Abstract
Several antibody-targeting cancer immunotherapies have been developed based on T cell activation at the target cells. One of the most potent activators of T cells are bacterial superantigens, which bind to major histocompatibility complex class II on antigen-presenting cells and activate T cells through T cell receptor. Strong T cell activation is also one of the main weaknesses of this strategy as it may lead to systemic T cell activation. To overcome the limitation of conventional antibody–superantigen fusion proteins, we have split a superantigen into two fragments, individually inactive, until both fragments came into close proximity and reassembled into a biologically active form capable of activating T cell response. A screening method based on fusion between SEA and coiled-coil heterodimers was developed that enabled detection of functional split SEA designs. The split SEA design that demonstrated efficacy in fusion with coiled-coil dimer forming polypeptides was fused to a single chain antibody specific for tumor antigen CD20. This design selectively activated T cells by split SEA–scFv fusion binding to target cells.
Keywords: cancer therapy, T cell, antibody engineering, protein chimera, protein engineering, protein splicing, cancer immunotherapy, split protein, superantigen, T cell activation
Introduction
Cancer therapy is experiencing great progress, especially based on application of biological drugs and cell immunotherapy. Nevertheless, cancer remains one of the leading causes of death in developed countries. In the last decade, immunotherapy became an important approach for fighting cancer, where the main goal is to activate the patient's own immune system to specifically recognize and kill tumor cells. Antibody-based therapeutics that target surface antigens expressed on tumor cells are successfully used for treatment of different types of cancer. Although unconjugated monoclonal antibodies are efficient, clinical studies showed that conjugating cytotoxic agents to monoclonal antibodies enhances their clinical utility (1). Antibody–drug conjugates are a class of highly potent biological drugs, composed of an antibody and an effector molecule. One of the immunotherapy approaches to potentiate the effects of monoclonal antibodies includes superantigens as effector molecules.
Superantigens (SAg)2 are potent activators of T lymphocytes and can activate up to 20% of T cells compared with conventional peptide antigens that activate only a small fraction of T cells (0.001% or less) (2, 3). The best characterized superantigens are a family of staphylococcal enterotoxins and streptococcal pyrogenic exotoxins secreted by the Gram-positive bacteria Staphylococcus aureus or Streptococcus pyogenes (3, 4). Superantigens do not need to be processed through antigen-presenting cells but can directly bind to class II major histocompatibility complex (MHC class II) expressed on antigen-presenting cells. Once bound to MHC class II, superantigen binds the T cell receptor (TCR) via the variable region of the TCR β chain (4). This results in activation of both cytotoxic T cells (CD8+) and helper T cells (CD4+), including massive release of cytokines, such as interleukin 2 (IL-2), interferon γ (IFN-γ), tumor necrosis factor α (TNFα), and perforins, which generate strong T cell cytotoxic capacity. The precondition for activating T cells is binding of superantigen to the MHC class II expressed on B cells, dendritic cells, and monocytes (5). SAg-directed T cells can lyse a variety of MHC class II–positive tumor cells. Because all tumor cells do not express MHC class II, to make superantigens selective for tumor antigens, Dohlsten et al. (6–8) exploited the conjugates between WT superantigen staphylococcal enterotoxin A (SEA) from S. aureus and antibody specific for tumor antigens. Because of the high affinity of SEA for MHC class II, a limitation of this approach was a retention of Ab-SEA fusion proteins in normal tissues expressing MHC class II, which caused systemic immune activation and dose-limiting toxicity (9). Therefore to lower the systemic effect of Ab-SAg fusion proteins, the Asp-227 to Ala (D227A) substitution was introduced into the SEA, reducing binding activity to MHC class II without affecting the TCR binding (10, 11). This point mutation lies in the SEA high-affinity MHC class II–binding site, which interacts with β chain of MHC class II complex in zinc-dependent manner. However SEA also contains a low-affinity MHC class II–binding site that interacts with α chain of MHC class II complex (12). Although D227A substitution in SEA reduced the binding affinity to MHC class II, the systemic cytotoxic effect on MHC class II expressing cells was only decreased, but not eliminated (13).
It has been shown that Ab-SEA fusion proteins are cytotoxic for target tumor cells irrespective of their MHC class II expression, in contrast to the induction of cytokine release from T cells which requires the presence of MHC class II–positive cells, such as monocytes. This might be explained by the low-affinity interaction of Ab-SEA fusion proteins and TCR β chain in the absence of MHC class II being sufficient to induce cytotoxic T cells to release granules, whereas induction of cytokine release requires a stronger TCR signal, as seen with the high-affinity interaction of the TCR with the SEA–MHC class II (14).
Here we present a novel approach to overcome the limitation of previous antibody–superantigen fusion proteins. A new generation of superantigens was designed with intact binding site for MHC class II that is able to activate T cell response only upon dimerization triggered by binding to cells expressing target antigen and does not affect MHC class II–positive healthy cells. To achieve this, SEA was split into two fragments, each individually inactive, until the fragments come into close proximity upon binding to target cells, where they reconstitute a biologically active form capable of activating T cell response. To detect functional split superantigen designs, a screening method was developed, where split SEA fragments were fused with interacting protein domains. The functional SEA regained its biologic activity only when split SEA fragments were fused with coiled-coil dimer forming polypeptides; meanwhile, split SEA fragments fused with noninteracting polypeptides did not regain their activity. The effective split SEA design was implemented for targeting B cells by fusion with single chain variable fragment against B cell antigen CD20 (scFv–CD20) for use in cancer immunotherapy.
Results
Design of split SEA
Design of split proteins is a challenging task, because it is difficult to predict which sites would ensure that the reassembled protein has the activity of a parent protein, while each split fragment individually remains inactive, and that the split fragments do not reassemble spontaneously. Split proteins often completely lose their biological activity (15). As superantigen we decided to use SEA, which is highly potent and among the most extensively characterized superantigens.
The split sites within the SEA have been selected based on the following requirements: a) at least one domain should comprise a sufficiently large independent folding domain containing several secondary structure elements, to form the nucleus of folding; b) split sites should preferentially be located in the loop regions, exposed to the solvent, which are less sensitive for the reconstitution of split proteins; and c) split site should inactivate the TCR-binding epitope, which prevents the constitutive T cell activation in the absence of complementation by the second split segment. Overlap of split segment amino acids has been on several occasions demonstrated as an effective strategy enabling the optimal positioning of the split segments (15, 16).
Therefore, to design an efficient split SEA we selected three different split sites based on the above-stated requirements, where each of the split constructs overlapped in nine amino acid residues (Fig. 1). A 3D crystal structure of SEA (17) was used as a guide for the design.
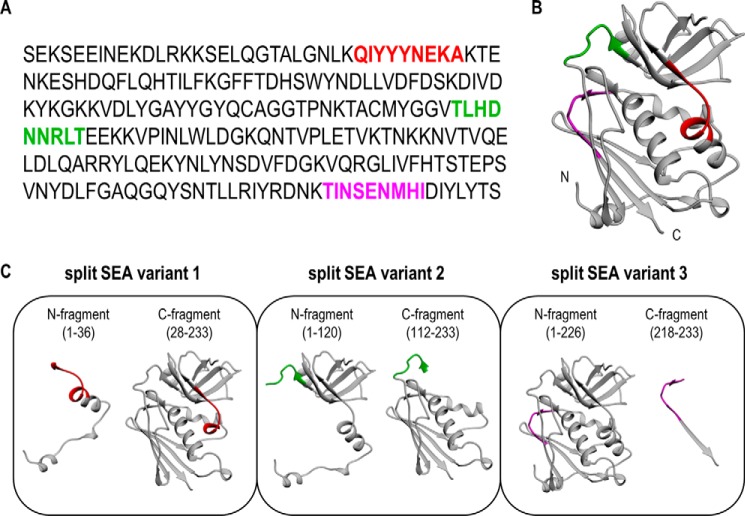
Figure 1.
Design of split SEA variants. A, amino acid sequence of SEA. B, crystal structure of SEA (PDB 1ESF (17)). C, ribbon representation of split SEA variants. Selected three different split sites with overlaps are colored red (split SEA variant 1), green (split SEA variant 2), and magenta (split SEA variant 3). All split designs comprise an overlap of regions of nine amino acid residues.
Screening method for the detection of effective split SEA designs
For the detection of functional split protein designs it is desirable to have an easily measurable readout, similar to the split GFP (18), where the readout is reconstituted fluorescence. Depending on the fluorescence intensity it can be determined which split protein construct exhibits the highest biological activity, whereas in the case of split superantigens, the readout is more complex. To solve this issue we developed a method for screening candidate superantigen split sites, based on split SEA fusion with heterodimerizing polypeptides (Fig. 2B) to select split SEA design which meets three criteria important for further application as cancer therapeutics. First, each split fragment individually should not trigger activation of T cells; second, the reassembled split SEA should be biologically active; and third, split fragments should not spontaneously reassemble into a biologically active form. To monitor the biological activity of split SEA, human peripheral blood mononuclear cells (PBMC) were stimulated with fusion proteins and the release of human cytokine IL-2, as the readout of T cell activation, was detected with an ELISA assay.
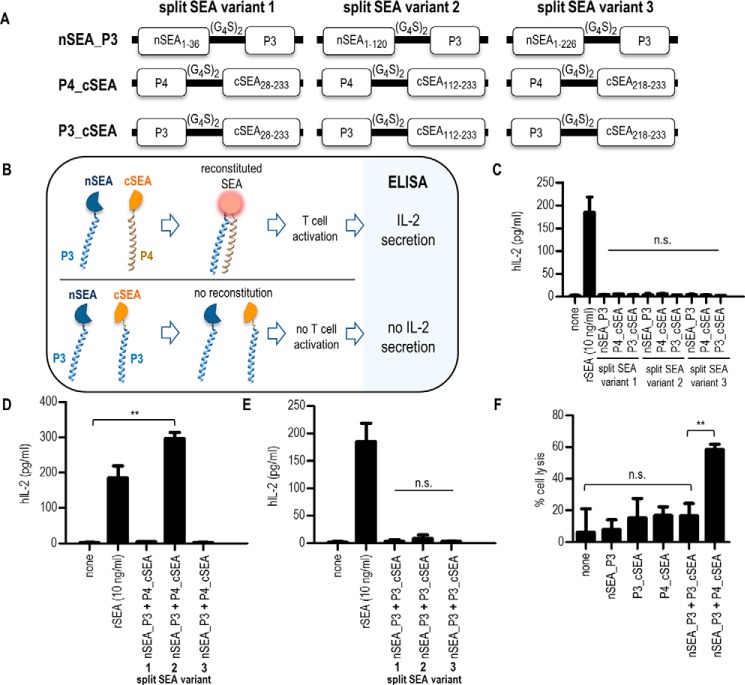
Figure 2.
Screening method for the detection of effective split SEA designs. A, a schematic diagram of split SEA–coiled-coil fusion constructs used in this study. Three different split SEA variant designs were fused with either P3 or P4 polypeptide through glycine–serine linker (10 amino acids). All constructs were cloned into pFLAG–CMV3 vector containing signal peptide for secretion and FLAG tag for detection of expression. B, schematic representation of newly developed screening method for detection of effective split SEA variant designs. Split SEA fragments are fused with P4 and/or P3 polypeptide and split SEA reassemble into an active form only when split SEA fragments are fused with coiled-coil forming P3 and P4 polypeptides, meanwhile split fragments fused with non–coiled-coil forming P3 polypeptides do not regain its biologic activity. For detection of effective split SEA designs, capable of activating T cells, IL-2 is measured by ELISA. C–E, stimulation assays. Briefly, PBMCs were stimulated with 10 ng/ml of recombinant SEA or with supernatant collected from HEK293T cells containing split SEA–coiled-coil fusion proteins. After 24 h incubation at 37 °C, supernatant was collected and the production of human IL-2, as an indicator of T-cell activation, was measured by commercially available ELISA. PBMCs were stimulated with each split SEA–coiled-coil fusion protein separately (50 μl of each) (C), with combination of split SEA fragments fused with P4 and P3 polypeptide (25 μl of each) (D), and with combination of split SEA fragments fused only with P3 polypeptide (25 μl of each) (E). F, cytotoxicity assay. In brief, the target cell line BCWM was incubated with 25 μl of HEK293T supernatant containing split SEA2–coiled-coil fusion proteins. SEA-reactive T cell line was used as effector cell line in ratio 1:10. After overnight incubation the percentage of cell lysis of the BCWM cell line was determined by IVIS Lumina. Data are presented as the mean ± S.D. (n = 3), statistical analysis with a two-tailed t test (**, p < 0.01; n.s., not significant).
Split SEA fragments were fused with coiled-coil dimer-forming P3 and P4 polypeptides (Fig. 2A) designed and tested previously in our lab (19, 20), that form a tight and specific heterodimer. Both polypeptide domains were genetically fused with SEA fragments by a 10 amino acid glycine-serine peptide linker. Coiled-coil dimers were selected because interactions that govern their dimerization specificity are well-understood and orthogonality of used coiled-coil forming polypeptides in mammalian cells ensures that each polypeptide binds preferably to its designated partner polypeptide and does not interfere with other cellular processes.
Our goal was not only to determine the effective split SEA design but also to establish a fast and simple screening method for the detection. Human embryonic kidney 293T (HEK293T) cells were transiently transfected with plasmids coding for fusion proteins that were cloned into a pFLAG–CMV3 vector containing signal sequence for secretion. To confirm that transiently transfected HEK293T cells secrete biologically active recombinant SEA into the media in the amount sufficient to activate T cell response, we first generated a construct with a WT SEA (Fig. S1A) and compared the biological activity of mammalian cell–produced protein with pure SEA (Fig. S1, B and C).
To select functional split SEA designs where each split fragment individually does not activate the T lymphocytes, following the example of recombinant WT SEA, PBMCs were stimulated with a HEK293T supernatant containing secreted split SEA fragments fused with P3 or P4 polypeptide. The presence of fusion proteins in the supernatant was confirmed by the Western blot analysis (Fig. S1D). None of the split SEA fragment fused to the P3 or P4 polypeptide from all three split variant designs caused T cell activation (Fig. 2C). PBMCs were therefore further stimulated with a combination of split SEA fragments fused to P3 or P4 polypeptide to determine whether any of the selected split variant pairs could reassemble into the biologically active form after coiled-coil dimer formation. Only split variant 2 efficiently reassembled, whereas split SEA variants 1 and 3 did not reassemble into a biologically active form (Fig. 2D). To rule out that activation of T cells is a result of the spontaneously reassembled split SEA, PBMCs were stimulated with HEK293T supernatant containing both split SEA fragments fused with a P3 polypeptide, so that the coiled-coil dimer should not form (Fig. 2E). We confirmed that in case of split SEA variant 2 (SEA2), the split fragments indeed have to be in the close proximity to reassemble into a biologically active form that activates T cell response. This is important for further application, because the spontaneously reassembled split SEA could cause T cell activation against nontarget cells expressing MHC class II. To investigate the cytotoxic activity induced by the split SEA2 reassembly, its ability to mediate killing of MHCII+ cells by human reactive T cells was measured. Efficient cell killing of target BCWM cells expressing MHC class II (21) was observed only when split SEA2 fragments were fused with P3 and P4 polypeptides (Fig. 2F), whereas no cytotoxicity was observed by each fusion protein alone. The expression of MHC class II on BCWM cells was confirmed by flow cytometry (Fig. S2).
As seen from the immunoblot analysis (Fig. S1D) fusion proteins P4_cSEA and P3_cSEA of the split SEA variant 3 were not observed in the HEK293T supernatant. To increase the yield of those variants both fusion proteins were fused with the maltose binding protein, which resulted in an improved production (Fig. S3A). Again, the stimulation assay revealed that split SEA variant 3 fragments fused with polypeptides P3 or P4 alone are not able to activate T cells. Activation of T cells did not occur even in case of stimulation with a combination of split SEA3 fragments fused to P3 and P4 polypeptide that form a heterodimer (Fig. S3B).
Because of the favorable results obtained with split SEA2, for which a 3D model of fusion proteins was prepared (Fig. S4), further work was focused on split SEA2.
The impact of linker length and orientation on the reconstitution of split SEA
We first decided to investigate the influence of a linker length on the functionality of reconstituted split SEA. It is known that linker length may strongly affect split protein reassembly (22). DNA constructs were prepared coding for split SEA2 linked to P3 and P4 polypeptides through glycine–serine containing linker of different lengths (Fig. S5). Fusion proteins were expressed in HEK293T cells and secreted into the media. Western blot analysis confirmed the presence of fusion proteins of the expected size in cell suspension (Fig. S5). Few additional bands most likely occur because of the proteolysis or glycosylation. Glycosylation is a frequent posttranslation modification of proteins, and the vast majority of secreted eukaryotic proteins are glycosylated. Because of the glycosylation, proteins migrate differently on SDS-PAGE (23). Therefore for the bands on Western blots with slower mobility than the expected size, we proposed that this is because of the glycosylation. Analysis of the N- and O-linked glycosylation sites revealed positive scores for the potential N-glycosylation sites within split SEA2–coiled-coil fusion proteins. All three potential N-glycosylation sites are present in split SEA fragments and are surface exposed according to the 3D molecular model (Fig. S6).
Stimulation assay revealed that PBMCs stimulated with HEK293T cell suspension containing both split fragments of SEA2 fused with P3 and P4 coiled-coil dimer forming polypeptides, activated T cells to the similar extent regardless of the peptide linker length (Fig. 3A), and were also cytotoxic for target cells expressing MHC class II (Fig. 3B). Cell stimulation and cytotoxicity assays with different amounts of HEK293T supernatant containing split SEA2 linked to P3 and P4 polypeptides alongside integral SEA, revealed a dose-dependent response (Fig. S7).
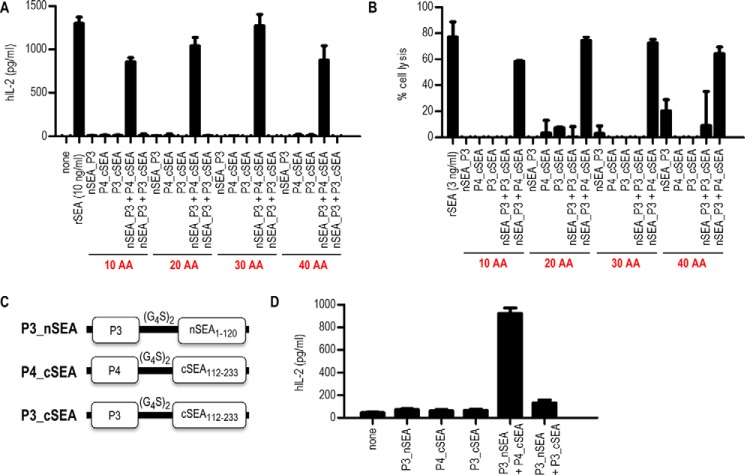
Figure 3.
Characterization of split SEA variant 2 coiled-coil fusion proteins. A, PBMCs were stimulated with 10 ng/ml of recombinant SEA (rSEA) or with 20 μl of supernatant collected from HEK293T cells containing split SEA–coiled-coil fusion proteins with different linker lengths. After 24 h incubation at 37 °C, supernatant was collected and the production of human IL-2, as an indicator of T-cell activation, was measured by commercially available ELISA. B, cytotoxicity assay. In brief, the target cell line BCWM was incubated with 25 μl of HEK293T supernatant containing split SEA2–coiled-coil fusion proteins. SEA-reactive T cell line was used as effector cell line in ratio 1:10. After overnight incubation the percentage of cell lysis of the BCWM cell line was determined by IVIS Lumina. C, a schematic diagram of DNA constructs with changed orientation, where split SEA variant 2 fragments are located on the N-terminal site and fused with either P3 or P4 polypeptide through glycine–serine linker (10 amino acids). D, PBMCs were stimulated with 50 μl of HEK293T supernatant containing split SEA–coiled-coil fusion proteins with changed orientation. After 24 h incubation at 37 °C, supernatant was collected and the production of human IL-2 was measured by commercially available ELISA assay. Data are presented as the mean ± S.D. (n = 3).
The importance of interacting domain orientation has also been reported (24, 25); therefore, we investigated how the orientation of split SEA2 fragments and polypeptides (P3 or P4) might influence the split SEA2 reassembly. DNA constructs were prepared (Fig. 3C) where the interacting domains were at the N terminus of the fusion proteins. Again stimulation assay showed that split SEA2 reassembled when the orientation was reversed, which is likely because of the sufficient length of the linker to enable reconstitution in case of pairing in both orientations (Fig. 3D).
Engineered human scFv–CD20 specifically targets the CD20 tumor antigen
Targeting of split SEA can be provided by fusion with antibodies or specificity determining single chain variable fragment (scFv) against target cell surface markers such as CD20, specific for B lymphocytes. To prepare split SEA2/scFv–CD20 fusion proteins that came into close proximity only upon binding of scFv–CD20 to target cells expressing CD20 at their surface and trigger T cell response, we generated humanized scFv–CD20, comprising humanized variable light (VL) and variable heavy (VH) chain (26), connected by a Whitlow peptide linker (27). Subsequently, a fluorescent protein Venus was fused to scFv–CD20 through a glycine–serine linker (Fig. S8A) to track the binding of scFv–CD20 to target cells. We selected the scFv–CD20 that targets a well-known B cell lymphoma CD20 tumor antigen as a targeting moiety, because it is expressed at high surface density on B cells from the pro-B phase through memory cells, is not rapidly internalized upon binding of antibody, and is not shed from cells (28). For fast characterization, fusion protein was expressed in HEK293T cells and secreted into the media (Fig. S8B), and cell supernatant was incubated with B cell lymphoma line Raji expressing target CD20 antigen. The efficient binding of scFv–CD20 to target CD20 antigen expressed on Raji cells was confirmed by flow cytometry and confocal imaging (Fig. S8C). The same design of scFv–CD20 was further used for fusion with split SEA2.
Design and production of split SEA2/scFv–CD20 fusion proteins
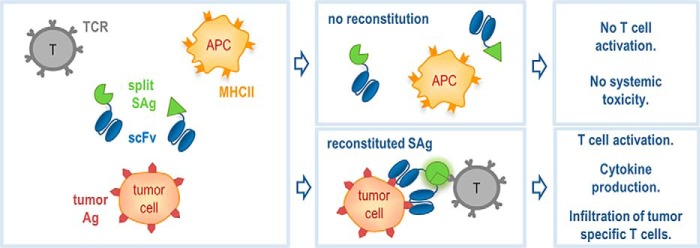
Proof-of-concept fusion proteins were designed where split SEA2 fragments were genetically fused to a scFv–CD20. Binding of two antibodies specific for the tumor antigens at the target cell membrane, fused with split superantigen fragments, were expected to bring the inactive split SEA fragments into close proximity so they could reassemble into the biologically active form that triggers the T cell response (Fig. 4). The main advantage of this approach is the specificity for tumor cells without affecting other cells expressing MHC class II, which should, unlike the standard approach with WT superantigen fused with antibody, decrease the toxic systemic effect of superantigen. A pair of fusion proteins for targeting CD20+ lymphomas was designed. Each fusion protein contained a humanized scFv–CD20, 10 amino acid linker and split SEA2 fragment, nSEA2 or cSEA2, respectively (Fig. 5A).
Figure 4.
Schematic representation of split SEA/scFv fusion proteins for potential use in cancer immunotherapy. Split superantigen can be fused with target seeking moiety, such as antibody, specific for tumor antigen which targets superantigen directly to tumor cells expressing the target antigen. The binding of two antibodies specific for tumor antigens fused with split superantigen fragments brings the inactive split fragments into close proximity so they can be reassembled into biologically active form that activates T cell response. The main advantage of the approach is the specificity only for tumor cells without affecting healthy cells expressing MHC class II, which completely eliminates the toxic systemic effect of superantigen. Moreover superantigen induces infiltration of tumor specific T cells to the tumor site, which is one of the main goals of cancer immunotherapy.
Figure 5.
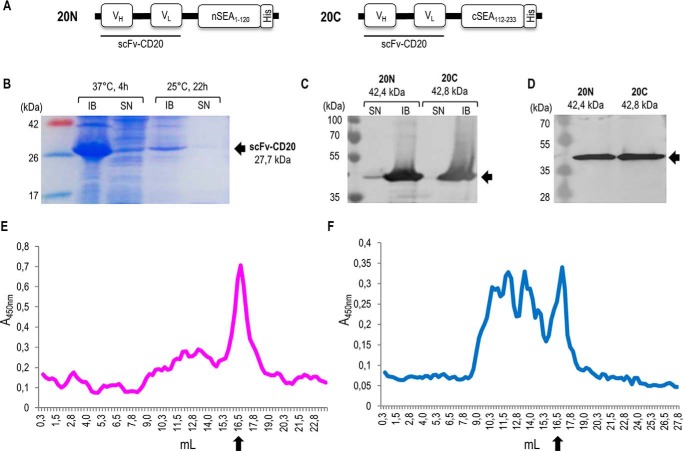
Design and production of split SEA2/scFv-CD20 fusion proteins. A, schematic representation of fusion proteins scFv–CD20/nSEA2 (20N) and scFv–CD20/cSEA2 (20C) in pET-19b vector. VH, variable region of heavy chain; VL, variable region of light chain; (G4S)2, glycine-serine linker; split SEA variant 2 fragments nSEA1–120 and cSEA112–233; His, histidine tag. B, SDS-PAGE analysis of scFv–CD20 produced in E. coli after induction with 1 mm IPTG. Representative soluble (SN) and insoluble (IB) fractions after production at different temperatures and times are shown. C, Western blot analysis of fusion proteins scFv-CD20/nSEA2 and scFv-CD20/cSEA2 produced in E. coli after induction with 0.2 mm IPTG. Soluble (SN) and insoluble (IB) fractions after 4 h production at 37 °C are shown. D, Western blot analysis of purified and refolded fusion proteins scFv–CD20/nSEA2 and scFv–CD20/cSEA2. E and F, analysis of fusion proteins scFv–CD20/nSEA2 (E) and scFv–CD20/cSEA2 (F) by size-exclusion chromatography. Arrows indicate the monomeric form of fusion proteins at the expected molecular mass of 42 kDa.
Both recombinant fusion proteins containing a histidine tag were produced in Escherichia coli and purified on a Ni-NTA column. We aimed to produce recombinant fusion proteins in the bacterial expression system in a soluble form, but although the WT SEA (Fig. S9A) was produced in the soluble form, both split SEA2 fragments (nSEA2 in cSEA2) were insoluble and were restricted to the insoluble fraction (Fig. S9, C and E). The inspection of a crystal structure of SEA with highlighted hydrophobic amino acid residues confirmed that both split SEA2 fragments expose additional hydrophobic surfaces compared with the WT SEA (Fig. S9, D and F). According to the model, splitting of the SEA produced additional solvent-accessible hydrophobic patches, as the total area of hydrophobic surface increased from 924 Å2 to 1988 Å2 corresponding to SEA and split SEA2 fragments combined, respectively (Table S2). Moreover, the portion of solvent-accessible hydrophobic surfaces also increased from 10.4% for WT SEA to 19.5% and 15.5% nSEA2 and cSEA2, respectively (Table S2). Increase in surface hydrophobic patches can be accounted to the emergence of new regions (shown in red in Fig. S9, D and F), which may account for the low solubility of split fusion proteins in aqueous buffers.
Also scFv–CD20 alone was insoluble and produced in the insoluble fraction (Fig. 5B) and similarly scFv–CD20/nSEA2 and scFv–CD20/cSEA2 (Fig. 5C). Several strategies, including strain selection, temperature, and level of induction were tested (data not shown) to produce both fusion proteins in the soluble fraction, however without success. Purification of the GdnHCl solubilized fusion proteins scFv–CD20/nSEA2 and scFv–CD20/cSEA2 was performed on a Ni-NTA column and refolding at low concentrations to hinder aggregation (Fig. 5D). Refolding buffer screening identified 0.5 m arginine as the best solubility enhancer, however that buffer was toxic for cell assays. Hence, although not optimal, 20 mm Tris-HCl, 200 mm NaCl was used to refold fusion proteins, which were then used for cell stimulation experiments without further concentration. Size-exclusion chromatography was performed for both fusion proteins and revealed elution of scFv–CD20/nSEA2 and scFv–CD20/cSEA2 at volumes corresponding to the size of monomers with some aggregates (Fig. 5, E and F).
Binding of split SEA2/scFv–CD20 fusion proteins to CD20 antigen
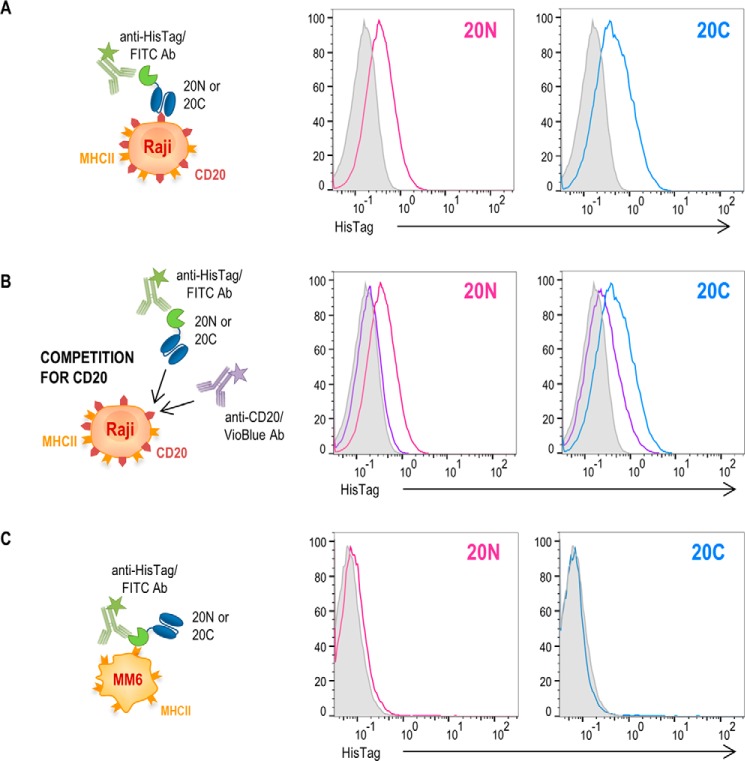
To examine the specificity of split SEA2/scFv–CD20 fusion proteins for tumor antigen CD20, Raji cells were incubated with both fusion proteins separately and binding was detected by anti-HisTag/FITC. As seen in Fig. 6A, both fusion proteins scFv–CD20/nSEA2 and scFv–CD20/cSEA2 efficiently bound to cells expressing CD20 antigen. In addition, we show that in the case of addition of anti-CD20/VioBlue antibody, binding of fusion proteins to the cell surface was strongly decreased because of the competition for the binding site (Fig. 6B). In contrast, both fusion proteins failed to bind to MHCII+CD20− Mono-Mac-6 cells (Fig. 6C), thereby confirming that the fusion proteins bound to target cells via scFv–CD20 and not via split SEA2 fragments interacting with MHC class II complex. The expression of antigen CD20 and/or MHC class II on Raji and Mono-Mac-6 cells was also confirmed by flow cytometry (Fig. S2).
Figure 6.
Target cell binding of split SEA2/scFv-CD20 fusion proteins. A and C, binding assays. Histograms demonstrate surface binding of the indicated fusion proteins at concentration of 200 nm, either scFv–CD20/nSEA2 (20N) and scFv–CD20/cSEA2 (20C), respectively, on target cells Raji (A) or Mono-Mac-6 (C). B, the binding of both fusion proteins, 20N and 20C, respectively, at 200 nm concentration was analyzed on Raji cells in the presence (purple curves) or absence (magenta and blue curves) of anti-CD20/VioBlue for competition. The shaded curves show the target cells stained only with anti-HisTag/FITC.
T cell activation induced by split SEA2/scFv–CD20 fusion proteins
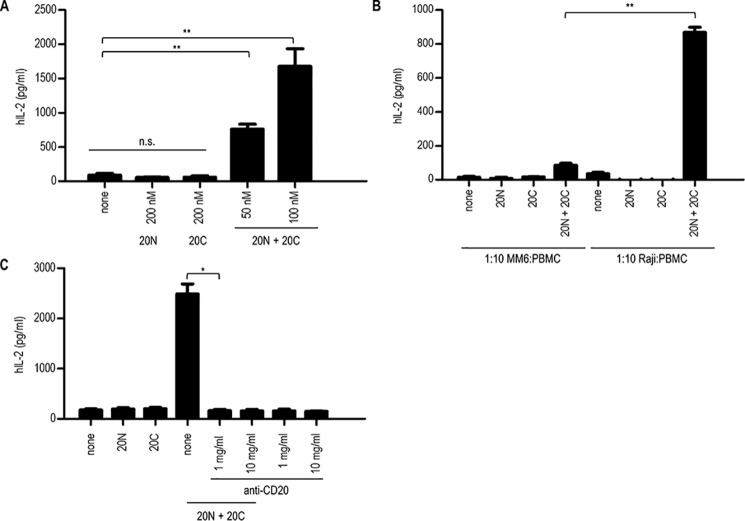
When split SEA2/scFv–CD20 fusion proteins bind to tumor antigens through the scFv domains, the split SEA2 fragments should reassemble into a biologically active form and trigger activation of T cells. To evaluate this, a stimulation assay was performed where B lymphoma Raji cell line (CD20+MHCII+) was used as a target tumor cell line and PBMCs as the source of effector cells. Fig. 7A shows that each fusion protein, scFv–CD20/nSEA2 or scFv–CD20/cSEA2, alone could not induce T cell response; meanwhile, when tumor cells expressing CD20 were incubated with both fusion proteins scFv–CD20/nSEA2 and scFv-CD20/cSEA2, a significant increase in IL-2 production occurred, which demonstrates selective activation of T cells. The main advantage of this approach is that none of the fusion proteins direct T cell activation against antigen-presenting cells expressing MHC class II. To confirm the specificity for tumor cells expressing CD20+, Mono-Mac-6 cell line (CD20−MHCII+) was used in the stimulation assay. As seen in Fig. 7B, when Mono-Mac-6 cells were incubated with both scFv–CD20/nSEA2 and scFv–CD20/cSEA2 fusion proteins, the activation of T cell response was significantly lower compared with activation when Raji cells were used as target cells, confirming increased selectivity of the split SEA2 design strategy. To additionally confirm that the presence of CD20 antigen is required for split SEA2 reconstitution into a bioactive form capable of activating T cell response, the target cells were incubated with anti-CD20 (MabThera) that shares the same epitope with scFv–CD20. When anti-CD20 antibody blocked the target tumor antigen CD20 split SEA2 could not reconstitute into the biological active form; therefore, no T cell activation occurred whereas in the absence of anti-CD20 antibody, split SEA2 fragments reconstituted and activated T cells (Fig. 7C).
Figure 7.
Stimulation assays. A–C, production of human cytokine IL-2 by stimulated PBMCs. Raji cells (A–C) or Mono-Mac-6 (B) were used as a target cell line and PBMCs as effector cells in ratio 1:10. Cells were stimulated with anti-CD20 antibody (C) and/or with 100 nm fusion proteins scFv–CD20/nSEA2 (20N) and/or scFv-CD20/cSEA2 (20C) (A–C). After 24 h incubation at 37 °C, supernatant was collected and the production of human IL-2, as an indicator of T-cell activation, was measured by commercially available ELISA assay. Data are presented as the mean ± S.D. (n = 3), statistical analysis with a two-tailed t test (*, p < 0.05; **, p < 0.01; n.s., not significant).
Discussion
Previous research of superantigen targeting was based on antibody–superantigen fusion proteins composed of target-seeking moieties, typically antibodies, fused to WT (6–8) or mutated SEA with reduced binding affinity to MHC class II (11, 13). Indeed, mutated SEA decreased binding affinity to MHC class II, nevertheless the systemic cytotoxic effect on healthy MHC class II–positive cells has not been eliminated (13). In this report we aimed to design a SEA that would not affect MHC class II expressing cells and would activate T cell response only when bound to a tumor antigen via a targeting moiety. The underlying idea was to design a split SEA where superantigen is split into two fragments, each by itself inactive. The fragments are brought into close proximity by binding to cell surface antigens, dimerize, and reconstitute into a biologically active form capable of activating T cell response.
To identify the effective split SEA designs as quickly and simply as possible, a screening method was developed (Fig. 2B), based on a split SEA fused to dimerizing domains, that enabled us to select and characterize a functional split SEA design using IL-2 release as a marker of T cell activation. The goal was to determine a split SEA site, which met criteria important for further application in a cancer immunotherapy. To additionally simplify the method, all SEA–coiled-coil fusion proteins were produced in HEK293T cells and without any further purification, using cell supernatants in a stimulation assay. This approach demonstrated sufficient production to activate T cell response. Among the three tested split SEA designs only split SEA2 was able to reconstitute and activate T cells when linked with dimerizing pair P3 and P4. It is likely that the reason for the successful reconstitution of SEA2 was because in this case both split domains formed several elements of a secondary structure forming a folding unit.
The controls based on nondimerizing domains demonstrated that activation of T cells is not a result of spontaneously reassembled split SEA2 and that a close proximity of split fragments through interacting domains is required. This screening method based on a split protein fused to heterodimerizing polypeptides can be easily adapted to other effector molecules potentially interesting for cancer immunotherapy. Somewhat surprisingly, the linker length and orientation had no effect on the reconstitution, probably because the smallest length was already sufficiently long to enable pairing in both orientations.
To trigger split SEA2 reconstitution at the target tumor cells, scFv–CD20 was used which targets a well-known CD20 tumor marker, abundantly expressed across the B cell lymphoma cells. Although the concentration of soluble isolated scFv–CD20/nSEA2 and scFv-CD20/cSEA2 fusion proteins was not sufficient for any extended biochemical characterization, because of the high hydrophobicity of split SEA, we were able to perform the biological assay of T cell activation which confirmed binding of both fusion proteins to target antigen CD20. The lack of T cell activation by MHC class II–positive cells confirms the split SEA concept. Target specificity was further confirmed by inhibition of T cell activation by anti-CD20 antibody.
Antibody–superantigen fusion proteins have been designed and used for cancer immunotherapy (29–31). Although mutated and chimeric SEA have lower affinity to MHC class II compared with WT SEA because of the D227A substitution, at higher dosages systemic immune activation and toxic side effects can occur, which may be critical for clinical applications as this limits the maximal tolerated dose. Therefore, split superantigen design is likely to cause a lower systemic cytokine release.
It has been reported that the presence of MHC class II–positive cells, such as monocytes, is required for full T cell activation (14); therefore, for the maximum therapeutic efficiency it would be desirable to have an intact MHC class II–binding site on SEA. Our approach in contrast meets two criteria at the same time: disrupted MHC class II binding, so it does not affect MHC class II–positive cells, whereas the reassembled SEA2 regains all features of the WT SEA.
CD20 antigen selected in this report is an established tumor antigen already used in cancer therapy (target for therapeutic mAb rituximab), and it is expressed at high density on cell membrane. Moreover, it forms a tetramer in the membrane (32), which is favorable for our design as the tetrameric arrangement should bring the split fusion proteins into the close proximity. Tetrameric nature of the target therefore eliminates the requirement for high surface density. For other monomeric surface antigens this could be an issue, limiting the approach to highly expressed surface markers and the need to extend the linker segment length.
One of the drawbacks of SEA in cancer immunotherapy is the preexisting antibodies against SEA, which might however be bypassed by engineering the immunogenic surface exposed residues that do not affect T cell activation (33). SEA–antibody fusion functionally resembles bispecific antibodies called BiTE (bispecific T-cell engager), where anti-CD3 is used to recruit T cells to the tumor site (34). In this case antibody may bind to T cells in the absence of target cells, whereas the split superantigen concentrates all therapeutic protein at target cells limiting T cell activation only to those sites. In addition antibody-targeted superantigen induces infiltration of effector T cells to the tumor site, which is one of the main goals of cancer immunotherapy (35).
In conclusion, we demonstrated that SEA can be split into two fragments, each of them biologically inactive, until they reassemble into a biologically active form upon dimerization. This strategy however opens an additional option by the possibility of linking split SEA fragments with two different antibodies, each specific for different tumor antigen, essentially performing an AND logical function, which could increase the selectivity for tumor cells.
Experimental procedures
Materials
Materials used were as follows: SEA (Sigma), human IL-2 (Roche), mitomycin C (Sigma), IPTG (Gold Bio), lysozyme (Sigma), Benzonase® Nuclease (Merck), CPI protease inhibitors (Sigma), luciferin (PerkinElmer), 5× ELISA/ELISPOT Diluent (eBioscience), 1× TMB solution (eBioscience), and anti-CD20 antibody (MabThera; Roche).
Antibodies used for immunodetection and ELISA were as follows: anti-FLAG antibody produced in rabbit (diluted 1:2000 for immunodetection; Sigma), goat anti-rabbit IgG H+L (HRP) (diluted 1:5000 for immunodetection; Abcam), tetra His antibody (diluted 1:1000 for immunodetection and ELISA; Qiagen), Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) (diluted 1:3000 for immunodetection; diluted 1:4000 for ELISA; Jackson ImmunoResearch).
Antibodies used in flow cytometry analysis and confocal microscopy were as follows: anti-CD20/VioBlue (clone LT20; Miltenyi Biotec), anti-HisTag/FITC (clone GG11-8F3.5.1; Miltenyi Biotec), and anti-HLA-DR, DP, DQ/APC (clone REA332; Miltenyi Biotec).
Design and cloning
The DNA fragments coding for SEA or split SEA (UniProt, A5A2I3), scFv–CD20 (26), Venus (GenBank, AAZ65844.1), maltose binding protein (UniProt, P0AEX9), and P3 and P4 polypeptides (19) were synthesized by Integrated DNA Technologies (gBlocks) and subcloned into mammalian (pFLAG–CMV3) or bacterial expression (pET-19b) vector using standard DNA technology. Amino acid sequences of all used constructs are listed in Table S1.
All cloning of the fusion proteins was performed using Gibson assembly (36) and the various linkers and tag sequences were inserted as part of PCR amplification primers and overlapping sequences for the Gibson assembly. Split SEA–coiled-coil fusion proteins, scFv–CD20/Venus fusion protein, and WT SEA were expressed in mammalian expression system from the CMV promoter of the pFLAG–CMV3 vector. Split SEA2/scFv–CD20 fusion proteins were cloned in pET-19b vector for inducible expression in bacterial expression system. All constructs were verified with DNA sequencing.
Cells
The cell lines used in these studies were the HEK293T cells (ATCC), human B cell lymphoma line Raji (a gift from N. Kopitar-Jerala), BCWM.1 cell line established from the bone marrow aspirate of a patient with Waldenström's macroglobulinemia (21), and human cell line Mono-Mac-6 (a gift from K. Triantafilou).
PBMCs were isolated from heparinized blood of healthy blood donors at the Blood Transfusion Centre of Slovenia, following their informed consent and based on the approval of the Slovenian National Medical Ethics Committee at the Ministry of Health (0120-413/2016-2 KME 38/08/16). Authors state that the study abides by the ethical principles for medical research in humans (Declaration of Helsinki, 2008). All participants signed agreement forms. The cells were isolated by density centrifugation over Ficoll-Paque cushion and incubated in RPMI (Invitrogen Life Technologies), supplemented with 10% (v/v) heat-inactivated FBS (Invitrogen Life Technologies). PBMCs were used for experiments or stored in a secure liquid nitrogen freezer until use.
SEA reactive human T cell line (SEA-T) was established as described (37). Briefly, PBMCs (3 × 106 cells/ml) were activated with mitomycin C–treated Raji cells preincubated with 100 ng/ml SEA (Sigma-Aldrich) in RPMI medium with 10% FBS. The T cell lines were restimulated biweekly with 20 units/ml IL-2, weekly with mitomycin C–treated SEA-coated Raji cells and cultivated for 4–12 weeks before being used in the assay. The viability of the effector cells, as determined by trypan blue exclusion, exceeded 50%. Cells were frozen in 90% FBS and 10% DMSO (Sigma). All cell lines were cultivated at 37 °C in 5% CO2.
Transient production of recombinant proteins in mammalian expression system
In preparation for transient production the HEK293T cells were grown to 50–75% confluence. HEK293T cells were transiently transfected with plasmid pFLAG–CMV containing DNA insert coding for the WT SEA, scFv–CD20/Venus, or the selected split SEA fragments fused with the P3 or P4 polypeptides. For transfection, commercially available transfection reagent polyethylenimine was used according to the manufacturers' protocol. 3–5 days after, transfection media was removed and saved at −20 °C for further analysis.
Immunoblotting
HEK293T supernatant containing fusion proteins was mixed with 4× Laemmli buffer (to the final concentration of 1× Laemmli buffer) and then heat denatured at 95 °C for 5 min. Proteins were separated by SDS-PAGE and transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare). Blots were incubated with appropriate antibodies by the use of iBind Western Systems (Thermo Fisher Scientific) according to the manufacturer's protocol. The immunoblots were visualized on G-box (Syngene) after they were developed using Pico or Femto Sensitivity Substrate (Thermo Fisher Scientific).
Prediction of glycosylation sites
N- and O-linked glycosylation sites were predicted using the NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/)3,4 and NetOGlyc (38) prediction programs.
Production of recombinant proteins in prokaryotic expression system
Recombinant fusion proteins were produced using an E. coli BL21 (DE3) or E. coli Nico21 (DE3) expression system. Cells were transformed with pET-19b plasmid coding for SEA, nSEA2, cSEA2, or scFv–CD20 and grown at 37 °C and 160 rpm in LB medium containing ampicillin (0.1 mg/ml). After induction of protein expression with 0.2 mm or 1 mm IPTG, different incubation times and temperatures were tested as indicated. Cell pellets were resuspended in the lysis buffer (20 mm Tris-HCl, 200 mm NaCl, 1 mg/ml lysozyme, 1 mm MgCl2, CPI protease inhibitors, 10 mm imidazole, 10% glycerol, 15 units/ml Benzonase® Nuclease, pH 7.0) and incubated at RT for 1 h with occasional mixing. After centrifugation at 4 °C at 8000 × g, bacterial inclusion bodies and soluble fraction were assessed by SDS-PAGE or Western blot analysis.
Recombinant fusion proteins were produced using an E. coli Nico21 (DE3) expression system. Cells were transformed with pET-19b plasmid coding for scFv–CD20/nSEA2 or scFv–CD20/cSEA2 and grown at 37 °C and 160 rpm in LB medium containing ampicillin (0.1 mg/ml). After induction of protein expression with 0.2 mm IPTG, incubation proceeded for additional 4 h at 37 °C. Cell pellets were resuspended in the lysis buffer (20 mm Tris-HCl, 200 mm NaCl, 1 mg/ml lysozyme, 1 mm MgCl2, CPI protease inhibitors, 10 mm imidazole, 10% glycerol, 15 units/ml Benzonase® Nuclease, pH 7.0) and incubated at RT for 1 h with occasional mixing. After centrifugation at 4 °C at 8000 × g, bacterial inclusion bodies were solubilized in 100 mm sodium phosphate buffer, pH 7.0, containing 6 m guanidine hydrochloride (6 m GdnHCl buffer) and purified by chelating chromatography (Ni-NTA agarose column; Qiagen, GE). The fusion proteins were eluted with 6 m GdnHCl buffer containing 500 mm imidazole and then refolded by dialysis as followed. The concentration of the affinity-purified denatured fusion proteins was adjusted to 10–100 μg/ml in 6 m GdnHCl buffer. The diluted and denatured fusion proteins were then dialyzed three times against 2 liters of 20 mm Tris-HCl, pH 7.0, 150 mm NaCl buffer and assessed for homogeneity by SDS-PAGE and Western blot analysis.
Size-exclusion chromatography coupled with ELISA
0.5 ml of sample was injected to a size-exclusion column (Superdex 200 Increase 10/300 GL) and separated at 0.5 ml/min (running buffer: 20 mm Tris buffer, pH 7.0, 150 mm NaCl). Fractions of 0.25 ml were collected and further analyzed with ELISA. Sample from each collected fraction was coated to the 96-well high-binding plates (overnight, 4 °C). After washing (with wash buffer: 1× PBS, 0.05% Tween 20) samples were incubated for 1 h at RT with 1× ELISA/ELISPOT Diluent. Then followed the steps of washing, detection with primary antibodies (anti-His antibody; 1 h at RT), washing, secondary antibodies (Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L); 1 h at RT), and washing. After the addition of substrate (TMB solution) the reaction was stopped with 0.16 m sulfuric acid. The plates were read on a microplate reader at 450 nm, and again at 630 nm for correction by subtraction of the reading at 630 nm from that at 450 nm.
Calculation of hydrophobic patches
Total surface area of all hydrophobic patches and the portions in WT SEA and split SEA2 fragments were calculated based on the predictions from SWISS-PDB viewer tool.
Surface molecule detection
Cell lines Raji, Mono-Mac-6, and BCWM were stained with anti-CD20/VioBlue and anti-HLA-DR, DP, DQ/APC according to manufacturers' protocol.
Stimulation assay
PBMCs were seeded in 96-well microplate (5 × 104 cells per well) and co-cultured with different concentrations of recombinant SEA and with HEK293T cell suspension containing WT SEA or split SEA–coiled-coil fusion proteins. The supernatant was harvested 24 h after incubation at 37 °C. Then the human IL-2 was measured using a standard ELISA according to manufacturer's protocol (Ready-Set-Go! ELISA; eBioscience).
Raji or Mono-Mac-6 cell lines were used as a target tumor cell line. 2 × 104 cells were seeded in 96-well microplate and human PBMCs as effector cells were added in ratio 1:10. The cells were co-cultured with recombinant SEA or anti-CD20 antibody (MabThera) and/or with split scFv-CD20/nSEA2 and/or scFv-CD20/cSEA2 fusion proteins, respectively. The supernatant was harvested 24 h after incubation at 37 °C. Then the human IL-2 was measured using a standard ELISA assay according to manufacturers' protocol (ELISA Ready-SET-Go!; eBioscience).
Binding assays
5 × 105 Raji were resuspended in 200 μl scFv–CD20/Venus. After 2 h at 37 °C in the dark, cells were washed and analyzed by flow cytometry or confocal microscopy using the Leica TCS SP5 inverted laser-scanning microscope.
5 × 105 Raji or Mono-Mac-6 cells were resuspended in 90 μl PBS and 10 μl of anti-CD20/VioBlue was added. After 10 min incubation at 2–8 °C in the dark, the cells were washed with PBS and 200 nm scFv–CD20/nSEA2 and scFv–CD20/cSEA2 fusion proteins were added, respectively. Mixture of cells and fusion proteins was incubated for 1–2 h at 37 °C, then washed with PBS, and incubated for 10 min at 2–8 °C in the dark with antibodies anti-HisTag/FITC. Binding of fusion proteins containing histidine tag on tumor antigen CD20 or MHC class II was analyzed by flow cytometry. Flow cytometry analysis was performed using a CyFlow space flow cytometer (Partec). The data were analyzed using FlowJo software (TreeStar).
Cytotoxicity assays
The cytotoxicity was determined by standard luciferase-based assay. In brief, BCWM (expressing firefly luciferase–GFP) cells served as target tumor cells and were co-cultured with effector SEA-T cells in triplicates at the indicated ratio using black-walled 96-well plates with 5 × 104 target cells per well. Recombinant SEA or fusion proteins were added at indicated concentrations. Target cells alone were plated at the same cell density to determine maximal luciferase expression (RLU max). After overnight incubation, d-luciferin at a final concentration 150 μg/ml was added directly to each well. Emitted light was detected with IVIS® Lumina Series III (PerkinElmer) and quantified using Living Image® 4.5.2 (PerkinElmer). Lysis was determined as (1− (RLUsample)/(RLUmax)) × 100.
Statistical analysis
Data are presented as mean ± S.D. Representative graphs and images are shown. The Student's unpaired two-tailed t test was used for the statistical comparison of the data.
Author contributions
A. G.-U. validation; A. G.-U., U. R., and Ž. S. investigation; A. G.-U. visualization; A. G.-U., U. R., and Ž. S. methodology; A. G.-U. and R. J. writing-original draft; A. G.-U. and R. J. writing-review and editing; R. J. conceptualization; R. J. resources; R. J. supervision; R. J. funding acquisition; R. J. project administration; R. J. discussion.
Supplementary Material
Acknowledgments
We are grateful to Fabio Lapenta for the help with optimizing the production of fusion proteins and for many insightful discussions. Our thanks to Jana Aupič for providing P3 and P4 structure models. We thank Bojana Stevović, Irena Škraba, Robert Bremšak, and Darija Oven for the technical support. We thank our generous blood donors for the contribution to this study.
This work was supported by Slovenian Research Agency Grant P4-0176 (to R. J.) and for the Ph.D. study (to A. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9 and Tables S1 and S2.
R. Gupta, E. Jung, and S. Brunak, unpublished work.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SAg
- superantigens
- MHC
- major histocompatibility complex
- TCR
- T cell receptor
- SEA
- staphylococcal enterotoxin A
- PBMC
- peripheral blood mononuclear cells
- GdnHCl
- guanidine hydrochloride
- Ni-NTA
- nickel-nitrilotriacetic acid
- scFv
- single chain variable fragment
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- RT
- room temperature
- RLU
- relative light units.
References
- 1. Kang J. S., and Lee M. H. (2009) Overview of therapeutic drug monitoring. Korean J. Intern. Med. 24, 1–10 10.3904/kjim.2009.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Proft T., and Fraser J. D. (2003) Bacterial superantigens. Clin. Exp. Immunol. 133, 299–306 10.1046/j.1365-2249.2003.02203.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fraser J., Arcus V., Kong P., Baker E., and Proft T. (2000) Superantigens—powerful modifiers of the immune system. Mol. Med. Today 6, 125–132 10.1016/S1357-4310(99)01657-3 [DOI] [PubMed] [Google Scholar]
- 4. Baker M. D., and Acharya K. R. (2004) Superantigens: Structure-function relationships. Int. J. Med. Microbiol. 293, 529–537 10.1078/1438-4221-00298 [DOI] [PubMed] [Google Scholar]
- 5. Hedlund G., Dohlsten M., Lando P.A., and Kalland T. (1990) Staphylococcal enterotoxins direct and trigger CTL killing of autologous HLA-DR+ mononuclear leukocytes and freshly prepared leukemia cells. Cell. Immunol. 129, 426–434 10.1016/0008-8749(90)90218-G [DOI] [PubMed] [Google Scholar]
- 6. Dohlsten M., Hedlund G., Akerblom E., Lando P. A., and Kalland T. (1991) Monoclonal antibody-targeted superantigens: A different class of anti-tumor agents. Proc. Natl. Acad. Sci. U.S.A. 88, 9287–9291 10.1073/pnas.88.20.9287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dohlsten M., Abrahmsén L., Björk P., Lando P. A., Hedlund G., Forsberg G., Brodin T., Gascoigne N. R., Förberg C., and Lind P. (1994) Monoclonal antibody-superantigen fusion proteins: Tumor-specific agents for T-cell-based tumor therapy. Proc. Natl. Acad. Sci. U.S.A. 91, 8945–8949 10.1073/pnas.91.19.8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dohlsten M., Lando P. A., Björk P., Abrahmsén L., Ohlsson L., Lind P. and Kalland T. (1995) Immunotherapy of human colon cancer by antibody-targeted superantigens. Cancer Immunol. Immunother. 41, 162–168 10.1007/BF01521342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brodin T. N., Persson R., Soegaard M., Ohlsson L., d'Argy R., Olsson J., Molander A., Antonsson P., Gunnarsson P. O., Kalland T., and Dohlsten M. (1998) Man-made superantigens: tumor-selective agents for T-cell-based therapy. Adv. Drug Deliv. Rev. 31, 131–142 10.1016/S0169-409X(97)00097-5 [DOI] [PubMed] [Google Scholar]
- 10. Gidlöf C., Dohlsten M., Lando P., Kalland T., Sundström C., and Tötterman T. H. (1997) A superantigen-antibody fusion protein for T-cell immunotherapy of human B-lineage malignancies. Blood 89, 2089–2097 [PubMed] [Google Scholar]
- 11. Forsberg G., Ohlsson L., Brodin T., Björk P., Lando P. A., Shaw D., Stern P. L., and Dohlsten M. (2001) Therapy of human non-small-cell lung carcinoma using antibody targeting of a modified superantigen. Br. J. Cancer 85, 129–136 10.1054/bjoc.2001.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abrahmsén L., Dohlsten M., Segrén S., Björk P., Jonsson E., and Kalland T. (1995) Characterization of two distinct MHC class II binding sites in the superantigen staphylococcal enterotoxin A. EMBO J. 14, 2978–2986 10.1002/j.1460-2075.1995.tb07300.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forsberg G., Skartved N. J., Wallén-Öhman M., Nyhlén H. C., Behm K., Hedlund G., and Nederman T. (2010) Naptumomab estafenatox, an engineered antibody-superantigen fusion protein with low toxicity and reduced antigenicity. J. Immunother. 33, 492–499 10.1097/CJI.0b013e3181d75820 [DOI] [PubMed] [Google Scholar]
- 14. Dohlsten M., Sundstedt A., Björklund M., Hedlund G., and Kalland T. (1993) Superantigen-induced cytokines suppress growth of human colon-carcinoma cells. Int. J. Cancer 54, 482–488 10.1002/ijc.2910540321 [DOI] [PubMed] [Google Scholar]
- 15. Shekhawat S. S., and Ghosh I. (2011) Split-protein systems: Beyond binary protein-protein interactions. Curr. Opin. Chem. Biol. 15, 789–797 10.1016/j.cbpa.2011.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y., Li S., Chen T., Hua H., and Lin Z. (2009) Random dissection to select for protein split sites and its application in protein fragment complementation. Protein Sci. 18, 399–409 10.1002/pro.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schad E. M., Zaitseva I., Zaitsev V. N., Dohlsten M., Kalland T., Schlievert P. M., Ohlendorf D. H., and Svensson L. A. (1995) Crystal structure of the superantigen staphylococcal enterotoxin type A. EMBO J. 14, 3292–301 10.1002/j.1460-2075.1995.tb07336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kent K. P., Childs W., and Boxer S. G. (2008) Deconstructing green fluorescent protein. J. Am. Chem. Soc. 130, 9664–9665 10.1021/ja803782x [DOI] [PubMed] [Google Scholar]
- 19. Gradišar H., Božič S., Doles T., Vengust D., Hafner-Bratkovič I., Mertelj A., Webb B., Šali A., Klavžar S., and Jerala R. (2013) Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 9, 362–366 10.1038/nchembio.1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drobnak I., Gradišar H., Ljubetič A., Merljak E., and Jerala R. (2017) Modulation of coiled-coil dimer stability through surface residues while preserving pairing specificity. J. Am. Chem. Soc. 139, 8229–8236 10.1021/jacs.7b01690 [DOI] [PubMed] [Google Scholar]
- 21. Ditzel Santos D., Ho A. W., Tournilhac O., Hatjiharissi E., Leleu X., Xu L., Tassone P., Neri P., Hunter Z. R., Chemaly M. A. Z., Branagan A. R., Manning R. J., Patterson C. J., Moreau A. S., Ciccarelli B., et al. (2007) Establishment of BCWM.1 cell line for Waldenström's macroglobulinemia with productive in vivo engraftment in SCID-hu mice. Exp. Hematol. 35, 1366–1375 10.1016/j.exphem.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 22. Kondo N., Miyauchi K., Meng F., Iwamoto A., and Matsuda Z. (2010) Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J. Biol. Chem. 285, 14681–14688 10.1074/jbc.M109.067090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Croset A., Delafosse L., Gaudry J.-P., Arod C., Glez L., Losberger C., Begue D., Krstanovic A., Robert F., Vilbois F., Chevalet L., and Antonsson B. (2012) Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. J. Biotechnol. 161, 336–348 10.1016/j.jbiotec.2012.06.038 [DOI] [PubMed] [Google Scholar]
- 24. Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., and Ohad N. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427 10.1111/j.1365-313X.2004.02206.x [DOI] [PubMed] [Google Scholar]
- 25. Paulmurugan R., and Gambhir S. S. (2007) Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal. Chem. 79, 2346–2353 10.1021/ac062053q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen H., Qu Z., and Goldenberg M. D. (December 19, 2006) Anti-CD20 antibodies and fusion proteins thereof and methods of use. U.S. Patent No. 7,151,164
- 27. Whitlow M., Bell B. A, Feng S. L., Filpula D., Hardman K. D., Hubert S. L., Rollence M. L., Wood J. F., Schott M. E., and Milenic D. E. (1993) An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng. 6, 989–995 10.1093/protein/6.8.989 [DOI] [PubMed] [Google Scholar]
- 28. Smith M. R. (2003) Rituximab (monoclonal anti-CD20 antibody): Mechanisms of action and resistance. Oncogene 22, 7359–7368 10.1038/sj.onc.1206939 [DOI] [PubMed] [Google Scholar]
- 29. Giantonio B. J., Alpaugh R. K., Schultz J., McAleer C., Newton D. W., Shannon B., Guedez Y., Kotb M., Vitek L., Persson R., Gunnarsson P. O., Kalland T., Dohlsten M., Persson B., and Weiner L. M. (1997) Superantigen-based immunotherapy: A phase I trial of PNU-214565, a monoclonal antibody-staphylococcal enterotoxin A recombinant fusion protein, in advanced pancreatic and colorectal cancer. J. Clin. Oncol. 15, 1994–2007 10.1200/JCO.1997.15.5.1994 [DOI] [PubMed] [Google Scholar]
- 30. Shaw D. M., Connolly N. B., Patel P. M., Kilany S., Hedlund G., Nordle O., Forsberg G., Zweit J., Stern P. L., and Hawkins R. E. (2007) A phase II study of a 5T4 oncofoetal antigen tumour-targeted superantigen (ABR-214936) therapy in patients with advanced renal cell carcinoma. Br. J. Cancer 96, 567–574 10.1038/sj.bjc.6603567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hawkins R. E., Gore M., Shparyk Y., Bondar V., Gladkov O., Ganev T., Harza M., Polenkov S., Bondarenko I., Karlov P., Karyakin O., Khasanov R., Hedlund G., Forsberg G., Nordle Ö., and Eisen T. (2016) A randomized phase II/III study of naptumomab estafenatox + IFNα versus IFNα in renal cell carcinoma: Final analysis with baseline biomarker subgroup and trend analysis. Clin. Cancer Res. 22, 3172–3181 10.1158/1078-0432.CCR-15-0580 [DOI] [PubMed] [Google Scholar]
- 32. Polyak M. J., Li H., Shariat N., and Deans J. P. (2008) CD20 homo-oligomers physically associate with the B cell antigen receptor: Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. J. Biol. Chem. 283, 18545–18552 10.1074/jbc.M800784200 [DOI] [PubMed] [Google Scholar]
- 33. Erlandsson E., Andersson K., Cavallin A., Nilsson A., Larsson-Lorek U., Niss U., Sjöberg A., Wallén-Öhman M., Antonsson P., Walse B., and Forsberg G. (2003) Identification of the antigenic epitopes in staphylococcal enterotoxins A and E and design of a superantigen for human cancer therapy. J. Mol. Biol. 333, 893–905 10.1016/j.jmb.2003.09.009 [DOI] [PubMed] [Google Scholar]
- 34. Baeuerle P. A., and Reinhardt C. (2009) Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res. 69, 4941–4944 10.1158/0008-5472.CAN-09-0547 [DOI] [PubMed] [Google Scholar]
- 35. Litton M. J., Dohlsten M., Hansson J., Rosendahl A., Ohlsson L., Kalland T., Andersson J., and Andersson U. (1997) Tumor therapy with an antibody-targeted superantigen generates a dichotomy between local and systemic immune responses. Am. J. Pathol. 150, 1607–1618 [PMC free article] [PubMed] [Google Scholar]
- 36. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 37. Rosendahl A., Kristensson K., Riesbeck K., and Dohlsten M. (2000) T-cell cytotoxicity assays for studying the functional interaction between the superantigen staphylococcal enterotoxin A and T-cell receptors. in Bacterial Toxins: Methods and Protocols. Methods in Molecular BiologyTM (Holst O., ed.) Vol. 145, Humana Press, Totowa, NJ: 10.1385/1-59259-052-7:241 [DOI] [PubMed] [Google Scholar]
- 38. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T. B. G., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., and Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.