SUMMARY
We developed a flexible toolkit for combinatorial screening in Saccharomyces cerevisiae that generates large libraries of cells, each uniquely barcoded to mark a combination of DNA elements. This interaction Sequencing platform (iSeq 2.0) includes genomic landing pads that assemble combinations through sequential integration of plasmids or yeast mating; 15 barcoded plasmid libraries containing split selectable markers (URA3AI, KanMXAI, HphMXAI, NatMXAI); and an array of ~24,000 “double-barcoder” strains that can make existing yeast libraries iSeq-compatible. Various DNA elements are compatible with iSeq: DNA introduced on integrating plasmids, engineered genomic modifications, or entire genetic backgrounds. DNA element libraries are modular and interchangeable, and any two libraries can be combined, making iSeq capable of performing many new combinatorial screens by short-read sequencing.
Keywords: Combinatorial screening, Saccharomyces cerevisiae, barcode sequencing, yeast, fitness, landing pad, interaction screening, iSeq, loxP, Cre
eTOC
A flexible toolkit for barcode-based combinatorial screening in Saccharomyces cerevisiae is developed. It includes genomic landing pads that assemble combinations through sequential integration of plasmids or yeast mating; 15 barcoded plasmid libraries containing split selectable markers (URA3AI, KanMXAI, HygMXAI, NatMXAI); and an array of ~24,000 “double-barcoder” strains. Various DNA elements are compatible: DNA introduced on integrating plasmids, engineered genomic modifications, or entire genetic backgrounds. DNA element libraries are modular and interchangeable, and any two libraries can be combined.
INTRODUCTION
Large-scale combinatorial screens of DNA elements have been used to define the genetic and protein-protein interaction networks of a cell (Bandyopadhyay et al, 2010; Collins et al, 2007; Costanzo et al, 2010; Díaz-Mejía et al, 2018; Frost et al, 2012; Han et al, 2017; Roguev et al, 2008; Shenet al, 2017) (Ito et al, 2001; Rolland et al, 2014; Tarassov et al, 2008; Uetz et al, 2000; Yachie et al, 2016), interrogate the genetic determinants and evolutionary constraints of protein interactions (Aakre et al, 2015; Diss & Lehner, 2018; Raman et al, 2016; Younger et al, 2017), and discover combinations of natural or synthetic small RNAs that underlie a phenotype (Wong et al, 2015; 2016). The earliest combinatorial screens to be developed, such as the Yeast Two-Hybrid (Y2H) assay, took advantage of pairwise yeast mating to build and assay combinatorial libraries in arrays on agar (Ito et al, 2001; Uetz et al, 2000). However, because combinations must be constructed and assayed one-at-a-time, throughput of these methods is limited.
More recently, a number of strategies have been developed that build and/or assay combinatorial libraries in pools using next generation sequencing of unique DNA barcodes (Díaz-Mejía et al, 2018; Diss & Lehner, 2018; Jaffe et al, 2017; Schlecht et al, 2017; Wong et al, 2015; 2016; Yachie et al, 2016; Younger et al, 2017). These methods allow large combinatorial libraries to be constructed quickly, stored in pools, and economically re-assayed under new conditions. Variations of two general strategies have been developed to build such combinatorial libraries: 1) two or more DNA elements are added to a uniquely-identifiable barcoded plasmid, and plasmid libraries are subsequently inserted into cells (Diss & Lehner, 2018; Han et al, 2017; Wong et al, 2015; 2016), or 2) two barcoded haploid yeast libraries are mated, and in vivo recombination fuses the two barcodes (Díaz- Mejía et al, 2018; Jaffe et al, 2017; Schlecht et al, 2017; Yachie et al, 2016; Younger et al, 2017). An example of the second strategy is our initial interaction Sequencing (iSeq) (Jaffe et al, 2017; Schlecht et al, 2017) platform. A DNA barcode and loxP recombination site is introduced at a common chromosomal location in each yeast mating type. Barcodes are placed on opposite sides of the loxP such that mating and loxP recombination between homologous chromosomes yield a barcode-loxP-barcode on one chromosome.
Increases in throughput resulting from pooled combinatorial screens such as iSeq could open new frontiers in systems biology and bioengineering by, for example, providing a means by which to assay how biological networks change across time, environment, or genotype (Celaj et al, 2017; Fischbach & Krogan, 2010). However, existing assays, including our initial iSeq platform, have generally been built for one or two use cases, and repurposing an assay to a novel use is not always possible. Ideally, a library of potential interaction partners, could be used with any other library. For maximal utility and flexibility, we envision that a potential interaction partner could be an exogenous DNA element that is added to a cell such as a guide RNA or open reading frame (ORF), a specific modification to a cell genome such as a gene knockout or gene fusion, or an entire genotype.
Here, we develop a flexible toolkit for combinatorial screening in the yeast Saccharomyces cerevisiae that we call iSeq 2.0. This toolkit can generate large barcoded libraries of cells, each of which contains a unique combination of any two potential interaction partners. A potential interaction partner library can be reused with any other library to develop a novel screen. We provide a set of 15 plasmid libraries each containing >120,000 barcodes, which are likely to allow iSeq to be performed in many experimental, industrial and wild yeast strains. To facilitate barcoding of existing yeast libraries for use with iSeq, we provide a library of ~24,000 arrayed barcoded yeast strains. Lastly, we demonstrate the utility of iSeq by screening for protein-protein interactions and by measuring the fitness effects of combinations of genes involved in amino acid synthesis.
RESULTS AND DISCUSSION
Overview of the iSeq 2.0 platform.
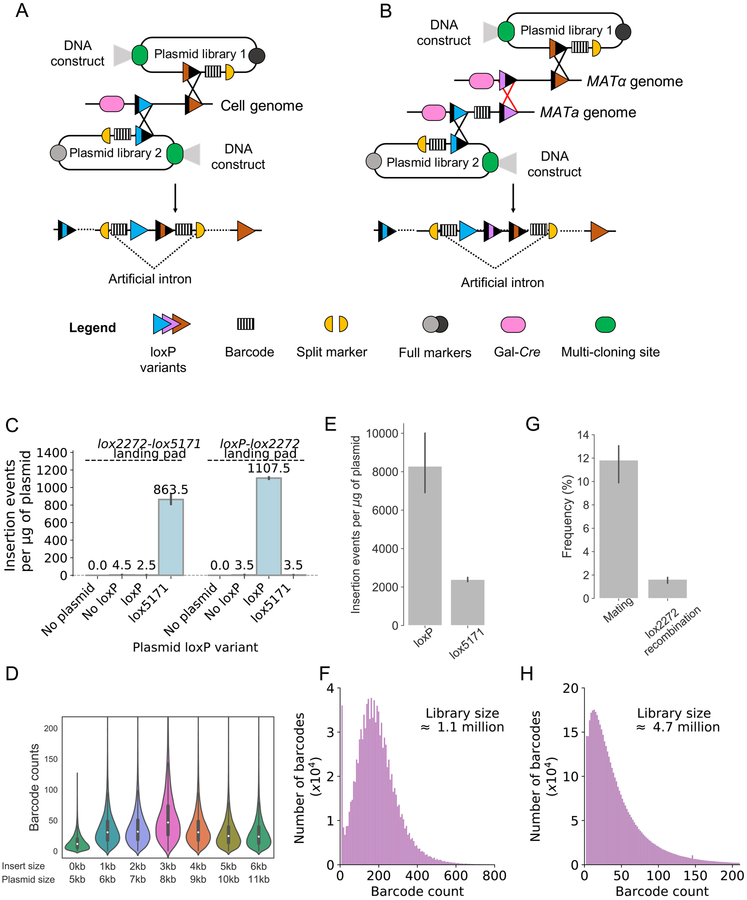
The iSeq 2.0 system functions by first introducing a “landing pad” at a defined genomic location in a yeast strain (STAR Methods, Figure S1A). The landing pad consists of a galactose inducible Cre recombinase and tandem loxP variants that are incompatible with each other (Figure S1B, Table S1). Each loxP variant serves as either a plasmid integration site or a recombination site between homologous chromosomes. The landing pad can build combinatorial libraries in two ways. In the sequential integration method, barcoded plasmid libraries are integrated in tandem, creating a 140 base-pair barcode-lox-lox-barcode (Figure 1A). In the mating method, plasmids are integrated into the genome of MATa and MATα haploids. Subsequent mating and recombination between homologous chromosomes creates a 195 base-pair barcode-lox-lox-lox-barcode (Figure 1B). In both methods, plasmids are integrated in their entirety by being looped in at a single loxP site. LoxP variants are used in configurations that result in a high rate of production of the desired double barcode products (Figure S1B) (Albert et al, 1995). Selection for double barcode constructs is achieved by reassembly of a split positive selectable marker, one half of which is on each plasmid. One of four split selectable markers can be used, three of which have been designed here (URA3AI (Yoshimatsu & Nagawa, 1989), KanMXAI, HphMXAI, NatMXAI). Barcoded plasmids can be engineered to contain additional DNA constructs or to serve as markers for specific genomic modifications or strain backgrounds. Once double barcode libraries are constructed, highly parallel quantitative screens can be performed by competitive growth and short read sequencing.
Figure 1. iSeq 2.0 mechanism, construction fidelity, bias and scalability.
(A and B) Plasmids are introduced into the yeast genome via Cre-loxP-mediated recombination. The black “X”s between loxP sites indicate separate yeast transformations, and can occur in either order. Each integrating plasmid contains a loxP variant (loxP, blue, or lox5171, brown), half of a split selectable marker (URA3AI, KanMXAI, HphMXAI, or NatMXAI, yellow), a full selectable marker (KanMX, HphMX, or NatMX, grey), a random barcode, and a multi-cloning site that can be used to insert genetic constructs prior to yeast transformation. Darkened regions in loxP variants indicate a mutation in one palindromic arm. Variants with mutations in both arms (2 darkened regions) recombine at a greatly reduced rate, driving formation of double barcode constructs (Zhang & Lutz, 2002). The recombination events also reconstruct a split selectable marker, separated by an artificial intron containing the barcodes and loxP variants. By the sequential integration method (A), integration of two plasmids forms a 140 base pair barcode-lox-lox-barcode. By the mating method (B), plasmids are integrated into MATa and MATα cells. MATa and MATα cells are mated and induced to recombine a third loxP variant (lox2272, purple, red “X”), bringing the integrated plasmids to the same chromosome and forming a 195 base pair barcode-lox-lox-lox-barcode.
(C) Number of insertion events from transformations into yeast containing two tandem loxP variants (lox2272-lox5171or loxP-lox2272). Transformations are performed with no plasmid, or plasmids containing no loxP variant, a loxP variant, or a lox5171 variant. Note that different selection markers are used for each landing pad (STAR Methods), so insertion numbers are not comparable between loxP variants.
(D) Pools of barcoded plasmids containing various sized inserts were integrated into yeast at the loxP variant in the landing pad. Violin plots show the distribution of barcode counts at each plasmid size. The width of each violin is fixed, and violin areas are not proportional to the number of barcodes.
(E) The number insertion events per μg plasmid DNA for plasmids containing a loxP or a lox5171 site. URA3AI reconstruction is selected for in both cases.
(F) A histogram of the number of reads per barcode for a yeast library containing ~1.1 million barcodes, constructed by the sequential insertion method. Median = 179, SD = 111.
(G) Lower bound of the rate of mating and reciprocal translocation between homologous chromosomes from a high-density mating protocol.
(H) A histogram of the number of reads per barcode for a yeast library containing ~4.7 million barcodes, constructed by the mating method. Median = 33, SD = 66.
iSeq 2.0 construction fidelity, bias and scalability.
A robust combinatorial screening platform requires high fidelity construction of libraries. Depending on the application, it may also require the ability to screen different sized exogenous DNA elements or extremely large interaction spaces, and to minimize variation in the initial frequency distribution of the library so that all combinations can be economically measured. To test the performance of iSeq 2.0 in these criteria, we performed a series of experiments that follow.
Construction fidelity.
A loxP recombination site consists of an 8-base pair “core” surrounded by two 13 base pair palindromic sequences. LoxP variants with different cores (lox5171 and lox2272) have been reported to recombine efficiently with themselves but not with each other or wild-type loxP (Lee & Saito, 1998). To test the loxP variant specificity in our platform, we integrated plasmids containing a single loxP variant into a landing pad strain that does or does not contain a compatible loxP variant (STAR Methods, Figure 1C). We find that a high rate of integration requires compatible loxP variants, with integration rates between incompatible loxP variants being on par with random genomic integration.
Plasmid integration bias.
iSeq could potentially be used to test combinations of different sized exogenous DNA constructs (e.g. ORFs). However, plasmid size may impact integration efficiency, resulting in frequency biases within the pool. Frequency biases are undesirable because they require deeper sequencing to resolve all genotypes. To test the impact of plasmid size on the genomic integration rate, we constructed various size iSeq-compatible plasmids, with each plasmid design represented by >6000 unique barcodes (STAR Methods). Plasmids were in vitro assembled, transformed into bacteria, isolated from bacterial pools, and integrated into the yeast genome as pools at the landing pad locus. Barcode sequencing revealed that insert size has a minor impact on integration efficiency, with most insert sizes having similar frequency distributions (Figures 1D and S2).
iSeq scalability.
Combinatorial screen size scales roughly exponentially with the number of individual constructs (N × N constructs = N2 combinations). Thus, combinatorial screens are unlikely limited by the number of individual constructs, but rather by how many combinations can be realistically generated and sequenced. For the sequential insertion method, the size generated is limited by the integration efficiency of the second plasmid library to be introduced (N × N double barcodes requires ≥N × N integration events). Using a modified lithium acetate yeast transformation protocol and the split URA3 marker (STAR Methods), we found that plasmids integrate at high efficiency (Figure 1E): a plasmid maxi-prep (1 mg of plasmid DNA) generates an estimated ~107 double barcodes. We validated this scale by integrating 0.9 mg of a complex plasmid library and sequencing one-tenth of recovered integrants (STAR Methods). We found ~1.1 × 106 double barcodes within a reasonably narrow frequency distribution (Figure 1F). For the mating method, the size is limited by both the number of mating events and the number of loxP-mediated chromosomal translocation events. Using a protocol that mates ~1010 cells on a standard agar plate, we found a lower bound mating efficiency of ~11.83% (Figure 1G and STAR Methods). By inducing recombination in mated diploids, we found a lower bound recombination efficiency of ~1.63% (Figure 1G and STAR Methods). Based on these estimates, we predict that a single mating plate can produce > 1.93 × 107 (1010 × 11.83% × 1.63%) double barcoding events. We validated this approximate scale by mating ~4200 barcoded MATa strains to ~1200 barcoded MATα strains and sequencing diploid recombinants. We found ~4.7 × 106 double barcodes (94% of expected), with a distribution that is somewhat broader than the library built by sequential insertion (Figure 1H). Continued selection of the split selectable marker assures that barcodes are not lost. However, some experimental scenarios may require growth in non-selectable media. Under these conditions, we find that the functional split URA3 is lost at a rate of 4.68 × 10−6 per cell per generation (STAR Methods).
Construction and validation of the iSeq 2.0 toolkit.
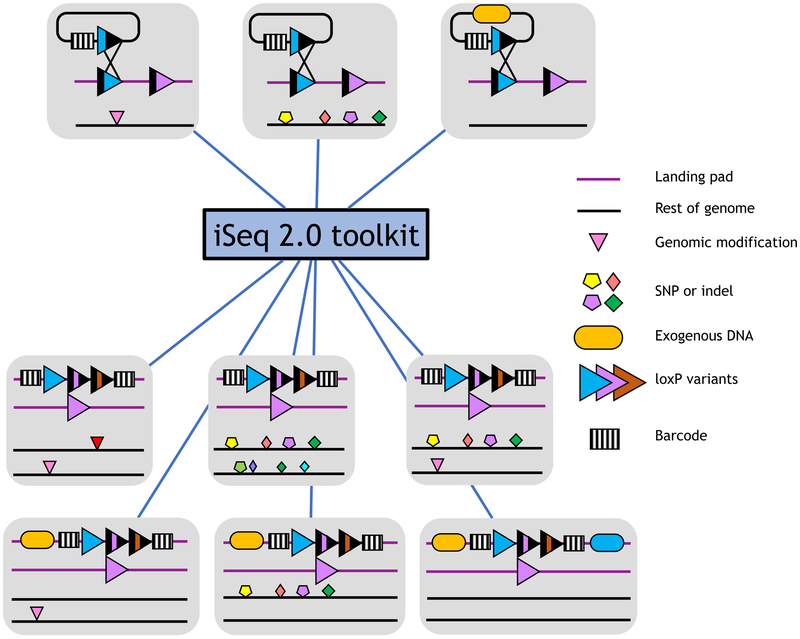
To make the iSeq 2.0 platform easily applicable to a wide variety of potential combinatorial screens in natural, engineered, or industrial yeasts (Figure 2), we next constructed a toolkit consisting of barcoded plasmid libraries with various split selectable markers and two arrays of barcoded yeast strains.
Figure 2. Potential combinatorial screens with iSeq 2.0.
Top: a barcode marks a potential interaction partner. A potential interaction partner can be an engineered genomic modification (left), a genotype (middle), or an exogenous DNA construct (right). Bottom: six potential combinatorial libraries generated using the iSeq mating method. All potential interaction partner libraries on plasmids or in yeast are intercompatible, meaning that constructed libraries can be reused for novel screens.
A versatile set of barcoded plasmid libraries with split drug resistance markers.
Our previous version of iSeq (Jaffe et al, 2017; Schlecht et al, 2017) relied on the split URA3 marker (URA3AI) (Yoshimatsu & Nagawa, 1989), a tool with diverse applications (Yoshimatsu & Nagawa, 1989), (Lee et al, 2008), (Curcio & Garfinkel, 1991), (Jaffe et al, 2017), (Schlecht et al, 2017). URA3AI requires a ura3 auxotroph in the target strain, which can limit its utility for natural, engineered, or industrial yeasts. To address this limitation, we designed a collection of split positive selection markers (HphMXAI, KanMXAI, and NatMXAI) that provide resistance to a drug (hygromycin, G418, and nourseothricin, respectively) and can be used in prototrophic yeast (Goldstein & McCusker, 1999). We found that plasmid integration and reconstruction of each split drug marker provides resistance to the drug, as expected (STAR Methods, Figure S2B–S2D). Using these new split markers in combination with full selectable markers, we constructed a set of 15 iSeq-compatible plasmid libraries, containing between 120,000 and 3,500,000 barcodes each (Table S2). Each library contains a multicloning site for integration of additional DNA constructs.
An array of ~24,000 double barcoder yeast strains.
Many existing yeast libraries could be potentially used with iSeq, such as the YKO (Winzeler, 1999), DaMP (Breslow et al, 2008), GFP (Huh et al, 2003), Tet-promoter (Mnaimneh et al, 2004), insertional mutant (Ross-Macdonald et al, 1999), protein interactome (Tarassov et al, 2008) and SWAp-Tag (Meurer et al, 2018) collections. To facilitate this, we constructed a collection of 11,582 MATa and 13,195 MATα arrayed yeast strains that each have a unique barcode integrated in the mating version of the iSeq landing pad (the “double-barcoder” collection, Table S8 and S9). Strains in this collection contain a dual SGA reporter (can1Δ::MFA1pr-HIS3::MFα1pr-LEU2) (Tong et al, 2004), which, following mating, provides a means by which to isolate haploids of either mating type without the need for tetrad dissections. Additionally, barcoded arrays can be used to partition large oligonucleotide libraries or strain collections into physically separated arrays (Smith et al, 2017) using a REcombinase-Directed Indexing (REDI) method (Figure S3).
Validation of the iSeq 2.0 platform.
To validate that iSeq can be used to carry out quantitative and reproducible combinatorial screens, we made a subset of strains from the protein interactome collection iSeq compatible, and used fitness-based measurements to screen for known protein-protein interactions (Figure S4) (Tarassov et al, 2008; Schlecht et al, 2017). We found that fitness measurements are highly reproducible between replicate growth cultures (Pearson’s r = 0.997) and double barcodes representing the same protein pair (Pearson’s r = 0.961). Additionally, our assay had a similar rate of false positives and false negatives as traditional yeast two hybrid or protein fragment complementation assays. We next tested if iSeq can be used to screen added DNA elements of various sizes. We added combinations of auxotrophic markers to yeast lacking those ORFs, and assayed for their effect on fitness in various environments (Figure S5). We recovered all ORF combinations, despite large differences in their lengths. Here too, fitness estimates correlated well across replicate cultures and double barcodes representing the same combination of ORFs.
iSeq 2.0 applications.
One element of the iSeq toolkit is a “double barcoder” yeast collection that can make existing yeast libraries iSeq compatible. As an extension, combinatorial screens could be performed with collections of natural isolates (Liti et al, 2009) or with segregants from a cross. For example, pooled double barcoder strains could be mated to a genetically divergent strain and sporulated to recover a library of barcoded segregants. Alternatively, iSeq landing pads could be integrated into strain backgrounds of interest, with subsequent integration of barcoded plasmid libraries serving as a genotype marker. One potential caveat of the second method is that transformation and/or integration efficiency may vary between natural isolates or segregants, making large screens more difficult.
While iSeq was initially developed for yeast, Cre-loxP has been demonstrated to function in a wide variety of organisms including mammalian cells (Lo et al, 2003; Morin et al, 2006; Yang et al, 2011). This raises the possibility that iSeq by sequential integration could be ported to function in other cell types. Additionally, introduction of other recombination sites on either the landing pads or the integrating plasmids could allow for additional rounds of plasmid integration, thereby extending the capability of iSeq to test for higher-order interactions. While the number of orthogonal loxP variants is currently limited (Lee & Saito, 1998), additional site-specific recombination systems, such as FLP/FRT, R/RS, or Dre/Rox could be employed (Araki et al, 1992; Chuang et al, 2016; Schlake & Bode, 2002).
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sasha Levy (sflevy@stanford.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast Strains
All the strains used in the study were in the S288c background. The genotypes of all the constructed strains are listed in Table S5.
METHOD DETAILS
Construction of unbarcoded plasmid backbones and complex barcode plasmid libraries
For each library, a plasmid backbone was constructed to contain one half of a split marker (HphMXAI, KanMXAI, NatMXAI, or URA3AI), a full selectable marker (KanMX, HphMX, or NatMX), a multicloning site, an ampR marker, a pBR322 bacterial origin, but no yeast replication origin). Plasmids pBAR1, pBAR6, and pBAR7 were cloned from the following sources by standard methods: 1) plasmid backbone/bacterial origin from pAG32, 2) NatMX, KanMX and HphMX from pAG25, pUG6 and pAG32 respectively, 3) URA3 from pSH47, 5) artificial intron, multiple cloning site (MCS), random barcodes and lox sites were synthesized de novo (IDT). Plasmids pBAR8, pBAR9, pBAR10, pBAR11, pBAR12, pBAR13, pBAR14, pBAR15, pBAR16, pBAR17, pBAR18, and pBAR19 (Table S4) were constructed by inserting de novo synthesized gblocks (IDT) containing 5’HphMX, 3’HphMX, 5’KanMX, 3’KanMX, 5’NatMX, or 3’NatMX (Table S3) into plasmid backbones pBAR6 or pBAR7 via restriction digestion and ligation.
To insert random barcodes into pBAR plasmids, a series of oligos (PXL005, PXL006, PXL296, PXL297), each containing a unique restriction site (KpnI or BamHI or BglII), a barcode region containing random 20 nucleotides, a loxP variant (lox66 or lox5171/66) and a region of homology to pBAR1, were ordered from IDT. PXL005, paired with P23, was used to generate barcodes and lox66 to insert into pBAR7, pBAR12, pBAR19, pBAR20. PXL006, paired with P23, was used to generate barcodes and lox5171/66 to insert into pBAR6, pBAR14, pBAR15, pBAR16, pBAR17. PXL296, paired with P23, was used to generate barcodes and lox66 into pBAR10, pBAR11, pBAR13, pBAR18. PXL297, paired with P23, was used to generate barcodes and lox5171/66 into pBAR8 and pBAR9. Random sequences were limited to 5 nucleotide stretches to prevent the inadvertent generation of restriction sites. For example, to generate a HphMX-lox66 barcode library (pBAR7–L1), the digested PCR product derived from PXL005 was ligated into digested pBAR7. To generate a KanMX-lox5171/66 barcode library (pBAR6–L1), the digested PCR product derived from PXL006 was ligated into digested pBAR6. For each ligation, ~12–15 ng of pooled ligation product was electroporated into 10-beta electrocompetent cells (NEB). Cells were allowed to recover from electroporation in liquid LB media for 30 minutes and plated onto 118 (pBAR7-L1) or 93 plates (pBAR6-L1). Because the oligonucleotide library is highly complex (~420 or ~1012 potential barcodes), it is unlikely that any two transformants contain the same barcode. We therefore estimate the complexity of each barcode library by counting the number of ampicillin-resistant colonies recovered. The pBAR7-L1 library was plated at a density of ~25,500 CFU/plate, for a total of ~3,000,000 colonies. The pBAR6-L1 library was plated at a density of ~17,000 CFU/plate, for a total of ~1,600,000 colonies. During the recovery period in liquid media, some fraction of the cells could have undergone a cell cycle, meaning that our true library complexity is likely to be less than the number of colonies we observed. Colonies of each library were scraped from plates and pooled in 500 ml LB+Carbenicillin. A fraction of each pool was used directly for plasmid preps to generate two plasmid libraries pBAR6-L1 and pBAR7-L1. For the rest of the pBAR plasmids, the primers used in the amplification of barcodes and loxP variants were described above. Following the same electroporation and recovery procedures, one barcode plasmid library was constructed for each pBAR backbone. The detail of these libraries was listed in Table S2.
iSeq toolkit strain construction
Yeast landing pad strains were constructed via four sequential gene replacements. All transformations were performed using a standard high-efficiency lithium acetate method (Gietz & Schiestl, 2007b). First, Gal-Cre-NatMX was amplified from the plasmid pBAR1 (Levy et al, 2015)using the primers (PEV8 and PEV9). This PCR product was then transformed into two S288C derivatives, BY4741 and BY4742 (Brachmann et al, 1998), creating the SHA333 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-NatMX) and SHA319 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ybr209w::GalCre-NatMX) strains (Table S5). Each strain was verified by PCR for successful integration. Second, the magic marker construct, MFA1pr-HIS3-MFα1pr-LEU2(Tong et al, 2004), was amplified from DNA extract from a haploid derivative of UCC8600 (Lindstrom & Gottschling, 2009)using the published primers (P14 and P15) (Tong et al, 2004). The resulting fragment was used to replace CAN1in SHA319 and SHA333 via homologous recombination. This insertion allows for selection of either MATa or MATα haploids via growth on synthetic complete (SC) media containing canavanine and lacking either histidine or leucine respectively. Correct integration was verified by PCR. Yeast strains following this replacement are SHA342 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-NatMX can1::MFA1pr-HIS3-MFα1pr-LEU2) and SHA349 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ybr209w::GalCre-NatMX can1::MFA1pr-HIS3-MFα1pr-LEU2). Third, the NatMX cassette in SHA342 and SHA349 strains was replaced with URA3. The URA3 cassette was amplified from pRS426 with the PXL003 an PXL004. The PCR product was inserted into the genome by homologous recombination to create the XLY001 strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-URA3 can1::MFA1pr-HIS3-MFα1pr-LEU2) and XLY009 strain (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ybr209w::GalCre-URA3 can1::MFA1pr-HIS3-MFα1pr-LEU2). Fourth, URA3 was replaced by homologous recombination with one of three duplex ultramers (PXL008, PXL043, PXL044), each containing a tandem loxP site. These oligos were transformed into XLY001 cells and integration was selected for via 5-Fluoroorotic Acid (5-FOA) counter selection of URA3. This replacement resulted in XLY003 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-lox71-lox5171/71 can1::MFA1pr-HIS3-MFα1pr-LEU2), XLY005 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-lox71-lox2272/71 can1::MFA1pr-HIS3-MFα1pr-LEU2) XLY011 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-lox2272/66-lox5171/71 can1::MFA1pr-HIS3-MFα1pr-LEU2). The sequence of all integrated tandem loxP variants was confirmed by PCR and Sanger sequencing.
To construct strains with multiple auxotrophies that also contain the necessary elements of our interaction sequencing platform, we mated the S288C derivative BY4727 (ATCC) (MATα his3Δ300 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0) (Brachmann et al, 1998), to XLY003, XLY005 and XLY011. Haploid segregants were selected to contain lys2Δ0, trp1Δ63, CAN1, the tandem loxP sites, and the correct mating type by standard methods. Selected segregants are XLY065 (MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ybr209w:: GalCre-lox71-lox5171/71), XLY058 (MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ybr209w:: GalCre-lox2272/66-lox5171/71) and XLY059 (MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ybr209w:: GalCre-lox71-lox2272/71).
iSeq integration specificity
To test the specificity of the recombination between different loxP variants in our barcoding platform, we performed duplicate transformations of two strains containing tandem loxP sites, XLY005 (lox71-lox2272/71) and XLY011 (lox2272/66-lox5171/71), with 700 ng of barcode plasmids that contain no loxP site, a compatible loxP site, or an incompatible loxP site. XLY005 was transformed with pBAR6 (no loxP), pBAR7-L1 (compatible), pBAR6-L1 (incompatible). XLY011 was transformed with pBAR7 (no loxP), pBAR6-L1 (compatible), pBAR7-L1 (incompatible). For each transformation, ~1×108 cells were used. Following transformation, cells were plated on YPG agar overnight. Cell lawns were replica-plated onto the appropriate selectable plates to form colonies. Colonies were counted by hand.
iSeq integration bias
For experiments that measure the impact of plasmid size on integration efficiency, we generated two barcoded libraries by inserting various sizes of non-expressing exogenous DNA fragments. Briefly, three different 1kb fragments were PCR amplified from human KCNIP4 intronic transcript (NCBI Reference Sequence: NR_002813.1). These three 1kb fragments were used to build 1kb, 2kb, 3kb, 4kb, 5kb, 6kb inserts in the multiple cloning sites region on the backbone of pBAR7 via a series of restriction digestion and ligation reactions. The resulting plasmids with six different insert sizes were restriction digested at XhoI and NotI sites. Each of the six inserts was ligated into the cut pBAR6-L1 and pBAR7-L1 plasmid libraries. For each insert size and empty barcode vector (12 in total), 10 to 12 colonies presumably containing a unique barcode were picked and Sanger sequenced to discover the barcode. Clones were arrayed in 96-well plate and grown in LB+Carbenicillin to saturation overnight. Saturated wells of each insert size with same loxP site were pooled together and inoculated into 100 ml LB+Carbenicillin for overnight growth. Saturated cultures of different insert size with the same loxP site were pooled together for plasmid preparation using Plasmid Plus Maxi Kit (QIAGEN) to generate pBAR6-L1-Insert and pBAR7-L1-Insert libraries. To examine the insertion bias resulting from the recombination between lox71 and lox66, ~5 μg of pBAR6-L1 plasmid library was inserted into XLY003, resulting in ~1,000 transformants, which were pooled and transformed with ~ 500 μg pBAR7-L1-Insert library using the large-scale transformation protocol. Cells were grown in liquid YPG + G418 for overnight galactose induction. Transformants were selected by plating cells on 62 SC-Ura plates at a density of ~29,600 CFU/plate for a total of ~1.84 ×106. To examine the insertion bias resulting from the recombination between lox5171/71 and lox5171/66, ~2 μg of pBAR7-L1 plasmid library were inserted into XLY003, resulting in ~350 transformants, which were pooled and transformed with ~300μg pBAR6-L1-Insert library. Cells were grown in liquid YPG + Hyg for overnight galactose induction. Transformants were selected on 59 SC-Ura plates at a density of ~28,000 CFU/plate for a total of ~1.65 ×106.
iSeq integration efficiency, mating efficiency, and recombination efficiency
The number of double barcodes that can be generated using the sequential insertion method depends on the integration efficiency via loxP recombination. To test this efficiency, we first generated two clonal single barcode yeast strains containing a single inserted barcoded plasmid. pBAR7-L1 and pBAR6-L1 were inserted into XLY003 to create XLY091 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-lox71/66-HphMX-Barcode-loxP-lox5171/71 can1::MFA1pr-HIS3-MFα1pr-LEU2) and XLY092 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0ybr209w::GalCre-lox71-lox5171/71/66-Barcode-KanMX-lox5171 can1:: MFA1pr-HIS3-MFα1pr-LEU2), respectively. XLY091 (~1×108cells) and XLY092 (~1×108 cells) was transformed with 1μg pBAR6-L1 and 1μg pBAR7-L1, respectively. After transformation, cells were incubated in 800μl YPG liquid media overnight. Cell density was measured after incubation to estimate the growth during galactose induction (~2 generations). The lower bound assumes growth of ~2 generations following integration, while the upper bound assumes no growth following integration. Transformants were selected by plating cells on SC-Ura plates and the colonies were counted.
The number of double barcodes that can be generated using the mating method depends on 1) the mating efficiency, and 2) the recombination efficiency between lox2272/71and lox2272/66. To estimate these two efficiencies, we first generated two clonal single barcode yeast strains containing a single inserted barcoded plasmid. We inserted pBAR7-L1 into MATa XLY005 to create XLY023 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-lox71/66-HphMXBarcode-loxP-lox2272/71 can1::MFA1pr-HIS3-MFα1pr-LEU2) and pBAR6-L1 into MATα XLY011 to create XLY024 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ybr209w::GalCrelox2272/66-lox5171/71/66-Barcode-KanMX-lox5171 can1:: MFA1pr-HIS3-MFα1pr-LEU2). The two clones were grown to saturation in YPD liquid, mixed in equal volumes, and plated overnight on YPD at a density of 1 × 1010 cells/plate. Cell lawns were scraped, and cells were counted using a Z2 particle counter (Beckman Coulter) to determine the number of cell divisions that occurred on the plate (~1.4 generations).
To estimate the mating efficiency, we plated ~1000, 2000, 3000, 4000 cells of the pool on YPD+G418+Hyg plates in triplicate. Only the mated diploids can grow on YPD+G418+Hyg. The number of colonies was then used to calculate the upper and lower bound of the mating efficiency. The lower bound assumes growth of 1.4 generations following mating, while the upper bound assumes no growth following mating. To test the recombination efficiency between lox2272/71 and lox2272/66, we isolated a single diploid from the above mating, grew this clone overnight in 5 ml YPD, and collected all the cells and inoculated in 5ml YPG overnight. Cells were counted to determine the number of cells divisions that occurred during the incubation (~2 generations). We then plated ~1000, 2000, 3000, 4000 cells from overnight cultures on SC-Ura in triplicate. The numbers of colonies on SC-Ura plates were used to calculate the recombination efficiency between the two lox2272 variants. The lower bound assumes growth of ~2 generations following recombination, while the upper bound assumes no growth following recombination.
Fluctuation test for loss of split URA3 function
A double barcoded clone containing the split URA3 marker was grown in 5mL SC-Ura overnight to saturation. The cell number at saturation (1.5 × 108) was determined on a Coulter counter. Cells were washed with SC media twice and diluted to ~400 cells/μl. Using a LabCyte Echo acoustic liquid dispensing system, we transferred 0.1μl diluted cultures (~40 cells) and 0.9μl SC media into wells of a 96-well plate (12 replicate wells). Cells were grown at 30°C for 2 days in a hydration chamber. Each well was resuspended in water, spotted on 5-FOA plates, and grown at 30°C for five days. FOA-resistant (ura3) colonies were counted by eye. The rate of reversion to ura3 was calculated using bz-rates (Gillet-Markowska et al. 2015), assuming a plating efficiency of 1 and an unknown relative fitness of ura3 vs. URA3 cells in SC (4.68 × 10−6).
Construction of a yeast random barcode library
To construct a yeast barcode library using the sequential insertion method, the pBAR6-L1 plasmid library (10μg) was transformed into ~1 × 108 XLY003 cells. After transformation, cells were plated on one YPG plates overnight. The lawns were replica-plated onto YPD+G418 plates and grown for 2 days. All the cells were pooled and ~1 × 109 of the pool was inoculated into 20 ml YPD+G418 liquid for overnight growth. Freshly made 2 × YPD (500 ml) was inoculated with 5.5 ×109 cells from the above culture and grown for 4 hours. Cells were harvested and transformed with 900 μg pBAR7-L1 using a large-scale high-efficiency transformation protocol (Gietz & Schiestl, 2007a). After transformation, cells were harvested and resuspended in 70 ml YPD+G418 liquid for ~16 hours of galactose induction. After dilution, 1/10th of the cells were plated on 192 SC-Ura plates at a density of ~6,000 CFU/plate, for a total of ~1,150,000 colonies. During the overnight galactose induction, the population underwent between 1 and 2 generations of growth, meaning that our true library complexity is likely to be less than the number of colonies we observe.
To construct a yeast barcode library using the mating method, the pBAR6-L1 plasmid library (4μg) and pBAR7-L1 (2μg) were transformed into ~1 ×108 of XLY011 and XLY005 cells, respectively. After transformation, cells were plated onto YPG plates and grown overnight. The XLY011 and XLY005 transformation lawns were replica-plated on YPD+G418 and YPD+Hyg plates, respectively, and grown for 2 days. The transformants (~1000 for XLY011and ~4000 for XLY005) were pooled and grown in 250 ml of liquid YPD+G418 or YPD+Hyg overnight. To perform the bulk mating, 5 ×1010 cells of each pool were collected and mixed. The mixture was split and plated on 10 YPD agar plates for overnight incubation. Mating pools were scraped and resuspended in YPD+G418+Hyg. The density of the pool was measured to estimate the growth of the population during the overnight mating. All the cells from the bulk mating (~1.75 × 1011) were inoculated into 500 ml YPG+G418+Hyg liquid for overnight shaking. The culture density was measured to estimate the growth during galactose induction. We plated ~11,000 cells from the culture on SC-Ura plates in duplicate to estimate the percentage of successful recombinants in the pool. The estimation of the percentage of URA3+ cells is used to calculate the number of unique mating and recombination events. However, because the population grew between 1 and 2 generations during both mating and galactose induction, the true number of unique mating and recombination events is likely to be less than the number we estimate. All the cells were then inoculated in 4 liters of SC-Ura liquid to enrich for double barcoded cells.
Construction of the double barcoder strain collection by REDI
We first constructed two large yeast pools with high barcode library complexities. Briefly, a single colony of XLY005 and XLY011 was grown in 50 ml YPD liquid overnight to saturation. Then 30 ml of saturated XLY005 cells (~6 × 109) and 45 ml of saturated XLY011 cells (~9 × 109) were inoculated into 1 liter and 1.5 liters of 2 × YPD liquid, respectively, and grown for ~4 hours at 30 °C. Cells were harvested and washed twice with 25 ml of sterile water. XLY005 and XLY011 were transformed with 1.2 mg of pBAR20-L1 and 2 mg of pBAR6-L1, respectively, using a large-scale transformation protocol (Gietz & Schiestl, 2007a). XLY005 and XLY011 transformations were washed twice with sterile water to remove the transformation mixture. Each pool was resuspended in liquid YPG at a density of ~2 × 108 cells/ml and incubated at 30 °C for ~20 hours. The resulting pools were named XLY005 + BC (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ybr209w::GalCre-lox71/66-KanMX-5’URA3-Barcode-loxP-lox2272/71 can1:: MFApr1-HIS3-MFα1pr-LEU2) and XLY011 + BC (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ybr209w::GalCre-lox2272/66-lox5171/71/66-Barcode-3’URA3-KanMX-lox5171 can1:: MFA1pr-HIS3-MFα1pr-LEU2). XLY005 + BC and XLY011 + BC were washed with water and frozen in aliquots at −80°C. A ROTOR HDA (SINGER instruments) was used to assist the colony picking and arraying. Briefly, a vial of frozen XLY005 + BC or XLY011 + BC transformed pools was dissolved in 1 ml water. After measuring the density, ~3 ×106 XLY005 + BC cells or ~6 ×106 XLY011 + BC cells were plated on one YPD + 2 x G418 square plate (400 μg/ml G418). A high concentration of G418 helps to reduce the background of nontransformants or non-integrants. After a ~48-hour incubation at 30°C, colonies were picked and arrayed into 96 or 384-well plates by ROTOR HDA using plate imaging on a flatbed scanner and a home-made MATLAB colony picking algorithm. Non-transformants, non-integrants and colonies in close proximity were filtered out by using colony size and shape thresholds. The above process was repeated to obtain ~14,000 and ~18,000 colonies for XLY005 + BC and XLY011 + BC, respectively. To identify the barcodes in each well, we applied the REDI strategy (Smith et al, 2017). We typically mated any unknown plate to two plates of the opposite mating type with “known” barcodes at each position. After 24-hours on YPD agar plates, colonies were replica-plated onto the selection plates (SC - Met - Lys - Ura + Galactose) to allow recombined diploids to grow. After a 96-hour incubation at 30°C, colonies on each plate were pooled with 5 ml water, and frozen in −20°C. Frozen cells were thawed, and DNA was purified using MasterPureTM Yeast DNA Purification Kit (Epicentre). Barcode PCR and sequencing was performed on each pool, as described below. Double barcodes from each mated pair were sequenced at >100x coverage, and first and second barcodes were independently clustered using Bartender (default settings) (Zhao et al, 2017). Because the physical location of all “known” barcodes on an array is known a priori, a double barcode read unambiguously identifies the position of an “unknown” barcode on the complementary array. To eliminate wells that could contain two or more “unknown” barcodes from the arrays, we selected wells that contained one and only one “unknown” barcode (a single Bartender cluster) across both mated plates (>200 reads total) to re-array. These re-arrayed colonies are the “double barcoder” collection (Table S8 and S9).
Construction of an iSeq library for protein-protein interaction screening
Overview: To demonstrate that the iSeq 2.0 toolkit can adapt existing yeast libraries for use with barcode-based combinatorial screens, we mated the double-barcoder strains to strains from the protein interactome collection (Tarassov et al, 2008). The protein-interactome collection has been previously used to perform large-scale Protein-fragment Complementation Assays (PCAs). These assays use an arrayed mating strategy to test for Protein-Protein Interactions (PPIs) via reconstruction of murine DiHydroFolate Reductase (mDHFR) marker fragments, which confer resistance to the drug methotrexate. Protein interactome strains were mated to the appropriate mating type from the double-barcoder collection, diploids were sporulated, and haploids with the same mating type as the original protein interactome strain were selected (Figure S4A). These barcoded versions of the protein interactome strains were then mated in pools to construct diploid double barcode strains. Using this method, we constructed a set of strains containing protein pairs that have been previously used to benchmark PPI assays in yeast (Yu et al, 2008). Among these are 70 well-documented yeast protein-protein interaction pairs (positive reference set or PRS) and 67 random pairs (random reference set or RRS), each represented by 2–8 unique double barcodes (898 total). Besides the double barcode at the iSeq locus, each strain contains a pair of complementary mDHFR fragments that are in frame and C-terminal to the two genes of interest. In addition, we constructed positive control strains that contain the complete mDHFR gene (10 barcodes) and negative control strains that lack any mDHFR fragment (100 barcodes). We pooled all strains and grew them for 9 generations in serial batch culture in synthetic complete media that does or does not contain 0.5 ug/ml methotrexate, diluting 1:8 every ~3 generations. We sequenced double barcode amplicons at each time point and used the double barcode trajectories to estimate the relative fitness of each strain in the pool (Li et al, 2018). Consistent with our previous version of iSeq (Schlecht et al, 2017), we could not detect any fitness differences between genotypes in the absence of methotrexate (all trajectories are flat, Figure S4E, F, G, H). However, in the presence of methotrexate, significant fitness differences between double barcodes were detected (Figures S4B, C, D). To determine if a protein pair physically interacted, we performed a Student’s t-test of all fitness measures of a protein pair against all fitness measures of the 100 negative control strains (-mDHFR) and defined a positive PPI as p < 10−10 (Bonferroni corrected). We found 14.3% and 1.5% positive PPIs in the PRS and RRS, respectively (Figure S4L). These values roughly correspond to the true positive and false positive rates, respectively, and are similar to traditional Y2H and PCA assays.
To generate positive control strains containing a full length of mDHFR, the DHFR[1,2] fragment was cloned from the plasmid of pAG25 linker-DHFR[1,2]-NatMX (Tarassov et al, 2008) with the primers of pZL071 and PZL072. The plasmid of pAG32 linker-DHFR[3]-HphMX (Tarassov et al, 2008) was linearized with primers of pZL073 and pZL074. Then, the two PCR products were assembled by Gibson assembly to form a new plasmid that contains the full mDHFR sequence (pAG32 linker-DHFR-HphMX). We then used primers pZL065 and pZL070 to PCR the fragment containing linker-mDHFR-HphMX from this plasmid. The linker-DHFR-HphMX cassette was then subcloned into pS413 TEF1pr-His3 between BamHI and XhoI restriction sites 5’ to the TEF1pr sequence, creating the pS413 TEF1pr-linker-DHFR-HphMX plasmid. This plasmid was used to create homologous recombination cassette for HO locus by PCR using 5’ and 3’ oligonucleotides consisting of 40-nucleotide sequences homologous to the upstream and downstream sequences of HO. The TEF1pr-linker-DHFR-HphMX cassette was placed at the ho locus in BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) via homologous recombination. We mated this strain with 10 different MATa barcode strains on YPD plates at 30°C for 24 h, selected for diploids on YPD+clonNat+Hyg+G418 plates at 30 °Cfor 48 h, replicated onto sporulation plates using the Singer ROTOR HDA (Baryshnikova et al, 2010), incubated at room temperature for 7 days, replicated onto SC-Met-Lys-Cys-Arg-Leu+Canavanine+G418+Hyg plates, incubated at 30 °C for a week, and replicated into SC-Met-Lys-Cys-Arg-Leu+Canavanine+G418+Hyg liquid media and incubated at 30 °C for 96 h. This procedure resulted in 10 barcoded MATα methotrexate-resistant strains (MATα his3Δ1 leu2Δ0 ura3Δ0 ybr209w::GalCre-lox71/66-KanMX-3’URA3-Barcode-loxP-lox2272/71
can1::MFApr1-HIS3-MFα1pr-LEU2 ho::mDHFR-HphMX). Each of these 10 barcoded MATα methotrexate-resistant strains was mated with one of 10 barcoded MATa strains (10 mating pairs). Matings were performed on YPD plates at 30 °C for 24 h, diploids were selected on YPD + Hyg + clonNat plates at 30 °C for 48 h, replicated onto SC-Met-Lys-Ura+Galactose plates, incubated at 30 °C for 48 h, replicated into SC-Ura+G418+Hyg+clonNat liquid media, incubated at 30 °C for 48 h, and pooled together to form the final pool of 10 barcoded methotrexate-resistant diploid strains.
To generate the negative control strains lacking mDHFR, the NatMX and HphMX were amplified from plasmids of pAG25-linker-DHFR[1,2]-NatMX and pAG32-linker-DHFR[3]-HphMX using primers containing 40-bp that are homologous to the upstream and downstream of HO. The resulting cassettes were integrated into BY4741 and BY4742, respectively. Using the mating and sporulation protocol described above, we mated BY4741-derived strain to 20 different barcoded MATα strains on YPD plates, selected diploids on YPD + Hyg + clonNat plates, induced meiosis, and selected haploid clones containing 20 different barcodes (MATaybr209w:: GalCre-lox2272/66-lox5171/71/66-Barcode-5’URA3-KanMX-lox5171 can1::MFApr1-HIS3-MFα1pr-LEU2 ho::NatMX his3Δ1 leu2Δ0 ura3Δ0)onSC-Met-Lys-Cys-Arg-His+Canavanine+G418+clonNat plates. Similarly, we mated BY4742-derived strain to 10 different MATa barcode strains on YPD plates, selected diploids on YPD + Hyg + clonNat plates, induced meiosis, and selected haploid clones containing 10 different barcodes (MATαybr209w:: GalCre-lox71/66-KanMX-3’URA3-Barcode-loxP-lox2272/71
can1::MFApr1-HIS3-MFα1pr-LEU2 Ho::HphMX his3Δ1 leu2Δ0 ura3Δ0) onSC-Met-Lys-Cys-Arg-Leu+Canavanine+G418+Hyg plates. Each of the 10 barcoded MATαmethotrexate-sensitive strains was mated with each of the 10 barcoded MATa methotrexate-sensitive strains (100 mating pairs) on YPD plates at 30 °C for 24 h, selected on YPD + Hyg + clonNat plates at 30 °C for 48 h, replicated onto SC-Met-Lys-Ura+Gal plates, incubated at 30 °C for 48 h, replicated into SCUra+G418+Hyg+clonNat liquid media, incubated at 30 °C for 48 h, and pooled together to form the final pool of 100 barcoded methotrexate-sensitive diploid strains
To benchmark the performance of our barcode-based PPI assay, we used a set of 70 likely yeast protein-protein interaction pairs [“positive reference set” (PRS)] and a set of 67 random pairs [“random reference set” (RRS)] that have been used previously for benchmarking. We picked bait and prey versions of ORF-DHFR fragment fusion strains involved in PRS and RRS from Yeast Protein Interactome Collection (Dharmacon, YSC5849) (Tarassov et al, 2008) with the Singer ROTOR HDA robot. We mated each bait and prey strain with two barcoded strains from our double barcoder library on YPD plates at 30 °C for 24 h, selected on YPD+clonNat+G418 plates at 30 °C for 48 h, replicated onto sporulation plates using the Singer ROTOR HDA, incubated at room temperature for 7 days, transferred onto SC-Met-Lys-Cys-Arg-His+Canavanine+G418+clonNat plates, incubated at 30 °C for 96 h, replicated into SC-Met-Lys-Cys-Arg-His+Canavanine+G418+clonNat liquid media, and incubated at 30 °C for 96 h. Similarly, each bait MATα ORF-DHFR[3] strain was mated with two MATa barcode strains on YPD plates at 30 °C for 24 h, selected on YPD+clonNat+Hyg+G418 plates at 30 °C for 48 h, replicated onto sporulation plates and incubated at room temperature for 7 days, transferred on to SC-Met-Lys-Cys-Arg-Leu+Canavanine+G418+Hyg plates and incubated at 30 °C for a week, and replicated into SC-Met-Lys-Cys-Arg-Leu+Canavanine+G418+Hyg liquid media and incubated at 30 °C for 96 h. The PRS, RRS, negative controls and positive controls were grown to saturation separately in SC-Ura media. These four pools were mixed at ~ 2 × 104: 2 × 104: 1 × 104: 1 ratio in cell numbers as a single pool. Cells of this pool (~6.25 × 107 cells) were inoculated into both SC+DMSO and SC+Methotrexate in triplicate. Cells were grown for five days by serial dilution, bottlenecking ~1:8 every 24 hours. Cells grew ~3 generations between each transfer for a total ~9 generations of growth.
Barcode-based combinatorial screening of yeast ORFs
Overview: To demonstrate that the iSeq toolkit can quantitatively screen added DNA elements of various sizes, we added pairs of plasmids, each of which contains an auxotrophic marker (HIS3, LEU2, MET15, LYS2, and TRP1), to yeast strains that lack all of these markers. To measure these fitness effects in both haploid and diploid yeast, we constructed barcoded auxotrophic rescue libraries using both the sequential integration method and the mating method (at least 56 unique barcode pairs per combination) and assayed the fitness of each strain in 12 different growth conditions that either include all amino acids (YPD and SC), or exclude one or two amino acids. We found that fitness estimates of double barcodes correlate well across replicate cultures (0.84 < r < 0.98, Pearson’s r, Figures S5A, B), double barcodes representing the same combination of ORFs integrated in the same orientation (Pearson’s 0.68 < r < 0.82, Figures S5E and S5G), and the opposite orientation (Pearson’s 0.66 < r < 0.79, Figures S5F and S5H). Fitness changes were as expected in both haploids and diploids, with diploid rescues generally causing greater fitness increases (Figures S5C and S5D).
We first generated two barcoded auxotrophic rescue libraries by inserting various ORFs that rescue common yeast auxotrophies into pBAR6-L1 and pBAR7-L1. The MET15, HIS3, TRP1, LEU2, LYS2 ORFs were PCR amplified from pRS421, pRS423, pRS424, pRS425, D1433 his3::LYS2 Disrupter Converter plasmids, respectively (Brachmann et al, 1998; Christianson et al, 1992; Voth et al, 2003). All five ORFs were inserted into pBAR6-L1 or pBAR7-L1 by Gibson Assembly. Briefly, ORFs were amplified with primers that extended the amplicon 20 base pairs at 5’ end and 21 base pairs at the 3’ end (Table S3). Extended 5’and 3’ regions are homologous to sequences in the destination plasmids flanking NheI and BclI restriction sites, respectively. Each library was linearized using the NheI and BclI restriction sites and plasmids were assembled to contain each ORF. Assembled plasmids were inserted into DH5α bacteria by chemical transformation. For each ORF insertion and for plasmids containing a barcode but no ORF, 8–10 clones were picked, and Sanger sequenced to discover the unique barcode. Clones were arrayed in 96-well plates and grown in 200 μl of LB+Carbenicillin to saturation overnight. Saturated cells containing clones with the same loxP site were combined and inoculated into 500 ml LB+Carbenicillin liquid for plasmid preparation using the Plasmid Plus Maxi Kit (QIAGEN). Final libraries, pBAR6-L1-AuxR and pBAR7-L1-AuxR, containing 53 and 52 barcodes, respectively, were subsequently used to generate yeast genomic double barcode libraries.
To insert the first barcoded auxotrophic rescue library into the genome of a haploid, ~60 μg of pBAR6-L1-AuxR plasmid library (53 barcodes) was inserted into XLY065, resulting in ~45,000 transformation events. Transformants were grown for 2 days on selectable media. All the cells were pooled and 1 ×109 of the pool was inoculated into 20 ml YPD+G418 liquid for overnight. Then, ~4.4 ×109 cells from the above culture were inoculated into freshly made 500 ml 2 ×YPD for 4 hours and transformed with ~1 mg of pBAR7-L1-AuxR using the same large-scale transformation protocol as above. Cells were grown in liquid YPG + G418 for galactose induction overnight. Transformants were selected by plating cells on 78 SC-Ura plates at a density of ~ 200,000 CFU/plate for a total of ~1.6 ×107 colonies. Colonies from ten plates were pooled together (~2 ×106) and stored in cryogenic vials with 25% glycerol (~1.68 ×109 cells/vial) at −80°C.
To construct a diploid double barcode library, we first transformed XLY059 (MATa) with pBAR7-L1-AuxR and XLY058 (MATα) with pBAR6-L1-AuxR, resulting in ~15,000 and ~10,000 transformants, respectively. XLY059 and XLY058 transformants were then pooled and mated on three YPD plates as described above, generating in excess of 7 × 105 mating events. All transformants were pooled together and stored cryogenic vials with 25% glycerol (~1.33 × 109 cells/vial) at −80°C.
One stock vial of each auxotrophic rescue yeast barcode library described above containing ~1.68 × 109 (haploid) and 1.33 × 109 (diploid) cells, respectively, was inoculated into 45 ml SCUra media and grown to saturation. Triplicate 5 ml cultures of media lacking zero (YPD and SC), one (SC-Lys, SC-Leu, SC-Met, SC-Trp, SC-His), or two (SC-Met-Leu, SC-Met-His, SC-His-Trp, SC-Lys-Trp, SC-His-Leu) amino acids were inoculated with 625 μl saturated culture (~ 1.2 × 108 (haploid) and ~1.4 × 108 (diploid) cells) of each library (Cysteine was also dropped out in media lacking methionine to select for MET15). Cells were grown for five days by serial dilution, bottlenecking ~1:8 every 24 hours. Cells grew ~3 generations between each transfer for a total ~12 generations of growth. We found that fitness differences caused by the MET15 ORF were extremely subtle. A likely explanation for this is that the MET15 ORF we used (Brachmann et al, 1998) has been reported to incompletely rescue a met15 auxotroph (Cost & Boeke, 1998). In support of this, we found that combinations that contain a Null plasmid (e.g. Null/TRP1) generally cluster closely with combinations that have replaced the Null with the MET15ORF (e.g. MET15/TRP1). Because of this problem with the MET15ORF, we excluded it from our dosage analysis.
Quantitative barcode PCR and amplicon sequencing
Genomic DNA from cells at each transfer was prepared using MasterPureTM Yeast DNA Purification Kit (Epicentre). A two-step PCR was performed, as described (Levy et al, 2015)with modifications. First, 2–3 cycles PCR with OneTaq polymerase (New England Biolabs) was performed. Generally, we amplified 100–500 ng of genomic DNA in a single 50 μl PCR reaction to ensure efficient amplification. Primers (Table S3 Primer list) for this reaction following these formats:
Forward:
ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNXXXXXTTAATATGGACTAAAGGAGGCTTTT
Reverse:
CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNNNNXXXTCGAATTCAAGCTTAGATCTGATA. The Ns in these sequences correspond to any random nucleotide and are used in the downstream analysis to remove skew in the counts caused by PCR jackpotting. The Xs correspond to a one of several multiplexing tags, which allows different samples to be distinguished when loaded on the same sequencing flow cell. The complete list of all primers used in barcode amplicon PCR can be found in Table S3. To minimize the sampling bias during the first PCR, we amplified a minimum of ~500 genome copies per barcode or double barcode in each sample. Therefore, the total amount of template per sample depended on the size of the library. For small libraries (<60,000 barcodes in haploid or <30,000 barcodes in diploid) such as the auxotrophic rescue library, 100 ng (haploid) or 200 ng (diploid) template was used in a single PCR reaction. For larger libraries, multiple PCR reactions were performed, each of which was following the rule of thumb described above. Depending on the size of the library, two different PCR cleanup protocols were used. For small ones, A simple magnetic bead solution with “SeraPure Beads” protocol from (DeAngelis et al, 1995; Rohland & Reich, 2012)and Sera-Mag SpeedBeads (Thermo) was used to enrich target barcode amplicons. Briefly, a 0.6x volume (30μl) of beads solution was added to the PCR tube to precipitate the large template DNA. Then, the resulting supernatant was collected and added with a 1.2x volume (60 μl) of beads solution to precipitate the small barcode amplicons. After size selection, PCR products were eluted into 33 μl of water. For larger ones, PCR products from multiple 50ul reactions were pooled and cleaned using NucleoSpin columns (Macherey-Nagel) and eluted into 33 μl of water. A second 23–25 cycles PCR was performed with PrimeStar HS polymerase (Takara), with 33 μl of cleaned product from the first PCR as template and 50 μl total volume per tube. Primers for this reaction were the standard Illumina paired-end ligation primers (PE1 and PE2). PCR products were cleaned using “SeraPure Beads” and quantitated by Bioanalyzer (Agilent) and Qubit fluorometry (Life Technologies). Cleaned amplicons were pooled and paired end sequenced on an Illumina MiSeq, HiSeq or NextSeq with 25% PhiX DNA spike-in. Sequencing reads were clustered into barcodes by using Bartender with default parameters (Zhao et al, 2017).
Fitness Estimation
Lineage trajectories for each double barcode were used to estimate the fitness using the Fit-Seq software, which uses a likelihood maximization strategy (Li et al, 2018). In the auxotrophic rescue experiment, the fitness of each double barcode was calculated relative to the fitness of strains containing two barcoded empty vectors (no auxotrophic rescue ORF, 100 double barcodes in total).
Validation of split MX cassettes
To test the functionality of all three split drug cassettes (HphMXAI, KanMXAI, NatMXAI), we performed a series of transformations. First, we first transformed XLY003 with three different plasmids (BXL121, BXL122 and BXL124), each of which contains a full drug marker and 3’half of a different drug marker. Single clones of the resulting transformants were isolated to generate XLY082 (3’HphMXAI), XLY083 (3’NatMXAI) and XLY085 (3’KanMXAI), respectively. To test the HphMXAI, XLY082 was transformed with 1 μg of BXL149 (5’HphMXAI-KanMX), or BXL147 (5’KanMXAI-HphMX) and incubated in liquid 800 μl YPG overnight. Transformants were selected by plating on YPD+Hyg plates. To test the NatMXAI, XLY083 was transformed with 1μg of BXL162 (5’NatMXAI-KanMX), or BXL169 (5’HphMXAI-NatMX) and incubated in liquid 800 μl YPG overnight. Transformants were selected by plating on YPD+clonNat plates. To test the KanMXAI, XLY085 was transformed with 1μg of BXL147 (5’KanMXAI-HphMX), or BXL149 (5’HphMXAI-KanMX) and incubated in liquid 800 μl YPG overnight. Transformants were selected by replica-plating on YPD+G418 plates. In addition, we tested the split markers by integrating 5’ half of the cassettes first by transforming XLY003 with three different plasmids (BXL164, BXL147 and BXL149). Isolated single clones of the resulting transformants form above were XLY088 (5’NatMXAI), XLY089 (5’HphMXAI) and XLY090 (5’KanMXAI), respectively. To test the NatMXAI, XLY088 was transformed with 1μg of BXL122 (3’NatMXAI-KanMX), or BXL160 (3’KanMXAI-NatMX) and incubated in liquid 800 μl YPG overnight. Transformants were selected by plating on YPD+clonNat plates. To test the HphMXAI, XLY089 was transformed with 1μg of BXL133 (3’HphMXAI-NatMX), or BXL123 (3’NatMXAI-HphMX) and incubated in liquid 800 μl YPG overnight. Transformants were selected by plating on YPD+Hyg plates. To test the KanMXAI, XLY090 was transformed with 1μg of BXL124 (3’KanMXAI-HphMX), or BXL122 (3’NatMXAI-KanMX) and incubated in liquid 800 μl YPG overnight. Transformants were selected on YPD+G418 plates. Colonies were counted by hand. Transformations that reconstruct drug resistance markers result in fewer integrants than those that reconstruct URA3AI, and integration rates are usually lower for split markers (e.g. HphMXAI) than for full markers (e.g. HphMX). Nevertheless, insertion rates for split drug markers are sufficient to generate >500,000 integrants from a typical plasmid maxiprep (1 mg).
Quantification and Statistical Analysis
Statistical correlations were computed by Pearson correlation tests, comparison between groups were computed by two-sided Student t-tests. All Pearson’s r values are indicated in the figures.
Data availability
The raw read counts, estimated fitness, and protein pair or genotype of each double barcode in PPI screen and ORF screen can be found in Table S6 and S7, respectively. The MATa and MATα barcoder strain collections can be in Table S8 and S9, respectively.
Supplementary Material
Table S6. Related to Figure 2. Data for the PPI screen, including raw read counts, estimated fitnesses, and genotypes.
Table S7. Related to Figure 2. Data for ORF screen including raw read counts, estimated fitnesses, and genotypes.
Table S8. Related to Figure 2. MATa barcoder strains.
Table S9. Related to Figures 2. MATα barcoder strains.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| DH5a Competent cells | Invitrogen | 18265017 |
| NEB 10-betaelectrocompetent cells | NEB | C3020K |
| Chemicals, Peptides, and Recombinant Proteins | ||
| G418 solution (50 mg/mL) | Sigma | 4727878001 |
| Hygromycin B | ThermoFisher | 10687010 |
| Nourseothricin | Jena Bioscience | AB-102XL |
| Carbenicillin disodium | Sigma | 4800-94-6 |
| 5-Fluoroorotic Acid Monohydrate (5-FOA) | US Biological | F5050 |
| Critical Commercial Assays | ||
| Gibson Assembly Cloning Kit | NEB | E5510S |
| PrimeStar HS DNA Polymerase | Clontech | R010 |
| Deposited Data | ||
| Protein-protein interaction barcode counts and estimated fitnesses | This paper | Table S6 |
| ORF rescue barcode counts and estimated fitnesses | This paper | Table S7 |
| MATa barcoder strain collection | This paper | Table S8 |
| MATα barcoder strain collection | This paper | Table S9 |
| Experimental Models: Organisms/Strains | ||
| See Table S5 for yeast strains and genotypes | This paper | Table S5 |
| Oligonucleotides | ||
| See Table S3 for primer sequences | This paper | Table S3 |
| Recombinant DNA | ||
| See Table S4 for plasmid sequences | This paper | Table S4 |
| Software and Algorithms | ||
| Bartender | Zhao et al., 2017 | https://github.com/LaoZZZZZ/bartender-1.1 |
| Fit-Seq | Li et al., 2018 | https://github.com/sashaflevy/Fit-Seq |
Highlights.
A modular and interchangeable platform for pooled interaction screening in yeast
Compatible with exogenous DNA, engineered genomic modifications, or cell genotypes
Includes 15 compatible barcoded plasmid libraries with various split markers
Includes an array of ~24,000 “double-barcoder” yeast strains
Acknowledgements
We are grateful to Ian Ehrenreich, Robert St. Onge, and Takeshi Matsui for suggestions on the manuscript. This work was supported by grants from US National Institute of Health (R01HG008354 and U01HL127522 to SL), The Louis and Beatrice Laufer Center, and the New York State Center for Biotechnology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
SL and XL have filed a patent application (WO2017075529A1) related to this manuscript.
References
- Aakre CD, Herrou J, Phung TN, Perchuk BS, Crosson S & Laub MT (2015) Evolving New Protein-Protein Interaction Specificity through Promiscuous Intermediates. Cell 163:594–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert H, Dale EC, Lee E & Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649–659 [DOI] [PubMed] [Google Scholar]
- Araki H, Nakanishi N, Evans BR, Matsuzaki H, Jayaram M & Oshima Y (1992) Site-specific recombinase, R, encoded by yeast plasmid pSR1. J. Mol. Biol 225:25–37 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, Fiedler D, et al. (2010) Rewiring of Genetic Networks in Response to DNA Damage. Science 330:1385–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Dixon S, Vizeacoumar FJ, Myers CL, Andrews B & Boone C (2010) Chapter 7 - Synthetic Genetic Array (SGA) Analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe In Laboratory Methods in Enzymology: Cell, Lipid and Carbohydratepp 145–179. Academic Press; [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P & Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ & Weissman JS (2008) A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celaj A, Schlecht U, Smith JD, Xu W, Suresh S, Miranda M, Aparicio AM, Proctor M, Davis RW, Roth FP, et al. (2017) Quantitative analysis of protein interaction network dynamics in yeast. Mol. Syst. Biol 13:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH & Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122 [DOI] [PubMed] [Google Scholar]
- Chuang K, Nguyen E, Sergeev Y & Badea TC (2016) Novel Heterotypic Rox Sites for Combinatorial Dre Recombination Strategies. G 36:559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446:806–810 [DOI] [PubMed] [Google Scholar]
- Cost GJ & Boeke JD (1998) A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12:939–941 [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. (2010) The Genetic Landscape of a Cell. Science 327:425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. (2016) A global genetic interaction network maps a wiring diagram of cellular function. Science 353:aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ & Garfinkel DJ (1991) Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis MM, Wang DG & Hawkins TL (1995) Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 23:4742–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diss G & Lehner B (2018) The genetic landscape of a physical interaction. eLife 7:e32472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Mejía JJ, Celaj A, Mellor JC, Coté A, Balint A, Ho B, Bansal P, Shaeri F, Gebbia M, Weile J, et al. (2018) Mapping DNA damage-dependent genetic interactions in yeast via party mating and barcode fusion genetics. Mol. Syst. Biol 14:e7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymond JS (2013) Chapter Twelve - Saccharomyces Cerevisiae Growth Media In Laboratory Methods in Enzymology: Cell, Lipid and Carbohydrate, Lorsch J (ed) pp 191–204. Academic Press; [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R & Marillonnet S (2008) A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA & Krogan NJ (2010) The next frontier of systems biology: higher-order and interspecies interactions. Genome Biol. 11:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Elgort MG, Brandman O, Ives C, Collins SR, Miller-Vedam L, Weibezahn J, Hein MY, Poser I, Mann M, et al. (2012) Functional Repurposing Revealed by Comparing S. pombe and S. cerevisiae Genetic Interactions. Cell 149:1339–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA & Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345 [DOI] [PubMed] [Google Scholar]
- Gietz RD & Schiestl RH (2007a) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc 2:38–41 [DOI] [PubMed] [Google Scholar]
- Gietz RD & Schiestl RH (2007b) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc 2:31–34 [DOI] [PubMed] [Google Scholar]
- Gillet-Markowska A, Louvel G and Fischer G (2015) bz-rates: A Web Tool to Estimate Mutation Rates from Fluctuation Analysis. G3 5:2323–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL & McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A & Bassik MC (2017) Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol 35:463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF & Brasch MA (2000) DNA Cloning Using In Vitro Site-Specific Recombination. Genome Res. 10:1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS & O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M & Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe M, Sherlock G & Levy SF (2017) iSeq: A New Double-Barcode Method for Detecting Dynamic Genetic Interactions in Yeast. G3 7:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G & Saito I (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216:55–65 [DOI] [PubMed] [Google Scholar]
- Lee K, Zhang Y & Lee SE (2008) Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454:543–546 [DOI] [PubMed] [Google Scholar]
- Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS & Sherlock G (2015) Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519:181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ & Elledge SJ (2005) MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat. Genet 37:311–319 [DOI] [PubMed] [Google Scholar]
- Li F, Salit M & Levy SF (2018) Fitness estimation of pooled barcode or amplicon sequencing studies. Cell Systems, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL & Gottschling DE (2009) The Mother Enrichment Program: A Genetic System for Facile Replicative Life Span Analysis in Saccharomyces cerevisiae. Genetics 183:413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. (2009) Population genomics of domestic and wild yeasts. Nature 458:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo J, Lee S, Xu M, Liu F, Ruan H, Eun A, He Y, Ma W, Wang W, Wen Z, et al. (2003) 15,000 Unique Zebrafish EST Clusters and Their Future Use in Microarray for Profiling Gene Expression Patterns During Embryogenesis. Genome Res. 13:455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer M, Duan Y, Sass E, Kats I, Herbst K, Buchmuller BC, Dederer V, Huber F, Kirrmaier D, Štefl M, et al. (2018) Genome-wide C-SWAT library for high-throughput yeast genome tagging. Nature Methods 15:598–600 [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng W-T, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. (2004) Exploration of Essential Gene Functions via Titratable Promoter Alleles. Cell 118:31–44 [DOI] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Kirkpatrick R, Butterfield YS, Young AC, Stott J, Barber S, et al. (2006) Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 16:796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK & Johnston JR (1986) Genealogy of principal strains of the yeast genetic stock center.Genetics 113:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman AS, White KI & Ranganathan R (2016) Origins of Allostery and Evolvability in Proteins: A Case Study. Cell 166:468–480 [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. (2008) Conservation and Rewiring of Functional Modules Revealed by an Epistasis Map in Fission Yeast. Science 322:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N & Reich D (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22:939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. (2014) A Proteome-Scale Map of the Human Interactome Network. Cell 159:1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, Coelho PSR, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K-H, Sheehan A, Symoniatis D, Umansky L, et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413–418 [DOI] [PubMed] [Google Scholar]
- Schlake T & Bode J (2002) Use of Mutated FLP Recognition Target (FRT) Sites for the Exchange of Expression Cassettes at Defined Chromosomal Loci. Biochemistry 33:12746–12751 [DOI] [PubMed] [Google Scholar]
- Schlecht U, Liu Z, Blundell JR, St Onge RP & Levy SF (2017) A scalable double-barcode sequencing platform for characterization of dynamic protein-protein interactions. Nat. Comms 8:15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. (2017) Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 14:573–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA & Waterston RH (2017) DNA sequencing at 40: past, present and future. Nature 550:345–353 [DOI] [PubMed] [Google Scholar]
- Smith JD, Schlecht U, Xu W, Suresh S, Horecka J, Proctor MJ, Aiyar RS, Bennett RAO, Chu A, Li YF, et al. (2017) A method for high-throughput production of sequence-verified DNA libraries and strain collections. Mol. Syst. Biol 13:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Molina MMS, Shames I, Malitskaya Y, Vogel J, Bussey H & Michnick SW (2008a) An in Vivo Map of the Yeast Protein Interactome. Science 320:1465–1470 [DOI] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. (2004) Global Mapping of the Yeast Genetic Interaction Network.Science 303:808–813 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403:623–627 [DOI] [PubMed] [Google Scholar]
- Voth WP, Wei Jiang Y & Stillman DJ (2003) New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20:985–993 [DOI] [PubMed] [Google Scholar]
- Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, Gu Z, Bruno D, Miranda M, Nguyen M, et al. (2007) Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104:12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA (1999) Functional Characterization of the S. cerevisiae Genome by Gene Deletion and Parallel Analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- Wong ASL, Choi GCG, Cheng AA, Purcell O & Lu TK (2015) Massively parallel high-order combinatorial genetics in human cells. Nat. Biotechnol 33:952–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ASL, Choi GCG, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M, et al. (2016) Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc. Natl. Acad. Sci. USA 113:2544–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachie N, Petsalaki E, Mellor JC, Weile J, Jacob Y, Verby M, Ozturk SB, Li S, Cote AG, Mosca R, et al. (2016) Pooled-matrix protein interaction screens using Barcode Fusion Genetics. Mol. Syst. Biol 12:863–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. (2011) A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8:659–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T & Nagawa F (1989) Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science 244:1346–1348 [DOI] [PubMed] [Google Scholar]
- Younger D, Berger S, Baker D & Klavins E (2017) High-throughput characterization of protein–protein interactions by reprogramming yeast mating. Proc. Natl. Acad. Sci. USA 114:12166–12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. (2008) High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science 322:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z & Lutz B (2002) Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein. Nucleic Acids Res. 30:90e–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Liu Z, Levy SF & Wu S (2017) Bartender: a fast and accurate clustering algorithm to count barcode reads. Bioinformatics 34:739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S6. Related to Figure 2. Data for the PPI screen, including raw read counts, estimated fitnesses, and genotypes.
Table S7. Related to Figure 2. Data for ORF screen including raw read counts, estimated fitnesses, and genotypes.
Table S8. Related to Figure 2. MATa barcoder strains.
Table S9. Related to Figures 2. MATα barcoder strains.
Data Availability Statement
The raw read counts, estimated fitness, and protein pair or genotype of each double barcode in PPI screen and ORF screen can be found in Table S6 and S7, respectively. The MATa and MATα barcoder strain collections can be in Table S8 and S9, respectively.