Abstract
Glucuronoyl esterases (GEs) catalyze the cleavage of ester linkages found between lignin and glucuronic acid moieties on glucuronoxylan in plant biomass. As such, GEs represent promising biochemical tools in industrial processing of these recalcitrant resources. However, details on how GEs interact with their natural substrates are sparse, calling for thorough structure-function studies. Presented here is the structure and biochemical characterization of a GE, TtCE15A, from the bacterium Teredinibacter turnerae, a symbiont of wood-boring shipworms. To gain deeper insight into enzyme–substrate interactions, inhibition studies were performed with both the WT TtCE15A and variants in which we, by using site-directed mutagenesis, substituted residues suggested to have key roles in binding to or interacting with the aromatic and carbohydrate structures of its uronic acid ester substrates. Our results support the hypothesis that two aromatic residues (Phe-174 and Trp-376), conserved in bacterial GEs, interact with aromatic and carbohydrate structures of these substrates in the enzyme active site, respectively. The solved crystal structure of TtCE15A revealed features previously not observed in either fungal or bacterial GEs, with a large inserted N-terminal region neighboring the active site and a differently positioned residue of the catalytic triad. The findings highlight key interactions between GEs and complex lignin-carbohydrate ester substrates and advance our understanding of the substrate specificities of these enzymes in biomass conversion.
Keywords: protein structure, enzyme kinetics, carbohydrate, plant cell wall, enzyme structure, enzyme mechanism, enzyme mutation, biotechnology, carbohydrate esterase, carbohydrate-active enzymes, CE15, glucuronoyl esterase, lignin-carbohydrate complexes, uronic acid
Introduction
Glucuronoyl esterases (GEs)3 are enzymes that act on esters of d-glucuronic acid (GlcA) (1) and belong to the carbohydrate esterase family 15 (CE15) in the carbohydrate-active enzymes database (CAZy; www.cazy.org4 (2)). They have been shown to cleave lignin-carbohydrate (LC) ester bonds between 4-O-methyl-glucuronoyl moieties in xylan and alcohol moieties in lignin (see Fig. 1A) and are therefore suggested as promising tools for reducing plant biomass recalcitrance in industrial settings (3–5). To date, 27 GEs of both fungal and bacterial origin have been biochemically characterized on ester-linked GlcA substrates with varying levels of substitutions and complexity (6–20).
Figure 1.
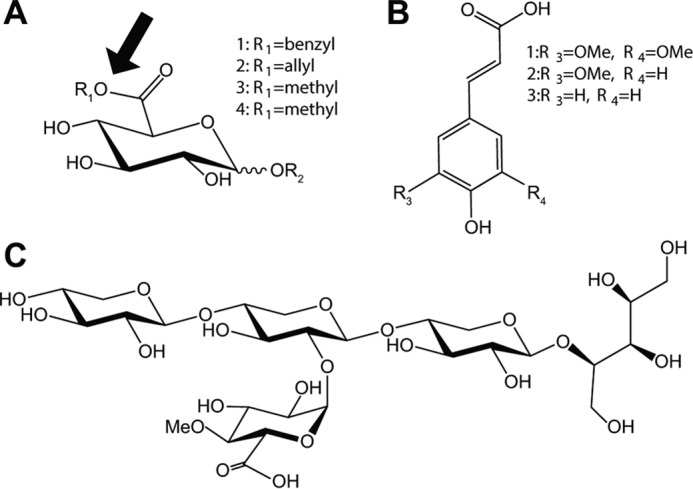
Substrates used for assaying GE activity and compounds used as inhibitors for GE reactions. Esters of uronic acids used for assaying enzyme activity are shown: 1, BnzGlcA; 2, AllylGlcA; 3, MeGlcA; 4, MeGalA (A). The position of the OH group linked to the fourth carbon is equatorial for esters of GlcA and axial for GalA. The fourth position of GlcA is often methylated in native lignocellulose. R2 represents H in the model substrates but indicates the position where 4-O-MeGlcA is α-1,2–linked to the xylan backbone. The site of GE attack is indicated with an arrow. Hydroxycinnamic acids used as GE inhibitors are shown: 1, SA; 2, FA; 3, pCoA (B) and XUXXR (C). B and C compounds were added in increasing concentrations to enzymatic assays with BnzGlcA to investigate any inhibitory effect on GE activity with aromatic and carbohydrate compounds.
Since the discovery of GEs over a decade ago, the majority of research has focused on fungal GEs, whereas the diversity of bacterial CE15 enzymes has only recently begun to be explored (17, 20, 21). We recently published a comprehensive study on bacterial GEs where 10 novel CE15 enzymes with as low as 25% sequence identity among them were systematically characterized on esters of uronic acids (20). The bacterial GEs across the CE15 family demonstrated similarities and differences in substrate specificity, correlating with their phylogenetic positioning. The study also produced two novel CE15 protein structures (OtCE15A from Opitutus terrae and SuCE15C from Solibacter usitatus), which together with the recent structural determination of MZ0003, a CE15 member from a marine arctic metagenome study (21), represent the only bacterial CE15 structures available thus far.
Three factors have been argued as important for GE substrate specificity regarding substitutions on the GlcA moiety: the length of the carbohydrate portion linked to the anomeric carbon (8, 16, 22), the properties of the alcohol (lignin-derived) portion of the ester, and the presence of a 4-O-methyl substituent on the glucuronoate moiety (see Fig. 1A). Although studies indicate that GEs have minimal specificity toward the presence or type of carbohydrates linked to the anomeric position of the GlcA, the enzymes tend to have increased specificity toward bulkier alcohol moieties and a methylation on the 4-OH position (8–10, 13, 14, 22). However, recent characterizations have revealed several bacterial CE15 enzymes with high activities on ester substrates that lack 4-O-methylation, indicating a degree of diversity across the family (20). For fungal GEs, the configuration of the C4 hydroxyl moiety has been suggested to be important due to the lack of activity on esters of d-galacturonic acid (GalA) (6, 8). Similar preferences have been reported for some bacterial GEs, although several bacterial CE15 enzymes are promiscuous and exhibit equal activities on both GlcA- and GalA-derived substrates (20). Additionally, two bacterial CE15 enzymes have significant acetyl esterase activity in addition to their GE activity, which again indicates a broader substrate specificity among bacterial CE15 enzymes (17, 23). The differences in substrate specificity among CE15 enzymes potentially point toward different biological roles, especially in cases where multiple CE15 enzymes are encoded by a single microorganism.
Five GE structures have been determined by X-ray crystallography (20, 21, 24, 25). All of them share a typical α/β-hydrolase fold, with a solvent-exposed active site containing the catalytic residues. The three bacterial structures differ from the two fungal structures (TrGE (Cip2) from Trichoderma reesei and StGE2 from Thermothelomyces thermophila (previously Myceliophthora thermophila and Sporotrichum thermophile)) by three inserted regions and key active site differences. Of the three regions, a prominent insertion of ∼45 residues creates a ridge on one side of the active site, resulting in a significantly deeper cleft compared with the fungal structures (20, 21). The active sites of the bacterial structures have further been proposed to be more flexible due to the absence of disulfide bridges that are present in the fungal structures (21). TrGE (Cip2), StGE2, and the bacterial OtCE15A and SuCE15C all share the same catalytic triad: serine-histidine-glutamate. The triad is, however, not fully conserved in all CE15 enzymes, as glutamate has been found to be replaced by other amino acids in a number of enzymes. In MZ0003, a cysteine is found at the equivalent position of the canonical glutamate, and a closely positioned aspartate is suggested to act as the acidic residue in the reaction (21). Interestingly, an aspartate is found at the equivalent noncanonical position in OtCE15A and SuCE15C in addition to the canonical glutamate.
Only a fraction of the CE15 members listed in CAZy have been characterized, and the properties of different GEs are currently difficult to compare due to use of various GlcA ester model substrates and different biochemical analyses and experimental conditions. In addition, a lack of detailed structure-function studies precludes understanding of the molecular interactions between these enzymes and their substrates, which is essential for understanding their potential role(s) in microbial biomass conversion.
The aim of the present study was to investigate structure-function relationships of TtCE15A, a novel CE15 enzyme from the Gram-negative bacterium Teredinibacter turnerae. T. turnerae has been isolated from the gills of a wood-boring shipworm, a mollusk that digests lignocellulose in marine environments (26). The bacterium encodes multiple CE15 enzymes (27), which were hypothesized to be novel GEs involved in the degradation of lignocellulose on the sea floor. In the present work, TtCE15A was biochemically characterized on uronic acid ester substrates, and its structure was solved by X-ray crystallography. Residues proposed to interact with both the xylan and lignin portions of natural substrates were investigated through comparative inhibition studies using both the WT enzyme and enzyme variants.
Results
Sequence analysis and enzyme production
T. turnerae encodes three CE15 enzymes (TtCE15A, -B, and -C; locus tags TERTU_0517, TERTU_3447, and TERTU_3514, respectively). The enzymes are relatively distant in terms of sequence (37% identity between A and B, 25% identity between A and C, and 51% identity between B and C). TtCE15A is phylogenetically more distant to the fungal CE15 enzymes than are the other two TtCE15 enzymes and the previously characterized bacterial members (20). The most similar characterized CE15 enzymes are the bacterial OtCE15D, SuCE15B, and SlCE15B and -C from Spirosoma linguale, with sequence identities between 39 and 43%. The sequence identities between TtCE15A and the five previously structurally characterized CE15 enzymes range between 30 and 34% (Table S1). TtCE15B and TtCE15C could not be expressed solubly in our experiments, whereas TtCE15A production yielded ∼80 mg of protein/liter of culture broth after nickel-affinity purification.
Biochemical characterization and substrate specificity of TtCE15A
Kinetic parameters were determined on esters of glucuronic and galacturonic acids as well as 4-nitrophenyl acetate (pNP-Ac) (Table 1 and Fig. 1A). Benzyl glucuronoate (BnzGlcA) was used to determine the pH optimum, which was shown to be alkaline (pH 8.5; Fig. S1). TtCE15A exhibited a remarkably high substrate turnover number on BnzGlcA, kcat = 132 s−1, which is more than 2-fold higher than previously reported (58 s−1 for SuCE15C (20)) on this substrate. The Km value for BnzGlcA of 3.5 mm was similar to several other characterized bacterial and fungal GEs (14, 20, 28). The substrate affinity decreased drastically for glucuronoate esters with smaller alcohol portions (allyl and methyl), with a Km of 50 mm for allyl glucuronoate (AllylGlcA) and an unsaturable reaction for methyl glucuronoate (MeGlcA; up to 50 mm), where the Km value consequently could not be determined. The kinetic data emphasize the apparent importance of a larger and bulkier alcohol part of the ester to reach full activity for TtCE15A. Although the kcat/Km was found to be high (between 1 and 40 s−1 mm−1) for TtCE15A on all three GlcA esters, it was 2–3 orders of magnitude lower using methyl galacturonoate (MeGalA) as a substrate (0.012 s−1 mm−1; Table 1). This demonstrates that TtCE15A is a true GE, with minimal promiscuous activity for GalA relative to GlcA. TtCE15A was additionally assayed on pNP-Ac; however, no significant acetyl esterase activity could be detected.
Table 1.
Kinetic parameters of TtCE15A WT and variants on model substrates
Esterase activity was assayed with benzyl, allyl, and methyl esters of GlcA and GalA and pNP-Ac (acetyl esterase activity).
| Substrate | TtCE15A | Km | kcat | kcat/Km |
|---|---|---|---|---|
| mm | s−1 | s−1 m−1 | ||
| BnzGlcA | WT | 3.47 ± 0.28 | 132 ± 3.6 | (3.83 ± 0.16) × 104 |
| S281A | 4.00 ± 0.30 | 0.360 ± 0.010 | (9.05 ± 0.06) × 101 | |
| F174A | 25.8 ± 1.4 | 136 ± 4.8 | (5.25 ± 0.35) × 103 | |
| F174D | 53.1 ± 3.9 | 43.1 ± 2.4 | (8.11 ± 0.75) × 102 | |
| W376A | 22.7 ± 3.8 | 18.3 ± 1.9 | (8.03 ± 1.58) × 102 | |
| W376D | 15.4 ± 4.0 | 12.8 ± 1.8 | (8.32 ± 2.44) × 102 | |
| E374A | 21.8 ± 2.1 | 4.12 ± 0.25 | (1.89 ± 0.22) × 102 | |
| S304E/E374A | 12.7 ± 2.0 | 2.83 ± 0.23 | (2.23 ± 0.20) × 102 | |
| S304E | 29.4 ± 9.6 | 1.92 ± 0.42 | (6.54 ± 2.57) × 101 | |
| AllylGlcA | WT | 49.0 ± 1.7 | 200 ± 4.3 | (4.08 ± 0.17) × 103 |
| F174A | Cannot be saturated up to 50 mm | (4.20 ± 0.02) × 102 | ||
| F174D | Cannot be saturated up to 50 mm | (1.22 ± 0.02) × 102 | ||
| MeGlcA | WT | Cannot be saturated up to 50 mm | (1.03 ± 0.01) × 103 | |
| F174A | Cannot be saturated up to 50 mm | (7.82 ± 0.15) × 101 | ||
| F174D | Cannot be saturated up to 50 mm | (3.17 ± 0.05) × 101 | ||
| MeGalA | WT | Cannot be saturated up to 50 mm | (1.20 ± 0.09) × 101 | |
| pNP-Ac | WT | Cannot be saturated up to 10 mm | 2.50 ± 0.40 | |
Structural determination
Overall structure
To gain deeper insight into the structure-function relationship of TtCE15A and the CE15 family as a whole, structural characterization was pursued. The structure of a selenomethionine-substituted TtCE15A was determined by X-ray crystallography using SAD phasing (Protein Data Bank (PDB) code 6hsw). TtCE15A has an α/β-hydrolase fold similar to previously determined CE15 structures, consisting of a three-layer sandwich with a solvent-exposed cleft comprising the active site with its catalytic triad. Three TtCE15A molecules were found in the asymmetric unit, with a Cα root mean square deviation below 0.2 Å (all atoms <1 Å), indicating a high degree of similarity between the protein chains.
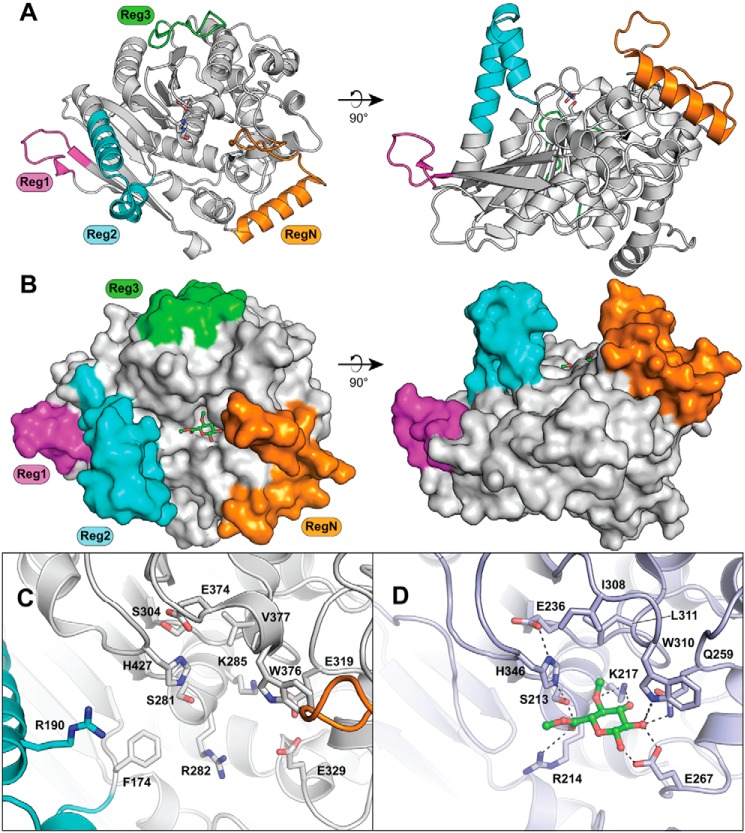
Like the previously determined bacterial structures (20, 21), TtCE15A has three inserted regions relative to the fungal CE15 structures (Reg1, residues 122–134; Reg2, 175–200; Reg3, 399–415) (Fig. 2, A and B). Reg1 and Reg3 have structures similar to the corresponding regions in MZ0003, OtCE15A, and SuCE15C, whereas Reg2 lacks both sequence and structural similarity with the others (Figs. S2 and S3). Although being shorter in sequence (26 versus 40–45 residues), Reg2 of TtCE15A forms a much longer helix compared with the Regs2 of OtCE15A, SuCE15C, and MZ0003. The longer Reg2 helix present in TtCE15A builds up a larger ridge around the previously proposed lignin-binding region and thus presents a deeper substrate-binding groove compared with previously solved structures (Fig. 2, A and B, and Fig. S2). On the ridge built up by Reg2, multiple hydrophobic residues are found on the side facing the active site (Phe-174, Ile-178, Trp-179, Phe-182, Met-186, and Ile-194), whereas the opposite face is more hydrophilic. At the N terminus, TtCE15A has an additional extension (RegN, residues 27–55) consisting of ∼30 residues neighboring the previously proposed xylan-binding region (20). RegN forms a further deepening of the active site pocket opposite Reg2, a feature not present in any previously determined CE15 structure (Fig. 2, A and B). RegN consists of several hydrophilic residues, which may be involved in binding of xylan or xylooligosaccharides.
Figure 2.
Structure of TtCE15A. The overall structure (A) and space-filling representation (B) with the methyl ester of 4-O-methyl glucuronoate substrate shown as green sticks in (B) were generated from structural alignment with the substrate complex structure of StGE2 (PDB code 4g4j). The inserted regions relative to the fungal CE15 enzymes, regions N, 1, 2, and 3, are colored orange, magenta, cyan, and green, respectively. Comparison of the active site organization of TtCE15A (C) and StGE2 (D) shows conservation of residues between the enzymes for binding of the glucuronoate moiety, whereas TtCE15A has portions of RegN and Reg2 additionally protruding into the active site.
Active site pocket
Although all CE15 members exhibit variability in the hydrophilic residues surrounding the active site, several key residues observed to interact with the methyl esterified 4-O-methyl glucuronoate ligand in the structurally solved complex with StGE2 (25) are conserved in TtCE15A: Arg-282, interacting with the glucuronoyl carboxylate moiety; Lys-285, hydrogen bonding to both the 3-OH and the oxygen of the 4-O-methyl group; and Trp-376, hydrogen bonding to the glucuronoyl 2-OH (Fig. 2, C and D). These residues are conserved across all structurally characterized CE15 members (Fig. S3). The conserved tryptophan has also been suggested to interact with the xylan portion of the substrate (20). The recent structural determinations of the first bacterial CE15 enzymes identified a phenylalanine close to the catalytic histidine and serine within the pocket of Reg2 that has been proposed to interact with aromatic components of the LC substrates (20, 21). Although absent in fungal CE15 enzymes, the residue is conserved among all bacterial CE15 members characterized to date, including TtCE15A (Phe-174; Fig. S3).
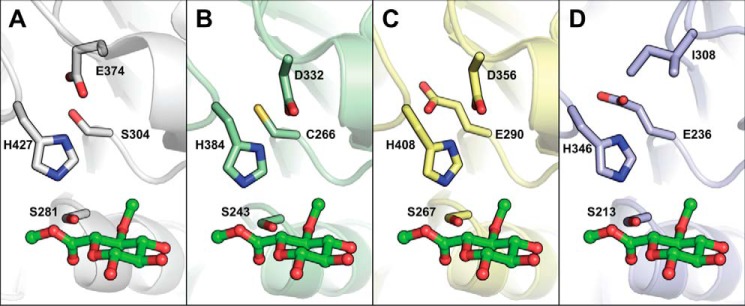
The serine and histidine residues of the catalytic triad found in all previously solved structures (fungal and bacterial) are conserved in TtCE15A (Ser-281 and His-427; Fig. 3). However, the proposed canonical glutamate of the catalytic triad is absent (similar to MZ0003), and this position is occupied by a serine residue (Ser-304) (Fig. 3A). In MZ0003, a closely positioned aspartate (Asp-332) has been proposed to act as the acidic residue of the catalytic triad in place of the missing canonical glutamate (confirmed by mutagenesis) (Fig. 3B). In TtCE15A, a glutamate, Glu-374, is found in an equivalent position, which would likely fulfill the same role. Interestingly, OtCE15A and SuCE15C both have an aspartate at the equivalent position of the Glu-374 of TtCE15A and the Asp-332 of MZ0003 in addition to having the glutamate of the canonical catalytic triad (Fig. 3C). The fungal StGE2 and Cip2 structures both have isoleucine residues in the equivalent position of the noncanonical glutamate/aspartate (Fig. 3D).
Figure 3.
Comparison of catalytic residues among selected structurally characterized CE15 members. The TtCE15A (A), MZ0003 (B; PDB code 6ehn), OtCE15A (C; PDB code 6gs0), and StGE2 (D; PDB code 4g4g) are shown in gray, green, yellow, and blue, respectively. The enzymes are shown relative to the methyl ester of 4-O-methyl glucuronoate substrate shown as green sticks, generated from structural alignment with the complex structure of StGE2 (PDB code 4g4j). The catalytic residues and equivalently placed residues are shown in sticks. Notably, the canonical acidic residue observed in StGE2 is absent in TtCE15A and MZ0003, and instead the acidic residue is found on a different loop, and in OtCE15A, acidic residues are found in both positions.
Investigation of the noncanonical catalytic triad acidic residue
Due to the absence of a canonical glutamate in TtCE15A and the positioning of Glu-374 in the active site, the latter residue was investigated as a potential noncanonical acidic residue in the catalytic triad. Consistent with the proposed role of the residue in the catalytic triad, an E374A variant had a 100-fold decrease in kcat (4.1 s−1) together with an increased Km (21.8 mm) for the model substrate BnzGlcA (Table 1). Similar to previous reports for other serine esterases, substituting the acidic residue may strongly reduce, but not eliminate, the activity of the enzyme as other residues or water molecules can partially substitute for the missing functional group (29). An S304E/E374A variant was produced to assess whether the native high turnover number could be restored by introduction of a glutamate at the position of the canonical acidic residue. However, the turnover number of the S304E/E374A variant on BnzGlcA was not recovered (kcat = 2.8 s−1; Km = 12.7 mm). Furthermore, a variant harboring solely the S304E substitution, i.e. possessing a glutamate in both the canonical and noncanonical positions (Glu-304 and Glu-374) was additionally catalytically crippled (kcat = 1.9 s−1; Km = 29.4 mm) (Table 1). Collectively, the results indicate that the TtCE15A is fine-tuned to utilize Glu-374 as the acidic residue in the catalytic mechanism, supporting the enzyme's high turnover rate, and that the utilization of the residue in this position is distinct from CE15 members exhibiting the canonical acidic residue.
Interaction of Trp-376 with the glucuronoate moiety
In the fungal StGE2, the conserved tryptophan in the active site (Trp-376 in TtCE15A) aids in binding GlcA through hydrogen bonding between the indole nitrogen and the 2-OH of the sugar (25). Enzyme variants of the TtCE15A tryptophan (W376A and W376D) were catalytically compromised, displaying 5-fold increased Km values and 10-fold decreased kcat values (Table 1). These results indicate that the tryptophan makes a considerable contribution to the interaction with GlcA, which when perturbed reduces catalytic efficiency on the model substrates.
Investigation of the putative lignin-binding pocket
Because the bulky benzyl moiety on the model substrate BnzGlcA appears to be crucial for substrate affinity, the possible inhibition of the action of TtCE15A on BnzGlcA by aromatic molecules was investigated. Benzyl alcohol and three different hydroxycinnamic acids, ferulic (FA), sinapic (SA), and p-coumaric acids (pCoA) (Fig. 1B), common phenolic compounds in lignocellulose-derived streams, were investigated as potential inhibitors of the BnzGlcA esterase reaction. We determined the IC50 values (amount of inhibitor required to reduce the reaction rate by 50%) for these reactions, where possible, to be able to estimate the extent of the inhibitory effect(s).
Inhibition was observed when FA, SA, and pCoA were added to the reactions individually. The IC50 values for these compounds were 0.55, 0.56, and 0.83 mm, respectively, in reactions containing 0.4 mm BnzGlcA (Table 2 and Fig. 4, A–C). No inhibition of TtCE15A activity was observed when adding benzyl alcohol (the product of the BnzGlcA reaction; up to 12 mm). The lower IC50 values of FA and SA as inhibitors compared with pCoA suggests that the methoxy groups of FA and SA may provide interactions with the active site residues. The overall results from the inhibition experiments using hydroxycinnamic acids indicated that there are interactions between the active site of TtCE15A and aromatic molecules. Following previous docking studies performed on OtCE15A using a benzyl esterified glucuronoxylotriose oligosaccharide (20), specific interactions of a phenylalanine residue with the aromatic portions of LC structures were postulated. Enzyme variants of the equivalently positioned Phe-174 of TtCE15A were therefore created to investigate interaction with the benzyl moiety.
Table 2.
IC50 values of different compounds on the TtCE15A WT enzyme and enzyme variants using BnzGlcA as substrate
NM, not measured; NI, no/minor inhibition detected (>87% retained enzyme activity after maximum addition of inhibitor).
| TtCE15A | FA | SA | pCoA | XUXXR |
|---|---|---|---|---|
| mm | mm | mm | mm | |
| WT | 0.55 ± 0.045 | 0.56 ± 0.030 | 0.83 ± 0.04 | 5.51 ± 0.28 |
| F174A | 0.084 ± 0.005 | 0.19 ± 0.022 | 4.84 ± 0.47 | NM |
| F174D | 0.40 ± 0.040 | 0.77 ± 0.064 | 26.0 ± 12.0 | NM |
| W376A | NM | NM | NM | NI |
Figure 4.
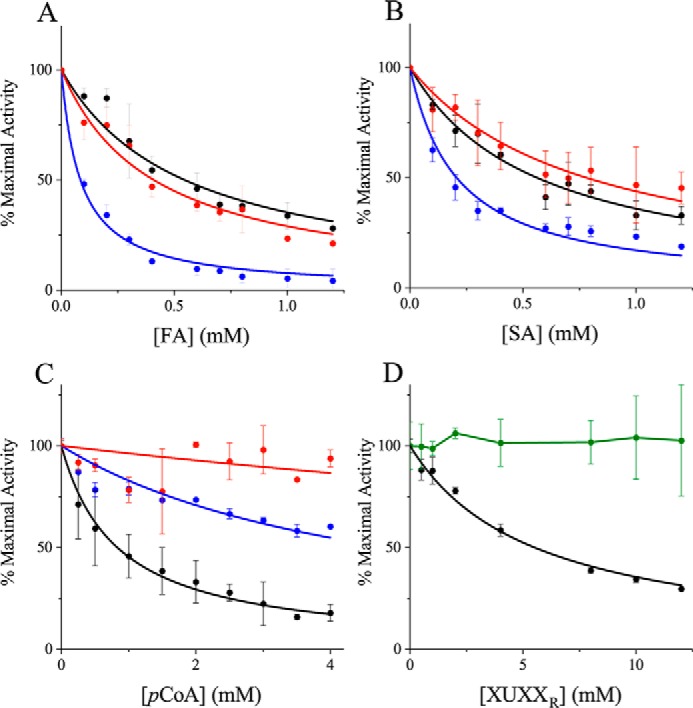
Inhibition of TtCE15A with aromatic compounds and xylooligosaccharides. Inhibition of the WT enzyme activity (black) compared with F174A (blue) and F174D (red) by FA (A), SA (B), and pCoA (C) and inhibition of TtCE15A WT activity (black) compared with W376A (green) by XUXXR (D) are shown. The inhibitors were added in increasing concentrations to 0.4 mm BnzGlcA, up to 1.2 mm FA or SA, 4 mm pCoA, and 12 mm XUXXR. The inhibitory effect of the compounds was calculated by nonlinear regression, fitting Equation 1 (see “Experimental procedures”) to the data (not possible for W376A with XUXXR). Error bars represent S.D. from the mean value of duplicate measurements. The data are normalized to facilitate comparison, where 100% maximal activity corresponds to the following rates (v/[E]t): 11.1 (WT), 1.7 (F174A), 0.3 (F174D), and 3.0 s−1 (W376A). Similar or even stronger inhibition by FA and SA was observed for the Phe-174 variants compared with the WT enzyme. However, the variants lacking Phe-174 or Trp-376 were less inhibited by pCoA and XUXXR than the WT enzyme.
Substitution of Phe-174 with alanine (F174A) resulted in a 10-fold higher Km (26 mm) for BnzGlcA, whereas the kcat value remained similar (136 s−1; Table 1). A second variant, F174D, was even more catalytically impaired, with a 15-fold increase in Km (53 mm) and a halved kcat value (43 s−1). Both variants exhibited lower kcat/Km values by an order of magnitude or more compared with the WT enzyme assayed using AllylGlcA or MeGlcA as substrates. Differential scanning fluorimetry confirmed similar melting temperatures (Tm) for the Phe-174 variants (52–54 °C) and the WT enzyme (52 °C), suggesting that the reduced catalytic efficiencies are not due to significantly compromised enzyme structures. Collectively, the results suggest that Phe-174 is important not only for binding of aromatic benzyl moieties, but it also affects the catalytic action on smaller substrates.
The inhibition studies using hydroxycinnamic acids were repeated for the two enzyme variants F174A and F174D (Table 2 and Fig. 4, A–C), with a hypothesis that the inhibitory effects would be reduced compared with the WT enzyme. Both variants were less inhibited by pCoA with higher IC50 values observed (6-fold for F174A and 30-fold for F174D; Table 2 and Fig. 4C). However, the IC50 values for FA and SA were similar (F174D) or significantly lowered (F174A; 6-fold decrease) compared with the reactions with the WT enzyme (Table 2 and Fig. 4, A and B). The results are puzzling and do not clearly indicate an involvement of Phe-174 in binding FA or SA. However, it is possible that the increased space resulting from replacing Phe-174 with smaller residues enables the methoxylated FA and SA to bind closer to the catalytic residues and thereby more easily perturb the catalytic action of the enzyme variants. To fully elucidate the role of Phe-174 in binding of aromatic structures, enzyme–ligand structures are required. Despite extensive efforts, we have thus far been unable to obtain ligand complexes with TtCE15A, but future experiments might illuminate which residues provide key contributions in substrate recognition.
Investigation of potential xylan-interacting residues
We previously postulated that GEs may interact with a larger carbohydrate portion of xylan than just the glucuronoate moiety (20). To examine how TtCE15A may interact with mono- and oligosaccharides, potential inhibition of the hydrolysis of BnzGlcA was assayed using a range of carbohydrates: arabinose, glucose, xylose, xylotriose, and the reduced 23-(4-O-methyl-d-glucuronyl)-α-d-xylotetraitol (XUXXR; Fig. 1C). XUXXR mimics the carbohydrate portion of native LC ester structures. None of the monosaccharides nor xylotriose (up to 12 mm) was able to inhibit the reaction of the enzyme with BnzGlcA (at 0.4 mm), but addition of XUXXR to the reactions reduced the activity of TtCE15A on BnzGlcA significantly (IC50 = 5.51 mm; Table 2 and Fig. 4D).
Interactions between the conserved active site tryptophan and the carbohydrate portion of LC structures have previously been proposed from our docking simulations on OtCE15A (20). In TtCE15A, to investigate whether the conserved tryptophan (Trp-376) may interact with the xylose moiety α-1,2–linked to 4-O-MeGlcA, we performed inhibition experiments with the previously described W376A enzyme variant. In contrast to the WT TtCE15A enzyme, addition of XUXXR did not inhibit the activity of W376A, which retained 100% activity even in the presence of 12 mm XUXXR (Fig. 4D). This observation supports the hypothesis that Trp-376 is involved in positioning and/or binding to the main chain xylan or xylooligosaccharides in native cell wall–derived substrates.
We attempted to obtain data for the binding of GlcA and benzyl alcohol, respectively, to TtCE15A using isothermal titration calorimetry and surface plasmon resonance measurements. Unfortunately, neither experimental setup yielded conclusive data. Possibly, this was a result of a relatively weak binding of TtCE15A to these molecules, as indicated by the relatively high Km value for BnzGlcA (Table 1).
Discussion
The proposed biological role of GEs is to hydrolyze ester bonds between lignin and glucuronoxylan in plant cell walls, which would greatly reduce the cell wall recalcitrance. However, information on how GEs interact with their native substrates is still lacking, primarily due to difficulties with substrate acquisition and suitable detection methods. The bacterial CE15 enzyme examined in the present work, TtCE15A, is the most dissimilar, in terms of primary structure, to the fungal enzymes characterized to date. Our investigations revealed a remarkably high substrate turnover rate of TtCE15A on the typical model substrate BnzGlcA as well as a strong preference for larger aromatic ester substituents on GlcA. The model substrates used in this study have been used to characterize the majority of previously studied bacterial GEs, which allows for comparisons of the kinetic behavior of TtCE15A with several closely and distantly related characterized CE15 enzymes. As expected, TtCE15A exhibited properties highly similar to those of the two GEs most similar in primary structure, OtCE15D and SlCE15B, by strongly preferring the BnzGlcA substrate over smaller substrates and displaying a strict preference for esters of GlcA (no activity detected for MeGalA) (20).
The crystal structure of TtCE15A revealed features previously not observed in either fungal or bacterial GEs. The lack of a canonical catalytic glutamate residue and the existence of a noncanonically positioned carboxylic acid in a catalytically favorable orientation have previously been observed in the MZ0003 enzyme (21). Here, we demonstrated through mutagenesis that the noncanonical Glu-274 in TtCE15A is the general acid of the catalytic triad. However, attempting to switch the position of the glutamate to the canonical position typically encountered in GEs (S304E/E374A) did not significantly increase the activity above that of the E374A variant, indicating that the position of the acidic residue is not interchangeable. Interestingly, there are aspartate residues present in the equivalent position of Glu-374 in both of the previously solved structures of OtCE15A and SuCE15C, but whether these residues (Asp-356 in OtCE15A and Asp-346 in SuCE15C) could participate in catalysis is not known. Because the activity is only reduced but not abolished in the absence of the catalytic carboxylate, this has likely allowed a later rescuing by change in a neighboring side chain and the shift of the positioning of the catalytic acid in GEs.
The inhibition studies performed for TtCE15A-catalyzed reactions revealed, for the first time, that small aromatic compounds as well as specific oligosaccharides can inhibit GE activity. The results strengthen the prevailing hypothesis that GEs interact with both aromatic lignin and hydrophilic carbohydrate parts of the substrate, although the exact nature of these interactions remains elusive. We have demonstrated that a tryptophan residue (Trp-376) is important for hydrolysis of small substrates, and the inhibition profiles are consistent with it also being important for binding GlcA-decorated xylotriose. The length of the xylan chain linked to the anomeric position of the GlcA may possibly be of less importance, as has previously been shown by the comparable kinetic values observed for the GE ScGE (from Schizophyllum commune) on esters comprising either xylose- or xylooligosaccharide structures (22). The most striking structural feature of TtCE15A was the presence of a large N-terminal extension opposite Reg2. An N-terminal loop of this size is unprecedented among characterized GEs to date. Residues of Reg2 have previously been proposed to interact with lignin or lignin fragments, and the positioning of RegN suggests a role in binding of carbohydrate moieties of LC complexes. Overall, the inserts of TtCE15A lead to a structure with a deep substrate-binding groove and possibly extensive interactions with both lignin and carbohydrates. What biological consequences this deepened active site topology may have, compared with the open-face active sites of structurally solved fungal enzymes, are currently not understood. As a symbiont of marine shipworms, T. turnerae is likely presented with partially solubilized wood-derived material, which may be rich also in LC fragments. Possibly, TtCE15A has evolved to target smaller LC fragments in solution that can be captured by the clamplike active site, whereas the studied fungal enzymes would be more adapted to act on larger, solid substrates that are more abundant in terrestrial environments.
The two additional CE15 enzymes encoded by T. turnerae, TtCE15B and TtCE15C, might complement TtCE15A in the biology of the bacterium. A multiple sequence alignment (Fig. S4) shows some noteworthy differences among the three enzymes. TtCE15B and TtCE15C possess, unlike TtCE15A, the canonically positioned glutamate, and TtCE15C additionally has an aspartate in the noncanonical position, similar to the active sites of OtCE15A and SuCE15C. Notably, the conserved tryptophan suggested to bind xylan chains in previously characterized GEs is not conserved in either TtCE15B or TtCE15C, and from the multiple sequence alignment, it appears that both TtCE15B and TtCE15C lack the putative lignin-binding phenylalanine residue conserved among characterized bacterial CE15 members. Moreover, the lack of inserted regions and the presence of possible disulfide bridges in both TtCE15B and TtCE15C are features shared with characterized fungal GEs. Previous investigations of S. linguale, a species also encoding three CE15 genes, showed that expression of each gene depends on the type of carbon source to which the host is exposed (20). Taken together, it is reasonable to assume that the different T. turnerae CE15 genes are nonredundant and exhibit unique substrate specificities, although further investigations are required to elucidate the roles of these enzymes in biomass decomposition.
In this work, we have provided a detailed structure–function study of a novel and highly active GE from CE15, with a proposed role in separation of carbohydrates from lignin. From structural analysis and biochemical characterization followed by site-directed mutagenesis of key active site residues, new insights into both the structural diversity and key residues involved in substrate binding in GEs were attained. In future studies, comparisons of active site residues of various GE enzymes could yield a more comprehensive understanding of the general interactions between GEs and their complex substrates consisting of both hydrophilic carbohydrates and hydrophobic lignin, as would synthesis of more varied model substrates with higher similarity to the proposed native LC structures. Structures of GEs in complex with biologically relevant ligands would further illuminate our understanding of how GEs interact with their natural substrates.
Experimental procedures
Cloning, expression, and purification of TtCE15A
The TtCE15A gene was amplified from genomic DNA of T. turnerae DSM 15142 (DSMZ) by PCR (primers are listed in Table S2). The product was cloned into a modified pET-28a vector (In-Fusion HD kit, Clontech), containing N-terminal His6 tags and tobacco etch virus protease cleavage sites, and gene expression was performed in Escherichia coli BL21(λDE3). Cells were grown in lysogeny broth (LB), at 37 °C with 200 rpm shaking, containing 50 μg/ml kanamycin. At A600 of ∼0.5, expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (0.2 mm final concentration), and the cells were incubated at 16 °C overnight. Cells were harvested by centrifugation (5000 × g, 10 min); resuspended in 20 mm Tris buffer (pH 8) containing 250 mm NaCl, 5 μg/ml lysozyme, and 10 μg/ml DNase; and disrupted by sonication. Cell debris was removed by centrifugation (18,000 × g, 15 min), and proteins were purified using immobilized metal ion affinity chromatography on an ÄKTA system (GE Healthcare) using 5-ml HisTrapTM Excel columns. 50 mm Tris (pH 8), 250 mm NaCl was used as binding buffer, and elution was performed in one step with 250 mm imidazole. The purified fraction was dialyzed into 20 mm Tris buffer (pH 8).
Site-directed mutagenesis
The plasmid containing TtCE15A was used as template to create TtCE15A variants according to the QuikChange II site-directed mutagenesis method (Agilent Technologies) using primers listed in Table S2. Mutant strand synthesis reaction (thermal cycling) with Phusion high-fidelity DNA polymerase (Thermo Fisher) was followed by DpnI digestion (Thermo Fisher) of the amplification products and transformation into StellarTM competent cells (Clontech). Gene expression was performed in E. coli BL21(λDE3), and the protein was purified as described for the WT enzyme. Differential scanning fluorimetry with SYPROTM Orange protein stain (Invitrogen) was performed to monitor the thermal transition of the TtCE15A enzyme variants. Measurements were performed in triplicates on a Stratagene MX3005P qPCR instrument (Agilent Technologies) with gradual heating of the sample plate (1 °C/min) from 25 to 90 °C.
Enzyme assays
Glucuronoyl esterase activity was assayed with the substrates BnzGlcA, AllylGlcA, MeGlcA, and MeGalA (Carbosynth) and monitored continuously with the K-URONIC kit (Megazyme) as reported previously (20). Although the pH optimum of TtCE15A was at pH 8.5, standard assays were kept at pH 7.5 due to instability of the ester substrates at the higher pH. The substrates were dissolved in 100% DMSO, and all reactions contained less than 10% DMSO. pH dependence profiles were generated with 2 mm BnzGlcA in a three-component buffer containing 25 mm acetic acid, 25 mm MES, and 50 mm Tris-HCl (pH 4.5–9.5) (30). Acetyl esterase activity was assayed using pNP-Ac (Sigma-Aldrich) as described previously (20). Curve fitting and kinetic parameters were calculated using OriginPro 2017 software (OriginLab).
Crystallization and data collection
TtCE15A was screened for crystallization in MRC two-drop crystallization plates (Molecular Dimensions) using an Oryx 8 robot (Douglas Instruments). Sitting drops (0.3 μl) were mixed with protein:reservoir volume ratios of 3:1 or 1:1 using 35 mg/ml TtCE15A in 20 mm Tris (pH 8.0). Hits from Morpheus screens (Molecular Dimensions) were optimized, and final crystallization conditions were as follows: 0.09 m halogens, 0.1 mbuffer system 1 (imidazole and MES), and 50% (v/v) precipitant mixture 2 (40% (v/v) ethylene glycol and 20% (w/v) PEG 8000) (31). Crystals were mounted using nylon loops and flash frozen in liquid N2 in the absence of additional cryoprotectant. An initial data set, which diffracted to ≈2.5 Å, was collected on ID30B at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Molecular replacement using the previously determined fungal and bacterial CE15 structures as templates (25–45% identity) was unsuccessful, and collection of anomalous data was pursued. The pET-28a construct was transformed into E. coli T7 Express Crystal (methionine auxotroph; New England Biolabs), and the protein was expressed in minimal medium containing seleno-l-methionine (SeMet) according to the supplier's recommendations (Molecular Dimensions). TtCE15A–SeMet was screened for crystallization, and crystals grew in the same conditions as the native protein. A data set of TtCE15A–SeMet was collected at beamline P11 of Petra III at 0.975 Å.
Data processing and structure determination
Diffraction data were processed with XDS (32) with good statistics to 2.1 Å (Table S3), significantly better than the previously collected native data set, and was solely pursued for structure determination. Structure solution was carried out in Phenix (33). AutoSol (33, 34) was used to find the selenium sites and generate the initial SAD map with three protein molecules in the asymmetric unit. The figure of merit values before and after density modification in AutoSol were 0.34 and 0.64, respectively. The structure was initially built with Phenix AutoBuild (35), rebuilt in Coot (36), and further refined with Phenix Refine (37) in alternating cycles. Ligand compounds were added in Coot. The model has >97% residues in the most favorable Ramachandran regions with <0.2% Ramachandran outliers (38). Table S3 lists the final model refinement statistics.
Inhibition studies
The aromatic compounds benzyl alcohol (BnzAlc), SA, FA, and pCoA (Sigma-Aldrich) as well as the carbohydrate compounds arabinose, glucose, xylose, xylotriose, and XUXXR (Megazyme) were added to GE reactions, and the effect on activity of TtCE15A (WT and variants) was monitored using the K-URONIC kit. Reactions with 400 μm BnzGlcA (significantly below the Km) were supplemented with the putative inhibitor in concentrations between 0 and 12 mm for carbohydrate compounds and BnzAlc, 0 and 4 mm for pCoA, or 0 and 1.2 mm for SA and FA. FA and SA concentrations above 1.2 mm resulted in too high background absorbance levels at the wavelength used in the coupled assay. Inhibition with only GlcA could not be performed due to the assay setup. IC50 values were obtained, where possible, by plotting the activity versus the concentration of inhibitor and fitting Equation 1 with nonlinear regression using OriginPro 2017 software.
| (Eq. 1) |
Author contributions
J. A. B., S. M., L. L. L., and J. L. conceptualization; J. A. B., S. M., J.-C. N. P., L. O., L. L. L., and J. L. data curation; J. A. B., S. M., J.-C. N. P., L. O., L. L. L., and J. L. formal analysis; J. A. B., S. M., J.-C. N. P., L. O., L. L. L., and J. L. validation; J. A. B., S. M., J.-C. N. P., L. L. L., and J. L. investigation; J. A. B., S. M., and J. L. visualization; J. A. B., S. M., J.-C. N. P., L. L. L., and J. L. methodology; J. A. B., S. M., and J. L. writing-original draft; J. A. B., S. M., L. O., L. L. L., and J. L. writing-review and editing; L. O., L. L. L., and J. L. resources; L. O., L. L. L., and J. L. supervision; L. O., L. L. L., and J. L. funding acquisition; L. L. L. and J. L. project administration.
Supplementary Material
Acknowledgments
We thank Nicole Koropatkin and Tiantai Ma from the University of Michigan Medical School for performing isothermal titration calorimetry measurements and data interpretation and Istvan Horvath from Chalmers University of Technology for assistance with surface plasmon resonance measurements and data interpretation.
The work performed at Chalmers University of Technology was supported by the Knut and Alice Wallenberg Foundation through the Wallenberg Wood Science Center. The collaborative work between Chalmers University of Technology and Copenhagen University was supported by MAX4ESSFUN: Cross Border Science and Society Program Grant CTH-003 and Novo Nordisk Foundation Grant NNF17OC0027698 (Biotechnology-based Synthesis and Production Research). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 6hsw) have been deposited in the Protein Data Bank (http://wwpdb.org/).
This article contains Figs. S1–S4 and Tables S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GE
- glucuronoyl esterase
- CE15
- carbohydrate esterase family 15
- LC
- lignin-carbohydrate
- GlcA
- d-glucuronic acid
- GalA
- d-galacturonic acid
- BnzGlcA
- benzyl glucuronoate
- AllylGlcA
- allyl glucuronoate
- MeGlcA
- methyl glucuronoate
- MeGalA
- methyl galacturonoate
- pNP-Ac
- 4-nitrophenyl acetate
- FA
- ferulic acid
- SA
- sinapic acid
- pCoA
- p-coumaric acid
- XUXXR
- 23-(4-O-methyl-d-glucuronyl)-α-d-xylotetraitol
- Tt
- T. turnerae
- Ot
- O. terrae
- Su
- S. usitatus
- Tr
- T. reesei
- St
- S. thermophile
- Sl
- S. linguale
- Sc
- S. commune
- SAD
- single-wavelength anomalous diffraction
- BnzAlc
- benzyl alcohol
- Reg
- region.
References
- 1. Monrad R. N., Eklöf J., Krogh K. B. R. M., and Biely P. (2018) Glucuronoyl esterases: diversity, properties and biotechnological potential. A review. Crit. Rev. Biotechnol. 38, 1121–1136 10.1080/07388551.2018.1468316 [DOI] [PubMed] [Google Scholar]
- 2. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnling Bååth J., Giummarella N., Klaubauf S., Lawoko M., and Olsson L. (2016) A glucuronoyl esterase from Acremonium alcalophilum cleaves native lignin-carbohydrate ester bonds. FEBS Lett. 590, 2611–2618 10.1002/1873-3468.12290 [DOI] [PubMed] [Google Scholar]
- 4. d'Errico C., Börjesson J., Ding H., Krogh K. B., Spodsberg N., Madsen R., and Monrad R. N. (2016) Improved biomass degradation using fungal glucuronoyl-esterases-hydrolysis of natural corn fiber substrate. J. Biotechnol. 219, 117–123 10.1016/j.jbiotec.2015.12.024 [DOI] [PubMed] [Google Scholar]
- 5. Mosbech C., Holck J., Meyer A. S., and Agger J. W. (2018) The natural catalytic function of CuGE glucuronoyl esterase in hydrolysis of genuine lignin-carbohydrate complexes from birch. Biotechnol. Biofuels 11, 71 10.1186/s13068-018-1075-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong D. W., Chan V. J., McCormack A. A., Hirsch J., and Biely P. (2012) Functional cloning and expression of the Schizophyllum commune glucuronoyl esterase gene and characterization of the recombinant enzyme. Biotechnol. Res. Int. 2012, 951267 10.1155/2012/951267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai A. Y., Canam T., Gorzsás A., Mellerowicz E. J., Campbell M. M., and Master E. R. (2012) Constitutive expression of a fungal glucuronoyl esterase in Arabidopsis reveals altered cell wall composition and structure. Plant Biotechnol. J. 10, 1077–1087 10.1111/j.1467-7652.2012.00735.x [DOI] [PubMed] [Google Scholar]
- 8. Duranová M., Hirsch J., and Kolenová K., and Biely P. (2009) Fungal glucuronoyl esterases and substrate uronic acid recognition. Biosci. Biotechnol. Biochem. 73, 2483–2487 10.1271/bbb.90486 [DOI] [PubMed] [Google Scholar]
- 9. d'Errico C., Jørgensen J. O., Krogh K. B., Spodsberg N., Madsen R., and Monrad R. N. (2015) Enzymatic degradation of lignin-carbohydrate complexes (LCCs): model studies using a fungal glucuronoyl esterase from Cerrena unicolor. Biotechnol. Bioeng. 112, 914–922 10.1002/bit.25508 [DOI] [PubMed] [Google Scholar]
- 10. Katsimpouras C., Bénarouche A., Navarro D., Karpusas M., Dimarogona M., Berrin J. G., Christakopoulos P., and Topakas E. (2014) Enzymatic synthesis of model substrates recognized by glucuronoyl esterases from Podospora anserina and Myceliophthora thermophila. Appl. Microbiol. Biotechnol. 98, 5507–5516 10.1007/s00253-014-5542-9 [DOI] [PubMed] [Google Scholar]
- 11. Vafiadi C., Topakas E., Biely P., and Christakopoulos P. (2009) Purification, characterization and mass spectrometric sequencing of a thermophilic glucuronoyl esterase from Sporotrichum thermophile. FEMS Microbiol. Lett. 296, 178–184 10.1111/j.1574-6968.2009.01631.x [DOI] [PubMed] [Google Scholar]
- 12. Huynh H. H., and Arioka M. (2016) Functional expression and characterization of a glucuronoyl esterase from the fungus Neurospora crassa: identification of novel consensus sequences containing the catalytic triad. J. Gen. Appl. Microbiol. 62, 217–224 10.2323/jgam.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 13. Huynh H. H., Ishii N., Matsuo I., and Arioka M. (2018) A novel glucuronoyl esterase from Aspergillus fumigatus-the role of conserved Lys residue in the preference for 4-O-methyl glucuronoyl esters. Appl. Microbiol. Biotechnol. 102, 2191–2201 10.1007/s00253-018-8739-5 [DOI] [PubMed] [Google Scholar]
- 14. Hüttner S., Klaubauf S., de Vries R. P., and Olsson L. (2017) Characterisation of three fungal glucuronoyl esterases on glucuronic acid ester model compounds. Appl. Microbiol. Biotechnol. 101, 5301–5311 10.1007/s00253-017-8266-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Topakas E., Moukouli M., Dimarogona M., Vafiadi C., and Christakopoulos P. (2010) Functional expression of a thermophilic glucuronoyl esterase from Sporotrichum thermophile: identification of the nucleophilic serine. Appl. Microbiol. Biotechnol. 87, 1765–1772 10.1007/s00253-010-2655-7 [DOI] [PubMed] [Google Scholar]
- 16. Biely P., Malovíková A., Uhliariková I., Li X. L., and Wong D. W. (2015) Glucuronoyl esterases are active on the polymeric substrate methyl esterified glucuronoxylan. FEBS Lett. 589, 2334–2339 10.1016/j.febslet.2015.07.019 [DOI] [PubMed] [Google Scholar]
- 17. De Santi C., Willassen N. P., and Williamson A. (2016) Biochemical characterization of a family 15 carbohydrate esterase from a bacterial marine arctic metagenome. PLoS One 11, e0159345 10.1371/journal.pone.0159345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spániková S, and Biely P. (2006) Glucuronoyl esterase—novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett. 580, 4597–4601 10.1016/j.febslet.2006.07.033 [DOI] [PubMed] [Google Scholar]
- 19. Lin M. I., Hiyama A., Kondo K., Nagata T., and Katahira M. (2018) Classification of fungal glucuronoyl esterases (FGEs) and characterization of two new FGEs from Ceriporiopsis subvermispora and Pleurotus eryngii. Appl. Microbiol. Biotechnol. 102, 9635–9645 10.1007/s00253-018-9318-5 [DOI] [PubMed] [Google Scholar]
- 20. Arnling Bååth J., Mazurkewich S., Knudsen R. M., Poulsen J. N., Olsson L., Lo Leggio L., and Larsbrink J. (2018) Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion. Biotechnol. Biofuels 11, 213 10.1186/s13068-018-1213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Santi C., Gani O. A., Helland R., and Williamson A. (2017) Structural insight into a CE15 esterase from the marine bacterial metagenome. Sci. Rep. 7, 17278 10.1038/s41598-017-17677-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spániková S, Poláková M., Joniak D., Hirsch J., and Biely P. (2007) Synthetic esters recognized by glucuronoyl esterase from Schizophyllum commune. Arch. Microbiol. 188, 185–189 10.1007/s00203-007-0241-x [DOI] [PubMed] [Google Scholar]
- 23. Aurilia V., Martin J. C., McCrae S. I., Scott K. P., Rincon M. T., and Flint H. J. (2000) Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology 146, 1391–1397 10.1099/00221287-146-6-1391 [DOI] [PubMed] [Google Scholar]
- 24. Pokkuluri P. R., Duke N. E., Wood S. J., Cotta M. A., Li X. L., Biely P., and Schiffer M. (2011) Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea jecorina. Proteins 79, 2588–2592 10.1002/prot.23088 [DOI] [PubMed] [Google Scholar]
- 25. Charavgi M. D., Dimarogona M., Topakas E., Christakopoulos P., and Chrysina E. D. (2013) The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst. Acta Crystallogr. D Biol. Crystallogr. 69, 63–73 10.1107/S0907444912042400 [DOI] [PubMed] [Google Scholar]
- 26. Distel D. L., Morrill W., MacLaren-Toussaint N., Franks D., and Waterbury J. (2002) Teredinibacter turnerae gen. nov., sp nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int. J. Syst. Evol. Microbiol. 52, 2261–2269 10.1099/00207713-52-6-2261 [DOI] [PubMed] [Google Scholar]
- 27. Yang J. C., Madupu R., Durkin A. S., Ekborg N. A., Pedamallu C. S., Hostetler J. B., Radune D., Toms B. S., Henrissat B., Coutinho P. M., Schwarz S., Field L., Trindade-Silva A. E., Soares C. A., Elshahawi S., et al. (2009) The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms). PLoS One 4, e6085 10.1371/journal.pone.0006085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sunner H., Charavgi M. D., Olsson L., Topakas E., and Christakopoulos P. (2015) Glucuronoyl esterase screening and characterization assays utilizing commercially available benzyl glucuronic acid ester. Molecules 20, 17807–17817 10.3390/molecules201017807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hedstrom L. (2002) Serine protease mechanism and specificity. Chem Rev 102, 4501–4524 10.1021/cr000033x [DOI] [PubMed] [Google Scholar]
- 30. Ellis K. J., and Morrison J. F. (1982) Buffers of constant ionic-strength for studying pH-dependent processes. Methods Enzymol. 87, 405–426 10.1016/S0076-6879(82)87025-0 [DOI] [PubMed] [Google Scholar]
- 31. Gorrec F. (2009) The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 42, 1035–1042 10.1107/S0021889809042022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925,10.1107/S0108768110007202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.