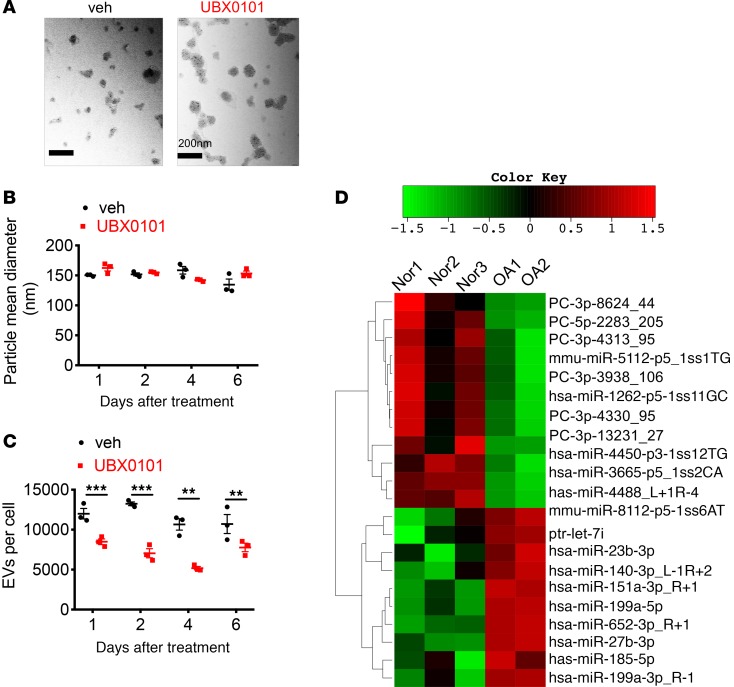
Figure 2. Decreased secretion of EVs from human OA chondrocytes after removing SnCs and alteration in miRs carried by synovial EVs from OA patients.
(A) Representative electron micrographs of EVs derived from OA chondrocytes treated with vehicle (veh) and UBX0101. Scale bar: 200 nm. (B and C) Mean size and concentration of EV enrichments released per cell in human OA chondrocytes at 1, 2, 4, and 6 days after incubation with veh or 43 μM UBX0101, measured by nanoparticle tracking analysis (n = 3 per data point). The experiment was performed 2 independent times. Data are shown as mean ± SEM. Statistical analysis was performed using 2-tailed t tests (unpaired). **P < 0.01, ***P < 0.001. (D) Heatmap and hierarchical clustering depicting statistically significant (P < 0.1 by t test) differentially expressed miRs. Synovial fluid was obtained from normal (Nor) individuals (age 77.3 ± 6.8 years; n = 3) and OA patients (age 64.5 ± 2.1 years; n = 2). PC, predicted candidate.
EVs from synovial fluid of aged normal and arthritic articular joints have different miR expression patterns.
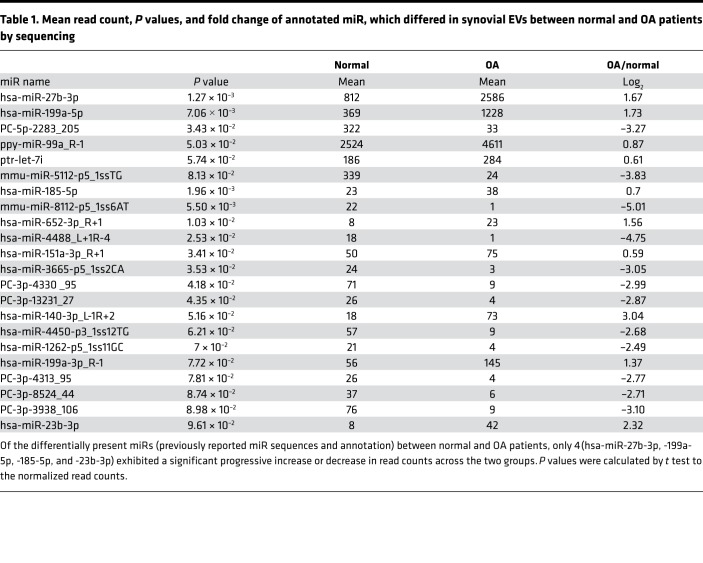
To determine whether EVs were also present in human disease, we isolated and evaluated EVs from synovial fluid from normal and OA patients at relatively advanced ages (70–80 years), when both groups are likely to harbor SnCs from aging and potentially previous trauma. The OA donors had clinical evidence of OA based on pain that led to total joint arthroplasty. EV size and concentration in synovial fluid from both OA and normal donors were similar. Sequencing identified a large number of known and potentially novel miRs that were differentially present in EVs isolated from OA patients compared with healthy controls (Supplemental Figure 3, A and B, and Supplemental Table 1). Twenty-two miRs were significantly upregulated or downregulated in OA synovial EV enrichments (Figure 2D and Table 1). Of the differentially expressed miRs, only 4 known miRs (hsa-miR-27b-3p, miR-199a-5p, miR-185-5p, and miR-23b-3p) exhibited changes in read counts when synovial EVs from OA patients were compared with those of normal patients. The predicted target genes for these differentially expressed EV-associated miRs and their biological function are shown in Supplemental Figure 3, C and D. It has been previously reported that pathways for mucin-type O-glycan biosynthesis and proteoglycans in cancer play important roles in the pathogenesis of OA (27). These data suggest that synovial fluid EV–derived miRs are significantly altered in OA compared with age-relate degeneration and can be used as potential biomarkers for OA diagnosis. Diseased human chondrocyte EVs changed with senolytic treatment that was previously demonstrated to increase cartilage production. The EVs isolated from synovial fluid were different than the cell-derived EVs. The content of the EVs produced by chondrocytes did not appear to overlap with EVs found in the synovial fluid, suggesting that EV production from cells may change with culture or EVs are produced by multiple different cell types in the articular joint, such as cells in the synovium.
Table 1. Mean read count, P values, and fold change of annotated miR, which differed in synovial EVs between normal and OA patients by sequencing.
Specific EV-associated miRs in synovial fluid correlate with response to senolytic therapy in a murine posttraumatic OA model.
We previously demonstrated that SnCs develop after PTOA created by anterior cruciate ligament transection (ACLT) and that their selective elimination reduced inflammation and pain in young and aged mice (4). Furthermore, in young mice, new cartilage formed on the articular surface after ACLT and senolytic treatment (Supplemental Figure 4A). We hypothesized that differences in the content of EV-associated miR and protein in joint fluids might correlate with OA and varying response to senolytics.
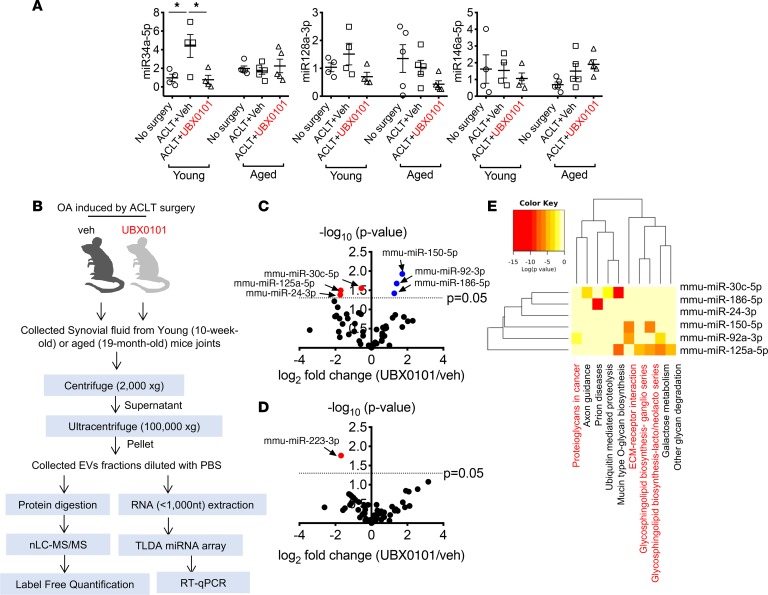
To determine whether EVs are present in the joint space after trauma, we isolated EVs from synovial fluid of mice after PTOA and examined the presence of common EV markers by mass spectrometry and FunRich analysis (28, 29). Comparison of the total proteins found in synovial EVs isolated from young and aged OA mice treated with vehicle or UBX0101 against records in Vesiclepedia revealed that there was robust enrichment for proteins annotated as EV associated (Supplemental Figures 4 and 5 and Supplemental Tables 2 and 3). We found that miR-34a, miR-146a, and miR-128a were more abundant in EVs enriched from the synovial fluid of young PTOA mice compared with no surgery controls. This miR profile is similar to that of EVs produced by human OA chondrocytes (Figure 3A). Clearance of SnCs by senolytic treatment significantly decreased miR-34a in synovial EVs from young OA joints. No differences in miR-34a, -146a, and -128a expression were observed in aged OA animals with or without SnCs removal; however, since these animals have SnCs before injury, accumulation of extracellular miRs may have masked any treatment-related differences.
Figure 3. miRs carried by EVs are differentially present in OA synovial fluid from young versus aged mice after clearance of SnCs.
(A) Quantification of miR-34a-5p, miR-128a-3p, and miR-146a-5p in young and aged PTOA mice treated with vehicle (veh) or the senolytic UBX0101, which can mediate senescence and the SASP, 28 days after ACLT surgery. All data are expressed as mean ± SEM, and each data point represents an individual mouse. One-way ANOVA with Tukey’s multiple-comparisons test was used for statistical analysis (young, n = 4; aged, n = 5). *P < 0.05. (B) Workflow of analysis of EVs from the synovial fluid of PTOA mice treated with veh or UBX0101. (C and D) Plots illustrating the fold change (UBX0101/veh; x axis) and significance level expressed as the log P value (y axis). The blue circles represent miRs that were upregulated and red circles represent miRs that were downregulated by UBX0101 compared with veh-treated PTOA young (C) and aged (D) mice (n = 3 per group). Significance was determined based on a P value cutoff of 0.05. (E) The heatmap reveals significant correlations among mmu-miR-30c-5p, -92a-3p, -24-3p, -186-5p, -125a-5p, and -150-5p, expression of which was significantly altered by UBX0101 treatment in young PTOA mice and the signaling pathways in which they are predicted by the DIANA-miRPath (v3.0) to participate.
To investigate potentially novel EV-associated miRs and proteins in young and old animals with PTOA treated with senolytic, we performed miR array and proteomic analyses on synovial fluid–derived EVs. The workflow for isolating and characterizing these EVs and the effects of senolytic treatment is illustrated in Figure 3B. Briefly, C57BL mice underwent ACLT of one rear limb to induce OA and were injected intra-articularly every other day with vehicle or UBX0101 (10 μl of a 1 mM solution) for 2 weeks starting 14 days after surgery. We collected synovial fluid on day 28 after surgery. EV enrichments were isolated through differential ultracentrifugation (30), followed by protein and RNA extraction for proteomics and miR analyses.
To identify EV-associated miRs, we performed TaqMan mouse miR Low-Density Array analysis. Several miRs showed distinct profiles when we compared young mice treated with drug with vehicle-treated controls. Levels of mouse miR-92a-3p (mmu-miR-92a-3p), -186-5p, and -150-5p increased after senolytic treatment, while mmu-miR-30c-5p, -24-3p, and -125a-5p decreased (Figure 3, C and D). In aged animals, only mmu-miR-223-3p levels decreased significantly in synovial EVs after drug treatment. These findings suggest that the levels of miR-30c, -24, -125a, -92a, -150, -186, and -223 in synovial fluid–derived EVs correlate with senolytic response in mice.
We next examined the target genes of the EV-derived miRs that are differentially expressed depending on the presence of SnCs. We applied the prediction algorithm DNA Intelligent Analysis (DIANA; DIANA-microT-CDS v5.0; ref. 31) to investigate the function of the extracellular miRs that are targets of the miRs. The analyses found that the increase in miR-92a-3p, -150-5p, and -186-5p combined with a decrease in miR-30c, -24, and -125a in the synovial EVs after senolytic treatment in young mice were enriched for target genes within pathways related to cartilage matrix formation (Figure 3E and Supplemental Figure 6). In contrast to that in young mice, analysis of EVs from aged mice identified only a decline in miR-223-3p after senolytic treatment. This miR participates in mucin-type O-glycan biosynthesis (Supplemental Table 4). These findings suggest that the miRs are differentially expressed in young and aged mice after UBX0101 treatment. These age-specific miRs identified target genes and signaling pathways that might explain the reduced cartilage regeneration observed in the older animals (4).
Synovial fluid EVs from young OA mice treated with UBX0101 contain proteins associated with cartilage growth and cartilage protease inhibitors.
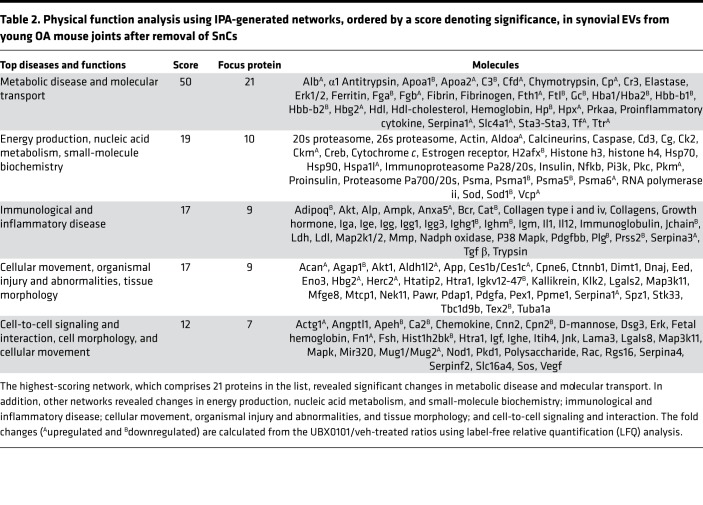
To relate EV miRs and their predicted pathways to protein expression, we performed label-free quantitative (LFQ) proteomics and ingenuity pathway analysis (IPA) on synovial fluid–derived EVs isolated from young and aged mice after injury and standard senolytic treatment. The analysis identified 59 proteins that were differentially expressed between young OA mouse joints with and without the senolytic. Specifically, Serpina 1 and Serpina 3 — subtypes of serine protease inhibitors that regulate proteases involved in the cartilage degradation (32) — were increased in young PTOA joints after senolytic treatment. In addition, SnC removal decreased the level of Prss2 — a protease, serine 2 that degrades type II collagen–rich cartilage ECM (Figure 4A). In addition, synovial EVs from treated young mice contained increased levels of aggrecan, one of the major ECM components in cartilage. Overall, the top 5 molecular networks predicted by the IPA analysis were (a) metabolic disease and molecular transport; (b) energy production, nucleic acid metabolism, and small-molecule biochemistry; (c) immunological and inflammatory disease; (d) cellular movement, organismal injury and abnormalities, and tissue morphology; and (5) cell-to-cell signaling and interaction (Table 2).
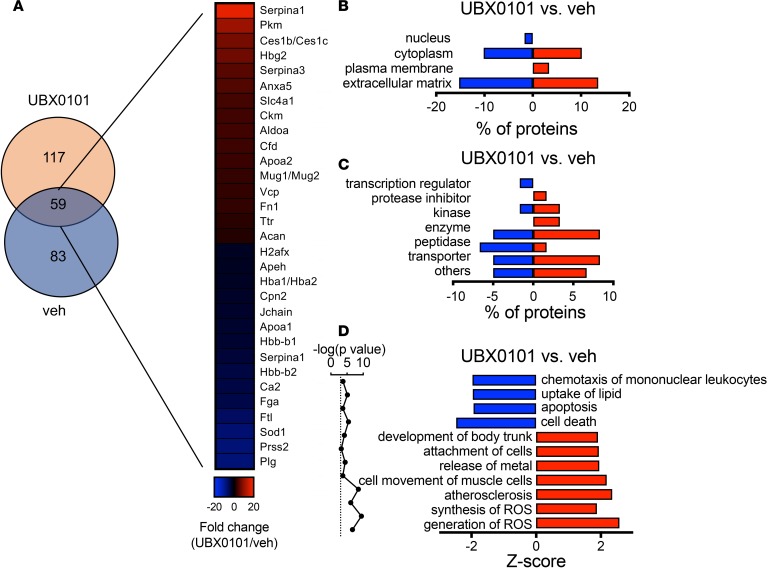
Figure 4. Identification of proteins with altered levels in synovial EVs derived from young OA mice after selective SnC clearance.
(A) Venn diagram of the number of proteins quantified by mass spectrometry and heatmap of upregulated or downregulated proteins (P < 0.05 calculated by a right-tailed Fisher’s exact test, fold change > 2, n = 3 per group). (B and C) Classification of the significantly regulated proteins according to their roles in cellular components and molecular functions. (D) Significant function and disease roles were analyzed by ingenuity pathway analysis (IPA) from upregulated and downregulated proteins after treatment with UBX0101. Bars with positive Z-scores indicate that functional activity is increased, whereas negative Z-scores indicate decreased activity.
Table 2. Physical function analysis using IPA-generated networks, ordered by a score denoting significance, in synovial EVs from young OA mouse joints after removal of SnCs.
To categorize the altered synovial EV–associated proteins after SnC clearance, we applied Gene Ontology analysis (Figure 4, B–D). As expected, the functions were predominantly in the ECM, protease inhibitor, and transporter categories (Figure 4C). The disease/functional analysis component of differentially expressed synovial EV–derived proteins in young OA mouse joints after SnC elimination showed increased development of body trunk, implicating cartilage growth (Figure 4D).
Senolytic treatment in aged mice induced primarily immunological changes in EV proteins, and age-related synovial EVs can transfer arthritic disease to young animals.
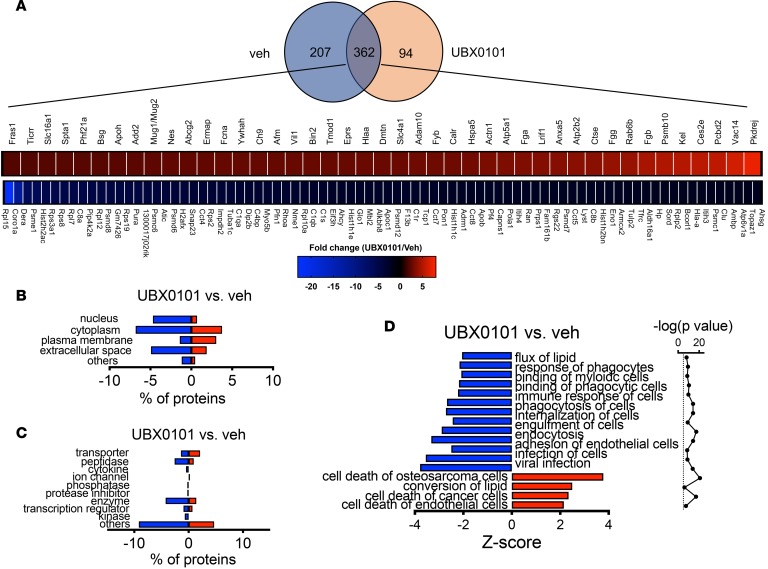
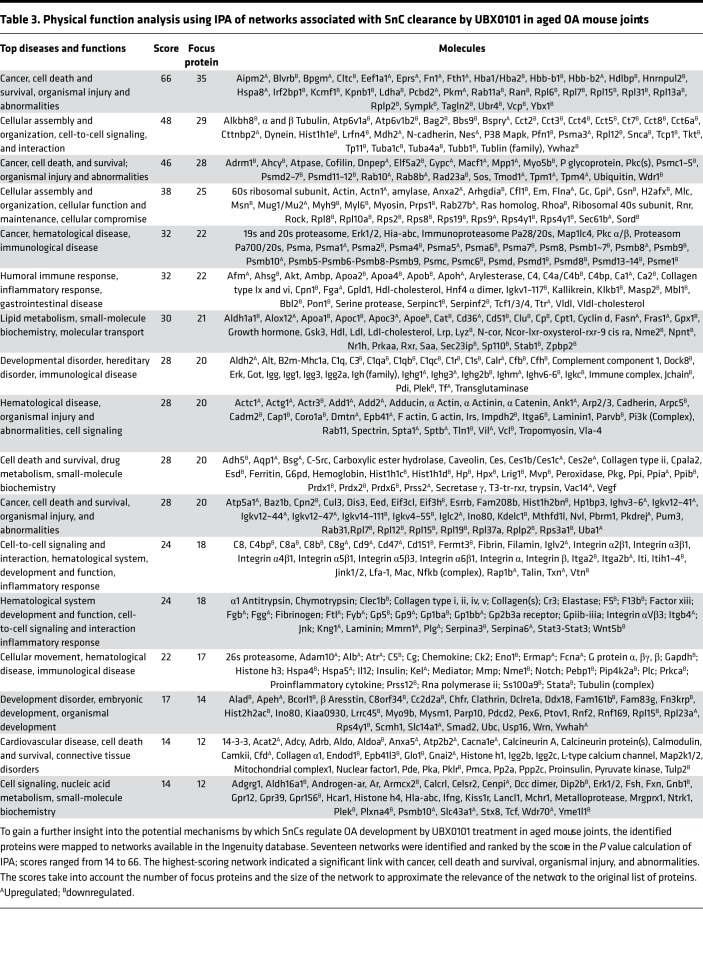
Synovial EVs from aged animals treated with only one round of senolytic therapy did not contain increased levels of cartilage ECM-related proteins, Serpins, or decreased serine proteases (Figure 5A). Gene Ontology analysis indicated that there were no significant changes in synovial EV–associated proteins after SnC clearance in aged animals (Figure 5, B and C). Most of the component changes after senolysis in aged animals were enriched in immunological responses, including the response of myeloid and phagocytic cells (Figure 5D). The top 5 molecular networks predicted by IPA were (a) cancer, cell death and survival, organismal injury, and abnormalities; (b) cellular assembly and organization, cell-to-cell signaling, and interaction; (c) cellular assembly and organization, cellular function and maintenance, cellular compromise, (d) cancer, hematological disease, immunological disease; and (e) humoral immune and inflammatory response (Table 3). These results are consistent with the in vivo observation of senolytic treatment reducing degeneration and pain in aged mice after traumatic injury. It also highlights the important of the immune system in disease and senolytic treatment in aged animals. Further studies on dosing and delivery of senolytics in aged mice exposed to articular trauma are needed.
Figure 5. Differential expression of synovial EV–derived proteins altered by selective clearance of SnCs in aged PTOA mice.
(A) Venn diagram of the number of proteins quantified by proteomics, and heatmap of upregulated and downregulated proteins that were present in PTOA joints and differentially present after UBX0101 treatment of 20-month-old mice (P < 0.05 calculated by a right-tailed Fisher’s exact test, fold change > 2, n = 3 per group). (B and C) Classification of the significantly differentially present proteins based on cellular components, molecular functions and cellular functions, and diseases. (D) Significant function and disease roles were analyzed by ingenuity pathway analysis (IPA) using quantitatively upregulated and downregulated EV proteins after treatment of aged PTOA mice with UBX0101.
Table 3. Physical function analysis using IPA of networks associated with SnC clearance by UBX0101 in aged OA mouse joints.
Even without significant trauma, aging contributes broadly to the development of chronic diseases such as OA. Aged mice experience cartilage degeneration and a more severe degenerative response after traumatic injury in the joint. To determine whether EVs from an aged animal can transfer and induce disease, we isolated EVs from the synovial fluid of aged mice and injected them into the articular space of young mice. Young mice that received the aged EVs developed marked cartilage degeneration after 84 days, as demonstrated by decreased Safranin-O staining for proteoglycans. The animals also exhibited increased pain, as demonstrated by decreased weight bearing on the EV-injected leg between days 56 and 84 compared with control animals injected with saline (Supplemental Figure 7). This work demonstrates the physiological and pathologic functions of senescence-associated synovial EVs and their ability to transfer a specific age-related disease to young animals.
Discussion
Multiple miRs in synovial fluids relevant to OA progression were differentially expressed, including miR-27b, -199a, -185, and -23b. These miRs have been suggested to play a role in bone sclerosis along with catabolic and inflammatory response (27, 33–36), pathways that are relevant in OA disease and diagnosis. The effect of SnC clearance on the synovial EV contents was defined in a murine model of PTOA. Differential expression of miR-34a after senolytic treatment occurred in both murine synovial EVs and human EVs from senescent chondrocytes. Therefore, expression of this miR may be used to evaluate disease progression and response to drugs that target SnCs. Further studies are needed to validate this observation, since there were low numbers of clinical samples due to limitations in collection of clinical OA samples.
Any disturbance of joint homeostasis is reflected in the levels of soluble factors (such as cytokines, enzymes, and growth factors) in the synovial fluid and possibly also in the number and content of EVs. Recent evidence suggests that synovial fluid–derived EV miRs vary with sex and disease state (27). miRs we found showing differential expression in OA (miR-27b, miR-199a, miR-185) were previously reported to contribute to OA progression by causing abnormal subchondral bone development, chondrogenesis, and catabolic and inflammatory gene expression (27, 33–36). Our studies revealed the presence of these OA-specific miRs in EVs, along with other unique miRs (e.g., hsa-miR-151a-3p_R+1, hsa-miR-652-3p_R+1, hsa-miR-4450-p3-1ss12TG, hsa-miR-3665-p5_1ss2CA, has-miR-4488_L+1R-4, and hsa-miR1262-p5-1ss11GC). We speculate that synovial EV–derived miRs contribute to dysregulated cartilage homeostasis and initiation/amplification of inflammation. miR expression profiling in human synovial fluid–derived EVs could provide a disease fingerprint. But future studies need to further examine EV profile differences at different timings and stages during the diseases process on a larger clinical study.
Transfer of age-related pathologies (or conversely the more popular transfer of youthfulness to aged animals) has been demonstrated using a number of modalities. Parabiosis studies that demonstrated the improvement of age-related pathology by introducing young factors, also found that exposure to the aged environment had a negative effect on the young animals. In a specific example of senescence transfer, transplantation of SnCs or human aged adipose tissue to young animals produced deleterious effects (37, 38). Our data show that EVs secreted from senescent chondrocytes isolated from human arthritic patients can transfer features of senescence to nonsenescent (from healthy donor) chondrocytes and suppress cartilage tissue formation. The increased secretion of EVs from highly enriched senescent chondrocyte populations — and especially their miR-34a cargo — can also provoke senescence in a paracrine manner (39). The phenotypic changes may correlate with the cellular senescence response during OA progression that also affects EV secretion, partially through p53 and one of its targets (17, 40, 41). miR‑140 is one of the few miRs that is highly expressed in nonsenescent chondrocytes (42), and its expression is lower in human OA cartilage as well as in IL-1B–induced inflammation in articular cartilage (24, 43). To validate physiological relevance and age-related pathology transference to young animals, SnC-associated EVs alone were able to induce symptoms of OA (pain) and induced tissue degeneration in the young joint.
There is mounting evidence that the immune system plays a role in the development of PTOA (44). The cellular sources of miRs that we found to be differentially expressed in synovial EVs after senolytic therapy have been defined previously (45). For example, miR-223 is significantly enriched in neutrophils and monocytes while miR-150 and -125 are exclusively expressed by lymphocytes. This suggests that clearance of SnCs modulates the response, recruitment, and activation of cells from the myeloid and lymphoid lineage. Just as the human chondrocyte EV production did not fully overlap with the EVs found in synovial fluid, these results provide evidence of other cell types that may be contributing to the EV population found in the synovial fluid. Future studies need to address the possibility of identifying the cellular origin of EVs and tissue-specific EV profiles in order to elucidate detailed mechanistic roles of EVs in the OA diseases. The findings also support the relevance of the immune system in PTOA, which when combined with age-related immune changes, may affect senolytic therapy design.
Methods
Cell isolation and culture.
Explanted OA articular cartilage and synovial fluid from human patients undergoing total knee arthroplasty were received from the National Disease Resource Institution (Philadelphia, Pennsylvania, USA). The cartilage tissue was cut into 1-mm3 pieces, washed 3 times with PBS supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Pen/Strep; Invitrogen, catalog 15140-122), and digested on a shaker for 16 hours at 37°C with 0.17% (w/v) type II collagenase (Worthington Biochemical, catalog 4176) in high-glucose DMEM (Gibco, catalog 11965-092) with 10% FBS (Hyclone, catalog SH30070.03). After the digestion, the filtrate was passed through a 70-μm strainer and cells were rinsed 3 times with growth media containing DMEM supplemented with 1% Pen/Strep and 10% FBS. For culture experiments in which EVs derived from SnCs were exposed to healthy chondrocytes, healthy chondrocytes were cultured with EVs (8 × 108) in growth medium that was precentrifuged at 100,000 rcf for 20 hours to remove FBS-associated EVs for 6 days.
Sorting senescent chondrocytes by flow cytometry.
Human primary osteoarthritic chondrocytes were trypsinized, collected in the chondrocyte growth medium, and immediately used for sorting in a FACSAria IIu Sorter (BD Biosciences). Signals were analyzed using FACS Diva Version 6.1.3 software. There are currently no known markers of SnCs for live sort; therefore, the sort was based on examining the enlarged size and accumulated autofluorescence of the age pigment lipofuscin in SnCs (20, 46). SnCs were sorted based upon size in FL1 and autofluorescence (488 nm) by FSC as previously described (31). The FSC/SSC dot plot of autofluorescence versus size was then generated and used to arbitrarily set up gates to sort the chondrocytes into 3 groups to generate populations with different proportions of SnCs. Live cells were sorted in buffer composed of PBS with 1% FBS and collected in buffer composed of chondrocyte medium with 2× Pen/Strep. We sorted populations containing high- (65%), medium- (45%), and low-senescent (20%) chondrocytes using the upper 5.9%, middle 10.8%, and lower 9.9% quartiles, respectively, with respect to both FSC-A and FITC-A, which was confirmed by SA-β-Gal assay (Supplemental Figure 1). We collected sorted populations of SnCs in chondrocyte growth medium and used these for coculture experiments.
Coculture of senescent chondrocytes with healthy chondrocytes.
A 24-well Corning Transwell plate with 6.5-mm inserts (MilliporeSigma, catalog 3470) was used for coculture experiments. Transwell inserts (0.4-μm pore size) containing varying concentrations of senescent chondrocytes at a cell density of 20,000/insert were fitted into the 24-well containing primary nonsenescent chondrocytes at a cell density of 20,000/well. The coculture was incubated at 37°C and 5% CO2 in chondrocyte growth medium for 7 days; control cultures contained only nonsenescent chondrocytes. For SnC-conditioned media, high- (65%), medium- (45%), and low-senescent (20%) chondrocytes were plated in a 24-well culture plate at a cell density of 20,000/well and collected the conditioned media after 7 days of culture. After centrifugation (300 g for 10 minutes at 4°C), we aliquoted the conditioned medium, which was stored at –80°C. Nonsenescent chondrocytes at a density of 20,000/well were cultured in the conditioned medium for 7 days. The conditioned medium was changed on alternate days.
Cytokine antibody array assay.
Cytokines in conditioned media were assessed using the human cytokine array kit (R&D Systems, catalog ARY005) according to the manufacture’s instructions. Briefly, conditioned media were prepared by washing approximately 0.2 × 105 cells once with PBS and incubating them in serum-free medium for 24 hours. The conditioned media were collected in 1.5-ml centrifuge tubes and clarified by centrifugation. The array membrane was incubated with 1.5 ml of 3-fold diluted conditioned media overnight at 4°C, washed, and incubated with biotin-conjugated antibody cocktail, washed, and then incubated with streptavidin-HRP conjugate. Cytokines were detected by Chemi reagent mix. The signals were developed on x-ray film and quantified relative to the average signal (pixel density) of a pair of duplicate spots representing each cytokine by ImageJ software (NIH). The experiments were performed in duplicate.
SA-β-Gal staining.
SA-β-Gal staining was done using a kit (BioVision Senescence Detection Kit, catalog K320-250) according to the manufacturer’s instructions. SnCs were identified as blue-stained cells under light microscopy. Total cells were counted using a nuclear DAPI counterstain in 10 random fields per culture dish to determine the percentage of SA-β-Gal–positive cells.
Surgically induced OA mouse model.
ACLT surgery was performed on 10-week-old or 19-month-old male C57BL/6 mice from Charles River. Mice were placed under general anesthesia with 3% isoflurane, and the hind limbs shaved and prepared for aseptic surgery. The knee joint was exposed following a medial capsular incision, and the ACL was transected with microscissors under a surgical microscope. After irrigation with saline to remove tissue debris, the skin incision was closed.
EV enrichment by ultracentrifugation.
EVs were isolated by differential ultracentrifugation according to established methods shown in Supplemental Figure 2A (30, 47). The 10,000 g step from that protocol was skipped because we did not see a need to focus on any particular class of EV in this initial study and thus did not try to deplete microvesicle-sized particles. For isolation of EVs from in vitro cultured cells, conditioned medium was collected and centrifuged at 2,000 rcf for 20 minutes at 4°C to remove cells and cell debris; the supernatant containing EVs was then centrifuged at 100,000 rcf for 70 minutes at 4°C. The EV pellet was suspended in PBS and centrifuged at 100,000 rcf again to eliminate contaminant proteins and nucleic acids. The pellet was then resuspended in PBS and store at –80°C until use. Isolation of EVs from synovial fluid was performed similarly, but synovial fluid was first diluted 1:4 with PBS prior to differential centrifugation.
EV characterization via NTA and transmission electron microscopy.
EVs isolated and resuspended in PBS were analyzed for size and concentration via NTA using a NanoSight NS300 (Malvern Panalytical) equipped with a 532-nm laser, low-volume flow-cell, syringe pump, and concentration upgrade. Thawed EV samples were further diluted in PBS to yield concentrations of between 20 and 100 particles/frame on NanoSight, and three 60-second videos were acquired per sample using the same camera settings and NTA 3.1 or 3.2 software. For transmission electron microscopy, EVs were incubated overnight on carbon film 400 square mesh transmission electron microscopy grids. Grids were briefly dipped in a droplet of ultrapure water, wicked dry, vacuum dried, and imaged on a Philips CM120 (Philips Research).
Protein extraction from synovial fluid–derived EVs from young and aged OA joints.
The final EV pellets were resuspended in 100 μl solubilization buffer (7 M urea, 2 M thiourea, in 30 mM Tris, pH 8.0). Aliquots from the solubilized EV extractions were used to determine the total protein concentration using the BCA method (Thermo Scientific).
miR qPCR array.
miR was harvested from EVs of conditioned media and synovial fluids of OA mouse joints using the miRCURY RNA isolation kit for biofluids (Exiqon, catalog 300112). A TaqMan low-density microRNA array A (TLDA; Thermo Fisher Scientific, catalog 4398967) was used for 375 miRs targets and for 6 common miRs as controls. Reverse transcription (TaqMan, PN 4366596), preamplification (TaqMan, PN 4391128), and TLDA card processing were done using manufacturer’s protocol. Data were extracted and processed as previously described (48). Normalization was performed to the geometric mean of 26 miRs detected in all samples.
In-solution trypsin/LysC/Glu-C digestion of EVs proteomes.
For proteomic analysis of proteins extracted from the EVs purified from synovial fluid from young and old OA joints of mice subjected to ACLT surgery, with or without UBX0101 treatment, equal aliquots (0.5–1 μg) were subjected to the in-solution reduction with 100 mM dithiothreitol for 50 minutes at 55°C followed by alkylation with 550 mM iodoacetamide for 1 hour at room temperature in the dark. The samples were further subjected to digestion for 18 hours at 37°C with endoproteinase Lys-C (sequencing grade, Promega) (1:50, protein/enzyme ratio) in 50 mM ammonium bicarbonate buffer, pH 8.5. Then, tryptic digestion was performed for 3 hours at 37°C, in 50 mM ammonium bicarbonate buffer, at pH 8.5 (1:50, protein/enzyme ratio). Finally, Glu-C was added (1:10, Glu-C/protein ratio) in ammonium 50 mM bicarbonate buffer (pH 7.5) at 37°C for 10 hours. Total peptides, extracted from all enzymatic digestions, were combined, desalted on C18 Prep clean columns, and further subjected to nanoLC/ESI/MS/MS on a Q Exactive HF quadrupole orbitrap mass spectrometer.
NanoLC-ESI-MS/MS analysis of peptides generated from the digestion with LysC/trypsin/Glu-C enzymes.
Each sample digest was analyzed by nano LC/MS/MS (liquid chromatography/mass spectrometry). For LFQ analysis, technical replicates (2 × 1μg) from each digested sample and corresponding EVs were analyzed on a Q Exactive HF quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific) coupled to an Easy nLC 1000 UHPLC (Thermo Fisher Scientific) through a nanoelectrospray ion source. The mass spectrometer was operated in the positive ion mode and data-dependent acquisition mode. The full MS scans were obtained with a m/z range of 300–1600 and a mass resolution of 120,000 at m/z 200. Higher-energy collision-induced dissociation was performed on the 15 most significant peaks, and tandem mass spectra were acquired at a mass resolution of 30,000 at m/z 200 and a target value of 1.00 × 105 with a maximum injection time of 100 ms.
Protein identification and label-free relative peptide quantification (LFQ analysis).
Raw files from each technical and biological replicate were filtered, de novo sequenced, and assigned with protein ID using PEAKS 7.0, 7.5, and 8.0 software (Bioinformatics Solutions) by searching against the mouse (Mus musculus) Swiss-Prot database (82,628 entries). The following search parameters were applied for LFQ analysis: trypsin, Lys-C, and GluC restriction for enzymes, with the allowance of one missing cleaved enzyme at one peptide end. The parent mass tolerance was set to 15–18 ppm using monoisotopic mass, and fragment ion mass tolerance was set to 0.05 Da. Carbamidomethyl cysteine (+57.0215 on C) was specified in PEAKS as a fixed modification. Methionine, lysine, proline, arginine, cysteine, and asparagine oxidations (+15.99 on CKMNPR) and deamidation of asparagine and glutamine (NQ-0.98) and pyro-Glu from glutamine (Q-18.01 N-term) were set as variable modifications. Data were validated using the FDR built into PEAKS 7.0–8.0, and protein identification was accepted at a confidence score (–10logP) > 15 for peptides and (–10logP) > 15 for proteins; a minimum of 1 peptide per protein after data was filtered for an FDR of less than 1.0% for peptides and less than 1.5% FDR for protein identifications (P < 0.05). LFQ analysis followed by quantitative IPA were performed as described previously (49).
Gene ontology, pathway enrichment, and protein analysis of proteomics data.
Networks, functional analyses, and biochemical and cellular pathways were generated by employing IPA (Ingenuity Systems). Specifically, experimentally determined protein ratios were used to calculate the fold changes by rescaling values using a log2 transformation, such that positive values reflected fold increases while negative values reflected fold decreases. For network generation, data sets containing gene identifiers (symbols) were uploaded into the IPA application together with their rescaled log2 transformation ratios. For all quantitative IPA, we used data sets that represent >2-fold expressed proteins across all analyzed samples. These molecules were overlaid onto a global molecular network in the Ingenuity Knowledge Base. The networks were then algorithmically generated based on their connectivity index using the IPA algorithm. The probability of having a relationship between each IPA-indexed function and the experimentally determined genes was calculated by a right-tailed Fisher’s exact test. The level of significance was set to P < 0.05. Accordingly, IPA identified the molecular and cellular pathways from the library of established biochemical pathways that were most significant to the data set (–log [P value] > 2.0).
miR target selection and validation.
To identify molecular pathways potentially altered by multiple miRs, we used DIANA-mirPath (v 3.0) (http://diana.imis.athena-innovation.gr/DianaTools/index.php), which performs an enrichment analysis of multiple miR target genes, comparing each set of miR targets to all known Kyoto encyclopedia of genes and genomes pathways.
Library prep and miR sequencing.
Sequencing was performed on the Illumina HiSeq 2500 platform in the 50 cycle SE configuration. In brief, single-end sequencing reads were cleaned with quality filter, adapter cutter, and length filter, using ACGT101_miR_v4.2g from LC Sciences. Considering that extremely low abundance might lead to false results, the known and new candidate miRs with less than 10 raw reads in the libraries were removed from the analysis. The cleaned reads were mapped to miRBase (miRBase v21) using Bowtie v1.1.1. The unmapped reads were mapped to multiple databases, including mRNA, Rfam11, and genomic DNA, using Bowtie v1.1.1. miR candidates we believe to be novel were identified from genome mapped reads based on the propensity of hairpin formation in corresponding genome positions using ACGT101_miR_v4.2g from LC Sciences. Data were normalized by dividing the sequence counts of individual samples by the corresponding normalization factors, which are the median values of the ratios between specific sample counts and geometric mean counts of all samples. The sequencing data presented in this study were submitted to the Gene Expression Omnibus (GSE126677).
Labeling EVs with a lipophilic and near-infrared fluorescent cyanine dye.
A 199-μl stock solution of DiR (Xenolight DiR, MW 1013.4, 25 mg, PerkinElmer, catalog 125964) was prepared by dissolving 25 mg in 3 ml 200 proof ethanol (~8.2 mM) and then diluting it in PBS to make a working solution of 320 μg/ml (0.31 mM). EVs from mouse synovial fluids were incubated with 320 μg/ml DiR at 37°C for 30 minutes. The EVs were then washed with 750 μl PBS and subjected to ultracentrifugation for 1 hour at 100,000 rcf. EVs were resuspend in 35 μl of 150 mM PBS. In vivo fluorescence imaging using the IVIS system was performed using 710 nm excitation and 760 nm emission at 10 minutes, 1 hour, 7 days, and 14 days after injecting DiR labeled EVs under the same imaging conditions.
Weight bearing.
Static incapacitance measurements were performed using an Incapacitance Tester (Columbus Instruments). Mice were first acclimated to the chamber at least 3 times before measurement. After acclimatization, mice were maneuvered inside the chamber such that they stood with 1 paw on each scale. Weight placed on each hind limb was measured over a 3-second interval for at least 3 separate measurements. Results are expressed as a percentage of weight placed on the operated limb versus contralateral control limb. The observer was blinded to the genotype and treatment of the mice.
Histology.
The mouse joints were fixed in 4% paraformaldehyde overnight, dehydrated in increasing concentrations of ethanol, and embedded in paraffin. Five-micrometer-thick sections were cut from the paraffin block and collected onto glass slides. The sections were stained for proteoglycans with aqueous Safranin-O (0.1%) for 5 minutes, and then specimens were mounted.
Statistics.
Data are expressed as mean ± SEM. All analyses were performed using Prism 8 (GraphPad software). Statistical tests used to calculate P values for each figure are indicated in the figure legends. One-way ANOVA and Tukey’s multiple-comparisons test were used for statistical analysis in Figures 1 and 3. Two-tailed t tests (unpaired) were performed for statistical analysis in Figure 2. A right-tailed Fisher’s exact test was performed to define statistical significance of functional analyses and biochemical and cellular pathways for proteomic studies in Figures 4 and 5. P < 0.05 was considered statistically significant (except P < 0.1 for differentially expressed miRs in synovial EVs between normal and OA patients in Figure 2D and Table 1).
Study approval.
All animal studies were performed with approval of and in accordance with the guidelines established by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine. Human cartilage and synovial fluid samples were received from the National Disease Resource Institution according to an IRB-approved protocol.
Author contributions
OHJ designed and carried out most of the experiments and analyzed data from experiments and wrote the manuscript with input from all coauthors. DRW, CCC, LS, and KWW designed experiments and analyzed and interpreted data from experiments. CC, SR, and BP performed experiments. JC and JJG analyzed and interpreted data and revised the manuscript. JHE conceived, designed, and supervised the study; analyzed and interpreted data; and wrote the manuscript. All authors discussed the results and commented on the manuscript. ZL and AMK carried out miR sequencing for human synovial EV samples.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge financial support from the Morton Goldberg Chair (to JHE), the Bloomberg-Kimmel Institute for Cancer Immunotherapy (to JHE), the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program (W81XWH-17-1-0627) (to JHE), the NIH (AG009909) (to JC), National Institute of Biomedical Imaging and Bioengineering (1P41EB021911-01) (to AMK), and a fellowship from the Glenn Foundation for Medical Research (to OHJ).
Version 1. 04/04/2019
Electronic publication
Funding Statement
the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program
to J.H.E
to J.H.E.
Footnotes
Conflict of interest: JHE, OHJ, and JC are inventors on patents owned by Unity Biotechnology, Buck Institute, Mayo Clinic, and Johns Hopkins and licensed to Unity Biotechnology (9,849,218 and 9,855,266). JC is a founder of Unity Biotechnology.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(7):e125019. https://doi.org/10.1172/jci.insight.125019.
Contributor Information
Ok Hee Jeon, Email: ojeon@buckinstitute.org.
David R. Wilson, Email: david.wilson@jhmi.edu.
Sona Rathod, Email: sona.rathod21@gmail.com.
Christopher Cherry, Email: ccherry.6574@gmail.com.
Zhenghong Lee, Email: zxl11@case.edu.
Jordan J. Green, Email: green@jhu.edu.
Judith Campisi, Email: jcampisi@buckinstitute.org.
Laura Santambrogio, Email: laura.santambrogio@einstein.yu.edu.
Jennifer H. Elisseeff, Email: jhe@jhu.edu.
References
- 1.Wieland HA, Michaelis M, Kirschbaum BJ, Rudolphi KA. Osteoarthritis - an untreatable disease? Nat Rev Drug Discov. 2005;4(4):331–344. doi: 10.1038/nrd1693. [DOI] [PubMed] [Google Scholar]
- 2.Loeser RF. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr Cartil. 2009;17(8):971–979. doi: 10.1016/j.joca.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin JA, Brown T, Heiner A, Buckwalter JA. Post-traumatic osteoarthritis: the role of accelerated chondrocyte senescence. Biorheology. 2004;41(3-4):479–491. [PubMed] [Google Scholar]
- 4.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23(6):775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price JS, et al. The role of chondrocyte senescence in osteoarthritis. Aging Cell. 2002;1(1):57–65. doi: 10.1046/j.1474-9728.2002.00008.x. [DOI] [PubMed] [Google Scholar]
- 6.Jeon OH, David N, Campisi J, Elisseeff JH. Senescent cells and osteoarthritis: a painful connection. J Clin Invest. 2018;128(4):1229–1237. doi: 10.1172/JCI95147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 9.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira-Gonzalez S, et al. Paracrine cellular senescence exacerbates biliary injury and impairs regeneration. Nat Commun. 2018;9(1):1020. doi: 10.1038/s41467-018-03299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15(8):978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Kato T, et al. Exosomes from IL-1β stimulated synovial fibroblasts induce osteoarthritic changes in articular chondrocytes. Arthritis Res Ther. 2014;16(4):R163. doi: 10.1186/ar4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Withrow J, Murphy C, Liu Y, Hunter M, Fulzele S, Hamrick MW. Extracellular vesicles in the pathogenesis of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2016;18(1):286. doi: 10.1186/s13075-016-1178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jubeck B, et al. Promotion of articular cartilage matrix vesicle mineralization by type I collagen. Arthritis Rheum. 2008;58(9):2809–2817. doi: 10.1002/art.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann BD, et al. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008;68(19):7864–7871. doi: 10.1158/0008-5472.CAN-07-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Effenberger T, et al. Senescence-associated release of transmembrane proteins involves proteolytic processing by ADAM17 and microvesicle shedding. FASEB J. 2014;28(11):4847–4856. doi: 10.1096/fj.14-254565. [DOI] [PubMed] [Google Scholar]
- 19.Overhoff MG, Garbe JC, Koh J, Stampfer MR, Beach DH, Bishop CL. Cellular senescence mediated by p16INK4A-coupled miRNA pathways. Nucleic Acids Res. 2014;42(3):1606–1618. doi: 10.1093/nar/gkt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Passos JF, von Zglinicki T. Methods for cell sorting of young and senescent cells. Methods Mol Biol. 2007;371:33–44. doi: 10.1007/978-1-59745-361-5_4. [DOI] [PubMed] [Google Scholar]
- 21.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahu J, Bustany S, Sola B. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 2012;3:e446. doi: 10.1038/cddis.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyaki S, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60(9):2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012;8(9):543–552. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feliciano A, Sánchez-Sendra B, Kondoh H, Lleonart ME. MicroRNAs regulate key effector pathways of senescence. J Aging Res. 2011;2011:205378. doi: 10.4061/2011/205378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolhe R, et al. Gender-specific differential expression of exosomal miRNA in synovial fluid of patients with osteoarthritis. Sci Rep. 2017;7(1):2029. doi: 10.1038/s41598-017-01905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathan M, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 29.Pathan M, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. 2017;6(1):1321455. doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 31.Paraskevopoulou MD, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakrishnan L, et al. Proteomic analysis of human osteoarthritis synovial fluid. Clin Proteomics. 2014;11(1):6. doi: 10.1186/1559-0275-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR, Haqqi TM. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62(5):1361–1371. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasadam I, Batra J, Perry S, Gu W, Crawford R, Xiao Y. Systematic identification, characterization and target gene analysis of microRNAs involved in osteoarthritis subchondral bone pathogenesis. Calcif Tissue Int. 2016;99(1):43–55. doi: 10.1007/s00223-016-0125-7. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Ning Y, Zhou B, Yang L, Wang Y, Guo X. Integrated bioinformatics analysis of the osteoarthritis‑associated microRNA expression signature. Mol Med Rep. 2018;17(1):1833–1838. doi: 10.3892/mmr.2017.8057. [DOI] [PubMed] [Google Scholar]
- 36.Ham O, et al. Upregulation of miR-23b enhances the autologous therapeutic potential for degenerative arthritis by targeting PRKACB in synovial fluid-derived mesenchymal stem cells from patients. Mol Cells. 2014;37(6):449–456. doi: 10.14348/molcells.2014.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433(7027):760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 38.Xu M, et al. Transplanted senescent cells induce an osteoarthritis-like condition in mice. J Gerontol A Biol Sci Med Sci. 2017;72(6):780–785. doi: 10.1093/gerona/glw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi A, et al. Publisher Correction: Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2018;9(1):4109. doi: 10.1038/s41467-018-06613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi A, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev. 2012;18(6):445–453. doi: 10.1089/ten.teb.2012.0116. [DOI] [PubMed] [Google Scholar]
- 43.Ntoumou E, et al. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics. 2017;9:127. doi: 10.1186/s13148-017-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Berg WB, Miossec P. IL-17 as a future therapeutic target for rheumatoid arthritis. Nat Rev Rheumatol. 2009;5(10):549–553. doi: 10.1038/nrrheum.2009.179. [DOI] [PubMed] [Google Scholar]
- 45.Witwer KW, Halushka MK. Toward the promise of microRNAs - Enhancing reproducibility and rigor in microRNA research. RNA Biol. 2016;13(11):1103–1116. doi: 10.1080/15476286.2016.1236172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hewitt G, von Zglinicki T, Passos JF. Cell sorting of young and senescent cells. Methods Mol Biol. 2013;1048:31–47. doi: 10.1007/978-1-62703-556-9_4. [DOI] [PubMed] [Google Scholar]
- 47.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Z, et al. Serum extracellular vesicle depletion processes affect release and infectivity of HIV-1 in culture. Sci Rep. 2017;7(1):2558. doi: 10.1038/s41598-017-02908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clement CC, et al. Protein expression profiles of human lymph and plasma mapped by 2D-DIGE and 1D SDS-PAGE coupled with nanoLC-ESI-MS/MS bottom-up proteomics. J Proteomics. 2013;78:172–187. doi: 10.1016/j.jprot.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.