Abstract
PTEN is a well-characterized tumour-suppressor gene that is lost or mutated in about half of metastatic castration-resistant prostate cancers and in many other human cancers. The restoration of functional PTEN as a treatment for prostate cancer has however proven difficult. Here, we show that PTEN mRNA can be reintroduced into PTEN-null prostate cancer cells in vitro and in vivo via its encapsulation in polymer-lipid hybrid nanoparticles coated with a poly(ethylene glycol) shell. The nanoparticles are stable in serum, elicit low toxicity, enable high PTEN mRNA transfection in prostate cancer cells, and lead to significant inhibition of tumour growth when delivered systemically in multiple mouse models of prostate cancer. We also show that the restoration of PTEN function in PTEN-null prostate cancer cells inhibits the PI3K-AKT pathway and enhances apoptosis. Our findings provide proof-of-principle evidence of the restoration of mRNA-based tumour suppression in vivo.
The onset and maintenance of cancer frequently involves gain of oncogenic function (via amplification or mutation of oncogenes) along with loss of tumour suppression. Phosphatase and tensin homolog deleted on chromosome ten (PTEN) is among the best-characterized tumour suppressors. PTEN mutations have been reported in a variety of human cancers1, including prostate cancer (PCa). Loss of one PTEN allele is seen in the majority of localized PCa, and homozygous deletion of PTEN is more common in metastatic castration-resistant PCa (mCRPC) (~50%) than in androgen-dependent primary tumours (~10%)2–8. Moreover, loss of PTEN protein expression is more frequent than genomic PTEN loss and has been correlated with high Gleason score and faster progression to metastasis9. PTEN encodes a dual phosphatase that acts on both lipid and protein substrates. By catalyzing phosphatidylinositol (3,4,5)-trisphosphate (PIP3) dephosphorylation, PTEN negatively regulates the phosphatidylinositol 3-kinase (PI3K)-AKT pathway10–14, a key signaling mediator of most receptor tyrosine kinases (RTKs)15. Recent integrative genomic profiling and whole-exosome sequencing analysis highlight the frequency of alterations of the PI3K-AKT pathway in PCa, which is associated with both primary (42%) and metastatic disease (~100%)16, 17. Since activation of the PI3K-AKT pathway upon PTEN loss enhances tumour cell survival, proliferation12, 18, migration19, 20, angiogenesis21, 22, and anti-apoptosis10, blocking this pathway has been proposed to inhibit tumour growth and sensitize tumour cells to apoptosis. While pharmacological inhibitors of the AKT-mTOR pathway are in clinical development, they cannot compensate fully for the loss of PTEN function, and show a poor toxicity profile.
Restoration of PTEN function via plasmid DNA transfection of tumour cells has been demonstrated in vitro 10; however, their in vivo delivery has been severely hindered by systemic instability. Recently, chemically modified mRNA has emerged as an intriguing alternative to DNA-based gene therapy, as its intrinsic qualities facilitate its ease of use as a genetic material that is independent of nuclear localization and genomic integration for transfection activity23, 24. mRNA also provides rapid protein expression even in non-dividing and hard-to-transfect cells (e.g., immune cells), as well as cancer cells. Moreover, mRNA offers more consistent and predictable protein expression kinetics than DNA, whose expression kinetics show random onset25–28. However, delivery of mRNA presents several potential challenges, including large size, highly negative charge, susceptibility to degradation, and suboptimal protein translation capacity if it is not effectively modified and delivered into cells29. Thus, safe and effective in vivo cytosolic delivery of mRNA to tumour tissues while retaining integrity and functional activity remains elusive. Although many nano-engineered formulations (e.g., polymer-based nanoparticles (NPs)30, 31, lipid-based NPs 32-35, and lipidoid NPs36, 37) designed for in vivo delivery of siRNA or other small oligonucleotides have shown promising results in laboratory or clinical settings38–41, they might not be efficient for mRNA delivery due to the inherent structural differences between small-size oligonucleotides (e.g., siRNA and microRNA) and mRNA in terms of length, charge density, and stability42–44. To overcome these issues, we employed polymer–lipid hybrid nanoparticles for the systemic delivery of modified PTEN mRNA to PCa tumours. Using this system, we successfully restored functional PTEN protein production, with consequent inhibition of tumour cell growth and induction of apoptosis both in vitro and in vivo. We expect that hybrid NP-mediated PTEN mRNA delivery could become a feasible approach for the treatment of cancers with PTEN deficiency.
RESULTS
Preparation and characterization of mRNA NP
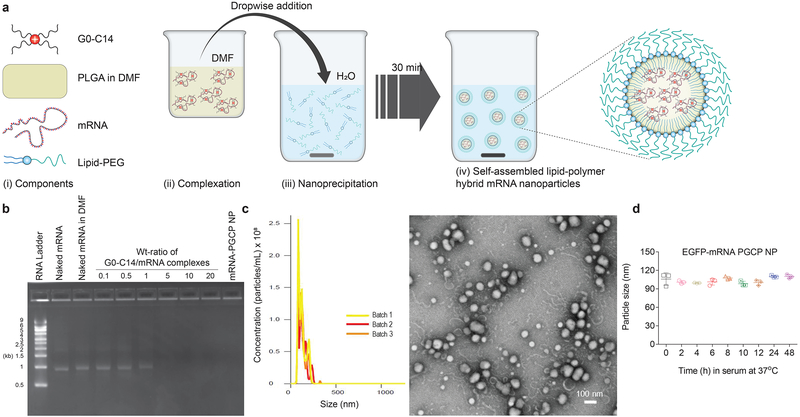
A robust self-assembly approach was employed to prepare the hybrid mRNA NPs using the cationic lipid-like compound G0-C14 and poly(lactic-co-glycolic acid) (PLGA) polymer coated with a lipid-poly(ethylene glycol) (lipid-PEG) shell45 (Fig. 1a). G0-C14 was used for mRNA complexation, and PLGA, a widely clinically used biodegradable and biocompatible polymer, was used to make a stable NP core. EGFP mRNA was used as a model mRNA, and the EGFP mRNA NP coated with ceramide-PEG is herein referred as to EGFP-mRNA-PGCP NP. We observed no effect of organic solvent (DMF) on the integrity or stability of EGFP mRNA, whether naked, complexed with G0-C14, or encapsulated in NPs (Fig. 1b). Fig. 1b also shows that G0-C14 effectively condensed EGFP mRNA at a weight ratio of 5 or above. The NPs were prepared at a G0-C14/mRNA weight ratio of 15, with no leaching of mRNA shown by electrophoresis, suggesting that most mRNA was encapsulated. The hybrid EGFP-mRNA-PGCP NPs were ~120 nm in size and spherical, as characterized by NanoSIGHT and transmission electron microscopy (TEM), respectively (Fig. 1c). Essential to the hybrid NP, the solid PLGA polymer core allowed formation of a stable and rigid nanostructure. The average surface charge measured by dynamic light scattering (DLS) was near neutral (5.96±0.76 mV), since the NPs have an outer lipid-PEG shell. In addition, the serum stability test showed no obvious changes in the particle size over 48h, suggesting the stability of the EGFP-mRNA-PGCP NP (Fig. 1d).
Figure 1. Preparation and characterization of mRNA NP.
(a) Self-assembly process of polymer-lipid hybrid mRNA NP and schematic representation of NP structure (i-iv). After self-assembly of cationic G0-C14 with anionic mRNA together with PLGA, the formulated polymer-lipid hybrid core was coated with lipid-PEG (EGFP-mRNA was used as reporter mRNA). (b) Agarose gel electrophoresis assay of mRNA stability in organic solvent, naked or complexed with cationic G0-C14, at various weight ratios (from 0.1 to 20). The formulated mRNA PLGA/G0-C14/ceramide-PEG (PGCP) NP was also run through gel to detect any mRNA leaching from the NP. About 0.125 μg of EGFP-mRNA was used for all groups in this assay. (c) The mRNA-PGCP NP was characterized with NanoSIGHT to check size distribution (n=3 batches, 121.5±2.3 nm), and transmission electron microscopy (TEM) to observe morphology. A weight ratio of 1:15 for mRNA: G0-C14 was used for the NP preparation. (d) Stability of mRNA-PGCP NP in 10% serum condition at 37°C was evaluated by measuring particle size changes determined with NanoSIGHT at various time points up to 48h. Error bars represent the s.d. of triplicate samples (n=3).
mRNA NPs exhibit low cytotoxicity and potent transfection efficiency in vitro, and protect mRNA activity from RNase degradation
To evaluate in vitro cytotoxicity, cells were treated with EGFP-mRNA-PGCP NP for 16h and further incubated with fresh culture medium for 24h; nearly 80% of PC3 cells were still viable at the highest EGFP mRNA concentration of 0.5 μg/ml (Fig. 2a). AlamarBlue toxicity assay was further extended for DU145 and LNCaP cells with no notable reduction in cell viability, maintaining ~90–100% viable cells at various EGFP mRNA concentrations from 0.062 to 0.5 μg/ml in DU145 cells and 100% cell viability at all concentrations in LNCaP cells (Supplementary Fig. 1a and Supplementary Fig. 2a, respectively).
Figure 2. In vitro toxicity and transfection efficiency of mRNA NPs in PC3 cells.
Cells were treated with various mRNA concentrations (at 0.062, 0.125, 0.250, 0.500 μg/mL) of EGFP-mRNA-PGCP NPs for 16h and further incubated for 24h in standard cell culture incubation conditions. (a) AlamarBlue cytotoxicity assay; cell viability was normalized with the untreated control group. Error bars represent the s.d. (n=6). (b) Transfection efficiency (%GFP positive cells) was determined using flow cytometry (****p<0.0001). Error bars represent the s.d. (n=4) and significance was determined using One-Way ANOVA test. (c) Transfection efficiency was also analyzed with the histograms for the respective groups using Flowjo software. (d) Fluorescence microscopy images of PC3 cells transfected with naked EGFP mRNA, EGFP-mRNA-PGCP NP, and Lipofectamine 2000 (L2K)-EGFP-mRNA (magnification at 20×). (e) Mechanism of cellular uptake and endosomal escape of mRNA NPs in PC3 cells. The cells were pre-incubated for 30 min in serum-free medium containing inhibitors (Filipin, CPZ, EIPA, and Baf A1 were used as the inhibitor for caveolae-mediated endocytosis, clathrin-mediated endocytosis, macropinocytosis, and intracellular proton pump effects, respectively) or combinations of inhibitors (Filipin, CPZ, or EIPA mixed with Baf A1) prior to transfection with EGFP-mRNA PGCP NPs at mRNA concentration of 0.250 μg/ml. Transfection efficiency (%GFP positive cells) was determined using flow cytometry (***p<0.001 and ****p<0.0001). Error bars represent the s.d. (n=5) and significance was determined using One-Way ANOVA test. (f) The mechanism of cellular uptake and intracellular transport of the hybrid mRNA NPs is schematically illustrated.
We next examined transfection efficacy in vitro. The EGFP-mRNA-PGCP NP mediated highly efficient transfection of PC3 cells, showing a dose-dependent linear increase of EGFP expression correlated with increasing EGFP mRNA concentrations (from 0.062 to 0.5 μg/ml) (Fig. 2b,c). The transfection efficacy (in terms of percentile of EGFP-positive cells) of the NPs at mRNA concentrations of 0.25 and 0.5 μg/ml was demonstrably greater than that mediated by the commercial transfection agent lipofectamine 2000 (L2K) at mRNA concentration of 0.5 μg/ml. This high transfection activity was confirmed by confocal microscopy (Fig. 2d), although L2K-mRNA-transfected PC3 cells exhibited higher fluorescent intensity. We found similar highly effective transfection activity of EGFP-mRNA-PGCP NP in two other PCa cell lines (DU145 and LNCaP); >98% and 86% efficiency at the 0.5 μg/ml concentration, respectively (Supplementary Fig. 1b,c and Supplementary Fig. 2b,c). While the transfection activity of our mRNA NP was comparable to that of L2K-mRNA in DU145 cells, the NP group showed significantly greater transfection efficacy relative to L2K-mRNA in LNCaP cells, especially at EGFP mRNA concentrations of 0.25 and 0.5 μg/ml.
To investigate the ability of the NPs to protect mRNA from RNase degradation, we incubated EGFP-mRNA-PGCP NP at two ratios of mRNA to RNase weight (1:1 and 1:10) for 30 min and then evaluated transfection activity in PC3 cells. A concentration of 0.250 μg/ml EGFP mRNA was used. Naked EGFP mRNA (without or with RNase incubation) complexed with L2K was used as a control. The transfection of naked EGFP mRNA (without RNAse incubation) complexed with L2K showed ~85% efficiency, whereas transfection in the RNase-treated groups was drastically reduced to the level similar to that of the untreated control. In contrast, EGFP-mRNA-PGCP NP notably maintained the integrity and activity of the mRNA at both RNase concentrations, consistently showing ~90% transfection capacity, comparable to the transfection in the absence of RNAse (Supplementary Fig. 3a,b).
Mechanisms of cellular uptake and endosomal escape of mRNA NPs
To evaluate the cellular uptake mechanisms and intracellular transport of mRNA NPs, we studied the transfection efficiency of EGFP-mRNA-PGCP NP in PC3 cells pre-treated with different inhibitors. NP transfection was not affected by either caveolae- or clathrin-mediated inhibitors (Filipin and CPZ, respectively). In contrast, uptake was significantly decreased from ~80% (without inhibitor) to ~40% in the presence of EIPA, a macropinocytosis inhibitor. Transfection of mRNA NPs was also markedly decreased from ~80% (without inhibitor) to ~58% in the presence of the proton-pump inhibitor Bafilomycin A1 (Baf A1)46, 47. We further tested transfection activity using combinations of Filipin, CPZ, or EIPA with Baf A1. Filipin+Baf A1 and CPZ+Baf A1 exhibited low transfection efficiency similar to Baf A1 alone, whereas EIPA+Baf A1 showed a combinatorial effect, exhibiting superior inhibition of transfection activity (Fig. 2e,f). These results suggest that cellular internalization of the EGFP-mRNA-PGCP NP is partly mediated by macropinocytosis, and after entering into the cells, the NPs were able to induce a proton-sponge effect to release the cargo cytosolically. All the above inhibitors are used commonly to investigate the cellular uptake/transport pathways of NPs45–47. The results thus suggest that this combinatorial cellular uptake and endosomal escape mechanism, together with mRNA’s inherent properties, could constitute one potential advantage of this mRNA delivery system.
Functional NP delivery of PTEN mRNA to PTEN-null PCa cells in vitro
We prepared PTEN mRNA by in vitro transcription (IVT) as previously described48, 49. PTEN mRNA was modified with ARCA capping and enzymatic polyadenylation and was fully substituted with Pseudo-UTP, 5’-Methyl-CTP, followed by DNase and phosphatase treatment. Substitution with Pseudo-UTP and 5’-Methyl-CTP in replacement of regular UTP and CTP was applied for reducing mRNA immunostimulation.43, 49, 50 The PTEN mRNA was also hemagglutinin (HA)-tagged to ensure easy detection and separation from endogenous message. We first transfected PTEN mRNA into PTEN-null PCa (PC3) cells using L2K to assess the facilitation of protein expression, diminish cancer cell viability, and suppress the PI3K-AKT pathway. PC3 cells transfected with L2K-PTEN-mRNA showed markedly higher HA-PTEN expression than pHAGE-PTEN WT (MSCV-N-Flag-HA-IRES-PURO gateway destination vector with long terminal repeat [LTR]-driven expression of PTEN wildtype) by immunofluorescence staining (Supplementary Fig. 4a). Western blotting also showed that HA-PTEN expression was significantly higher than plasmid PTEN transfection. Furthermore, L2K-PTEN-mRNA treatment downregulated the PI3K-AKT pathway, showing decreased phosphorylation of 4E-BP1-Ser65, PARS40-Thr246, and Foxo3a-Ser318/321 as determined by western blotting (Supplementary Fig. 4b). Accordingly, L2K-PTEN mRNA treatment dramatically decreased cell viability as measured by CyQUANT assay (Supplementary Fig. 4c).
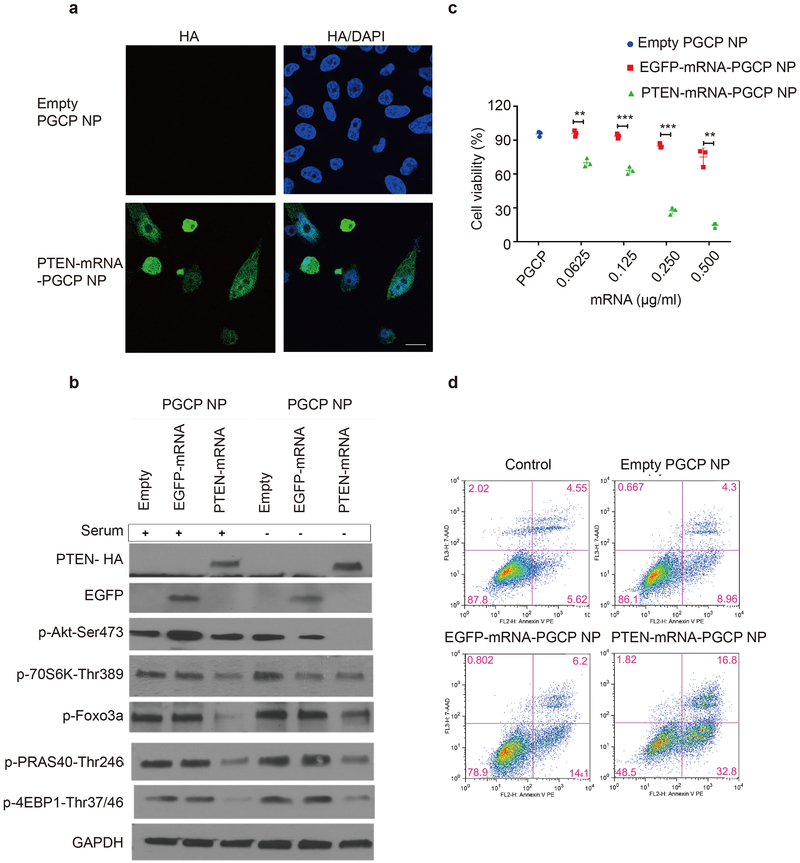
Next, we applied the NP system optimized above to determine whether our PTEN-mRNA-PGCP NP could restore the therapeutic functionality of tumour-suppressor PTEN to PCa cells. Both immunofluorescence staining (Fig. 3a, and see Supplementary Fig. 5 for bright field image) and western blot (Fig. 3b) confirmed the restoration of HA-PTEN expression transduced by PTEN-mRNA-PGCP NP treatment. It is worth noting that since PTEN-mRNA-PGCP NP treatment reduced cell viability, the cell density of this group was considerably lower than that of the group treated with control empty PGCP NP (Fig. 3a). Next, PTEN-mRNA-PGCP NP treatment significantly decreased cell viability in a dose-dependent manner compared to both empty PGCP NP and EGFP-mRNA-PGCP NP groups, as measured by MTT assay (Fig. 3c). We further found that after 48h treatment, PTEN-mRNA-PGCP NP efficiently inhibited PI3K-AKT signaling as indicated by the greater decrease in phosphorylation of Akt-Ser473, p70S6K-Thr389, 4E-BP1-Thr37/46, PARS40-Thr246, and Foxo3a-Ser318/321. PTEN-mRNA-PGCP NP treatment also further reduced basal phosphorylation of the above proteins under serum-starvation conditions (Fig. 3b). Moreover, early apoptosis was increased after treatment with PTEN-mRNA-PGCP NP, as indicated by elevated numbers of Annexin-V-positive cells via flow cytometry (Fig. 3d). A ~4-fold increase in cell death was noted in PC3 cells after PTEN-mRNA-PGCP NP treatment relative to control empty PGCP NP. We also noticed a slight increase in apoptosis in the EGFP-mRNA-PGCP NP-treated group; this was in part a possible consequence of inducing exogenous RNA into PC3 cells. However, this effect was very modest in comparison to the increase in apoptosis observed with the PTEN-mRNA-PGCP NP group. These results indicate that our hybrid NP system has the potential to effectively deliver PTEN mRNA and restore functional PTEN activity to tumour cells.
Figure 3. In vitro mechanism of PTEN-mRNA-PGCP NP treatment in PC3 cells and its therapeutic effect.
(a) Immunofluorescence staining of HA-PTEN after empty PGCP NP and PTEN-mRNA-PGCP NP treatment. (b) Western blot analysis of PI3K-AKT pathway signaling after mRNA-PGCP NP treatment in the presence (+) or absence (−) of serum. (c) The cell viability of PC3 cells after treatment with empty PGCP NP, EGFP-mRNA-PGCP NP, or PTEN-mRNA-PGCP NP measured by MTT assay (**p<0.01 and ***p<0.001). Error bars represent the s.d. of triplicate samples (n=3) and significance was determined using two-sided t-test. (d) Apoptosis was determined by flow cytometry after empty PGCP NP, EGFP-mRNA-PGCP NP, or PTEN-mRNA-PGCP NP treatment of PC3 cells.
Consistent with the above results, PTEN-mRNA-PGCP NP treatment remarkably reduced the cell viability of androgen receptor (AR)-positive PCa LNCaP cells as well as the invasive LNCaP LN3 subclone, both PTEN-deficient (Supplementary Fig. 6a). Cell apoptosis was also increased in LNCaP cells by PTEN-mRNA-PGCP NP treatment relative to empty PGCP NP or EGFP-mRNA-PGCP NP control groups (Supplementary Fig. 6b). In contrast, delivery of PTEN-mRNA with our PGCP NP into normal prostate epithelial cells (PreC) or to PTEN-competent DU145 cells did not significantly affect cell viability (Supplementary Fig. 7a), nor was there any significant change in PI3K-AKT signaling (Supplementary Fig. 7b). This is consistent with earlier results51 showing that PTEN+/− DU145 was refractory to conventional transfection with PTEN plasmid and suggests that restoration of PTEN may most effectively suppress growth and survival of tumour cells with defective PTEN expression. Similar to the results with PCa cells, PTEN-mRNA-PGCP NP treatment restored PTEN protein and growth-suppressive activity in PTEN-null MDA-MB-468 breast cancer cells, with treated cells showing reduced cell viability and PI3K-AKT signaling as well as induced apoptosis via PARP cleavage and Annexin V staining (Supplementary Fig. 8a-d). PTEN-competent MDA-MB-231 cells showed no effect on those phenotypes after PTEN-mRNA-PGCP NP treatment. Together, our results indicate the potential of PTEN-mRNA-PGCP NP to restore the tumour-suppressive activity of PTEN in PTEN-defective cells of different tumour origins.
In vivo pharmacokinetics and biodistribution of mRNA NPs
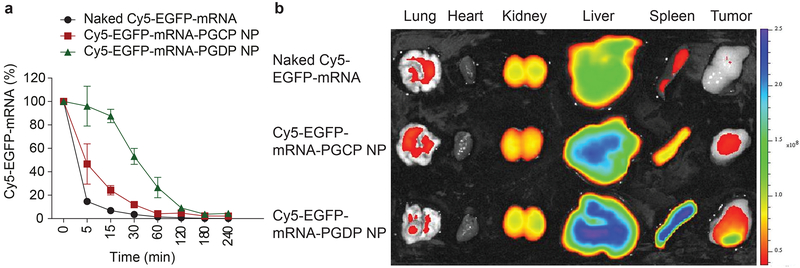
To predict the in vivo performance of our mRNA NPs for systemic delivery, we first evaluated pharmacokinetics (PK) by administering Cy5-EGFP-mRNA NPs prepared with two different lipid-PEGs (ceramide-PEG and DSPE-PEG, termed PGCP and PGDP, respectively) into healthy BALB/c mice via tail-vein intravenous (i.v.) injection and comparing the PK with that of naked Cy5-EGFP-mRNA. The EGFP-mRNA-PGDP NP possessed excellent physicochemical properties, with particle size and surface charge of 112.7±1.3 nm and 5.22±0.43 mV, respectively (Supplementary Fig. 9a) along with good stability in serum conditions and stable particle size over 48h at 37°C (Supplementary Fig. 9b). Notably, the in vitro transfection efficiency in PC3 cells was greater than 80% at an mRNA concentration of 0.5 μg/ml as measured by GFP fluorescence, which is comparable to that observed with L2K administration of GFP mRNA in vitro (Supplementary Fig. 10). The PK results showed that naked mRNA was cleared rapidly with a dramatic decrease to ~10% after 5 min. Cy5-EGFP-mRNA-PGCP NP slightly extended the circulation of Cy5-EGFP-mRNA at various time points compared to that of naked mRNA with a half-life (t½) of >5 min, whereas Cy5-EGFP-mRNA-PGDP NP had an even longer circulation profile (t½ >30 min). Moreover, ~30% of the Cy5-EGFP-mRNA PGDP NP was still circulating after 60 min, while naked mRNA and Cy5-mRNA-PGCP NP dropped to 1% and 4%, respectively. At 240 min, 5% of the Cy5-EGFP-mRNA-PGDP NP could still be detected (Fig. 4a). To evaluate biodistribution (BioD) and tumour accumulation, athymic nude mice carrying human PC3 xenograft tumour were injected with naked Cy5-EGFP-mRNA and Cy5-EGFP-mRNA NPs (both PGCP and PGDP) via tail vein. A high percentage of NPs accumulated in spleen and liver after i.v. administration. However, most importantly, Cy5-EGFP-mRNA-PGDP NP exhibited high tumour accumulation in the PC3-xenograft, whereas no or minimal signals in tumour were detected for naked Cy5-EGFP-mRNA or Cy5-EGFP-mRNA-PGCP NP (Fig. 4b). We also examined the BioD of PGCP or PGDP NP encapsulating Cy5-tagged PTEN mRNA to specifically assess the distribution of PTEN mRNA in vivo. The result explicitly reproduced similar BioD, exhibiting higher Cy5-PTEN-mRNA distribution in tumour by the PGDP NP, compared to naked Cy5-PTEN-mRNA or Cy5-PTEN-mRNA-PGCP NP (Supplementary Fig. 11a,b). This high tumour accumulation led us to advance this strategy into in vivo efficacy studies with tumour-bearing mice.
Figure 4. Effect of lipid-PEG on pharmacokinetics and biodistribution of mRNA NP.
(a) Circulation profile of naked Cy5 EGFP mRNA, and two different mRNA NP formulations with ceramide-PEG (termed as Cy5-EGFP-mRNA-PGCP NP) and DSPE-PEG (termed as Cy5-EGFP-mRNA-PGDP NP) in normal Balb/c male mice after injection (i.v., tail vein). Error bars represent the s.e.m. of triplicate samples (n=3). (b) Biodistribution of naked Cy5 EGFP mRNA, Cy5-EGFP-mRNA-PGCP NP, and Cy5-EGFP-mRNA-PGDP NP in different organs including tumours in athymic nude mice bearing PC3-xenograft tumour 24h post-injection.
In vivo therapeutic efficacy and mechanism of PTEN mRNA NP in PCa xenograft model
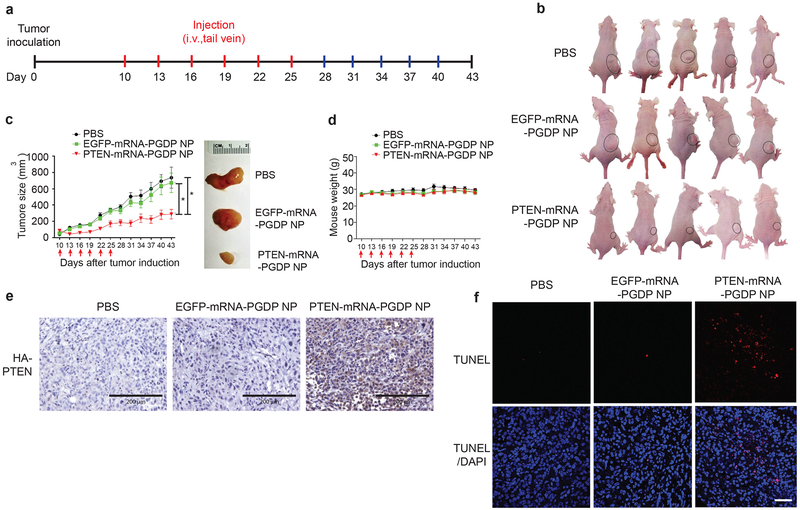
To validate the in vivo therapeutic efficacy of PTEN mRNA NP in PCa xenograft model, we systemically (i.v. via tail vein) injected PTEN-mRNA-PGDP NP every three days for six injections (Fig. 5a) in immunocompromised athymic nude mice bearing subcutaneous PC3 xenograft tumours. Tumour-bearing mice injected with PBS and EGFP-mRNA-PGDP NP were used as controls. Both PBS and EGFP-mRNA-PGDP NP groups showed rapid tumour growth, while PTEN-mRNA-PGDP NP treatment notably suppressed tumour growth compared to controls (Fig. 5b,c). The average tumour sizes rapidly increased to ~674 mm3 and ~738 mm3 for the controls, EGFP-mRNA-PGDP NP and PBS, respectively, which were significantly higher compared to ~288 mm3 for PTEN-mRNA-PGDP NP treatment at day 43 post tumour induction (Fig. 5c). Moreover, the average tumour weight for the PTEN-mRNA-PGDP NP treatment group was also significantly lower than that of control groups (Supplementary Fig. 12). No treatment group underwent significant changes in body weight, suggesting minimal toxicity (Fig. 5d). These results demonstrate the feasibility of using systemic mRNA NP delivery to reverse the effects of tumour suppressor loss in prostate tumours in vivo.
Figure 5. In vivo therapeutic validation of PTEN restoration using PTEN-mRNA NP in PCa xenograft model.
(a) Scheme of tumour inoculation and systemic injection (i.v., tail-vein) of PBS, EGFP-mRNA-PGDP NP, or PTEN-mRNA-PGDP NP in PC3 tumour-bearing male athymic nude mice. Mice were injected at day 10 post tumour inoculation. Injections were performed every three days for 6 times. (b) Whole-body images of mice bearing PC3-xenograft tumours treated with PBS, EGFP-mRNA-PGDP NP, and PTEN-mRNA-PGDP NP (at day 35). (c) Tumour growth to show in vivo therapeutic efficacy of PTEN-mRNA-PGDP NP (n=8) as compared to PBS (n=7) and EGFP-mRNA-PGDP NP (n=9) (*p<0.05 vs. PBS or EGFP-mRNA-PGDP NP). Error bars represent the s.e.m of the mean and significance was determined using One-Way ANOVA test. The arrows indicate i.v., tail vein injections. Tumour size measurement began on day 10 and continued every three days until day 43. The representative excised tumour images are also shown on the right. (d) The average body weight of the PC3 tumour-bearing xenograft mice over the course of therapy (mean±SEM). The effect of PTEN-mRNA-PGDP NP treatment in PC3 tumour-bearing xenograft mice was evaluated by (e) Immunohistochemistry staining of HA-PTEN (blue: nucleus; brown: HA-PTEN; bar indicates 200 μm) and (f) TUNEL assay on fixed tumour tissue after PBS, EGFP-mRNA-PGDP NP, or PTEN-mRNA-PGDP NP treatment (scale bar: 200 μm) (blue: nucleus; red: apoptotic cells and purple: merge of blue and red).
To further understand the mechanisms underlying the therapeutic activity of PTEN-mRNA-PGDP NP, we assessed HA-PTEN expression in tumour sections obtained on the third day after the last injection by immunohistochemistry analysis using HA antibody. PTEN-mRNA-PGDP NP treatment resulted in HA-PTEN protein expression in tumour, whereas PBS and EGFP-mRNA-PGDP NP controls did not show any background HA-PTEN expression (Fig. 5e). Next, TUNEL assay in tumour sections revealed that PTEN-mRNA-PGDP NP increased tumour cell apoptosis significantly more than either EGFP-mRNA-PGDP NP or PBS (Fig. 5f). These results suggest that the effective systemic restoration and efficient expression of PTEN in tumours mediated by NP delivery of PTEN mRNA leads to enhanced tumour-cell apoptosis and decreased tumour cell survival. Consequently, this approach may represent a viable treatment strategy for restoring tumour-growth suppression of PCa in vivo.
In vivo therapeutic efficacy of PTEN mRNA NPs in advanced PCa models
To validate the in vivo therapeutic efficacy of PTEN mRNA NP in advanced PCa models, we first established a disseminated PC3 metastatic model by injecting luciferase-tagged PC3 (PC3-luc) cells into the tail vein of immunocompromised, male athymic nude mice. Tumour metastases were detected in the lung and other organs of the mice, by bioluminescence imaging (Bruker Xtreme). Four weeks post tumour challenge, we systemically (i.v. via tail vein) injected PTEN or EGFP mRNA-loaded PGDP NPs every three days for five doses and compared to PBS. Based on the fold change of bioluminescence of PC3-luc cells in mice, it was found that the PTEN-mRNA-PGDP NP significantly prevented the progression of metastatic cancer when compared to PBS and EGFP-mRNA-PGDP NP treatment groups (Fig. 6a). Quantitative analysis demonstrated a significant difference of the fold change in average radiance at the experimental endpoint (day 15) in the PTEN-mRNA-PGDP NP vs. the PBS cohort (*p=0.0289), as well as the PTEN-mRNA-PGDP NP vs. the EGFP-mRNA-PGDP NP cohort (*p=0.0469) (Fig. 6b, 6c, Supplementary Fig. 13a). These results demonstrate the efficacy of the systemic delivery of PTEN mRNA-loaded NPs in a disseminated metastatic PCa model.
Figure 6. In vivo therapeutic validation of PTEN restoration using PTEN-mRNA NPs in disseminated metastatic PCa model.
(a) Scheme of i.v. tumour inoculation and systemic injection (i.v., tail-vein) of PBS, EGFP-mRNA-PGDP NP, or PTEN-mRNA-PGDP NP (n=8 mice per cohort) in disseminated PC3-luc metastatic male athymic nude mice. (b) Bioluminescent imaging for disseminated PC3-luc metastatic tumours at different time points post treatment. White color indicates signal >75×104 photons/s/cm2/sr. (c) Average radiance of tumour burden as normalized per mouse signal at treatment initiation (day 28) determined by bioluminescent imaging; inset: normalized average radiance per mouse at experimental endpoint (day 43) for each treatment group. Error bars represent the s.e.m. (n=8) and significance was determined using Mann-Whitney test. Each red arrow indicates treatment administration.
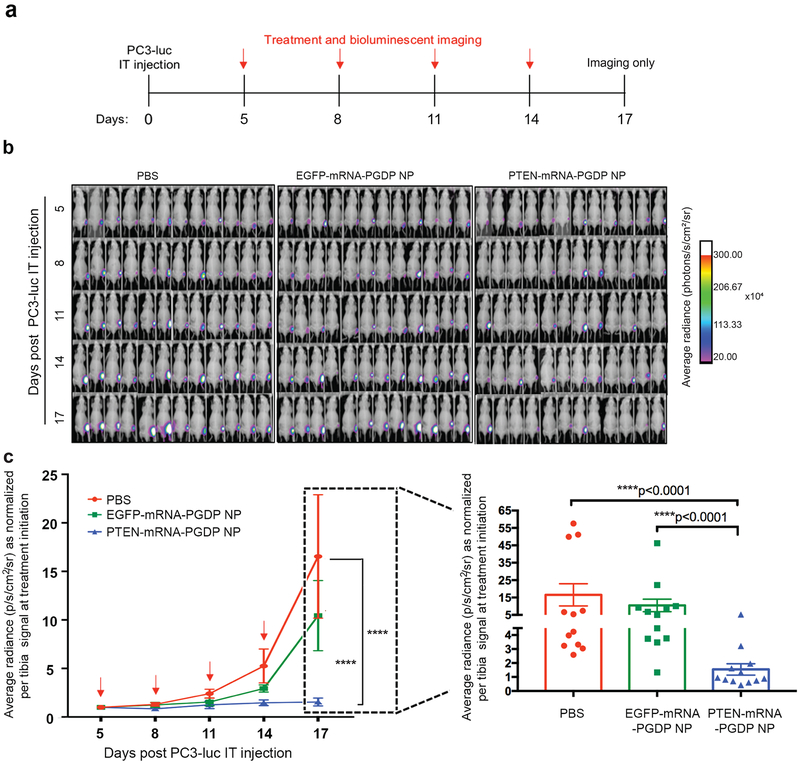
We also tested the mRNA NPs in a PCa bone model given that bone is the most common site of PCa metastasis52. We performed the orthotopic, intratibial (IT) injections with the PC3-luc cells in immunocompromised, male athymic nude mice, and systemically injected PTEN-mRNA-PGDP NPs five days post tumour inoculation and every three days for four injections in total (Fig. 7a). Both PBS and EGFP-mRNA-PGDP NP control groups demonstrated rapid tumour growth (Fig. 7b, c), and mice receiving PTEN-mRNA-PGDP NP as compared to PBS or EGFP-mRNA-PGDP NP treatment showed a significant decrease in fold change in average radiance at day 17 post tumour inoculation, which was the experimental endpoint (****p<0.0001 for both comparisons) (Fig. 7c, Supplementary Fig. 13b). For the IT orthotopic model experimental endpoint (day 17), cohorts which received PTEN-mRNA-PGDP NP treatment experienced a 90.7% reduction compared to cohorts which received PBS and a 85.23% reduction compared to cohorts which received EGFP-mRNA-PGDP NP (****p<0.0001 for both comparisons). These findings indicate an ability of PTEN-mRNA-PGDP NPs to reduce tibia tumour burden (Fig. 7c, Supplementary Fig. 13b). These results are further indicative of the ability of PTEN mRNA NP delivery to decrease tumour outgrowth in an orthotopic site of PCa metastasis. Notably, no treatment group showed any significant changes of body weight in both the disseminated metastatic and IT orthotopic PCa models, suggesting safety of this therapeutic system (Supplementary Fig. 14). Additionally, quantitative real-time PCR analysis of the bone marrow lysates and IHC analysis of the IT tumour sections conclusively show the expression of HA-PTEN in the bones, but no obvious expression was observed in tumours treated with either PBS or EGFP-mRNA-PGDP NP (Supplementary Fig. 15).
Figure 7. In vivo therapeutic validation of PTEN restoration using PTEN-mRNA NPs in IT orthotopic PCa model.
(a) Scheme of IT tumour inoculation and systemic injection (i.v., tail-vein) of PBS, EGFP-mRNA-PGDP NP, or PTEN-mRNA-PGDP NP (n=12 mice or 12 tibiae per cohort) in IT PC3-luc-bearing male athymic nude mice. (b) Bioluminescent imaging of total radiance of PC3-luc at time points post IT injections and treatments. White color indicates signal >300×104 photons/s/cm2/sr. (c) Average radiance of tumour burden as normalized per mouse signal at treatment initiation (day 5) determined by bioluminescent imaging; inset: normalized average radiance per mouse at experimental endpoint (day 17) for each treatment group. Error bars represent the s.e.m. (n=12) and significance was determined using Mann-Whitney test. Each red arrow indicates treatment administration.
In vivo safety profile of PTEN mRNA NP
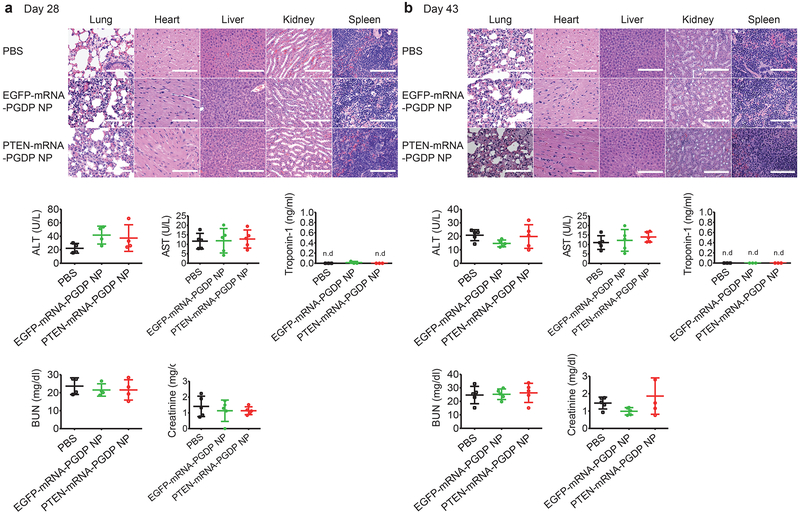
To evaluate the in vivo side effects of mRNA NPs, various organs and blood serum were harvested three days after the last injection (day 28) and at the end point (day 43) of the PCa xenograft experiment (Fig. 8a,b). Organs were sectioned and H&E stained. We found no histological differences in the tissues from lung, heart, liver, spleen, or kidney between PBS and NP treatment groups, suggesting no notable toxicity. For hematological analysis (days 28 and 43), we checked parameters including aspartate aminotransferase (AST) & alanine aminotransferase (ALT) to assess liver function, creatinine and blood urea nitrogen (BUN) to evaluate kidney activity, and troponin-1 to assess cardiac function using appropriate assay kits. We found no obvious changes in these parameters in serum from mice after treatment with the mRNA NPs, further indicating negligible side effects.
Figure 8. In vivo toxicity studies by histopathological and hematological analysis after treatment of mRNA NPs vs. PBS.
For histopathological assay, H&E staining of tissue sections of major organs was analyzed three days after the last injection of PBS, EGFP-mRNA-PGDP NP, and PTEN-mRNA-PGDP NP at day 28 (a), and at the endpoint of day 43 (b) post tumour inoculation as shown in the scheme of Figure 6a. Scale bars: 100 μm for lung, heart and spleen; 200 μm for kidney and liver. For hematological assay, the levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), blood urea nitrogen (BUN), creatinine, and troponin-1 in serum were measured at day 28 (a) and at day 43 (b) post tumour inoculation. Error bars represent the s.d. (n=5), and n.d.: not detectable).
We also investigated whether mRNA NPs mediated any in vivo immune response in immunocompetent mice to exclude the possibility that the anti-tumour efficacy of PTEN mRNA NP might be caused by an immunostimulatory effect. There was a similarly modest increase in levels of proinflammatory cytokine TNF-α for both the empty PGDP NP and the PTEN-mRNA-PGDP NP at 6h post-injection, suggesting that the cytokine response may be attributable to the NP itself rather than the effect of encapsulated PTEN-mRNA. PBS and naked PTEN mRNA stimulated minimal TNF-α response as expected (Supplementary Fig. 16). TNF-α levels for both empty PGDP NP and PTEN-mRNA-PGDP NP returned to baseline PBS levels 24h post-injection, suggesting that immune stimulation (e.g., TNF-α) by the NP itself was transient and that the PTEN mRNA had no lasting adverse immune/inflammatory activity. In sum, our results demonstrate effective reversal of the PTEN-null phenotype in PCa and in breast cancer cells in vitro and in vivo after systemic delivery of PTEN-mRNA-PGDP NP. The inhibitory effect is dependent on the presence of PTEN mRNA delivered by the NP and is not mediated by non-specific host responses to either the NP itself or to the introduction of other mRNA species.
DISCUSSION
Loss and/or mutation of tumour-suppressor genes is a dominant force in tumour development and clinical resistance to a variety of therapies53. Reversal of the phenotype induced by loss of tumour suppressors has long proven an elusive goal. Two major strategies have been employed for suppressor restoration: restoring a functional copy of a given tumour-suppressor gene via transfection; and the use of small-molecule agents to reactivate tumour-suppressor function via conformational change in the mutated molecule54. The latter approach has met with little success and is destined to be ineffective when the suppressor gene has been deleted. The major limitations of restoring suppressor gene in tumours include inefficient delivery to tumour cells, poor transfection efficacy, insufficient expression, and possible insertional mutagenesis. Consequently, restoring tumour-suppressor activity in cancer cells is highly challenging and requires the design of a functionally improved tumour suppressor with unique therapeutic modality that can withstand the rigors of systemic delivery, especially in the metastatic setting where the tumour burden is widely distributed. The modified PTEN mRNA NP technology reported herein is an early example of such an approach. To our knowledge, tumour inhibition following direct systemic restoration of a tumour-suppressor gene using an mRNA delivery strategy in vivo has not been shown before. Use of an mRNA delivery system allows rapid gene expression with controlled and predictable expression kinetics, higher transfection efficiency, and (most importantly) can eliminate genomic complexation and mutagenesis due to the inherent cytoplasmic and diminished nuclear expression of the desired protein 29. In addition, we report a delivery system using a new-generation polymer-lipid hybrid NP system that provides effective i.v. delivery of the suppressor mRNA to tumour xenografts. The end result is restoration of PTEN function as illustrated by inhibited both primary and advanced tumour growth, increased apoptosis, and blockade of the PI3K-AKT signaling pathway. Because PTEN loss is frequent in late-stage PCa, we suggest this approach may have feasibility in this patient population.
In previous reports, the most widely investigated non-viral gene delivery carriers such as polyethylenimine (PEI), DOTAP/cholesterol liposome, and the DOTAP/DOPE system49, 55–57 provided only suboptimal and variable mRNA transfection efficacy (40–80%) in cancer cells and fibroblasts (e.g., HeLa and NIH 3T3 cells). In addition, in PCa cells (e.g., PC3) it was reported that mRNA transfection efficacy of the PEI/mRNA complex was only 30%, although a high in vitro mRNA transfection activity in PC3 cells was found using a histidine-rich reducible polycation system58. Our polymer-lipid hybrid mRNA NP prepared using a robust self-assembly strategy provided 86–98.2% mRNA transfection capacity with minimal toxicity in various PCa cell lines (e.g., LNCaP, PC3, and DU145), a new standard for in vitro delivery of mRNA to tumours. Potential reasons for this effective mRNA delivery to tumours may be the relatively small size of the NPs (~120 nm), as well as their very high (95–100%) mRNA encapsulation efficiency, compact shape, and good stability. Whereas smaller particle size could be achieved with small oligonucleotides such as siRNA, it is more difficult with larger payloads such as mRNA. It is worth noting that such small NPs may more efficiently permeate the leaky tumour microvasculature and achieve greater tumour accumulation and deeper tissue penetration compared to larger NPs59–62.
For systemic in vivo application, PEGylation of NPs is a well-documented strategy to prevent rapid elimination from the circulation, since it reduces the interaction between NPs and serum proteins following recognition by mononuclear phagocytic system-mediated clearance mechanisms63, 64. Simultaneously, PEGylation could also hinder NP interactions with the target cell membrane, which may decrease tumour cell-mediated uptake. Therefore, proper dissociation of lipid-PEG molecules is necessary for optimal systemic circulation and extravasation of our mRNA NPs at the tumour site as well as effective uptake by tumour cells. We have demonstrated that the dissociation kinetics depend on the structure of the lipid-PEG molecules in the hybrid NPs and may be controlled by the length and/or saturation of lipophilic tails45. In that context, DSPE-PEG exhibited relatively slow de-PEGylation profile compared to ceramide-PEG, as we described earlier from our measurements of lipid-PEG’s dissociation kinetics from NPs in the presence of serum albumin, which is the most abundant plasma protein and binds with diacyl lipids65. Moreover, we observed a quicker release of ceramide-PEG than DSPE-PEG from NPs and also found that the surface charge of the NPs changed over time after lipid-PEG dissociation: the charge of ceramide-PEG NPs rapidly increased from 2.2 to 31.4 mV in 3h, but slowly for DSPE-PEG NPs from −4.0 to 11.9 mV over 24h, although both NPs were initially near neutral. This slow de-PEGylation profile conferred better PK, tumour biodistribution, and therapeutic efficacy for DSPE-PEG NPs45. Thus our hybrid mRNA NP coated with DSPE-PEG showed higher stability, longer circulation, and increased tumour accumulation compared to ceramide-PEG NP, indicating more efficient systemic restoration of mRNA to tumours and implications beyond any particular tumour suppressor and cancer type.
PTEN loss has been recognized for two decades as being involved in PCa progression66–68; surprisingly, there has been no progress on direct restoration of PTEN function in PTEN-null PCa cells, presumably due to the inefficient delivery and insufficient expression of PTEN at the tumour site. Recently, a secreted form of PTEN called PTEN-long (PTEN-L) that can penetrate cells has been discovered and shown to rescue PTEN function in PTEN-null U87-MG glioblastoma cells and MDA-MB-468 breast cancer cells in vitro and in vivo69. Our approach is complementary to these findings and is perhaps more generalizable to a wide variety of tumour-suppressive molecules. Recent protein structure studies have reported that PTEN and PTEN-L have different properties relating to conformational change, membrane binding, and substrate specificity70. Future studies will be necessary to explore the relative efficacy of these two approaches. It is further likely that delivery of mRNA NPs will achieve greater tumour-specific distribution than free protein. It was also recently reported that PTEN loss promotes resistance to clinical therapy as well as T-cell-mediated immunotherapy71, 72, suggesting that restoring functional PTEN via our PTEN mRNA NP technology could also be useful in immunotherapeutic applications and in rescuing chemosensitivity in resistant tumours. Therefore, delivery of mRNA NPs also holds the potential to potentiate other therapeutic approaches29. Our findings provide significant support for the proof-of-concept that the systemic restoration of PTEN rescues PTEN function in PTEN-null PCa and effectively suppresses tumour progression with negligible side effects.
In summary, restoration of PTEN represents a new approach to PI3K-AKT pathway inhibition, with the potential to significantly improve the treatment of cancer patients with loss of PTEN function. Moreover, considering the strong potential of mRNA therapy and the lack of systemic studies of in vivo mRNA transfection of tumours, this study sheds light on the useful application of NP-mediated mRNA delivery for validating tumour suppressors (e.g., PTEN and p53) as therapeutic targets in the treatment of cancer.
METHODS
Materials.
Ester-terminated poly(D,L-lactide-co-glycolide) (PLGA, viscosity 0.26–0.54 dL/g) was purchased from Durect Corporation. Cationic ethylenediamine core-poly(amidoamine) (PAMAM) generation 0 dendrimer (G0), bafilomycin A1 (Baf A1) were purchased from Sigma-Aldrich. Filipin III, chlorpromazine (CPZ) and 5-(N-ethyl-N-isopropyl)-amiloride (EIPA) were purchased from Cayman Chemicals (Ann Arbor, MI, USA). DSPE-PEG (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy{polyethylene glycol}]) with PEG molecular weight (MW) 5000, and ceramide-PEG (N-palmitoyl-sphingosine-1-(succinyl{methoxy[polyethylene glycol]}) with PEG MW of 2000 were obtained from Avanti Polar Lipids. Lipofectamine 2000 (L2K) was purchased from Invitrogen. EGFP mRNA (EGFP mRNA; modified with 5-methylcytidine and pseudouridine) and Cyanine 5 fluorescent dye-labeled EGFP mRNA (Cy5 EGFP mRNA; modified with 5-methylcytidine and pseudouridine modification) were purchased from TriLink Biotechnologies. Sequence-verified human PTEN wildtype, G129E, and G129R open reading frame were cloned into pENTR223 followed by recombination into the Gateway destination vector pHAGE (MSCV-N-Flag-HA-IRES-PURO, long terminal repeat [LTR]-driven expression) using λ recombinase. pLenti CMV Puro LUC (W168) was a gift from Eric Campeau (Addgene plasmid #17477)73. ViraPower Lentiviral packaging mix was purchased from Thermo Fisher Scientific. D-luciferin-K+ salt bioluminescent substrate (#122799) was obtained from PerkinElmer. Primary antibodies used in this work included the following: anti-PTEN (138G6), anti-p-Akt-ser473 (#9271), anti-p-70S6K-Thr389 (108D2), anti-p-FOXO3a-Ser318/321 (#9465), anti-p-PARS40-Thr246 (D4D2), p-4E-BP1-Thr37/46 (236B4), p-4E-BP1-Ser65 (#9451), anti-HA antibody (C29F4) and anti-PARP (#9542) antibodies (rabbit, Cell Signaling); anti-HA antibody (3F10) (rat, Roche); anti-HA-HRP conjugated antibody (A00169) (goat, GenScript); Anti-GFP antibody (A-6455) (rabbit, Life Technologies).
Preparation of modified PTEN mRNA.
Vector carrying open-reading frame (ORF) of PTEN was a gift from William Sellers74 (pSG5L HA PTEN wt; Addgene#10750). The vector was linearized by ApaI/EcoRI digestion and purified. HA-PTEN ORF under the regulation of T7 promoter was then amplified by PCR reaction. The amplicons were further purified and used as templates for in vitro transcription (IVT). The modified PTEN-mRNA was synthesized as described previously48, 49. In brief, IVT was conducted using MEGAscript T7 kit (Ambion) with 1–2 μg template and 7.5 mM ATP, 1.5 mM GTP, 7.5 mM 5-methyl-CTP, 7.5 mM pseudo-UTP (TriLink Biotechnologies), and 6 mM 3′−0-Me-m7G(5′)ppp(5′)G (anti-reverse cap analog, ARCA) (TriLink Biotechnologies). Reactions were incubated at 37°C for 4 hours, followed by Turbo DNase treatment for 15 min. 3´ poly(A)-tails were further added to IVT RNA products using a poly(A) tailing kit (Ambion). mRNA was purified by using the MEGAclear kit (Ambion), then treated with Antarctic Phosphatase (New England Biolab) at 37°C for 30 min, and further purified. Large-scale PTEN mRNA was custom-prepared by TriLink Biotechnologies as above (ARCA capped and enzymatically polyadenylated; fully substituted with Pseudo-U and 5’-Methyl-C; DNase and phosphatase treatment; Silica membrane purification) using 100–150 μg template containing T7 promoter and HA-PTEN ORF.
Synthesis of cationic lipid compound (G0-C14).
The cationic lipid-like compound (G0-C14) was synthesized from ethylenediamine core-poly(amidoamine) (PAMAM) generation 0 dendrimer (G0) using a ring-opening reaction by reacting with 1,2 epoxytetradecane according to previously described procedure45, 75. Briefly, 1,2 epoxytetradecane was mixed with PAMAM dendrimers G0 at a molar ratio of 7:1, where substoichiometric amounts of 1,2 epoxytetradecane were added to increase the proportion of products with one less tail than the total possible for a given amine monomer. The reaction was carried out for 2 days under vigorous stirring, and the crude mixture was separated on silica with gradient elution from CH2Cl2 to 75:22:3 CH2Cl2/MeOH/NH4OH using chromatography.
mRNA complexation ability of G0-C14 and its stability in organic solvent.
To assess the mRNA complexation ability of G0-C14 and its stability in organic solvent (DMF), naked EGFP-mRNA or EGFP-mRNA complexed with G0-C14 (in varying weight ratios from 1 to 20) were incubated with or without DMF for 30 min. For mRNA samples in DMF, electrophoresis was run without extracting mRNA from DMF into aqueous solution. The volumes of samples were then adjusted with loading dye (Invitrogen) and run into an E-Gel® 2% agarose (Invitrogen) gel for 30 min at 50 V. The ambion® millennium™ markers-formamide (Thermo Fisher Scientific) was used as a ladder. Finally, the gel was imaged under UV and the bands were analyzed.
Preparation of mRNA NP.
We employed a robust, self-assembly method to prepare mRNA-encapsulated polymer-lipid hybrid NPs as we previously described45, but with significant modification and optimization in ratios of reagents used in NP formulation. In brief, PLGA and G0-C14 were dissolved separately in dimethylformamide (DMF) at concentrations of 5 mg/ml and 2.5 mg/ml, respectively. Then PLGA (250μg in 50 μl) and G0-C14 (250μg in 100 μl) were mixed at a weight ratio of 1:1 in a small glass vial. mRNA (16 μg at 1 mg/ml concentration) in aqueous solution was mixed into the PLGA/G0-C14 organic solution (weight ratio of mRNA:PLGA:G0-C14 was 1:15:15) to form cationic lipid/mRNA nanocomplexes. This solution was then quickly nanoprecipitated into 10 ml of lipid-PEG (e.g., ceramide-PEG or DSPE-PEG) aqueous solution (0.1mg/ml concentration in DNase/RNase-free Hypure water) for ~20 seconds. The weight ratio of lipid-PEG to PLGA was 4:1. Upon nanoprecipitation, NPs formed instantly and were kept for 30 min at 600 rpm stirring at room temperature to stabilize. The NPs were then washed three times with ice-cold Hypure water using Amicon tubes (MWCO 100 kDa; Millipore) to remove organic solvent and free compounds and finally concentrated into 1 ml PBS solution. The NPs were used fresh or kept at −80°C to use later for various in vitro and in vivo studies. The mRNA NPs were run through gel electrophoresis as described above to check for any unencapsulated mRNA leaching. The NPs prepared with ceramide-PEG and DSPE-PEG were termed PGCP and PGDP NPs, respectively.
Physicochemical characterization and stability of mRNA NPs in serum condition.
mRNA NPs were characterized by assessing their size, surface charge, and morphology. Sizes were measured by NanoSIGHT (Malvern, NS300) at 20°C and analyzed using Nanoparticle Tracking Analysis (NTA), which utilizes the properties of both light scattering and Brownian motion to determine the particle size distribution of samples in liquid suspension. The surface charge of the NPs was determined by dynamic light scattering (DLS) with 15-mW laser and an incident beam of 676 nm (Brookhaven Instrument Corporation). A transmission electron microscope (TEM) was used to assess the NPs’ morphology and shape. For TEM, NPs were stained with 1% uranyl acetate and imaged using a Tecnai G2 Spirit BioTWIN microscope (FEI Company) at 80 kV. To check the in vitro stability of polymer-lipid hybrid mRNA NPs in serum as a means to mimic in vivo conditions, mRNA NPs were incubated in 10% serum containing PBS solution at 37°C in triplicate for various time periods (0, 2, 4, 8, 12, 24, and 48 h) with 100 rpm shaking. At each time point, an aliquot of NPs solution was taken for particle size measurement using NanoSIGHT and analyzed as described above to evaluate any change in size distribution at various time intervals. EGFP mRNA NPs were used in this study.
Cell culture.
Human PCa cell lines (PC3, DU145, LNCaP and its invasive subclone LNCaP LN3) and prostate epithelial cells (PreC) along with two breast cancer cell lines (MDA-MB-468 and MDA-MB-231) were used in various in vitro studies. All cells were purchased from American Type Culture Collection (ATCC). Cells were maintained in F-12K (ATCC), Eagle’s Minimum Essential Medium (EMEM; ATCC), Roswell Park Memorial Institute (RPMI) 1640 (ATCC), or Leibovitz’s L-15 (ATCC) cell-culture medium, according to the culture method for each cell type per the instructions from ATCC, supplemented with high-glucose, 10% fetal bovine serum (FBS; Gibco®) and 1% penicillin/streptomycin antibiotic (Thermo-Fisher Scientific). Cell culture and all biological experiments were performed at 37°C in 5% CO2 conditions in a cell-culture incubator. All cells were authenticated (using the “DDC Biomedical” or “Genetica DNA Laboratories” cell line authentication test) and checked for mycoplasma contamination before in vitro cell experiments and in vivo xenograft tumour model preparation.
Generation of luciferase-tagged PC3 cells.
The lentiviral vector pLenti CMV Puro LUC encoding the firefly Luciferase was transfected with Virapower Lentiviral packing mix to 293T cells using L2K. After 48 h, lentiviral supernatant was collected and added into 20–40% confluent PC3 cells. Polybrene (8 μg/ml) was added during the transduction. Two days after transduction, PC3 cells were selected by puromycin at 2 μg/ml concentration. Luciferase expression was analyzed by immunofluorescence staining and western blot. PC3-luc cells were maintained in media containing 1 μg/ml puromycin.
In vitro cytotoxicity and transfection activity of mRNA NP.
Cells were seeded at a density of 3~5×104 cells per well on 24-well plate and allowed to attach and grow until ~80% confluence. Cells were transfected with mRNA NPs at various mRNA concentrations (0.062, 0.125, 0.250, and 0.500 μg/ml) for 16h followed by washing with fresh complete medium and further incubated for 24h to check cytotoxicity as well as transfection efficiency. L2K was used as a standard transfection reagent (according to manufacturer’s protocol) to form L2K-mRNA complexes for comparison with the mRNA NPs. Cytotoxicity was measured by AlamarBlue® assay according to the manufacturer’s protocol using a microplate reader (TECAN, Infinite M200 Pro). AlamarBlue is a non-toxic assay that allowed us to continuously check real-time cell proliferation. For transection efficiency measurement, cells were harvested with 25% EDTA trypsin and washed two times and resuspended in PBS followed by measuring GFP expression using flow cytometry. The percentages of GFP-positive cells were calculated, and histograms were prepared using Flowjo software.
RNase Protection Assay.
To test whether the NPs protected the mRNA from RNase, naked EGFP mRNA and EGFP-mRNA PGCP NPs were incubated in RNase at two mRNA-to-RNase weight ratios (1:1 and 1:10) for 30 min at 37°C, shaking at 100 rpm. A concentration of 0.250 μg/ml EGFP mRNA was used in this study. After incubation, RNase was separated from the EGFP-mRNA PGCP NPs and naked mRNA by washing with water via centrifugation in 100 kDa Amicon filter tubes at 1300 rcf for 10 min. The post RNase-treated EGFP-mRNA PGCP NPs were then diluted in media, while the naked mRNA was complexed with L2K, and the PC3 cells were then transfected as described above and incubated for 16h. The medium was replaced and incubated for an additional 24h. The naked EGFP mRNA and EGFP-mRNA PGCP NPs without RNase treatment were used as negative controls. Cells were then harvested to measure EGFP expression by flow cytometry and analyzed using Flowjo as described above.
Mechanism of cellular uptake and endosomal escape of mRNA NPs.
To determine the mRNA NPs’ uptake and intracellular transport mechanism, 24-well plates were used to seed PC3 cells at an initial density of 5х104 cells/ml in 1 ml of growth medium and incubated for 24h at 37°C in 5% CO2 to allow the cells to attach. The cells were then pre-incubated for 30 min in serum-free medium containing inhibitors (Filipin at 1 μg/ml, CPZ at 10 μg/ml, EIPA at 10 μg/ml, and Baf A1 at 200 nM was used to block caveolae-mediated endocytosis, clathrin-mediated endocytosis, macropinocytosis, and intracellular proton pump inhibitor effects, respectively, alone and in combination). The cells were then transfected with EGFP-mRNA PGCP NPs at mRNA concentration of 0.250 μg/ml. After 16h, the old medium was replaced with fresh complete medium and incubated for an additional 24h. The cells were then harvested to check EGFP expression by flow cytometry and analyzed by Flowjo as described above.
Cell growth inhibition assay with PTEN mRNA NP.
Cell growth inhibition was determined by CyQUANT assay in 96-well plates. First, 3~5×103 cells per well per 100 μl were seeded in 96-well plates. The next day, cells were treated with mRNA NP or the L2K-mRNA mixture. After 72h incubation under standard cell-culture conditions, the culture medium was removed and plates were kept at −80°C for >24h. Cells were counted using the CyQUANT kit (Life Technologies) per the manufacturer’s instructions. Fluorescence measurements were made using a microplate reader with excitation at 485 nm and emission detection at 530 nm. Cell-growth inhibition was also determined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) bromide assay. Briefly, 3~5×103 cells were plated in a 96-well plate and treated with NPs the next day. After 16h, NPs were removed and fresh medium was added. After 72h of incubation, 10% culture volume of MTT (Sigma Chemicals, St. Louis, MO; 5 mg/ml) was added to each well. After incubation for an additional 4h, 200 μl of isopropanol-HCl solution was added to each well to dissolve the cell pellets. Absorbance was determined using a 96-well SpectraMax plate reader (Molecular Devices, Sunnyvale, CA) at 560 nm and 650 nm (background).
Apoptosis assay in vitro.
Cells were seeded in 6-well plates until ~80% confluence and then treated with PTEN mRNA NPs. The next day, the NPs were removed (16h post treatment) and kept in culture for another 24h. The supernatant and the cell monolayer were collected, washed with PBS, and processed for detection of apoptotic cells using the Annexin V-PE/7AAD apoptosis detection kit (BD Biosciences) according to the manufacturer’s instructions.
Western blot assay.
Protein extracts were prepared using NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 0.5% NP-40 substitute, 150 mM NaCl, and 12.5 mM NaF) supplemented with Complete Mini EDTA-free protease inhibitor tablets (Roche). Equal amounts of protein, as determined with a bicinchoninic acid (BCA) protein assay kit (Pierce/Thermo Scientific) according to the manufacturer’s instructions, were separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were blocked with 5% non-fat dry milk in TBST (50 mM Tris-HCl at pH 7.4 and 150 mM NaCl, and 0.1% Tween 20) and then incubated with appropriate primary antibodies. Signals were detected with horseradish peroxidase-conjugated secondary antibodies and an enhanced chemiluminescence (ECL) detection system (Amersham/GE Healthcare). When indicated, membranes were subsequently stripped for reprobing.
Immunofluorescent staining and microscopy.
For immunofluorescent staining, cells were plated onto coverslips in 6-well plates and grown overnight to 60–70% confluence. Cells were washed with ice-cold PBS and fixed with 4% paraformaldehyde (PFA, Electron Microscopy Sciences) in PBS for 15 min at room temperature (RT). Cells were then permeabilized by incubation in 0.2% Triton X-100-PBS for 8 min followed by blocking with PBS blocking buffer containing 2% normal goat serum, 2% BSA, and 0.2% gelatin for 1h at RT. Then the samples were incubated in primary antibody (1:200 anti-HA rat antibody) for 1h at RT, washed with PBS, and incubated in goat anti-rat-Alexa Fluor 488 (Molecular Probes) at 1:500 dilution in blocking buffer for 30 min at RT. Finally, stained cells were washed with PBS, counterstained with 500 nM DAPI, and mounted on slides with Prolong Gold antifade mounting medium (Life Technologies).
Animals.
Six-week-old BALB/c male normal mice were used for pharmacokinetics (PK) and immune response studies. To evaluate the biodistribution (BioD) of mRNA NP in various organs including tumours and test the therapeutic efficacy of PTEN mRNA NP to suppress tumour growth, male athymic nude mice (6 weeks old) were obtained from Charles River Laboratories. All animal studies were performed under strict regulations and pathogen-free conditions in the animal facility of Brigham and Women’s Hospital and in accordance with National Institutes of Health animal care guidelines. The animals had free access to sterile food pellets and water and were kept in the laboratory animal facility with temperature and relative humidity maintained at 23±2°C and 50±20%, respectively, under a 12-h light/dark cycle. Mice were kept for at least one week to acclimatize them to the food and environment of the animal facility. The animal protocol was approved by the Institutional Animal Care and Use Committees at Harvard Medical School.
Pharmacokinetic (PK) study.
For in vivo PK study, healthy BALB/c male mice (6 weeks) were divided into three groups (n=3 per group) and intravenously administered (i) naked Cy5 EGFP mRNA, (ii) Cy5-EGFP-mRNA-PGCP NP or (iii) Cy5-EGFP-mRNA-PGDP NP through the tail vein at a mRNA dose of 700 μg per kg of animal weight. At various predetermined time intervals (0, 5, 15, 30, 60, 120, 180, and 240 min), retro-orbital vein blood was withdrawn using a heparin-coated capillary tube, and the wound was gently pressed for a few seconds to stop the bleeding. Fluorescence intensity of Cy5 was measured at emission and excitation wavelengths of 640 and 670 nm, respectively, using a microplate reader. PK was calculated by calculating the percentage of Cy5 EGFP mRNA in blood at various time periods, normalized with the initial (0 min) time point.
PC3-xenograft tumour model preparation.
To prepare the PC3-xenograft tumour mice model, about 4×106 cells in 100 μL of culture medium mixed with 100 μL of matrigel (BD Biosciences) were implanted subcutaneously on the right flank of 6-week-old male athymic nude mice. Mice were monitored for tumour growth every other day according to the animal protocol.
Biodistribution (BioD) of mRNA NP in PCa xenograft tumour model.
For the BioD study, PC3 xenograft-bearing male athymic nude mice received intravenous injection of naked Cy5 EGFP mRNA, Cy5-EGFP-mRNA-PGCP NP, and Cy5-EGFP-mRNA-PGDP NP via tail vein injection at an mRNA dose of 700 μg per kg of animal weight. Twenty-four hours later, organs and tumours were harvested and imaged with the IVIS Lumina III In Vivo Imaging System (Perkin Elmer). To evaluate in vivo BioD specifically for PTEN mRNA in a tumour xenograft model, Cy5-tagged PTEN mRNA was prepared by substitution of 25% psedu-UTP with Cy5-UTP when conducting IVT reactions. The Cy5-PTEN-mRNA and its PGCP and PGDP NPs were then injected (i.v. via tail vein) for BioD analysis as described above.
In vivo therapeutic efficacy of PTEN mRNA NP in PCa xenograft tumour model.
For in vivo therapeutic efficacy, PGDP NP was used as a delivery system for PTEN mRNA and EGFP mRNA as a negative control. PC3 xenograft-bearing athymic nude mice were treated when the tumours were first palpable. The mice were randomly divided into three groups, which received (i) PBS (n=7), (ii) EGFP-mRNA-PGDP NP (n=9), or (iii) PTEN-mRNA-PGDP NP (n=8). Mice were injected with the above samples via tail vein at a mRNA dose of 700 μg per kg of animal weight at days 10, 13, 16, 19, 22, and 25 after tumour induction. Tumour size was measured using a caliper every three days through day 43, and average tumour volume (mm3) was calculated as: ½(length × width × height). The body weights of the mice were also determined. At day 28 (three days after the last injection), mice (1 mouse for PBS and 2 mice for each EGFP-mRNA-PGDP NP and PTEN-mRNA-PGDP NP group) were selected randomly for harvest of tumours to monitor PTEN expression and tumour cell apoptosis, and various organs to examine in vivo toxicity. The mice were imaged at day 35, and the image backgrounds were removed using Adobe Photoshop software. At day 43, mice were sacrificed and various organs (lung, heart, liver, kidney, and spleen) were collected to assess toxicity by immunohistochemical analysis. Blood serum was also collected at the two time points of days 28 and 43 for hematological assays.
In vivo therapeutic efficacy of PTEN mRNA NPs in advanced PCa models.
To assess the in vivo therapeutic efficacy of PTEN mRNA NP in advanced PCa, we prepared two different advanced PCa mice models: (1) an experimental metastasis model employing intravenous inoculation of luciferase-expressing PC3 (PC3-luc) prostate cancer (PCa) cells, and (2) a bone colonization of intratibial (IT) inoculation of PC3-luc cells as an orthotopic model of PCa established metastases. For experimental metastatic PCa model, 2.5×106 PC3-luc cells in 100 μL of PBS were implanted through i.v. tail vein injection into immunocompromised, male athymic nude mice (78 in total). Two weeks after implantation, mice were monitored for tumour growth every three days using an In-Vivo Xtreme imaging system (Bruker) for initial screening of PC3-luc disseminated mice for treatments. Four weeks post tumour inoculation, 24 mice (incidence rate of ~30%) with i.v. disseminated PCa (detected using an In-Vivo Xtreme imaging system [Bruker] after mice were injected intraperitoneally with 150 mg/kg luciferin substrate [PerkinElmer, Catalog# 122799]) were randomly divided into three groups (n=8 per treatment group), which received (i) PBS, (ii) EGFP-mRNA-PGDP NP, or (iii) PTEN-mRNA-PGDP NP. Treatments were performed via i.v. tail vein injection at mRNA dose of 700 μg per kg of animal weight. Initial treatment was performed at day 28 post tumour inoculation, followed by another four injections at day 31, 34, 37 and 40 in every three days (in total, five injections). Tumour images were also obtained every three days from the day of initial treatment (at day 28 post tumour inoculation) till day 43 post tumour inoculation using an In-Vivo Xtreme imaging system (Bruker) with a charge-coupled device (CCD) camera (exposure time 30 sec, binning of 1, field of vision [FOV] of 19cm, f/stop of 1.10, and no filter). Regions of interest (ROI) were quantified as average radiance (photon/sec/cm2/sr) using Bruker MI SE software, and the fold change of ROI in each measurement (at day 28, 31, 34, 37, 40 and 43) was normalized with the ROI at the day of treatment initiation (at day 28) and plotted using GraphPad software (Version 7).
To prepare the IT orthotopic PCa model, immunocompromised, male athymic nude mice were anesthetized with isoflurane, and 5×104 PC3-luc cells in 10 μL of PBS were injected into the right tibiae of each mouse (72 mice in total). After the cells were given time to establish in the bone (5 days post tumour inoculation in the bone), the mice with efficient IT injections and successful tumour models (36 mice with 50% success rate screened through established visual image and similar photon counts) were randomly divided into the above three groups (n=12 mice per treatment group) and treatments were initiated. Treatments were performed via i.v. tail vein injection at mRNA dose of 700 μg per kg of animal weight. Initial treatment was performed at day 5 post tumour inoculation, followed by another three injections at day 8, 11, and 14 in every three days (in total, four injections). Tumour images were also obtained every three days from the day of initial treatment (at day 5 post tumour inoculation) till day 17 post tumour inoculation using an In-Vivo Xtreme imaging system (Bruker) as mentioned above. Regions of interest (ROI) were quantified as average radiance (photon/sec/cm2/sr) using Bruker MI SE software, and the fold change of ROI in each measurement (at day 5, 8, 11, 14 and 17) was normalized with the ROI at the day of treatment initiation (at day 5) and plotted using GraphPad software (Version 7). The body weights of the mice were also determined every three days and plotted using GraphPad software (Version 7). At the terminal day 17 of IT tumour study, specimens of the 12 mice of each group were randomly divided into 2 sets to perform immunohistochemistry (n=6 mice) and quantitative real-time PCR (n=6 mice) assays to detect PTEN expression in the bone with tumour tissues.
Immunohistochemical staining to detect in vivo PTEN expression.
The expression of HA-PTEN protein in tumour tissue section was assessed by immunohistochemistry. Sections (5 μm thick) were obtained from tumours treated with PBS, PTEN-mRNA-PGDP NP, or EGFP-mRNA-PGDP NP. Paraffin-embedded sections were deparaffinized, rehydrated, and washed in distilled water. Samples were then incubated for 20 min with 0.3% hydrogen peroxide (H2O2) at room temperature to quench endogenous peroxidase activity followed by antigen retrieval in citrate buffer (pH 6.0) using a microwave for 10 min (2 times, each time 5 min). After washing with PBS (pH 7.4), the samples were treated with the Avidin/Biotin Blocking kit (Vector) to quench endogenous biotin, and then immersed in blocking buffer (1% BSA, 5% normal goat serum) for 60 min. Tissue sections were then incubated with primary rabbit anti-HA antibody at 4°C overnight in a humid chamber. After being rinsed with PBS, the samples were incubated with biotinylated secondary antibody for 30 min at room temperature, followed by incubation with the avidin–biotin–horseradish peroxidase complex (ABC kit, Vector Laboratories, Inc). Staining was developed with the diaminobenzidine peroxidase substrate kit (Impact DAB, Vector Laboratories, Inc) for 3 min. Sections were then counterstained with hematoxylin (Sigma), dehydrated, and mounted.
Quantitative real-time PCR assay.
Total RNA was extracted from bone marrow collected from mouse tibias using RNeasy mini kit (Qiagen, Valencia, CA). RNA was then reverse transcribed into cDNA using the high capacity cDNA reverse transcription kit (Thermo Fisher Scientific, Waltham, MA) and qRT-PCR was conducted with Syber Green master mix (Thermo Fisher Scientific). HA-PTEN mRNA expression was normalized to beta actin.
TUNEL apoptosis assay.
Tumours were extracted and fixed in formalin, embedded in paraffin, and sectioned at a thickness of 5 μm. Tumour cell apoptosis was determined by terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) assay (In Situ Cell Death Detection Kit, TMR red; Roche, #12–156-792–910) according to the manufacturer’s protocol. DAPI stain was used to assess total cell number.
In vivo toxicity evaluation: hematologic examination, histology, and immune response.
To evaluate in vivo toxicity, blood was drawn retro-orbitally and serum was isolated from PC3 xenograft athymic nude mice three days after the final injection (day 28), and at the end of the efficacy experiment (day 43). Aspartate aminotransferase (AST), alanine aminotransferase (ALT), blood urea nitrogen (BUN), creatinine, and troponin-1 were measured using assay kits for AST (BioVision), ALT and Creatinine (Cayman Chemical), BUN (Arbor Assays), and troponin-1 (Life Diagnostics) according to the manufacturers’ protocols. For histological examination, various organs (lung, heart, liver, kidney, and spleen) were also collected three days after the final NP injection (day 28), and at the end point of the experiment (day 43). The organs were then fixed with 4% paraformaldehyde and embedded in paraffin followed by sectioning (3~4μm) and staining with H&E. The slides were assessed using an EVOS Cell Imaging System (Thermo Fisher Scientific). Next, to check immunological response, male Balb/c immunocompetent male mice (n=3) received intravenous injection of PBS, naked PTEN mRNA (700 μg mRNA per kg), empty PGDP NP, and PTEN-mRNA PGDP NP. Six or twenty-four hours post injection, serum samples were collected and processed to measure the representative cytokine (i.e., TNF-α) by enzyme-linked immunosorbent assay (ELISA) (Affymetric eBioscience) according to the manufacturer’s protocol.
Statistical analysis.
All graphs were prepared using GraphPad Prism 7 software, and statistical analysis was also carried out using GraphPad Prism 7 software to perform One-Way ANOVA, two-sided t-test, or Mann-Whitney test. Mann-Whitney tests were performed for experiments in which the data was determined to be non-parametric by the normality test76–78 (i.e., for both disseminated metastatic and IT orthotopic experiments). All experiments were performed in triplicate unless otherwise stated. Error bars indicate standard deviation (s.d.), unless otherwise noted specifically as standard error means (s.e.m.). A p<0.05 value is considered statistically significant, where all statistically significant values shown in various Figures are indicated as: *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Data Availability.
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.
Supplementary Material
ACKNOWLEDGEMENTS
This work was in part supported by the Prostate Cancer Foundation (PCF) Young Investigator Award (J.S.); the David Koch-PCF Award in Nanotherapeutics (O.C.F., R.L. and P.W.K.); the US National Institutes of Health (NIH) grants CA200900 (J.S. and B.R.Z.), HL127464 (O.C.F.) and R00CA160350 (J.S.); the National Research Foundation of Korea (K1A1A2048701) (O.C.F.); and the DoD Prostate Cancer Research Program Postdoctoral Training Award (W81XWH-14–1-0268) (Y.X.). The authors would also like to thank Emma Reesor, Simon Guillemette, Jamie Rice, Yujing Li, Michela Zaffagni, Diane Bielenberg and Junqing Wang for their assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
COMPETING INTERESTS
O.C.F. and R.L. declare financial interests in Selecta Biosciences, Tarveda Therapeutics, Placon Therapeutics and Seer Biosciences. R.L. declares financial interests in Moderna Therapeutics.
REFERENCES
- 1.Song MS, Salmena L & Pandolfi PP The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 (2012). [DOI] [PubMed] [Google Scholar]
- 2.McCall P, Witton CJ, Grimsley S, Nielsen KV & Edwards J Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br. J. Cancer 99, 1296–1301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshimoto M et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet. Cytogenet. 169, 128–137 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Han B et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod. Pathol. 22, 1083–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhagen PC et al. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J. Pathol. 208, 699–707 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Yoshimoto M et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br. J. Cancer. 97, 678–685 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sircar K et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J. Pathol. 218, 505–513 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Schmitz M et al. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int. J. Cancer 120, 1284–1292 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Lotan TL et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin. Cancer Res. 17, 6563–6573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stambolic V et al. Negative regulation of PKB/Akt-dependent cell survival by the tumour suppressor PTEN. Cell 95, 29–39 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Furnari FB, Lin H, Huang HS & Cavenee WK Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc. Natl. Acad. Sci. USA. 94, 12479–12484 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA. 96, 6199–6204 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki A et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumour suppressor gene in mice. Curr. Biol. 8, 1169–1178 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Maehama T & Dixon JE The tumour suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273, 13375–13378 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Engelman JA, Luo J & Cantley LC The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Taylor BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasso CS et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backman SA et al. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29, 396–403 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Liliental J et al. Genetic deletion of the Pten tumour suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 10, 401–404 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Tamura M et al. Inhibition of cell migration, spreading, and focal adhesions by tumour suppressor PTEN. Science 280, 1614–1617 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Hamada K et al. The PTEN/PI3K pathway governs normal vascular development and tumour angiogenesis. Genes Dev. 19, 2054–2065 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang BH & Liu LZ PI3K/PTEN signaling in angiogenesis and tumourigenesis. Adv. Cancer Res. 102, 19–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Quabius ES & Krupp G Synthetic mRNAs for manipulating cellular phenotypes: an overview. Nat. Biotechnol. 32, 229–235 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Boczkowski D & Nair S Programming human dendritic cells with mRNA. Methods Mol. Biol. 969, 111–125 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto A, Kormann M, Rosenecker J & Rudolph C Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 71, 484–489 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Leonhardt C et al. Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine 10, 679–688 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Ligon TS, Leonhardt C & Radler JO Multi-level kinetic model of mRNA delivery via transfection of lipoplexes. PloS one 9, e107148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam MA et al. Biomaterials for mRNA delivery. Biomater. Sci 3, 1519–1533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis ME et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuckerman JE et al. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc. Natl. Acad. Sci. USA. 111, 11449–11454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabernero J et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 3, 406–417 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Strumberg D et al. Phase I clinical development of Atu027, a siRNA formulation targeting PKN3 in patients with advanced solid tumours. Int. J. Clin. Pharmacol. Ther 50, 76 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Schultheis B et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumours. J. Clin. Oncol 32, 4141–4148 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Tolcher AW et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumours. Cancer Chemother. Pharmacol. 73, 363–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akinc A et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotech. 26, 561–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehead KA et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehead KA, Langer R & Anderson DG Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8, 129–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuckerman JE & Davis ME Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov. 14, 843–856 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Yin H et al. Non-viral vectors for gene-based therapy. Nature Reviews Genetics 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Conde J, Oliva N, Atilano M, Song HS & Artzi N Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat. Mater. 15, 353–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kauffman KJ et al. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 15, 7300–7306 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Kormann MS et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotech. 29, 154–157 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Li B et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Letters 15, 8099–8107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu X et al. Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc. Natl. Acad. Sci. USA. 112, 7779–7784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Islam MA et al. The role of osmotic polysorbitol-based transporter in RNAi silencing via caveolae-mediated endocytosis and COX-2 expression. Biomaterials 33, 8868–8880 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Islam MA et al. Accelerated gene transfer through a polysorbitol-based transporter mechanism. Biomaterials 32, 9908–9924 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Warren L et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y et al. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 21, 358–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo X et al. Dual-functional lipid-like nanoparticles for delivery of mRNA and MRI contrast agents. Nanoscale 9, 1575–1579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H et al. PTEN induces chemosensitivity in PTEN-mutated prostate cancer cells by suppression of Bcl-2 expression. J. Biol. Chem. 276, 38830–38836 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Sturge J, Caley MP & Waxman J Bone metastasis in prostate cancer: emerging therapeutic strategies. Nature reviews. Clinical oncology 8, 357–368 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Smukste I & Stockwell BR Restoring functions of tumour suppressors with small molecules. Cancer Cell 4, 419–420 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Guo XE, Ngo B, Modrek AS & Lee WH Targeting tumour suppressor networks for cancer therapeutics. Curr. Drug Targets 15, 2–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bettinger T, Carlisle RC, Read ML, Ogris M & Seymour LW Peptide-mediated RNA delivery: a novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Res. 29, 3882–3891 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rejman J, Tavernier G, Bavarsad N, Demeester J & De Smedt SC mRNA transfection of cervical carcinoma and mesenchymal stem cells mediated by cationic carriers. J. Control. Release 147, 385–391 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Zou S, Scarfo K, Nantz MH & Hecker JG Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells. Int. J. Pharm. 389, 232–243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Read ML et al. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 33, e86 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong G, Braun RD & Dewhirst MW Hyperthermia enables tumour-specific nanoparticle delivery: effect of particle size. Cancer Res. 60, 4440–4445 (2000). [PubMed] [Google Scholar]
- 60.Alexis F, Pridgen E, Molnar LK & Farokhzad OC Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 5, 505–515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi J, Kantoff PW, Wooster R & Farokhzad OC Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer 17, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertrand N, Wu J, Xu X, Kamaly N & Farokhzad OC Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 66, 2–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knop K, Hoogenboom R, Fischer D & Schubert US Poly(ethylene glycol) in drug delivery: pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. Engl. 49, 6288–6308 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Guo X & Huang L Recent advances in nonviral vectors for gene delivery. Acc. Chem. Res. 45, 971–979 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu H et al. Structure-based programming of lymph-node targeting in molecular vaccines. Nature 507, 519–522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997). [DOI] [PubMed] [Google Scholar]
- 67.Steck PA et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 15, 356–362 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C & Pandolfi PP Pten and p27KIP1 cooperate in prostate cancer tumour suppression in the mouse. Nat. Genet. 27, 222–224 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Hopkins BD et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341, 399–402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masson GR, Perisic O, Burke JE & Williams RL The intrinsically disordered tails of PTEN and PTEN-L have distinct roles in regulating substrate specificity and membrane activity. Biochem. J. 473, 135–144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Juric D et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 518, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng W et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 6, 202–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campeau E et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS one 4, e6529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramaswamy S et al. Regulation of G1 progression by the PTEN tumour suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA. 96, 2110–2115 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu X et al. Enhancing tumour cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc. Natl. Acad. Sci. USA. 110, 18638–18643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cox TR et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature 522, 106–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Krzywinski M & Altman N Points of significance: Nonparametric tests. Nature methods 11, 467–468 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Tammela T et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.