Abstract
In the chloroplast psbD light-responsive promoter (LRP), a highly conserved sequence exists upstream from the bacterial −10/−35 elements. Multiple sequence-specific DNA binding proteins are predicted to bind to the conserved sequence as transcription factors. Using yeast one-hybrid screening of an Arabidopsis cDNA library, a possible DNA binding protein of the psbD LRP upstream sequence was identified. The protein, designated PTF1, is a novel protein of 355 amino acids (estimated molecular weight of 39.6) that contains a basic helix-loop-helix DNA binding motif in the predicted N-terminal region of the mature protein. Transient expression assay of PTF1-GFP fusion protein showed that PTF1 was localized in chloroplasts. Using the modified DNA sequence in the one-hybrid system, the ACC repeat was shown to be essential for PTF1 binding. The rate of psbD LRP mRNA accumulation was reduced in a T-DNA-inserted Arabidopsis ptf1 mutant. Compared with wild-type plants, the mutant had pale green cotyledons and its growth was inhibited under short-day conditions. These results suggest that PTF1 is a trans-acting factor of the psbD LRP.
In the early stage of light-induced chloroplast development, the transcription activity of chloroplast genes increases, leading to increased mRNA accumulation (Klein and Mullet, 1990; Rapp et al., 1992; DuBell and Mullet, 1995). Transcription rates vary among promoters, and in most cases, they reflect the amount of transcripts and the stoichiometric composition of proteins (Rapp et al., 1992). Although mRNA stability is an important regulatory factor in mature chloroplasts (Kawaguchi et al., 1992; Kim et al., 1993; Staub and Maliga, 1993; Shiina et al., 1998), the transcription rate plays a primary role in controlling gene expression in developing chloroplasts (for review, see Mullet, 1993; Mayfield et al., 1995; Stern et al., 1997).
Recent studies have clarified many of the molecular mechanisms of transcriptional regulation in plastids by successive cloning of RNA polymerase core and accessory subunits for plastid transcription (for review, see Maliga, 1998; Hess and Börner, 1999). One is nuclear-encoded bacteriophage-type RNA polymerase (Hedtke et al., 1997), which functions in transcription from the nuclear-encoded bacteriophage-type RNA polymerase promoters that exist in most non-photosynthetic genes (Hajdukiewicz et al., 1997; Kapoor et al., 1997). On the other hand, many plastid genes have eubacterial ς70-type promoters, which are preceded by “−10” and “−35” elements (consensus TATAAT and TTGACA, respectively; Hanley-Bowdoin and Chua, 1987; Igloi and Kössel, 1992). These promoters are recognized by plastid-encoded RNA polymerase (PEP), which is composed of plastid-encoded catalytic core subunits associated with nuclear-encoded subunits (for review, see Link, 1996; Maliga, 1998; and Hess and Börner, 1999). Many plastid-ς factors have been cloned in higher plants (Isono et al., 1997; Tanaka et al., 1997; Kestermann et al., 1998; Tozawa et al., 1998). As in bacterial RNA polymerase, plastid-ς factors are thought to be responsible for characteristic promoter recognition of holoenzymes. These achievements and relevant biochemical evidence support the idea that a change in the composition of multiple RNA polymerases with distinct sequence selectivity accounts for the developmental or environmental regulation of plastid transcription (Eisermann et al., 1990; Tiller and Link, 1993a; Nakahira et al., 1998; Satoh et al., 1999). In addition, plastid transcription is regulated by general DNA binding proteins (Nakano et al., 1997; Sato et al., 1998) and sequence-specific DNA binding proteins. The genes for the proteins that serve as trans-acting factors of plastid transcription have not been cloned so far.
psbD light-responsive promoter (LRP) is one of the best-characterized plastid promoters that is regulated by sequence-specific DNA binding proteins (Allison and Maliga, 1995; Kim and Mullet, 1995; To et al., 1996; Nakahira et al., 1998; Kim et al., 1999b). psbD encodes the D2 protein of photosystem II and is cotranscribed with psbC for CP43. LRP is one of several promoters (three–four) of the psbD/C operon, and the promoter sequence is conserved among higher plants (Christopher et al., 1992). It is not active in the dark and is activated by blue and UV-A light (Sexton et al., 1990; Christopher and Mullet, 1994; Satoh et al., 1997), dramatically increasing the accumulation of psbD/C mRNA (Kawaguchi et al., 1992). Light activation of LRP is thought to compensate for the photo-induced degradation of D2 and CP43 proteins, which are sensitive to blue and UV-A light (Christopher and Mullet, 1994). The LRP is an unusual ς70-type promoter that requires the “−10” element for transcription, but does not require the “−35” element (Nakahira et al., 1998; Kim et al., 1999b). A series of well-conserved repeat sequences upstream from the “−35” position act as a strong enhancer for the light-induced transcription (Allison and Maliga, 1995; Kim and Mullet, 1995; To et al., 1996; Nakahira et al., 1998). Although gel retardation assay has revealed that these repeat sequences bind sequence-specific DNA binding proteins, none of the proteins have been isolated or cloned. Identification of the trans-acting factors is essential to elucidate the unique mechanism of transcriptional regulation of the psbD LRP. In this study, we performed a one-hybrid screening of an Arabidopsis cDNA library using the psbD LRP upstream sequence. A cDNA of a novel protein that specifically binds to the psbD LRP sequence was cloned and the protein was proved necessary for sufficient transcription from psbD LRP for normal growth of plants.
RESULTS
One-Hybrid Screening of the psbD LRP Binding Factor
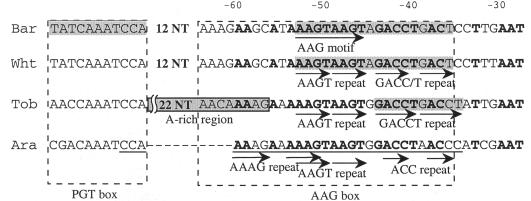
In Arabidopsis, multiple psbD mRNAs that vary at their 5′ are found. The 5′ ends are located 950, 550, and 190 bp upstream from the initiation codon, hereafter referred to as the −950, −550, and −190 ends, respectively (Hoffer and Christopher, 1997). The latter two 5′ ends result from either differential promoter utilization or processing of the −950 mRNA. Upstream of the −950 position exists the conserved sequence specific for the LRP of psbD of higher plants. The amount of mRNA with the −950 end increases in response to light (Hoffer and Christopher, 1997). Therefore, the mRNA with a −950 end is supposed to derive from primary transcription. The cis-acting element of the psbD LRP has been defined in tobacco (Allison and Maliga, 1995), barley (Kim and Mullet, 1995; Kim et al., 1999b), rice (To et al., 1996), wheat (Nakahira et al., 1998), and Arabidopsis (Christopher et al., 1999). According to these studies, the protein binding sites are found in the conserved plastid GT (PGT) box and around the AAGT and GACC/T repeats in the AAG box (Fig. 1). In barley, DNA binding protein complexes that bind to the PGT box and the AAG box are designated PGTF and AGF, respectively. The PGT box functions as a negative regulator, whereas the AAG box functions as an enhancer of light-dependent transcription (Kim and Mullet, 1995; Christopher et al., 1999). Similar results for the AAG box were obtained using wheat (Nakahira et al., 1998) and tobacco (Allison and Maliga, 1995). Our aim was to isolate a transcription activator of psbD LRP. Therefore, the highly conserved 29-bp sequence of the AAG box of Arabidopsis psbD LRP was used as the target element for the one-hybrid screening.
Figure 1.
Characteristics of the psbD LRP upstream sequences of four higher plants. Sequences from barley (Bar), wheat (Wht), tobacco (Tob), and Arabidopsis (Ara) are shown. The numbers on top represent the distance from the psbD LRP transcription start site. The nucleotides that are conserved among the four species are indicated with bold letters. The protein binding sequences are shadowed. The sequence that was used as the target sequence in the one-hybrid system is underlined.
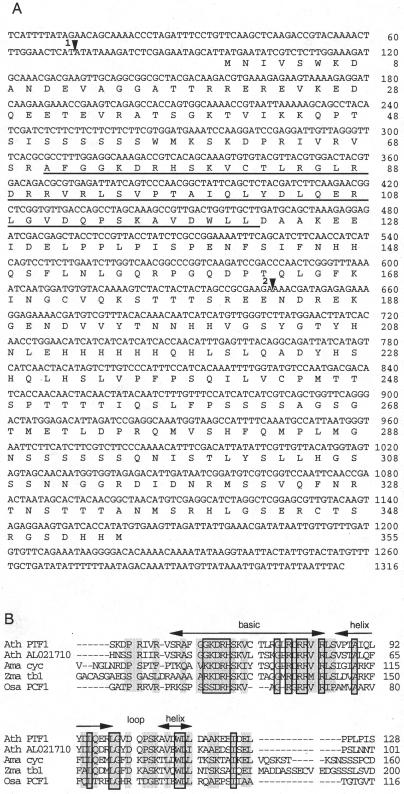
Four repeats of the target element were inserted into EcoRI sites of two reporter vectors. The EcoRI sites are located upstream from HIS3 and LacZ reporter genes with minimal promoters (PminHIS3 and PCYC1, respectively). The vectors were integrated into yeast strain YM4271 to make a target-reporter strain. Then an Arabidopsis cDNA activation domain (AD) fusion library was introduced into the strain. Approximately 4.5 × 105 transformants were screened on a Leu- and His-free medium. After 6 d, 103 to 104 colonies appeared on the plate and were tested for β-galactosidase activity. Three independent positive colonies that contained cDNA clones with identical sequences were found by the assay. Because no stop codon was found upstream from the first Met, these cDNA sequences did not seem to include the 5′ end. To obtain the full-length cDNA, PCR was carried out using a 5′ primer complementary to the library vector sequence, a cDNA-specific 3′ primer, and the cDNA-AD fusion library as a template (see “Materials and Methods”). A putative initiation codon was found after a 5′-untranslated region of 96 bp with stop codons (Fig. 2A).
Figure 2.
Sequence analysis of Arabidopsis PTF1 cDNA. A, Nucleotide sequence and deduced amino acid sequence of Arabidopsis PTF1 cDNA. The conserved bHLH region is underlined. Arrowhead 1, The position of the intron in 5′-untranslated region found by comparison with the genomic sequence; arrowhead 2, the T-DNA-insertion position of the ptf1 mutant. The GenBank accession number is AB014465. B, Sequence alignment of the conserved regions of PTF1 and four other proteins: Ath AL021710, Arabidopsis teosinte branched1-like protein; Ama cyc, Antirrhinum majus cycloidea protein; Zma tb1, Zea mays branched1 protein; and Osa PCF1, Oryza sativa PCF1. Residues conserved in all five proteins and in more than three proteins are boxed and shadowed, respectively.
The Protein Encoded by the Cloned cDNA Is a Chloroplast Protein That Contains a DNA- Binding Motif
The nucleotide sequence and the deduced amino acid sequence of the positive cDNA clone are shown in Figure 2A. The genomic sequence that corresponds to the clone (plastid transcription factor 1 [PTF1]) was located near marker mi74 at the top of chromosome III of Arabidopsis. Its predicted amino acid sequence contained a helix-loop-helix motif accompanied by a basic region (bHLH). The bHLH motif is known to be a bipartite sequence-specific DNA binding motif (Sun and Baltimore, 1991). This domain is conserved in PCF1 and PCF2 of rice (Kosugi and Ohashi, 1997), in the Arabidopsis expressed sequence tag clone of a predicted protein named teosinte branched1-like protein (EMBL accession no. AL021710), in the A. majus cycloidea protein (Luo et al., 1995), and in the Z. mays teosinte branched1 protein (Doebley et al., 1997; Fig. 2B). In PCF1 and PCF2, the conserved motif is essential for specific DNA binding as well as dimerization with each other or with themselves. The cycloidea and teosinte branched1 proteins have a putative nuclear localization signal and participate in the regulation of morphogenesis. The other part of the PTF1 sequence did not show significant similarity with any previously identified genes. The nuclear localization signal was not found in PTF1 by the sequence analysis.
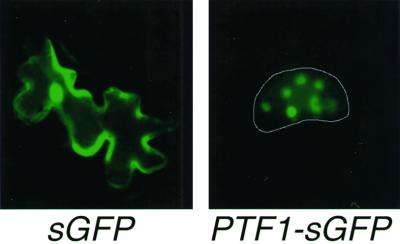
The N-terminal region of approximately 20 amino acids was 15% Ser and Thr and 25% Ala and Val. These amino acid residues are found in similar ratios in known transit peptides (Keegstra et al., 1989). To determine the subcellular localization of PTF1, its coding region was fused with the synthetic green fluorescent protein gene, sGFP (S65T; Chiu et al., 1996), and introduced into 3-week-old tobacco leaves by particle bombardment. Tobacco was used because the leaves are more robust than Arabidopsis leaves and clear pictures are easily obtained. After a 24-h incubation, green fluorescence was observed in the chloroplasts of transformed cells (Fig. 3). These results indicate that PTF1 is a chloroplast DNA binding protein. However, green fluorescence was also observed in the nucleus of some cells. The same result was obtained with bombarded Arabidopsis leaves (data not shown). Rare nuclear localization is feasible in a transient assay if the introduced gene accidentally picks up a nuclear localization signal after integration into a nuclear gene. Fluorescence in the nucleus alternatively could be an artifact.
Figure 3.
Chloroplast localization of the PTF1-sGFP (S65T) fusion protein in tobacco leaf epidermal cells. Detached tobacco leaves were bombarded with constructs carrying 35S-sGFP(S65T) (left) or 35S-PTF1-sGFP(S65T) (right). The epidermal cells were observed by fluorescence microscopy after a 24-h incubation.
PTF1 Specifically Binds to the ACC Repeat of psbD LRP
To test the sequence-specific DNA binding activity of PTF1, we performed a one-hybrid assay with a modified LRP sequence (Table I). In the yeast strain carrying the target sequence with a disrupted AAAG or AAGT repeat, the β-galactosidase activity was comparable to that of a strain with the wild-type sequence. However, in the ACC repeat-disrupted strain, the PTF1-AD fusion protein failed to induce β-galactosidase activity. These results strongly suggest that PTF1 specifically binds to the ACC repeat region. This result agrees well with the results obtained by other research groups using different species (Fig. 1). Although we did not succeed in producing enough recombinant PTF1 in Escherichia coli to directly prove the binding of the protein to the LRP sequence in vitro, this agreement supports that the PTF1 is one of the trans-acting factors of the psbD LRP.
Table I.
Binding assay of PTF1 to the wild-type and modified psbD LRP sequences
| Strain | Target Sequencea | Phenotype |
|---|---|---|
| Wild type | AAAAGAAAAAGTAAGTGGACCTAACCC | LacZ+b |
| mt-1 | CCCCTCCCAAGTAAGTGGACCTAACCC | LacZ+ |
| mt-2 | AAAAGAAACCTGCCTGGGACCTAACCC | LacZ+ |
| mt-3 | AAAAGAAAAAGTAAGTGGTAGTATGAC | LacZ−b |
Yeast strains that carry three (mt-1) or four (others) repeats of target sequences followed by lacZ under the control of PCYC1 in their genome DNA were transformed with the PTF1-AD fusion vector and tested for β-galactosidase activity.
Replaced nucleotides are indicated with bold letters.
LacZ+ or LacZ− represents the phenotype of transformants according to the β-galactosidase assay.
Correlated Expression of psbD LRP and PTF1 mRNA in Wild-Type Plants
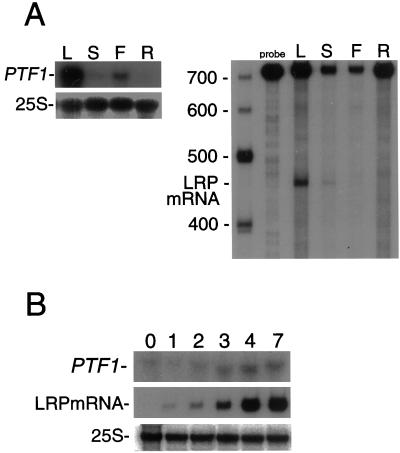
The tissue-specific expression of PTF1 was examined by RNA gel-blot analysis with total RNA isolated from leaves, stems, flowers, and roots of 32-d-old Arabidopsis plants. As shown in Figure 4A, PTF1 mRNA was most abundant in leaves, slightly detectable in stems and flowers, and traces were found in roots. Using the same RNA preparations, the accumulation of psbD LRP mRNA was examined by an S1 protection assay. The amount of psbD LRP mRNA was again highest in the leaves. Therefore, the tissue-specific expression pattern of PTF1 coincides with that of psbD LRP.
Figure 4.
Tissue-specific and light-dependent expression of PTF1 and psbD LRP mRNA. A, Accumulation of PTF1 and 25S rRNA detected by RNA gel-blot analysis (left) and psbD LRP mRNA detected by S1 protection assay (right) in different tissues. L, leaves; S, stems; F, flowers; R, roots. B, Accumulation of PTF1 and 25S rRNA (RNA gel-blot analysis) and psbD LRP mRNA (S1 protection assay) in seedlings grown in the dark for 3 d (0) and further illuminated for the indicated number of days (1–7).
The light dependency of PTF1 mRNA accumulation was examined in seedlings (Fig. 4B). Traces of PTF1 mRNA were found in 3-d-old dark-grown Arabidopsis seedlings. After 2 d of illumination, PTF1 mRNA content started to increase. On the other hand, no psbD LRP mRNA was detected in 3-d-old dark-grown seedlings. Then after 2 d of illumination, the psbD LRP mRNA increased rapidly and reached a maximum within 4 d. In conclusion, in etiolated seedlings, PTF1 expression precedes the psbD LRP activation, and both mRNA levels increase in parallel with illumination.
ptf1 Mutant Has Reduced psbD LRP Activity
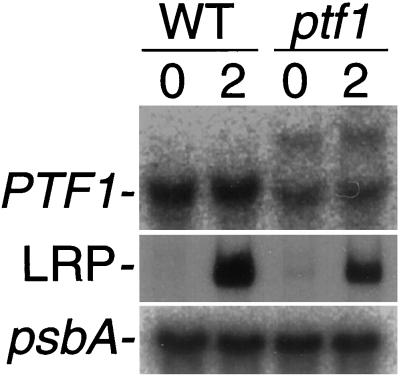
By means of PCR screening, we isolated a ptf1 mutant derived from a population of Arabidopsis lines (ecotype Wassilewskija) mutagenized with T-DNA insertions (Feldmann and Marks, 1987). Before screening, DNA gel-blot analysis was performed and PTF1 was identified as a single-copy gene (data not shown). T-DNA was inserted 535 bp from the initiation codon (Fig. 2A), and the PTF1 mRNA was split into two bands (Fig. 5). These results indicate that PTF1 is successfully destructed in the ptf1 mutant. Because we did not succeed in getting enough recombinant PTF1 to develop antibodies, the presence of the PTF1 could not be checked in the ptf1 mutant.
Figure 5.
Light response of PTF1 and psbD LRP mRNA in dark-adapted mature plants. Wild-type (WT) and ptf1 mutant (ptf1) Arabidopsis plants were grown for 14 d under continuous light and harvested after 2 d of dark adaptation (0) or following illumination for 2 h (2). PTF1 and psbA mRNAs were detected by RNA gel-blot analysis, and psbD LRP mRNA was detected by S1 protection assay.
No significant effect on the level of psbD LRP mRNA accumulation was observed in ptf1 plants grown under continuous light (data not shown); subsequently, the light response of psbD LRP in mature ptf1 and wild-type plants was examined (Fig. 5). When 2-week-old light-grown plants were dark adapted for 2 d, mRNA from the psbD LRP decreased to undetectable levels in both wild-type and ptf1 plants. After 2 h of illumination, both strains accumulated psbD LRP mRNA, but ptf1 accumulated only half as much as the wild type, although psbA mRNA was accumulated at a comparative level to the wild type in the ptf1 mutant. These results clearly show that disruption of the PTF1 gene causes reduced psbD LRP activity. However, no effect of light after dark adaptation was observed in PTF1 mRNA accumulation in wild-type plants (Fig. 5).
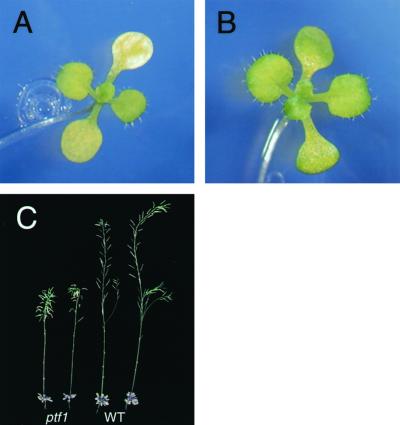
The ptf1 mutant did not show obvious phenotypic alterations when it was grown under continuous light (data not shown). However, under an 8-h short-day condition, ptf1 mutant cotyledons underwent early bleaching, whereas the cotyledons of wild-type plants remained green (Fig. 6, A and B). The ptf1 plants also displayed late flowering and dwarfism under the short-day condition compared with wild type (Fig. 6C). A shortage of psbD mRNA in the ptf1 mutant induced by limited light period may be responsible for these phenotypes.
Figure 6.
Phenotype of the ptf1 mutant grown under short-day condition. A, Twenty-one-day-old wild-type plant. B, Twenty-one-day-old ptf1 mutant with pale green cotyledons. C, Seventy-five-day-old mature wild-type and ptf1 mutant plants.
DISCUSSION
Correspondence between PTF1 and the AAG Box-Binding Proteins of Other Species
This is the first report on cloning of a plastid sequence-specific and nuclear-encoded transcription factor to our knowledge. PTF1 is a novel plastid protein encoded by the nuclear genome that has a conserved bHLH motif. The bHLH motif is a well-characterized domain that is involved in protein dimerization and DNA binding. This motif is found in a group of transcription factors that have been identified in eukaryotes ranging from yeast to mammals and plants (Sun and Baltimore, 1991; Toyama et al., 1999). Neither a PTF1 homolog nor the conserved psbD LRP sequence exists in cyanobacterium (data not shown). Because bHLH proteins have only been reported as eukaryotic transcription factors, the machinery for the light response of the psbD LRP may have been acquired after the endosymbiosis of the prokaryotic ancestor of chloroplasts.
PTF1 was shown to have sequence-specific DNA binding activity. It strictly recognizes the ACC repeat of the psbD LRP upstream sequence in yeast (Table I). The psbD LRP has two conserved upstream sequences: the PGT motif and the AAG box (Fig. 1; Christopher et al., 1999). At least two proteins have been detected to bind to the LRP upstream sequence. Protein complexes of barley binding to the PGT box and the AAG box are designated PGTF and AGF, respectively (Kim and Mullet, 1995). Proteins binding to the AAG box are also found in tobacco and wheat. In any of the species, the AAG box is required for the full light response, and the GACC/T repeat in the AAG box is involved in binding the protein(s) (Allison and Maliga, 1995; Nakahira et al., 1998; Kim et al., 1999b). The Arabidopsis ACC repeat that is specifically recognized by PTF1 corresponds to the GACC/T repeats of the three above-mentioned species (Fig. 1). Therefore, PTF1 with a predicted Mr of 39.6 may correspond to one of the AAG box binding proteins observed in other species. It is interesting that in barley, binding of 38-kD and 60-kD proteins to the PGT box was detected by southwestern analysis (Christopher et al., 1999). Although the PGT box is supposed to be a negative regulator, the 38-kD protein might correspond to PTF1, which may also interact with the PGT box under certain conditions and regulate transcription. Though high expression of PTF1 protein in E. Coli has not been successful yet, a detailed binding assay as well as the in vitro transcription assay using the recombinant PTF1 to the LRP upstream sequence will be required to investigate its function in the future.
PTF1 Enhances the Light-Induced Transcription of the psbD LRP
Mutation of the PTF1 gene caused a lower rate of psbD LRP mRNA accumulation than in the wild type when dark-adapted mature seedlings were illuminated (Fig. 5). Because the rate of psbA mRNA accumulation was not affected in the ptf1 mutant, disruption of the PTF1 gene seems to affect psbD LRP activity exclusively. When mutant plants were grown under short-day conditions, the ptf1 mutant plants produced yellow-green cotyledons (Fig. 6, A and B) and their growth was suppressed (Fig. 6C). These phenotypes were presumably caused by a shortage of psbD mRNA as a result of low psbD LRP activity and the short illumination period. The phenotypes of the ptf1 mutants also suggest that the psbD LRP plays an important role in maintaining plant vigor.
PTF1 mRNA was detectable in the 3-d-old dark-grown seedlings and the dark-adapted mature plants, although psbD LRP was not active (Figs. 4 and 5). This result indicates that PTF1 expression is not regulated by light at the transcriptional level. This observation is consistent with the observation that psbD LRP upstream sequences may act as a general enhancer regardless of the light condition (Nakahira et al., 1998). However, it does not exclude the possibility that PTF1 undergoes posttranslational modification when plants are exposed to light, as reported for PGTF, the PGT binding complex of barley: Its DNA binding activity is inhibited by ADP-dependent phosphorylation (Kim et al., 1999a). Furthermore, phosphorylation/dephosphorylation is known to regulate the function of PEP (Tiller and Link, 1993b). Putative phosphorylation sites are found in the PTF1 amino acid sequence (data not shown). Further study is needed to investigate the possible posttranslational regulation of the PTF1 function.
Suppression of the psbD LRP activity in the ptf1 mutant was incomplete, considering the fact that expression of the psbD LRP without the upstream sequence was observed at very low levels both in vivo (Allison and Maliga, 1995) and in vitro (Nakahira et al., 1998) under illumination. It is likely due to the existence of the additional higher Mr DNA binding factors that have been observed in tobacco A-rich region (see Fig. 1), barley (PGTF), and wheat (Allison and Maliga, 1995; Kim and Mullet, 1995; Nakahira et al., 1998). PTF1 alternatively might not be completely inactivated by the T-DNA insertion. The T-DNA is inserted 535 bp from the initiation codon, which is downstream from the conserved bHLH region (Fig. 2A). It is possible that the mutant PTF1 partially retains its function.
Possibility of Additional Trans-Acting Factors of the psbD LRP
Because transcription factors with a bHLH domain, like PTF1, bind DNA in the form of homo- or heterodimers (Sun and Baltimore, 1991), additional chloroplast proteins that interact with PTF1 may exist. Moreover, it is very likely that PTF1 needs hitherto unidentified factors to interact with the PEP that originates from the eubacterial RNA polymerase. PEP is composed of a plastid-encoded catalytic core complex and nuclear-encoded sigma factors (Isono et al., 1997; Tanaka et al., 1997; Kestermann et al., 1998; Tozawa et al., 1998). Two forms of PEP, designated enzyme A and enzyme B, have been identified in mustard (Pfannschmidt and Link, 1994). Enzymes A and B are dominant in chloroplasts and etioplasts, respectively. Enzyme B has features of eubacterial RNA polymerase and is sensitive to rifampicin, which inhibits prokaryotic gene transcription. On the other hand, enzyme A is rifampicin resistant (Pfannschmidt and Link, 1997) and is considered an eukaryote-like enzyme. Enzyme A consists of multiple subunits, including a protein kinase (Baginsky et al., 1997) and unidentified proteins in addition to the core enzyme (for review, see Link, 1996; Maliga, 1998). Investigation of the protein-protein interaction between PTF1 and the PEP enzyme A complex will provide further information about a novel mechanism of transcriptional regulation in plastids.
MATERIALS AND METHODS
The Yeast One-Hybrid Screening
Screening was performed using the MATCHMAKER one-hybrid system (CLONTECH, Palo Alto, CA). To make a target-reporter construct, four tandem repeats of the putative cis-acting element of the Arabidopsis psbD LRP (from −1010 to −982 where the psbD initiation codon is +1) were inserted into the EcoRI sites of pHISi and pLacZi plasmids. Both of the constructs were used to transform the yeast strain YM4271. The Arabidopsis MATCHMAKER cDNA expression library (ecotype Columbia) was purchased from CLONTECH. A 300-mL yeast culture was transformed using 20 μg of the cDNA and plated on synthetic minimal medium containing 30 mm 3-aminotriazole, but lacking His and Leu. After incubation at 30°C for 6 d, the colonies were transferred to filter paper and tested for β-galactosidase activity. Plasmids were extracted from the positive yeast colonies, amplified in Escherichia coli cells, and purified for sequencing. To obtain the 5′-end of the cDNA clone, the first PCR was carried out with 1 pmol μL−1 of vector primer 1 (5′-GGACGGACCAAACTGCGTATAACGCG-3′), 0.2 pmol μL−1 of cDNA-specific primer 2(5′-GCTGCATCAAGCAACCAGTCAACGGC-3′), and 6 ng μL−1 of DNA from the MATCHMAKER expression library as a template. The first PCR product was electrophoresed on an agarose gel, and the bands longer than 505 bp were collected and purified with a gel extraction kit (Qiagen, Hilden, Germany). Using the resulting DNA fragments as a template, a second PCR was performed with 1 pmol μL−1 each of vector primer 3 (5′-CGATGATGAAGATACCCCACCAAACCC-3′) and cDNA-specific primer 4 (5′-CAAAGGCGCGTGAAACCCTAACAATCC-3′). The products were ligated to pCR2.1 vector (Invitrogen, Groningen, The Netherlands) and the lengths of the inserts were checked by PCR. The clones with longer inserts were subsequently sequenced.
Transient Expression of GFP
The full length of the coding region of the positive cDNA clone was amplified by PCR and inserted between the SalI and NcoI sites of the 35S-sGFP(S65T) plasmid (Chiu et al., 1996). Detached leaves of 3-week-old tobacco plants were placed abaxial surface up on 0.8% (w/v) agar plates. Gold particles (1 μm in diameter; Bio-Rad, Hercules, CA) coated with the GFP plasmids were shot into the leaves with a particle gun (PDS 1000/He, Bio-Rad) under a vacuum of 28 inches Hg and a helium pressure of 1,350 psi (pound-force per square inch). After bombardment, the leaves were incubated at 25°C for 24 h and observed using a fluorescent microscope (AX70, Olympus, Tokyo). The light from a mercury lamp was filtered through an U-MNIBA filter for excitation to observe the emission from GFP.
Plant Growth
For RNA extraction, Arabidopsis seeds were sterilized with 70% (v/v) ethanol and 1% (w/v) sodium hypochlorite before sowing on Murashige and Skoog agar medium (Murashige and Skoog, 1962). After stratification at 4°C for 2 d, the seeds were grown at 22°C under continuous light for 2 to 3 weeks.
RNA Gel-Blot Analysis
Extraction of RNA was performed with an RNA isolation kit (TRI reagent, Molecular Research Center, Inc., Cincinnati) according to the manufacturer's protocol. Five to 10 μg of Arabidopsis total RNA was electrophoresed on a 1.2% (w/v) agarose gel containing 20 mm MOPS [3-(N-morpholino)propanesulfonic acid] (pH 7.0) and 5% (v/v) formaldehyde. Then, the RNA was transferred to a nylon membrane (Hybond-XL, Amersham Pharmacia Biotech, Buckinghamshire, UK) and prehybridized for from 3 h to overnight at 68°C in a buffer containing 5 × sodium chloride/sodium dihydrogen phosphate/EDTA buffer (SSPE), 5 × Denhardt's solution, 0.5% (w/v) SDS, and 60 ng mL−1 of denatured salmon sperm DNA. A radiolabeled DNA or RNA probe subsequently was added to the buffer and further hybridized at 68°C overnight. The membrane was washed three times with a buffer containing 1 × SSPE and 1% (w/v) SDS, and once with a buffer containing 0.1 × SSPE and 1% (w/v) SDS at 68°C before autoradiography.
S1 Protection Assay
A 5′-radiolabeled DNA probe (from −267 to +457 of the psbD LRP) was hybridized with 10 μg of total RNA at 37°C for 16 h in 10 μL of hybridization buffer containing 40 mm PIPES (1,4-piperazinediethanesulfonic acid) (pH 6.4), 1 mm EDTA (pH 8.0), 0.4 m NaCl, and 80% (v/v) formamide. The solution was diluted with 100 μL of ice-cold S1 nuclease mixture containing 0.28 m NaCl, 0.05 m sodiumacetate (pH 4.5), 4.5 mm ZnSO4, and 500 units mL−1 of S1 nuclease, and subsequently incubated at 20°C for 1 h. The protected DNA was extracted with phenol/chloroform, precipitated in 2-propanol, and electrophoresed on a denaturing gel containing 4% (w/v) acrylamide (mono:bis = 19:1), Tris-borate/EDTA buffer, and 7 m urea.
DNA Gel Blot Analysis
Arabidopsis plants grown on Murashige and Skoog medium were frozen and ground in liquid N2 with a mortar and pestle. Genomic DNA was extracted from the powdered tissue with a Nucleon DNA Extraction Kit (Amersham Pharmacia Biotech). The resulting DNA was extracted with chloroform and treated with RNase A. Ten micrograms of purified DNA were digested overnight with BamHI, EcoRI, or HindIII and precipitated with ethanol. Then approximately 1 μg of the digested DNA was electrophoresed on a 0.8% (w/v) agarose gel, denatured in 0.5 m NaOH and 1.5 m NaCl, neutralized in 0.5 m Tris-HCl (pH 7.4) and 1.5 m NaCl, and transferred to a nylon membrane. After denaturation in 0.4 m NaOH and neutralization in 0.2 m Tris (pH 7.4) and 2 × SSPE (1 × SSPE is 0.15 m NaCl, 20 mm NaH2PO4, and 2 mm EDTA [pH7.4]), the membrane was dried and hybridized with 25 ng of the radiolabeled PTF1 cDNA fragment (1,068 bp) at 65°C in a buffer containing 5 × SSPE, 5 × Denhardt's solution, 1% (w/v) SDS, and 1 mg mL−1 salmon sperm DNA. The membrane was subsequently washed in 1 × SSPE three times and 0.1% (w/v) SDS once and autoradiographed.
Screening of the ptf1 Mutant
The ptf1 mutant (stock no. CS15566) was screened as described by McKinney et al. (1995). Pooled DNA of T-DNA lines (6,000 lines, pool name CD5-7) and the T-DNA mutant seeds (ecotype Wassilewskija) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The PCR primers used were a combination of either left border (5′-GATGCAATCGATATCAGCCAATTTTAGAC-3′) or right border (5′-GTCCAGGATCCGATTGTCGTTTCCCGCCTT-3′) with either PTF1 5′-end primer (5′-GTCAGAGCCACCAGTGGCAAAACCGTAATT-3′) or PTF1 3′-end primer (5′-GGACCGACGACATCCGATTATCAATGTCTC-3′). For comparison, PCR reactions using only the LB or RB primer were performed. The PCR products were analyzed by DNA gel-blot hybridization with the PTF1 probe to confirm the specific amplification.
ACKNOWLEDGMENTS
The DNA sequence of the Arabidopsis chloroplast genome was provided by Dr. Satoshi Tabata (Kazusa DNA Research Institute, Kisarazu, Japan). The sGFP(S65T) plasmid was provided by Dr. Yasuo Niwa (University of Shizuoka, Shizuoka, Japan). We thank Dr. Rishikesh Bhalerao (Swedish University of Agricultural Sciences, Umeå, Sweden) for critical review of the manuscript.
Footnotes
This research was supported by a Grant-in-Aid from the Ministry of Science, Education and Culture of Japan (no. 11151230 to T.N. and K.B.). K.B. and K.Y. were supported by the Special Postdoctoral Researchers' Program of RIKEN.
LITERATURE CITED
- Allison LA, Maliga P. Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 1995;14:3721–3730. doi: 10.1002/j.1460-2075.1995.tb00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Link G. Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba) Plant Mol Biol. 1997;34:181–189. doi: 10.1023/a:1005802909902. [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Christopher DA, Kim M, Mullet JE. A novel light-regulated promoter is conserved in cereal and dicot chloroplasts. Plant Cell. 1992;4:785–798. doi: 10.1105/tpc.4.7.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Mullet JE. Separate photosensory pathways co-regulate blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Shen Y, Dudley P, Tsinoremas NF. Expression of a higher-plant chloroplast psbD promoter in a cyanobacterium (Synechococcus sp. Strain PCC7942) reveals a conserved cis element, designated PGT, that differentially interacts with sequence-specific binding factors during leaf development. Curr Genet. 1999;35:657–666. doi: 10.1007/s002940050465. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- DuBell AN, Mullet JE. Differential transcription of pea chloroplast genes during light-induced leaf development. Plant Physiol. 1995;109:105–112. doi: 10.1104/pp.109.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisermann A, Tiller K, Link G. In vitro transcription and DNA binding characteristics of chloroplast and etioplast extracts from mustard (Sinapis alba) indicate differential usage of the psbA promoter. EMBO J. 1990;9:3981–3987. doi: 10.1002/j.1460-2075.1990.tb07619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA, Marks MD. Agrobacterium-mediated transformation of germinating-seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol Gen Genet. 1987;208:1–9. [Google Scholar]
- Hajdukiewicz PTJ, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Chua N-H. Chloroplast promoters. Trends Biochem Sci. 1987;12:67–70. [Google Scholar]
- Hedtke B, Börner T, Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- Hess WR, Börner T. Organellar RNA polymerases of higher plants. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- Hoffer PH, Christopher DA. Structure and blue-light-responsive transcription of a chloroplast psbD promoter from Arabidopsis. Plant Physiol. 1997;115:213–222. doi: 10.1104/pp.115.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL, Kössel H. The transcriptional apparatus of chloroplasts. Crit Rev Plant Sci. 1992;10:525–558. [Google Scholar]
- Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H. Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains for ς70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:14948–14953. doi: 10.1073/pnas.94.26.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Suzuki JY, Sugiura M. Identification and functional significance of a new class of non-consensus-type plastid promoters. Plant J. 1997;11:327–337. doi: 10.1046/j.1365-313x.1997.11020327.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi H, Fukuda I, Shiina T, Toyoshima Y. Dynamical behavior of psb gene transcripts in greening wheat seedlings: I. Time course of accumulation of the psbA through psbN gene transcripts during light-induced greening. Plant Mol Biol. 1992;20:695–704. doi: 10.1007/BF00046454. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- Kestermann M, Neukirchen S, Kloppstech K, Link G. Sequence and expression characteristics of a nuclear-encoded chloroplast sigma factor from mustard (Sinapis alba) Nucleic Acids Res. 1998;26:2747–2753. doi: 10.1093/nar/26.11.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE. Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol. 1993;22:447–463. doi: 10.1007/BF00015975. [DOI] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE. ADP-dependent phosphorylation regulates association of a DNA-binding complex with the barley chloroplast psbD blue-light-responsive promoter. Plant Physiol. 1999a;119:663–670. doi: 10.1104/pp.119.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Mullet JE. Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light-responsive psbD-psbC promoter. Plant Cell. 1995;7:1445–1457. doi: 10.1105/tpc.7.9.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Thum KE, Morishige DT, Mullet JE. Detailed architecture of the barley chloroplast psbD-psbC blue light-responsive promoter. J Biol Chem. 1999b;274:4684–4692. doi: 10.1074/jbc.274.8.4684. [DOI] [PubMed] [Google Scholar]
- Klein RR, Mullet JE. Light-induced transcription of chloroplast genes. psbA transcription is differentially enhanced in illuminated barley. J Biol Chem. 1990;265:1895–1902. [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G. Green life: control of chloroplast gene transcription. BioEssays. 1996;18:465–471. [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1995;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- Maliga P. Two plastid RNA polymerases of higher plants: an evolving story. Trends Plant Sci. 1998;3:4–6. [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2–1 and act4–1. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- Mullet JE. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nakahira Y, Baba K, Yoneda A, Shiina T, Toyoshima Y. Circadian-regulated transcription of the psbD light-responsive promoter in wheat chloroplasts. Plant Physiol. 1998;118:1079–1088. doi: 10.1104/pp.118.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Murakami S, Shoji T, Yoshida S, Yamada Y, Sato F. A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell. 1997;9:1673–1682. doi: 10.1105/tpc.9.9.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol. 1994;25:69–81. doi: 10.1007/BF00024199. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol Gen Genet. 1997;257:35–44. doi: 10.1007/s004380050621. [DOI] [PubMed] [Google Scholar]
- Rapp JC, Baumgartner BJ, Mullet J. Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes. J Biol Chem. 1992;267:21404–21411. [PubMed] [Google Scholar]
- Sato N, Ohshima K, Watanabe A, Ohta N, Nishiyama Y, Joyard J, Douce R. Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell. 1998;10:859–872. doi: 10.1105/tpc.10.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J, Baba K, Nakahira Y, Shiina T, Toyoshima Y. Characterization of dynamics of the psbD light-induced transcription in mature wheat chloroplasts. Plant Mol Biol. 1997;33:267–278. doi: 10.1023/a:1005799001271. [DOI] [PubMed] [Google Scholar]
- Satoh J, Baba K, Nakahira Y, Tsunoyama Y, Shiina T, Toyoshima Y. Developmental stage-specific multi-subunit plastid RNA polymerases (PEP) in wheat. Plant J. 1999;18:407–415. doi: 10.1046/j.1365-313x.1999.00465.x. [DOI] [PubMed] [Google Scholar]
- Sexton TB, Christopher DA, Mullet JE. Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 1990;9:4485–4494. doi: 10.1002/j.1460-2075.1990.tb07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Allison L, Maliga P. rbcL transcript levels in tobacco plastids are independent of light: reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell. 1998;10:1713–1722. doi: 10.1105/tpc.10.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 1993;12:601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Higgs DC, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- Sun X-H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Tozawa Y, Mochizuki N, Shinozaki K, Nagatani A, Wakasa K, Takahashi H. Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 1997;413:309–313. doi: 10.1016/s0014-5793(97)00906-x. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Sigma-like transcription factors from mustard (Sinapis alba L.) etioplast are similar in size to, but functionally distinct from, their chloroplast counterparts. Plant Mol Biol. 1993a;21:503–513. doi: 10.1007/BF00028807. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993b;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K, Cheng M, Suen D, Mon D, Chen LO, Chen SG. Characterization of the light-responsive promoter of rice chloroplast psbD-C operon and the sequence-specific DNA binding factor. Plant Cell Physiol. 1996;37:660–666. doi: 10.1093/oxfordjournals.pcp.a028995. [DOI] [PubMed] [Google Scholar]
- Toyama T, Teramoto H, Ishiguro S, Tanaka A, Okada K, Takeba G. A cytokinin-repressed gene in cucumber for a bHLH protein homologue is regulated by light. Plant Cell Physiol. 1999;40:1087–1092. doi: 10.1093/oxfordjournals.pcp.a029491. [DOI] [PubMed] [Google Scholar]
- Tozawa Y, Tanaka K, Takahashi H, Wakasa K. Nuclear encoding of a plastid sigma factor in rice and its tissue- and light-dependent expression. Nucleic Acids Res. 1998;26:415–419. doi: 10.1093/nar/26.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]