Abstract
Microphthalmia-associated transcription factor (MiT) family translocation renal cell carcinoma harbors variable gene fusions involving either TFE3 or TFEB genes. Multiple 5′ fusion partners for TFE3 have been reported, including ASPSCR1, CLTC, DVL2, LUC7L3, KHSRP, PRCC, PARP14, NONO, SFPQ1, MED15, and RBM10. Each of these fusion genes activates TFE3 transcription which can be detected by immunostaining. Using targeted RNA-sequencing, TFE3 fusion gene partners were identified in 5 cases of TFE3 immunohistochemistry positive translocation renal cell carcinoma. Three cases demonstrated known fusions: ASPSCR1-TFE3, MED15-TFE3 and RBM10-TFE3. However, two cases showed unreported NEAT1-TFE3 and KAT6A-TFE3 fusion transcripts. The NEAT1-TFE3 RCC arose in a 59-year-old male; which demonstrated overlapping morphological features seen in NEAT2(MALAT1)-TFEB t(6;11) renal cell carcinoma, including biphasic alveolar/nested tumor cells with eosinophilic cytoplasm. The KAT6A-TFE3 renal cell carcinoma demonstrated typical morphological features of TFE3/Xp11 renal cell carcinoma including papillae, eosinophilic cytoplasm with focal clearing and abundant psammoma bodies. KAT6A gene fusion was reported in some cases of acute myeloid leukemia, which has not been previously reported in solid tumors. This report highlights the genetic complexity of TFE3 translocation renal cell carcinoma; and RNA-sequencing is a powerful approach for elucidating the underlying genetic alterations.
Introduction
Xp11 translocation renal cell carcinoma was renamed as MiT (microphthalmia-associated transcription factor) family translocation renal cell carcinoma in the 2016 World Health Organization classification [1]. This tumor type harbors variable gene fusions involving either TFE3 or TFEB genes [1]. TFE3 is a transcription factor gene located at chromosome band Xp11.23. Multiple 5′-fusion partners for TFE3 have been reported, including ASPSCR1, CLTC, DVL2, LUC7L3, KHSRP, PRCC, PARP14, NONO, SFPQ1, MED15, and RBM10 [2–11]. Each of these fusion genes activates TFE3 transcription, which results in the activation of downstream genes including targets normally activated by MiT family transcription factors (Cathepsin K and melanotic markers) [1, 12, 13]. TEF3/Xp11 renal cell carcinoma generally has papillary architecture, clear cells and psammoma bodies. The tumor cells are often positive for melanocytic markers, and negative or weakly positive for cytokeratins in contrast to classic clear cell and papillary renal cell carcinomas [6, 14].
Strong nuclear TFE3 stains using an antibody targeting the C-terminal portion of TFE3 is a sensitive and specific marker for Xp11 translocation renal cell carcinoma [15, 16]. However, it is difficult to optimize immunostaining for TFE3, as it is affected by variable fixation conditions. In clinical practice, it is not uncommon to encounter equivocal findings leading to false positive or false negative results. TFE3 break-apart florescence in situ hybridization (FISH) assay has been demonstrated to be more reliable to detect the presence of the translocation [5, 14, 15]. However, some renal cell carcinomas with morphological features of translocation renal cell carcinoma remain negative by immunostaining and FISH studies [5]. With the recent advancement of next-generation sequencing technology, RNA sequencing not only can identify cases not detected by other means, but also can identify various specific translocations in a single assay. In this study, we used targeted RNA-sequencing to identify the fusion genes in five cases of MiT family translocation renal cell carcinoma which had been diagnosed through TFE3 immunohistochemistry and/or FISH analysis for TFE3 fusions.
Materials and methods
Patients and samples
Five archived cases of renal cell carcinoma with expression of TFE3 by immunohistochemistry were retrieved from the Department of Pathology, Fox Chase Cancer Center (Table 1). Medical record review provided pertinent clinical information. Adequate tumor sampling for each case had been performed (case 1 (9 sections), case 2 (6 sections), case 3 (12 sections), case 4 (14 sections), case 5 (13 sections)). This study was approved by the Institutional Review Board.
Table 1.
Clinicopathological characteristics of five cases of renal cell carcinoma
| No. | Age | Sex | Fusion | Tumor size | Stage | Biphasic | Cytoplasm | Furhman grade | Psammoma body | Pigment | Necrosis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 59 | M | NEAT1-TFE3 | 7 cm | pT1b | Y | Clear/eosinophilic | 2 | Y | Y | N |
| 2 | 23 | F | ASPSCR1-TFE3 | 4 cm | pT1a | N | Clear | 3 | Y | N | N |
| 3 | 22 | F | MED15-TFE3 | 2 cm | pT1a | N | Clear/granular | 2 | N | N | N |
| 4 | 54 | F | RBM10-TFE3 | 10 cm | pT3a | Y | Clear/granular | 4 | N | N | FOCAL |
| 5 | 73 | M | KAT6A-TFE3 | 5 cm | pT1b | N | Clear/eosinophilic | 3 | Y | N | Y |
M, male; F, female; Y, yes; N, no; FOCAL, focally present
Immunohistochemistry
Immunohistochemistry was performed on 4-μm sections from formalin-fixed, paraffin-embedded (FFPE) tissue using a Ventana Benchmark XT automated stainer (Ventana Medical Systems, Tucson, AZ). The following antibodies were used: Melan-A (monoclonal mouse anti-human anti-body, A103; DAKO; 1:100), TFE3 (monoclonal mouse anti-human antibody, MRQ37, Cell Marque; prediluted), PAX8 (monoclonal mouse anti-human antibody, MRQ50, Cell Marque; prediluted), cytokeratin AE1/AE3 (monoclonal mouse anti-human antibody, AE1/AE3, Ventana; prediluted), cytokeratin 7 (monoclonal mouse anti-human antibody, OV-TL 12/30, Dako; 1:800), CAM 5.2 (monoclonal mouse anti-human antibody, BD, 1:100), Cathepsin K (monoclonal mouse anti-human antibody, 3F9, Cell marque, prediluted) and HBM45 (monoclonal mouse antihuman antibody, Dako; 1:60). The corresponding positive and negative controls were shown to be adequate.
RNA-sequencing and data analysis
RNA was isolated from FFPE samples using five 10 μm thick tissue sections. A High Pure FFPE RNA Isolation Kit (Roche, Indianapolis, IN) was used according to the manufacturer’s protocol. RNA was quantified using a Nanodrop apparatus and evaluated with an Agilent 2100 bioanalyzer.
Next-generation sequencing-based targeted RNA-sequencing analysis was performed using the Illumina TruSight RNA Fusion Panel and a MiniSeq sequencer according to the manufacturer’ s recommendation (Illumina, San Diego, CA). This targeted RNA fusion panel consists of 507 of the most well-known cancer-related fusion genes (including TFE3), and the gene list is available at www.illumina.com. The Trusight RNA fusion panel covers 7690 exonic regions that are targeted with a total of 21,283 probes.
FISH analysis
FISH/TFE3 analysis for case 1 and 2 were performed at the Hospital of the University of Pennsylvania (Philadelphia, PA) using dual-color DNA break-apart probes (KREATECH Repeat-Free Poseidon TFE3 (Xp11) Break probes, Leica Biosystems). FISH/TFE3 analysis on case 5 was performed at the Mayo Clinic (Rochester, MN) using break-apart probes Xp11.23 (3′TFE3, 5′TFE3) and probe XCEN (DXZ1). FISH for TFEB on case 1 was done at Mayo Clinic (Rochester, MN) using break-apart probes 6p21.1 (3′TFEB, 5′TFEB).
Results
Clinical characteristics
The patients consisted of three females and two males ranging in age from 22 to 73 (mean, 46 years) (Table 1). Three patients underwent partial nephrectomies, and two patients had radical nephrectomies.
Pathological findings
The pathological features are summarized in Table 1. Three cases demonstrated known fusions: ASPSCR1-TFE3, MED15-TFE3, and RBM10-TFE3. However, case 1 showed a NEAT1-TFE3 fusion gene due to a t(X;11)(p11.23; q13.1). Morphologically, the distinct features observed in this case include abundant psammoma bodies and pre-dominantly alveolar/nested growth pattern as opposed to the papillary architecture observed in the other four cases. This case also demonstrated biphasic tumor cells, i.e., larger epithelioid cells and small lymphocyte-like cells (Fig. 1a–c). Distinct basement membrane globules were not identified, as typically found in TFEB/t (6;11) renal cell carcinoma. The large epithelioid cells had eosinophilic cytoplasm with focal clearing (Fig. 1c), which were only focally positive for cytokeratins, including AE1/3, CAM5.2, and CK7. Melanin pigment was noted focally (Fig. 1c); and Melan A immunostaining was focally and weakly positive. These tumor cells were negative for HMB45 and Cathepsin K. Since TFE3 immunohistochemistry was focally positive (Fig. 1d), FISH analysis with TFE3 (Xp11) break-apart probes was performed, and this study confirmed the presence of a TFE3 translocation. FISH for TFEB translocation was negative.
Fig. 1.
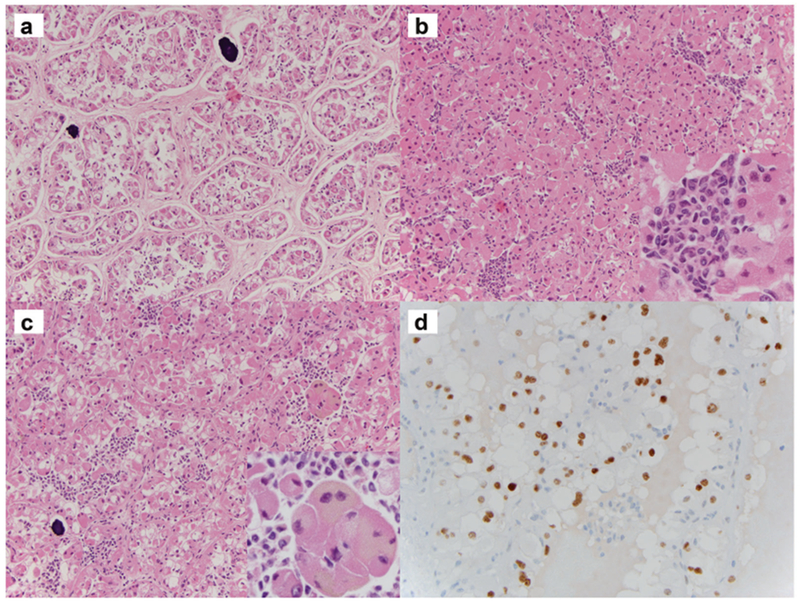
NEAT1-TFE3 renal cell carcinoma with alveolar/nested growth pattern and psammoma bodies (a). Biphasic morphology with both larger epithelial cells and smaller cells (inset) (b). The larger cells have eosinophilic/granular appearance with focal vacuolation/clearing. Some of the cells have brown pigments (inset) (c). Expression of TFE3 (d)
Another case had a KAT6A-TFE3 fusion gene due to a t (X;8)(p11.23; p11.21). This tumor featured typical TFE3/Xp11 morphology including papillae and abundant psammoma bodies. The tumor cells had eosinophilic cytoplasm with focal clearing (Fig. 2a, b). The tumor cells were positive for CAM5.2, and only focally positive for AE1/3, CK7 and Melan A. Similar to the NEAT1-TFE3 renal cell carcinoma, the TFE3 immunohistochemistry was only focally positive. FISH analysis with TFE3 (Xp11) break-apart probes was positive for a TFE3 translocation (Fig. 2c).
Fig. 2.
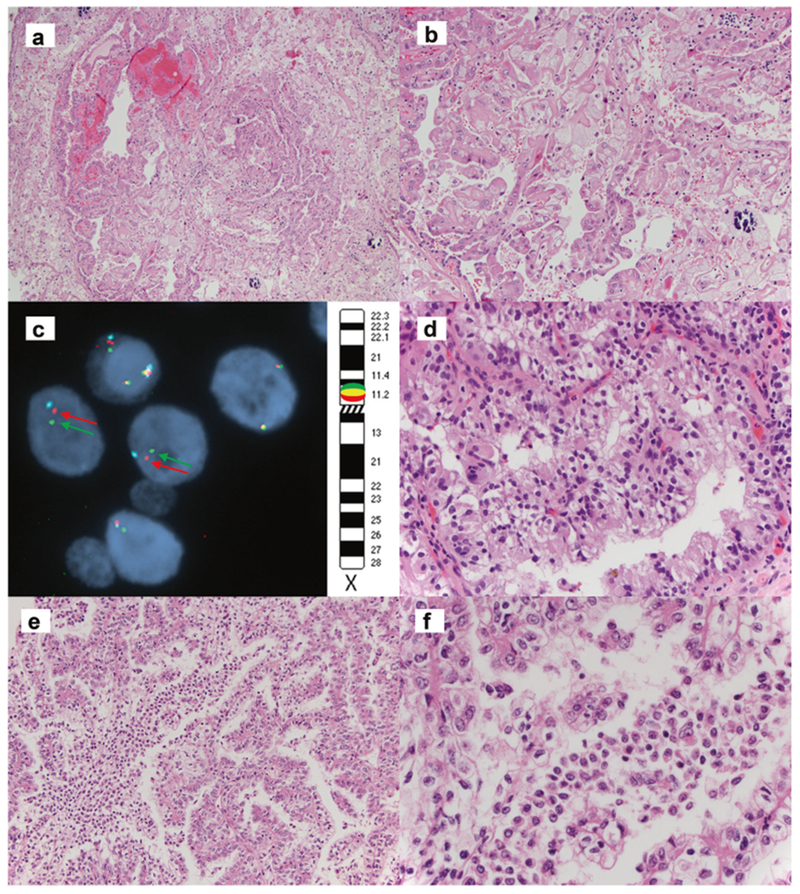
KAT6A-TFE3 renal cell carcinoma shows psammoma bodies, papillae and eosinophilic cytoplasm with clearing (a). High power of Fig a (b). FISH for KAT6A-TFE3 renal cell carcinoma shows TFE3 break-apart signals (green: 3′TFE3[Xp11.23], red: 5′TFE3[Xp11.23], azure: DXZ1[Xcen]) (c). MED15-TFE3 renal cell carcinoma (d). RBM10-TFE3 renal cell carcinoma (e). High power of Fig e (f)
Interestingly, the RBM10-TFE3 renal cell carcinoma (case 4, Fig. 2e, f) also showed biphasic tumor cells without basement membrane globules. However, unlike NEAT1-TFE3 renal cell carcinoma, the RBM10-TFE3 renal cell carcinoma did not feature eosinophilic cytoplasm. These tumor cells were positive for Melan A and Cathepsin K, negative for AE1/3, CAM5.2, CK7, and PAX8. The ASPSCR1-TFE3 renal cell carcinoma (case 2) case had typical TFE3/Xp11 morphology including papillae with clear cytoplasm and abundant psammoma bodies. The MED15-TFE3 renal cell carcinoma (case 3, Fig. 2d) showed papillae and granular cytoplasm with clearing; however, no psammoma bodies were identified. The tumor cells were focally positive for CAM5.2, Cathepsin K and Melan A, negative for AE1/3 and CK7, and they had no melanin pigments.
Discussion
Xp11 translocation renal cell carcinoma was first recognized as a specific category of renal cell carcinoma in the 2004 WHO classification [17]. In 2016, This group of renal cell carcinoma was renamed MiT family translocation renal cell carcinoma in the new World Health Organization classification [1]. This tumor type harbors variable gene fusions involving either TFE3 or TFEB genes [1]. The first fusion/translocation was cloned in 1996, which was found to fuse the TFE3 transcription factor gene on Xp11.2 to the PRCC gene located at 1q21.2 [18, 19]. Since that time, other various TFE3 fusion gene partners have been described, including ASPSCR1, CLTC, DVL2, LUC7L3, KHSRP, PARP14, NONO, SFPQ1, MED15, and RBM10 [2–11].
The predominant histological pattern of TFE3/Xp11 renal cell carcinoma is papillary architecture with clear cells and psammoma bodies. The tumor cells are frequently positive for melanocytic markers with negative or low expression of cytokeratins, which distinguishes them from other renal cell carcinoma subtypes [6, 14]. Renal cell carcinomas with MALAT1 (Alpha)-TFEB /t(6;11) are biphasic neoplasms composed of nested larger epithelioid cells with eosinophilic and vacuolated/clear cytoplasm and smaller cells surrounding basement membrane material [1, 20, 21]. The NEAT1-TFE3 renal cell carcinoma case reported here demonstrated a similar phenotype with an alveolar/nested growth pattern and only focal papillary architecture. The cytoplasm showed eosinophilic appearance with clearing. Our example lacks basement membrane material [21]. Interestingly, the MALAT1(metastasis-associated lung adenocarcinoma transcript 1) is also known as NEAT2 (nuclear-enriched abundant transcript 2) [22].
The Nuclear Enriched Abundant Transcript 1 (NEAT1) gene, located at 11q13.1, produces a long non-coding RNA (lncRNA), a class of intergenic non-coding RNAs longer than 200 nucleotides that can regulate specific gene expression at multiple steps. NEAT1 is a lncRNA localizing to a nuclear structure called paraspeckles [23]. NEAT1 is aberrantly up-regulated in various types of cancer including carcinomas of the ovary, colon and genitourinary system [24–26]. NEAT1 has been shown to be overexpressed in clear cell renal cell carcinoma, and upregulation of NEAT1 was positively correlated with tumor size, higher Fuhrman grade, and lymph node metastasis [26]. Interestingly, NEAT1 is involved in organizing a paraspeckle protein known as NONO (Non-POU domain-containing octamer-binding protein), which is one of the TFE3 fusion gene partners [6, 8, 27]. Similar to NEAT1, the TFEB/t(6;11) fusion partner MALAT1/NEAT2 is a lncRNA; and the fusion of NEAT2 and TFEB results in overexpression of TFEB [1, 21, 22, 28]. Although the NEAT2 promoter drives expression of the NEAT2-TEFB fusion gene, the NEAT2 gene does not contribute to the open reading frame [28, 29].
Except for the NEAT1-TFE3 renal cell carcinoma, the other four Xp11 renal cell carcinoma cases reported here had predominantly papillary architecture, cytoplasmic clearing and low/negative cytokeratin expression; and three of four cases were positive for Melan A. Two cases had psammoma bodies (Tables 1, 2). The KAT6A-TFE3 renal cell carcinoma case demonstrated typical morphological features of TFE3/Xp11 renal cell carcinoma including papillae and abundant psammoma bodies. In addition, the tumor cells had eosinophilic cytoplasm with focal clearing. Lysine acetyltransferase 6A (KAT6A) belongs to the histone acetyltransferase (HAT) family, which was reported to fuse to CREB-binding protein (CBP) in some cases of acute myeloid leukemia [30–32]. These proteins have histone acetyltransferase activity and are involved in transcriptional regulation and cell cycle control [33, 34]. Notably, gene fusions involving KAT6A have not been previously reported in solid tumors.
Table 2.
Immunohistochemistry profile of 5 cases of renal cell carcinoma
| No. | Age | Sex | Fusion | TFE3/FISH | TFE3 | PAX8 | Melan A | HMB45 | AE1/3 | CAM5.2 | CK7 | Cathepsin K |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 59 | M | NEAT1-TFE3 | >10%+ | 20%+ | 20%+ | 5%+ | NEG | 5%+ | 5%+ | 5%+ | NEG |
| 2 | 23 | F | ASPSCR1-TFE3 | >30%+ | 95%+ | 90%+ | NEG | 5%+ | 90%+ | 20%+ | 30%+ | NEG |
| 3 | 22 | F | MED15-TFE3 | N/A | 80%+ | 90%+ | 25%+ | NEG | NEG | 20%+ | NEG | 95%+ |
| 4 | 54 | F | RBM10-TFE3 | N/A | 10%+ | NEG | 70%+ | NEG | NEG | NEG | NEG | 95%+ |
| 5 | 73 | M | KAT6A-TFE3 | >50%+ | 50%+ | 70%+ | 10%+ | NEG | 15%+ | 75%+ | 25%+ | NEG |
NEG, negative;+, positive
Only ten cases of RBM10-TFE3 renal cell carcinoma have been reported. These cases can have sheets of small cells with “pseudo-rosette-like” architectures, mimicking the typical morphology of TFEB/t(6;11) renal cell carcinoma [5, 7]. The RBM10-TFE3 renal cell carcinoma case reported here not only was completely negative for AE1/3, CAM5.2, and CK7, it also did not express PAX8. However, it was positive for Melan A and Cathepsin K. This tumor might belong to melanotic Xp11 translocation renal cell carcinoma/perivascular epithelioid cell neoplasms, which can be keratin(−)PAX8(−)Cathepsin K(+) Melan A(+), although the most reported melanotic Xp11 translocation renal cell carcinoma/perivascular epithelioid cell neoplasms had SFPQ-TFE3 fusion. It may contain variable amounts of finely brown melanin pigment which was not identified in this case [6, 35, 36]. It should be mentioned that both TFE3 and RBM10 are located on the same chromosome sub-band (Xp11.23), only 1.82 Mb apart, so a rearrangement involving these two genes is very unlikely to be detected by conventional FISH analysis [5].
As noted by Argani et al., it is difficult to optimize TFE3 immunostaining due to variable fixation conditions [5, 16]. In our study, three of five cases only had focal or weak positivity for TFE3 staining. Therefore, one should interpret TFE3 immunostaining cautiously. Furthermore, even the use of FISH testing with break-apart probes can sometimes be problematic, as it may be difficult or impossible to detect an inversion or translocation of chromosome X, such as NONO-TFE3 and RBM10-TFE3 rearrangements [5–7, 37]. Thus, RNA-sequencing or targeted multiplex RT-PCR would be a better option for identifying such cases [37].
Conclusion
In this study, two new members of MiT family translocation renal cell carcinoma, NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, are reported. The renal cell carcinoma bearing a t(X;11)(p11.2;q13.1)/NEAT1-TFE3 gene fusion demonstrates overlapping morphological features with TFEB/t(6;11) renal cell carcinoma. The KAT6A-TFE3 renal cell carcinoma demonstrated typical features of TFE3/Xp11 renal cell carcinoma. This study further emphasizes the genetic complexity of this unusual group of renal tumors.
Acknowledgements
The authors would like to thank Wendy Davis, Magdalena Potoczek and Michele Pizzo for excellent technical support. This work was supported by NCI grant P30 CA006927.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Argani P, Cheville J, Ladanyi M. MiT family translocation renal cell carcinoma In: Moch H, Humphrey PA, Ulbright TM, et al. editors. WHO Classification of Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press, 2016. p. 33–34. [Google Scholar]
- 2.Argani P, Antonescu CR, Couturier J, et al. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2; q21). Am J Surg Pathol. 2002;26:1553–66. [DOI] [PubMed] [Google Scholar]
- 3.Argani P, Antonescu CR, Illei PB, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argani P, Lui MY, Couturier J, et al. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2; q23). Oncogene. 2003;22:5374–8. [DOI] [PubMed] [Google Scholar]
- 5.Argani P, Zhang L, Reuter VE, et al. RBM10-TFE3 renal cell carcinoma: a potential diagnostic pitfall due to cryptic intrachro-mosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am J Surg Pathol. 2017;41:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argani P, Zhong M, Reuter VE, et al. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol. 2016;40:723–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia QY, Wang XT, Zhan XM, et al. Xp11 translocation renal cell carcinomas (RCCs) with RBM10-TFE3 gene fusion demonstrating melanotic features and overlapping morphology with t(6;11) RCC: interest and diagnostic pitfall in detecting a paracentric inversion of TFE3. Am J Surg Pathol. 2017;41:663–76. [DOI] [PubMed] [Google Scholar]
- 8.Clark J, Lu YJ, Sidhar SK, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Goldfischer M, Babyeva S, et al. Identification of a novel PARP14-TFE3 gene fusion from 10-year-old FFPE tissue by RNA-seq. Genes Chromosomes Cancer. 2015;54:500–5. [DOI] [PubMed] [Google Scholar]
- 10.Malouf GG, Monzon FA, Couturier J, et al. Genomic heterogeneity of translocation renal cell carcinoma. Clin Cancer Res. 2013;19:4673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Classe M, Malouf GG, Su X, et al. Incidence, clinicopathological features and fusion transcript landscape of translocation renal cell carcinomas. Histopathology. 2017;70:1089–97. [DOI] [PubMed] [Google Scholar]
- 12.Kobos R, Nagai M, Tsuda M, et al. Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein. J Pathol. 2013;229:743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuda M, Davis IJ, Argani P, et al. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007;67:919–29. [DOI] [PubMed] [Google Scholar]
- 14.Green WM, Yonescu R, Morsberger L, et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–63. [DOI] [PubMed] [Google Scholar]
- 15.Calio A, Grignon DJ, Stohr BA, et al. Renal cell carcinoma with TFE3 translocation and succinate dehydrogenase B mutation. Mod Pathol. 2017;30:407–15. [DOI] [PubMed] [Google Scholar]
- 16.Argani P, Lal P, Hutchinson B, et al. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–61. [DOI] [PubMed] [Google Scholar]
- 17.Argani P, Ladanyi M. Renal carcinomas associated with Xp11.2 translocations/TFE3 gene fusions In: Eble JN, Sauter G, Epstein JI, et al. editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press, 2004. p. 37–38. [Google Scholar]
- 18.Sidhar SK, Clark J, Gill S, et al. The t(X;1)(p11.2; q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet. 1996;5:1333–8. [DOI] [PubMed] [Google Scholar]
- 19.Weterman MA, Wilbrink M, Geurts van Kessel A. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1) (p11; q21)-positive papillary renal cell carcinomas. Proc Natl Acad Sci USA. 1996;93:15294–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calio A, Brunelli M, Segala D, et al. t(6;11) renal cell carcinoma: a study of seven cases including two with aggressive behavior, and utility of CD68 (PG-M1) in the differential diagnosis with pure epithelioid PEComa/epithelioid angiomyolipoma. Mod Pathol. 2018;31:474–87. [DOI] [PubMed] [Google Scholar]
- 21.Argani P, Hawkins A, Griffin CA, et al. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1; q12) chromosome translocation. Am J Pathol. 2001;158:2089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seles M, Hutterer GC, Kiesslich T, et al. Current insights into long non-coding RNAs in renal cell carcinoma. Int J Mol Sci. 2016;17:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunwoo H, Dinger ME, Wilusz JE, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C, Li Z, Li Y, et al. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: a systematic review and meta-analysis. Oncotarget. 2017; 8:2672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang J, Qiao F, Tu J, et al. High expression of long non-coding RNA NEAT1 indicates poor prognosis of human cancer. Onco-target. 2017;8:45918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning L, Li Z, Wei D, et al. LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial-mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomark. 2017;19:75–83. [DOI] [PubMed] [Google Scholar]
- 27.Jiang L, Shao C, Wu QJ, et al. NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017;24:816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis IJ, Hsi BL, Arroyo JD, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21; q13) chromosome translocation. Proc Natl Acad Sci USA. 2003;100:6051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan HQ, Li ST, Shu Y, et al. Alpha gene upregulates TFEB expression in renal cell carcinoma with t(6;11) translocation, which promotes cell canceration. Int J Oncol. 2018;52:933–44. [DOI] [PubMed] [Google Scholar]
- 30.Borrow J, Stanton VP Jr, Andresen JM, et al. The translocation t (8;16)(p11; p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. [DOI] [PubMed] [Google Scholar]
- 31.Roberts I, Fordham NJ, Rao A, et al. Neonatal leukaemia. Br J Haematol. 2018;182:170–84 [DOI] [PubMed] [Google Scholar]
- 32.Gervais C, Murati A, Helias C, et al. Acute myeloid leukaemia with 8p11 (MYST3) rearrangement: an integrated cytologic, cytogenetic and molecular study by the groupe francophone de cytogenetique hematologique. Leukemia. 2008;22:1567–75. [DOI] [PubMed] [Google Scholar]
- 33.Camos M, Esteve J, Jares P, et al. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11; p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–54. [DOI] [PubMed] [Google Scholar]
- 34.Coenen EA, Zwaan CM, Reinhardt D, et al. Pediatric acute myeloid leukemia with t(8;16)(p11; p13), a distinct clinical and biological entity: a collaborative study by the International-Berlin-Frankfurt-Munster AML-study group. Blood. 2013;122:2704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argani P, Aulmann S, Karanjawala Z, et al. Melanotic Xp11 translocation renal cancers: a distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am J Surg Pathol. 2009;33:609–19. [DOI] [PubMed] [Google Scholar]
- 36.Argani P, Hicks J, De Marzo AM, et al. Xp11 translocation renal cell carcinoma (RCC): extended immunohistochemical profile emphasizing novel RCC markers. Am J Surg Pathol. 2010;34:1295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia QY, Wang Z, Chen N, et al. Xp11.2 translocation renal cell carcinoma with NONO-TFE3 gene fusion: morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod Pathol. 2017;30:416–26. [DOI] [PubMed] [Google Scholar]


