Abstract
Salmonella enterica is an intracellular bacterial pathogen that replicates within membrane-bound vacuoles through the action of effector proteins translocated into host cells. Salmonella vacuoles have characteristics of lysosomes but are reduced in hydrolytic enzymes transported by mannose-6-phosphate receptors (MPRs). We found that the effector SifA subverted Rab9-dependent retrograde trafficking of MPRs, thereby attenuating lysosome function. This required binding of SifA to its host cell target SKIP/PLEKHM2. Furthermore, SKIP regulated retrograde trafficking of MPRs in noninfected cells. Translocated SifA formed a stable complex with SKIP and Rab9 in infected cells. Sequestration of Rab9 by SifA-SKIP accounted for the effect of SifA on MPR transport and lysosome function. Growth of Salmonella increased in cells with reduced lysosomal activity and decreased in cells with higher lysosomal activity. These results suggest that Salmonella vacuoles undergo fusion with lysosomes whose potency has been reduced by SifA.
Lysosomes are membrane-bound acidic organelles that contain over 50 different hydrolytic enzymes. In the trans-Golgi network (TGN), the majority of newly synthesized hydrolytic enzymes bind the cation-dependent (CD-) and cation-independent (CI-) mannose phosphate receptors (MPRs), which then deliver the hydrolases to endosomes. Endosome acidification dissociates the hydrolases from their MPRs, which recycle back to the TGN. As early endosomes mature into late endosomes, the hydrolases are converted into their active forms and transported to lysosomes (1). Lysosomes fuse with vacuoles containing microbes and other exogenous material, causing their degradation.
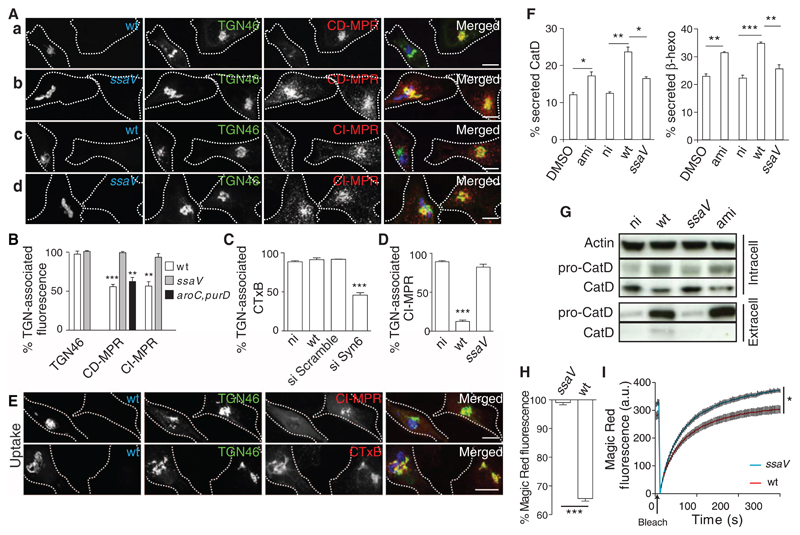
Salmonella enterica proliferates within mammalian cells in membrane-bound compartments: Salmonella-containing vacuoles (SCVs) (2). SCVs interact extensively with the endocytic pathway, resulting in acidic compartments (3) whose membranes are enriched in lysosomal membrane glycoproteins (4, 5) and which remain accessible to late endosomal and lysosomal content (6). However, the CD- and CI-MPRs, as well as their associated hydrolases, do not accumulate in SCVs (4, 5, 7, 8). To investigate this paradox, we studied the consequences of Salmonella infection on the distribution of MPRs. Human epithelial (HeLa) cells were infected with wild-type S. enterica serovar Typhimurium (Salmonella), and the amounts of TGN-localized CD-MPR and CI-MPR were quantified by three-dimensional confocal microscopy. In uninfected cells, the proteins accumulated as expected at the TGN (Fig. 1A). In infected cells, the CD- and CI-MPRs were distributed diffusely throughout the cell (Fig. 1, A and B), whereas the localization of other TGN-associated proteins was unaffected (fig. S1, A and B).
Fig 1. The SPI-2 T3SS interferes with Syn10-dependent retrograde trafficking of MPRs and reduces lysosome function.
(A and B) HeLa cells were infected for 14 hours with green fluorescent protein (GFP)–expressing wild-type (wt) (Aa and c) or ssaV mutant (Ab and d) Salmonella (blue) or (B) a replication-deficient aroC,purD mutant (23) that hasa functional SPI-2 T3SS (fig. S1C). Cells were immunolabeled for TGN46 (green) and CD-MPR (red) (a and b) or CI-MPR (red) (c and d). Scale bars, 10 μm. (B) Quantitation of mean fluorescent signals of TGN-associated TGN46 (24), CD-MPR, and CI-MPR, analyzed by three-dimensional (3D) confocal microscopy and normalized to signals in uninfected cells from the same sample. Error bars in all figures indicate SEM. (C to E) Plasma membrane–TGN transport assays. HeLa cells either were subjected to siRNA to deplete Syn6 or infected with wt or ssaV mutant Salmonella (blue in E) for 14 hours, then pulse-chased with either fluorescent CTxB [red in (E)] or CI-MPR antibody [red in (E)]. Fixed cells were immunolabeled for CI-MPR and TGN46 [green in (E)] or Rab6 (to identify the TGN in si Syn6– and si Scramble–treated cells), and TGN-associated CI-MPR or CTxB was quantified. Scale bars, 10 μm. (F and G) Rerouting of lysosomal enzymes. (F) HeLa cells were exposed to dimethyl sulfoxide (DMSO) or amiodarone (ami) [which redistributes MPRs (13)] and were not infected (ni) or infected with wt or ssaV mutant Salmonella for 14 hours. Secreted levels of cathepsin D (CatD) or β-hexosaminadase (β-hexo) are a percentage of total levels. (G) Intra- and extracellular proteins were analyzed by immunoblotting, using actin as a loading control. (H and I) Cathepsin B (CatB) activity was quantified by measuring fluorescent cleavage product of Magic Red-RR in living cells. (H) HeLa cells were infected for 14 hours with wt or ssaV mutant Salmonella and exposed to Magic Red-RR, and steady-state fluorescent product intensity was measured by flow cytometry. Bars represent the percentage of fluorescence of infected compared with uninfected cells in the same sample. (I) Cells as in (H) were photobleached and analyzed for new fluorescent product. Data were normalized to the mean fluorescence of uninfected cells. Vertical lines represent SEM for each of 100 time points. Statistically significant relationships are denoted (*P < 0.05; **P < 0.01; ***P < 0.005).
Salmonella translocates effector proteins across the SCV membrane via the Salmonella pathogenicity island 2 (SPI-2)–encoded type III secretion system (T3SS). SPI-2 T3SS effectors mediate intravacuolar bacterial replication (2). HeLa cells infected with an ssaV mutant strain that has a nonfunctional T3SS (9) were indistinguishable from uninfected cells in terms of the distribution of MPRs (Fig. 1, A and B). The reduction of MPR from the TGN began 6 to 8 hours after bacterial invasion (fig. S1D), when other effects of the SPI-2 T3SS become visible (10). Thus, the SPI-2 T3SS is required for the redistribution of MPRs from the TGN. Two endocytic-TGN recycling pathways have been described: One depends on the Q-soluble N-ethylmaleimide-sensitive-factor attachment protein receptor (Q-SNARE) syntaxin 6 (Syn6) and transports TGN46 and the cholera toxin B subunit (CTxB); the other depends on the Q-SNARE syntaxin 10 (Syn10) and involves retrograde transport of MPRs (11). Occasionally the CI-MPR is transported to the plasma membrane from the TGN (12). To determine whether intracellular Salmonella interferes with these pathways, we assayed the transport of fluorescent CTxB or an antibody against CI-MPR from the plasma membrane to the TGN in infected cells. Knockdown of Syn6 inhibited transport of the CTxB to the TGN, but there was no detectable effect of Salmonella on this pathway (Fig. 1C). In contrast, trafficking of CI-MPR antibody was dramatically reduced in cells infected with wild-type Salmonella (Fig. 1, D and E, and fig. S2A), which depended on a functional SPI-2 T3SS (Fig. 1D). Thus, the SPI-2 T3SS specifically interferes with Syn10-dependent retrograde trafficking of MPRs from endosomes to the TGN.
Interference of MPR recycling leads to misrouted secretion of newly synthesized lysosomal enzymes (13). We examined the effect of intracellular Salmonella on the secretion of two lysosomal enzymes: cathepsin D (CatD) and β-hexosaminidase (β-hexo). Infection of HeLa cells with wild-type Salmonella significantly increased extracellular activities of both enzymes compared with uninfected or untreated cells. The SPI-2 T3SS accounted for a major proportion of the Salmonella effect (Fig. 1F). Infection of cells by wild-type bacteria caused the appearance in the extracellular medium of large amounts of immature CatD (Fig. 1G), showing that it had not been processed in an endosomal environment. This was accompanied by a reduction in the level of intracellular mature enzyme and required an intact SPI-2 T3SS (Fig. 1G).
A misrouting of lysosomal enzymes would be expected to cause a reduction of lysosome function. We tested the effect of Salmonella infection and the SPI-2 T3SS on the activity of lysosomal cathepsin B (CatB) in living HeLa cells by using a substrate (Magic Red-RR, ImmunoChemistry Technologies, Bloomington, Minnesota) that becomes fluorescent upon cleavage (14). Cells infected with ssaV mutant bacteria had higher steady-state levels of fluorescence (Fig. 1H) and recovered to higher levels after photobleaching compared with cells infected with wild-type Salmonella (Fig. 1I and movies S1 and S2). A reduction in lysosomal CatB activity was also observed after small interfering RNA (siRNA)–mediated depletion of Syn10 in uninfected HeLa cells (fig. S3, A and B), whereas siRNA-mediated depletion of Syn6 enhanced CatB activity (fig. S3, A and B), which correlated with reduced and enhanced MPR localization at the TGN, respectively (fig. S3C).
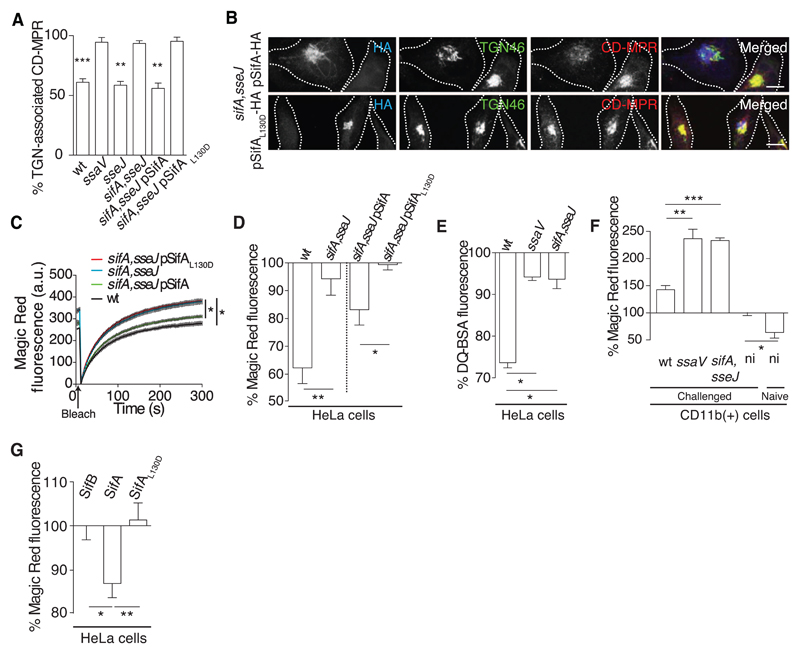
SifA is a Salmonella effector required to maintain the SCV membrane (10). Loss of the vacuolar membrane around a sifA mutant is inhibited by absence of another effector, SseJ (15). TGN-associated CD-MPR was reduced by infection with the sseJ mutant or wild-type strain but not by the sifA,sseJ or ssaV mutant strains (Fig. 2A). SifA interacts with a host protein, SifA and kinesin-interacting protein (SKIP)/pleckstrin homology domain containing, family M (with RUN domain) member 2 (PLEKHM2), to regulate kinesin activity and vacuolar membrane dynamics (16, 17). This interaction is dependent on leucine at position 130 (L130) of SifA (18). HeLa cells were infected with the sifA,sseJ mutant strain harboring a plasmid encoding either a hemagglutinin epitope–tagged wild-type SifA (SifA-HA) or a point mutant (SifAL130D-HA). Cells infected with Salmonella expressing wild-type SifA but not SifAL130D showed redistribution of MPR (Fig. 2, A and B). Similarly, lysosomal CatB activity was consistently lower in cells infected with either wild-type bacteria or with the sifA,sseJ mutant expressing SifA-HA compared with cells infected with the sifA,sseJ mutant or with this strain expressing SifAL130D-HA (Fig. 2, C and D). Cells infected with the sifA,sseJ mutant also had significantly higher lysosomal activity than wild-type–infected cells (Fig. 2E). SPI-2 T3SS-dependent inhibition of lysosome function was also observed in mouse-derived macrophages (fig. S4) and in CD11b(+) splenocytes from infected mice (Fig. 2F). Ectopic expression of SifA in HeLa cells significantly reduced TGN-localized CD-MPR (fig. S5A) and lysosomal activity when compared with cells expressing similar levels of SifAL130D or another effector, SifB (Fig. 2G and fig. S5B). Thus, SifA accounts for the inhibition of lysosomal function by Salmonella and the requirement for L130 implicates SKIP in this process.
Fig 2. SifA inhibits lysosome function.
(A) Quantitation from 3D confocal microscopy of the mean fluorescence signals of TGN-associated CD-MPR in HeLa cells infected with the indicated strains. (B) HeLa cells infected for 14 hours with sifA, sseJ double mutant Salmonella expressing SifA-HA or SifAL130D-HA were immunolabeled for HA (blue), TGN46 (green), and CD-MPR (red). (C) Magic Red-RR recovery assay of CatB activity in HeLa cells infected for 14 hours with the indicated strains. (D) HeLa cells were infected for 14 hours with the indicated Salmonella strains and exposed to Magic Red-RR, and the percentage of fluorescent product intensity of infected cells compared with uninfected cells from the same sample was measured at steady state by flow cytometry. (E) HeLa cells were infected as above and pulse chased with DQ-bovine serum albumin (DQ-BSA, Life Technologies, Paisley, UK), and the percentage of fluorescence between infected and uninfected cells from the same sample was measured by flow cytometry. (F) BALB/c mice were inoculated orally with the indicated Salmonella strains. After 48 hours, CD11b(+) splenocytes were harvested and subjected to Magic Red-RR assay. Bars represent the percentage of steady-state fluorescence of infected cells compared with uninfected cells from the same sample or of cells from uninfected mice compared with uninfected cells from infected animals. (G) HeLa cells were transfected with vectors encoding GFP-SifB, GFP-SifA, or GFP-SifAL130D, and 24 hours later cells were subjected to Magic Red-RR assay. In each case, fluorescence intensity in cells expressing similar levels of the indicated protein was normalized to that of untransfected control cells within the same sample and then to fluorescence caused by expression of SifB. Statistically significant relationships are denoted (*P < 0.05; **P < 0.01; ***P < 0.005).
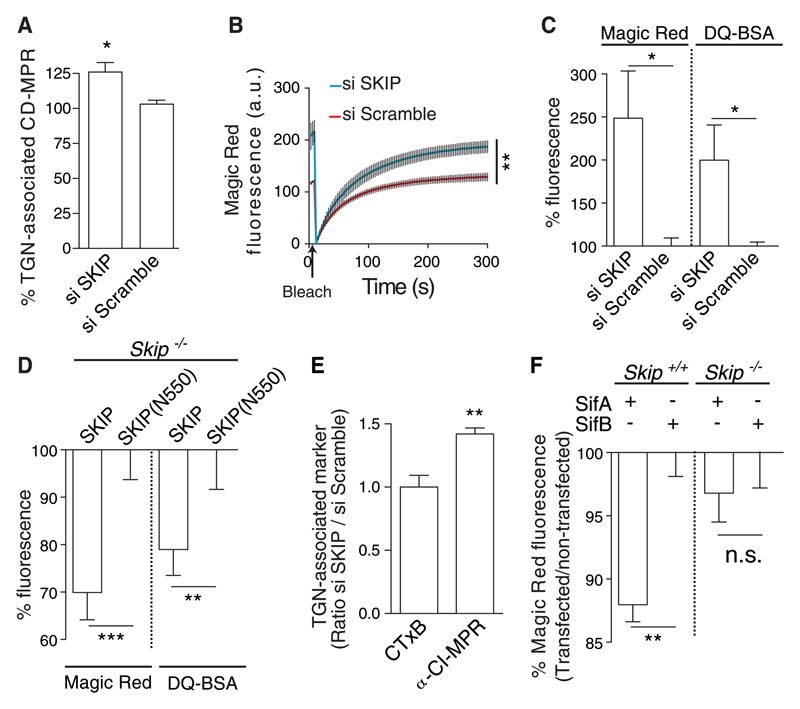
siRNA-mediated knockdown of SKIP (fig. S6A) resulted in an increase in both localization of the CD-MPR at the TGN (Fig. 3A) and CatB activity (Fig. 3, B and C). Immortalized mouse embryonic fibroblasts (MEFs) from SKIP knockout mice (fig. S6B) displayed significantly reduced CatB activity when transfected with a vector encoding wild-type SKIP compared with cells expressing a SKIP variant lacking its SifA-binding pleckstrin homology (PH) domain (N-550) (Fig. 3D and fig. S6, C and D). To investigate the possible involvement of SKIP in endosome-TGN trafficking, we subjected HeLa cells depleted of SKIP to retrograde transport assays. Depletion of SKIP had no detectable effect on traffic of CTxB from the plasma membrane to the TGN (Fig. 3E), but there was a significant increase in the amount of CI-MPR transported to the TGN (Fig. 3E and fig. S2B), indicating that SKIP negatively regulates the Syn10-dependent retrograde pathway and lysosome function. To determine whether the effect of SifA on lysosome function required SKIP, wild-type and Skip−/− knockout MEFs expressing either SifA or SifB were assayed for CatB activity. Compared with SifB, SifA reduced enzyme activity in wild-type MEFs but not in Skip−/− knockout cells (Fig. 3F). Thus, the inhibitory activity of SifA can be attributed to a pathway requiring SKIP.
Fig 3. SKIP negatively regulates lysosome function.
(A and B) HeLa cells depleted of SKIP by siRNA or exposed to scrambled siRNA were analyzed for TGN-associated CD-MPR (A) and by Magic Red-RR assay of recovery of CatB activity after photobleaching (B). (C) Steady-state levels of CatB activity and DQ-BSA hydrolysis analysed by flow cytometry. For each substrate, the data were normalized to that obtained by using scrambled siRNA. (D) Skip−/− MEFs expressing either an N-terminal domain of SKIP (N550) or full-length protein were analyzed for steady-state levels of CatB by Magic Red-RR assay. Data are normalized to SKIP (N550) samples. (E) HeLa cells were depleted of SKIP by siRNA (si SKIP) or exposed to scrambled siRNA and subjected to plasma membrane–TGN transport assays using fluorescent CTxB or CI-MPR antibody. Data were analyzed by 3D confocal microscopy and were normalized to that obtained by using scrambled siRNA. (F) Wild-type (Skip+/+) or Skip−/− MEFs were transfected with vectors encoding either GFP-SifA or GFP-SifB and assayed for CatB activity by Magic Red-RR. Bars represent the percentage of fluorescence of transfected cells compared with untransfected cells from the same samples, normalized to GFP-SifB in either Skip+/+ or Skip−/− cells. Statistically significant relationships are denoted (*P < 0.05; **P < 0.01; ***P < 0.005).
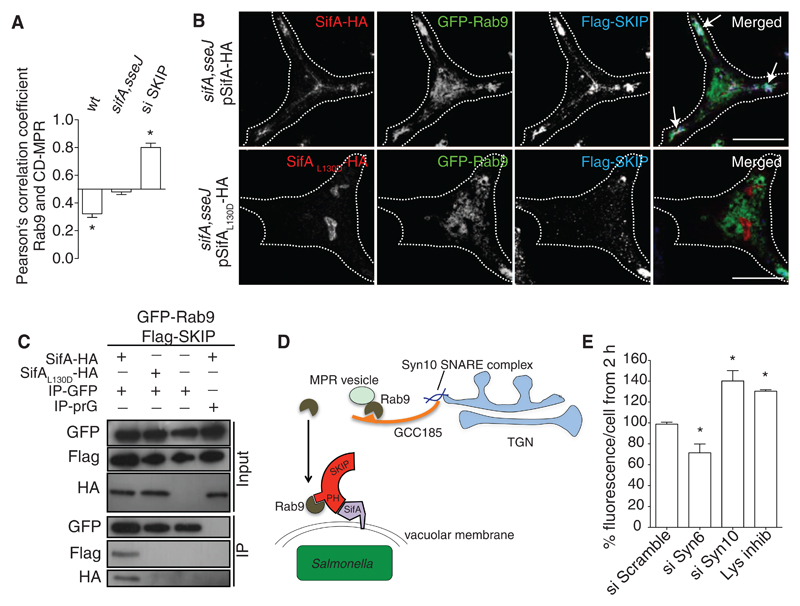
MPR levels were reduced in Salmonella-infected HeLa cells (fig. S7A), and a significant proportion of the remaining protein relocalized to structures containing Vps26 (fig. S7B), a component of the retromer complex that mediates sorting of cargo from endosomes before trafficking to the TGN (11, 19). Because there was no effect on the steady-state localization of Syn10 at the TGN (fig. S8), these observations suggested that Salmonella interferes with the Syn10 pathway downstream of retromer sorting and upstream of the Syn10-containing SNARE complex that mediates vesicle fusion at the TGN (11). The small guanosine triphosphatase (GTPase) Rab9 is required for late endosome-TGN transport of MPRs (20, 21). It also binds to the Golgi-associated protein GCC185, which tethers MPR-containing vesicles and enables their fusion at the TGN (11). The proportion of Rab9 colocalizing with CD-MPR was unaffected in HeLa cells infected with the sifA,sseJ mutant but significantly reduced in cells infected with wild-type bacteria. In contrast, there was significantly greater colocalization between these proteins in uninfected cells depleted of SKIP (Fig. 4A). Thus, SKIP appears to negatively regulate interactions between compartments containing MPR and Rab9, and SifA might enhance this by binding to SKIP.
Fig 4. Interactions between SifA, SKIP, and Rab9.
(A) HeLa cells depleted of SKIP (si SKIP) or infected for 14 hours with wt or sifA,sseJ double mutant Salmonella were fixed, labeled, and analyzed by quantitative confocal immunofluorescence microscopy. Pearson’s correlation coefficient was calculated between Rab9 and CD-MPR and compared to noninfected or si Scramble–treated cells. (B) HeLa cells stably expressing Flag-SKIP and GFP-Rab9 were infected for 14 hours with sifA,sseJ mutant Salmonella expressing either SifAwt-HA or SifAL130D-HA. Cells were fixed, labeled, and analyzed by confocal immunofluorescence microscopy. Arrows indicate co-localization of the three proteins. (C) Cells infected as in (B) were lysed and proteins were immuno-precipitated with GRP antibody–conjugated beads, or protein G (prG)–beads as a control. GFP-Rab9, SifA-HA, SifAL130D-HA, and Flag-SKIP were detected in input samples (input) and after immunoprecipitation (output) by means of SDS–polyacrylamide gel electrophoresis and immunoblotting. (D) Model depicting interference of Rab9-mediated retrograde transport of MPR to the TGN by sequestration of Rab9 by SifA and SKIP. (E) HeLa cells treated with scramble, Syn6, or Syn10 oligos or with lysosome inhibitor were infected with GFP-expressing wild-type Salmonella. Bacterial load, measured as GFP fluorescence per HeLa cell, was measured by flow cytometry (n > 10,000). The % fluorescence/cell represents the ratio between the median GFP fluorescence at 2 and 8 hours after infection, normalized to mock-transfected or DMSO-treated cells. Statistically significant relationships are denoted (*P < 0.05).
To investigate possible interactions between SifA, SKIP and Rab9 in infected cells, we infected stable cell lines expressing GFP-Rab9 and Flag-SKIP with Salmonella expressing SifA-HA or SifAL130D-HA. There was strong L130-dependent co-localization between SifA, SKIP, and Rab9 (Fig. 4B). Furthermore, SifA, SKIP, and Rab9 formed a stable complex in infected cells (fig. S9) that was dependent on L130 of SifA (Fig. 4C). Thus, SifA and SKIP appear to act as a “sink” for Rab9, thereby inhibiting MPR traffic and lysosome function (Fig. 4D). Indeed, overexpression of constitutively active Rab9 significantly impaired the ability of Salmonella to reduce lysosome function (fig. S10).
This work provides an explanation for the long-standing paradox that SCVs are relatively devoid of hydrolytic lysosomal enzymes (4) and yet retain lysosomal membrane glycoproteins in the vacuolar membrane and interact dynamically with the endolysosomal system (6). If Salmonella were exposed to lysosomal content, then its intracellular growth would be predicted to be sensitive to lysosome potency. Indeed, depletion of Syn6 (which augmented lysosome function; fig. S3B) resulted in reduced bacterial growth, whereas depletion of Syn10 (which decreased lysosome function; fig. S3B) or pharmacological inhibition of cathepsins resulted in increased bacterial growth (Fig. 4E and fig. S11). The actions of SPI-2 T3SS effectors that regulate SCV membrane dynamics (22) could promote interactions between SCVs and enzyme-depleted lysosomes to provide a source of vacuolar membrane and possibly nutrients to enable bacterial growth.
Supplementary Material
Acknowledgments
We thank C. Tang and members of the Holden laboratory for comments on the manuscript, S. Méresse for providing MEFs, S. Méresse and F. Randow for plasmids, and the European Conditional Mouse Mutagenesis Program Consortium for SkipPlekhm2−/− mice. We thank I. Dikic and D. McEwan for advice and discussions. This research was supported by a Wellcome Trust funded studentship to K.M. and grants to D.W.H. from the Medical Research Council and Wellcome Trust. The data are included in the main and the supplementary figures.
References and Notes
- 1.Saftig P, Klumperman J. Nat Rev Mol Cell Biol. 2009;10:623. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 2.Figueira R, Holden DW. Microbiology. 2012;158:1147. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 3.Rathman M, Sjaastad MD, Falkow S. Infect Immun. 1996;64:2765. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-del Portillo F, Finlay BB. J Cell Biol. 1995;129:81. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rathman M, Barker LP, Falkow S. Infect Immun. 1997;65:1475. doi: 10.1128/iai.65.4.1475-1485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drecktrah D, Knodler LA, Howe D, Steele-Mortimer O. Traffic. 2007;8:212. doi: 10.1111/j.1600-0854.2006.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Méresse S, Steele-Mortimer O, Finlay BB, Gorvel JP. EMBO J. 1999;18:4394. doi: 10.1093/emboj/18.16.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hang HC, et al. ACS Chem Biol. 2006;1:713. doi: 10.1021/cb600431a. [DOI] [PubMed] [Google Scholar]
- 9.Deiwick J, et al. J Bacteriol. 1998;180:4775. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beuzón CR, et al. EMBO J. 2000;19:3235. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganley IG, Espinosa E, Pfeffer SR. J Cell Biol. 2008;180:159. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh P, Dahms NM, Kornfeld S. Nat Rev Mol Cell Biol. 2003;4:202. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda K, et al. Biochem Biophys Res Commun. 2008;377:268. doi: 10.1016/j.bbrc.2008.09.121. [DOI] [PubMed] [Google Scholar]
- 14.Metcalf DJ, Calvi AA, Seaman MNj, Mitchison HM, Cutler DF. Traffic. 2008;9:1905. doi: 10.1111/j.1600-0854.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz-Albert J, et al. Mol Microbiol. 2002;44:645. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- 16.Boucrot E, Henry T, Borg J-P, Gorvel J-P, Méresse S. Science. 2005;308:1174. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 17.Jackson LK, Nawabi P, Hentea C, Roark EA, Haldar K. Proc Natl Acad Sci USA. 2008;105:14141. doi: 10.1073/pnas.0801872105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diacovich L, et al. J Biol Chem. 2009;284:33151. doi: 10.1074/jbc.M109.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seaman MNJ. J Cell Biol. 2004;165:111. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro AD, Riederer MA, Pfeffer SR. J Biol Chem. 1993;268:6925. [PubMed] [Google Scholar]
- 21.Barbero P, Bittova L, Pfeffer SR. J Cell Biol. 2002;156:511. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroeder N, et al. PLoS Pathog. 2010;6:e1001002. doi: 10.1371/journal.ppat.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beuzón CR, Salcedo SP, Holden DW. Microbiology. 2002;148:2705. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- 24.Salcedo SP, Holden DW. EMBO J. 2003;22:5003. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.