Abstract
Protein misfolding and overloaded proteostasis networks underlie a range of neurodegenerative diseases. No cures exist for these diseases, but developing effective therapeutic agents targeting the toxic, misfolded protein species in disease is one promising strategy. AAA+ (ATPases associated with diverse cellular activities) protein translocases, which naturally unfold and translocate substrate proteins, could be potent therapeutic agents to disassemble toxic protein conformers in neurodegenerative disease. Here, we discuss repurposing AAA+ protein translocases Hsp104 and proteasome-activating nucleotidase (PAN) to alleviate the toxicity from protein misfolding in neurodegenerative disease. Hsp104 effectively protects various animal models from neurodegeneration underpinned by protein misfolding, and enhanced Hsp104 variants strongly counter neurodegenerative disease-associated protein misfolding toxicity in yeast, Caenorhabditis elegans, and mammalian cells. Similarly, a recently engineered PAN variant (PANet) mitigates photoreceptor degeneration instigated by protein misfolding in a mouse model of retinopathy. Further study and engineering of AAA+ translocases like Hsp104 and PAN will reveal promising agents to combat protein misfolding toxicity in neurodegenerative disease.
Introduction
Proper folding of proteins is essential for their functions (1). However, protein folding in vivo is challenging because of high macromolecular concentrations (∼300–400 mg/mL), fluctuating microenvironmental conditions, and constraints imposed by cotranslational folding (1). To ward against these challenges, cells have evolved various molecular chaperones to help proteins achieve and maintain their native conformations and several protein-degradation machineries to remove terminally misfolded protein (2, 3). Collectively, these protein quality control factors comprise the protein homeostasis (proteostasis) network (2, 3).
The efficacy of the proteostasis network declines with age, and misfolded and aggregated proteins form and persist (2, 3). Misfolded proteins can gain toxicity by impeding the ubiquitin-proteasome system and have even been shown to directly inhibit proteasomes (4). This aspect is especially pronounced for postmitotic cells such as neurons, which cannot dilute cytoplasmic aggregates through cell division. Indeed, compromised proteostasis is now widely recognized as a unifying feature of several degenerative disorders (5). Diseases such as Parkinson’s disease and amyotrophic lateral sclerosis, despite having diverse clinical symptoms, are united in that the most prominent pathological feature of both disorders is the accumulation of misfolded protein (6). The situation is further complicated by the fact that proteins implicated in each disease adopt a range of misfolded conformations (7). Thus, in Parkinson’s disease, α-synuclein accumulates in toxic soluble oligomers and amyloid fibers that are the major component of cytoplasmic Lewy bodies in degenerating dopaminergic neurons (8, 9, 10, 11, 12). Likewise, in amyotrophic lateral sclerosis, the normally nuclear RNA-binding proteins TDP-43 and FUS accumulate in toxic oligomeric structures and cytoplasmic inclusions (13, 14, 15, 16, 17, 18, 19, 20). Therefore, effective therapeutic strategies must be able to recognize and eradicate each of these misfolded structures (21).
Protein translocases from the AAA+ (ATPases associated with diverse cellular activities) superfamily have emerged as interesting candidates to antagonize protein misfolding. AAA+ proteins are defined by the presence of conserved AAA+ domains containing Walker A and B motifs necessary for ATP binding and hydrolysis (22). AAA+ proteins deploy the power generated from ATP binding and hydrolysis to effect a diverse array of biological functions, including substrate protein unfolding and translocation (22, 23, 24, 25). This protein unfolding mechanism has fueled hypotheses that AAA+ protein translocases may be applied to counteract protein misfolding and the aggregation that underpins neurodegenerative disease (21, 26, 27, 28, 29, 30, 31). In this perspective, we highlight efforts to apply two AAA+ protein translocases, Hsp104 from yeast and proteasome-activating nucleotidase (PAN) from archaea, to mitigate protein misfolding in animal models of neurodegenerative disease.
Hsp104 protects dopaminergic neurons from α-synuclein toxicity in worms and rats
Hsp104 is an AAA+ protein translocase (32, 33, 34) first discovered in yeast as an essential factor for yeast survival during thermal stress (35). In addition, Hsp104 plays a role in the inheritance and maintenance of several yeast prions (26, 36, 37, 38, 39, 40). These activities are due to ATPase-driven protein unfolding and disassembly of protein aggregates, amyloids, and preamyloid oligomers (26, 27, 38, 41, 42), which it can perform on its own (26, 27, 38, 43) or in collaboration with Hsp70 and Hsp40 (41, 44). Indeed, Hsp104 was the first protein factor discovered to have these unique activities (26, 38, 41, 42). Typically, proteins processed by Hsp104 are restored to active soluble states and are not degraded (42, 45, 46, 47). However, Hsp104 can also promote the proteasomal degradation of some select clients (48, 49). Although Hsp104 is conserved among eubacteria, nonmetazoan eukaryotes, and some archaea, it is lost in metazoa (50). Perhaps unsurprisingly then, much speculation has arisen over why Hsp104 was lost and whether it might be added back to enhance the proteostasis network in mammalian lineages (21, 50, 51). Indeed, Hsp104 is well tolerated in mammalian cells, enhances their thermal tolerance, disaggregates heat-denatured luciferase aggregates, and reduces the aggregation and toxicity of various disease-linked proteins, suggesting a gain-of-function in the proteostasis network of these cells (51, 52, 53). Hsp104 also collaborates effectively with human chaperones Hsp70, Hsp40, Hsp110, and small Hsps (28, 54, 55, 56).
Of particular interest is the possibility that the ability of Hsp104 to disassemble amorphous aggregates, preamyloid oligomers, and amyloid fibers might be useful in combating neurodegenerative diseases, in which formation of these protein conformers are widely believed to underpin disease (5). In vitro, Hsp104 can dissolve fibrils composed of tau, amylin, polyglutamine, amyloid β, prion protein, and α-synuclein that are associated with human diseases (27, 55, 57). Hsp104 has also been applied in several animal disease models. Hsp104 expression reduces polyglutamine toxicity in Caenorhabditis elegans, fly, and rodent models (29, 58, 59, 60). Hsp104 reduces dopaminergic neuron degeneration in a rat model of Parkinson’s disease (55). Indeed, Hsp104 is the only protein factor known to eliminate α-synuclein oligomers and fibrils and exhibit protective effects in the rat substantia nigra (55). Furthermore, the substantia nigra of rats expressing Hsp104 and α-synuclein have a decreased load of phosphorylated α-synuclein inclusions, suggesting a link between the protein-disaggregase activity of Hsp104 and the protection of dopaminergic neurons (55). Thus, Hsp104 can indeed buffer proteotoxicity in diverse animal systems and is a therapeutic candidate for several neurodegenerative disorders.
However, Hsp104 activity against neurodegenerative disease proteins can be limited, and typically high concentrations of Hsp104 are required for modest disaggregation (27). Thus, strategies to enhance the disaggregase activity of Hsp104 against non-native substrates have been investigated (61). We engineered potentiated Hsp104 variants to potently suppress toxicity associated with TDP-43, FUS, and α-synuclein in yeast (28, 62, 63, 64, 65, 66, 67, 68). Potentiated Hsp104 variants have a substantially increased intrinsic disaggregase activity and, like wild-type Hsp104, can synergize with human Hsp70 and Hsp40 (28, 43, 62, 63, 65). Potentiated Hsp104s also efficiently eradicate preformed fibrils composed of TDP-43, FUS, and α-synuclein (28, 62); protect dopaminergic neurons in C. elegans (28); and disassemble FUS inclusions in mammalian cells (69). Recently, we also discovered an Hsp104 homolog, from the thermophilic fungus Calcarisporiella thermophila, that naturally suppresses proteotoxicity arising from TDP-43, α-synuclein, and polyglutamine misfolding (70). This finding raises the possibility that natural sequence variation among Hsp104 homologs may be harnessed for therapeutic protein-disaggregase modalities (70). Going forward, it will be very interesting to discover whether additional Hsp104 homologs have therapeutic activities, to understand the basis of these activities, and to apply these and other potentiated Hsp104 variants to more sophisticated animal neurodegenerative disease models to determine their effects.
PAN ameliorates proteotoxicity in rod photoreceptors that causes blindness
PAN is another interesting AAA+ protein unfoldase that has recently been applied to counteract toxic protein misfolding. PAN is homologous to the six AAA+ proteins that comprise the 19S regulatory particle in the eukaryotic proteasome (71). The subunit diversification that occurred during evolution from the archaeal PAN to the eukaryotic 19S proteasome may reflect the adaptation to a more complex proteasome, in analogy with putative adaptive diversification that has been proposed for the evolution of the eukaryotic chaperonin TRiC from the ancestral GroEL (72). Like the 19S particle, PAN associates with the 20S catalytic particle and unfolds substrates before their degradation (71). In archaea, proteins marked for proteasomal degradation are conjugated with ubiquitin-like small archaeal modifier proteins, which are engaged by PAN and shuttled to the proteasome (73). PAN can also recognize substrates with surface-exposed hydrophobic residues and translocate them into the proteasome for degradation (71, 74, 75). Moreover, PAN displays chaperone activity and prevents the aggregation of certain model substrates (76). These characteristics, in addition to its AAA+ architecture, draw parallels between PAN and Hsp104, including the possibility that PAN may counter protein misfolding in neurodegenerative disease. Indeed, a PAN variant was recently developed by adding a C-terminal FLAG epitope tag (PANet), which impedes PAN interaction with the 20S proteasome by obstructing a conserved HbYX proteasome-interacting motif (77). Thus, PANet retained the ability to unfold GFP tagged with an unstructured ssrA tag (GFP-ssrA), but lost the ability to stimulate gate opening of the 20S proteasome (77). PANet could be functionally expressed in mouse rod photoreceptors and did not affect retinal morphology, number of rod photoreceptors, or visual response to light compared to wild-type mice (77). Thus, PANet decouples substrate unfolding from proteasome degradation and is unlikely to have widespread off-target effects and aberrantly unfold essential endogenous proteins in rod photoreceptors.
Having established that PANet is well tolerated in mice, PANet was deployed in a mouse model of inherited blindness (77). In this model, mice lacking the γ subunit of the G-protein coupled receptor transducin (Gɣ1−/− mice) fail to form functional transducin complexes in rod photoreceptors, which in turn overwhelms the proteasome with mistargeted β (Gβ1) and α transducin subunits and ultimately causes severe toxicity and loss of photoreceptors (78, 79, 80). Excitingly, Gɣ1−/− mice that expressed PANet were protected against rod photoreceptor degeneration. Whereas almost all rods in Gɣ1−/− mice degenerated by 7 months, over half of the rods in Gɣ1−/− mice expressing the PANet transgene survived (77). Furthermore, Gɣ1−/−;PANet(+) mice maintained strong visual responses at 7 months as measured by the a-wave amplitude from electroretinogram analysis (77). Indeed, 7-month-old Gɣ1−/−;PANet(+) mice showed similar magnitudes in visual response to 1-month-old Gɣ1−/−;PANet(−) mice (77). Taken together, these studies establish that the PANet unfoldase is an effective tool to mitigate protein misfolding-mediated photoreceptor degeneration.
Although the phenotypic effects of PANet expression in Gɣ1−/− mice are undoubtedly promising, the molecular basis of how PANet interacts with the photoreceptor proteome to achieve those effects is unclear. We suggest four possible mechanisms here. One possibility is that PANet expression mitigates the rod cell degeneration chiefly by limiting nonproductive interactions of Gβ1 with other members of the proteostasis network. For instance, previous work reported that in Gɣ1−/− mice, more Gβ1 copurified with the chaperonin TRiC than in wild-type mice (80). Thus, Gβ1 may exert proteotoxicity by competitively inhibiting the folding of other TRiC clients. It will be interesting to determine whether this elevation in TRiC-associated Gβ1 is alleviated upon PANet expression. A second possibility is that Gβ1 may also form soluble oligomers that inhibit the proteasome, as has been determined previously (4), and PANet may prevent or reverse this oligomerization. This possibility could be explored by generating Gβ1 oligomers from a recombinant protein and determining whether PANet prevents or reverses oligomer formation. A third possibility is that PANet could facilitate the formation of hybrid transducin complexes. Indeed, a recent study demonstrated that in Gɣ1−/− mice, Gβ1 can associate with Gɣ2 and Gɣ3 (81). Thus, it would be interesting to determine whether the formation of noncanonical Gβɣ complexes is more efficient in PANet(+) mice. That Gɣ1−/−;PANet(+) mice have robust visual responses suggests that this may be possible. It would be interesting to measure the abundance of the transducin α1 and β1 subunits in PANet(+) mice, which previous studies showed are depleted in Gɣ1−/− mice (79), to assess whether they are protected from degradation in a PANet-dependent manner. A fourth possibility is that endogenous quality control factors fail to recognize orphan Gβ1 subunits in Gɣ1−/− mice (possibly because of aberrant chaperone interactions or the occlusion of degrons within oligomers) and target them for degradation. However, PANet-mediated unfolding of Gβ1 may facilitate recognition via downstream quality control pathways for protein orphans and enable Gβ1 degradation (82). These studies raise exciting questions for future studies concerning how PANet averts rod degeneration at the molecular level.
Another interesting avenue for future work includes determining the substrate repertoire of PANet. PANet may be deployed in diverse neurodegenerative diseases, similarly to the broad utility seen with Hsp104 (27). Similarly, PAN variants with augmented unfoldase activity may be engineered or discovered among naturally occurring homologs to more robustly antagonize proteotoxic misfolding (61, 83). Indeed, PAN could be deployed against many other neurodegenerative diseases linked to protein misfolding.
Conclusions and future opportunities
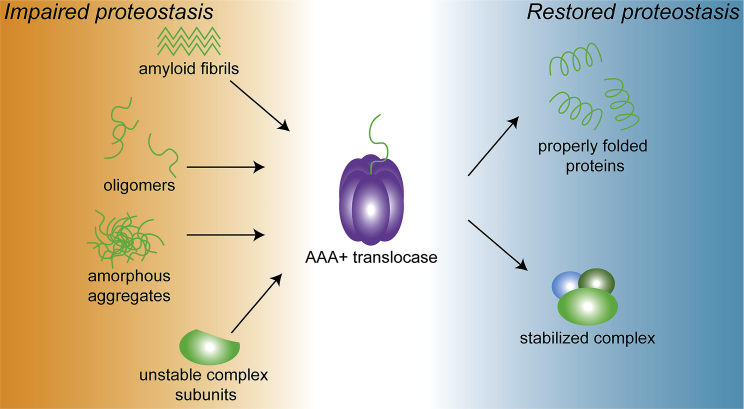
AAA+ protein translocases such as PAN and Hsp104 are unique hubs within the proteostasis network that can transform deleterious conformers back to their native form and function (Fig. 1), simultaneously eliminating loss-of-function phenotypes associated with the depletion of functional proteins and any toxic gain-of-function phenotypes associated with the accumulation of misfolded conformers. The foregoing studies establish the potential therapeutic utility of this protein remodeling in antagonizing protein misfolding to impede neurodegeneration. Yet, significant questions remain to be answered. Hsp104 and PAN are both from nonmetazoan lineages and are likely finely adapted to remodel the proteomes of their respective hosts; although the beneficial phenotypes seen upon expressing Hsp104 or PAN in diseased animals is auspicious, there is also undoubtedly room for improvement. Significant opportunity likely lies in protein engineering efforts to augment the therapeutic activities of Hsp104 or PAN to more efficiently detangle misfolded substrates. At the same time, attention must be paid to averting deleterious off-target effects that may arise while engineering these enhanced AAA+ proteins. For instance, Hsp104 expression in Drosophila is subject to some dose-dependent toxicity (29), and some potentiated Hsp104 variants are mildly toxic in some yeast genetic backgrounds (28, 63). Thus, an important future goal is understanding how AAA+ protein translocases such as Hsp104 and PAN discriminate substrates, and how that might be tailored to create “designer” disaggregases (30, 61, 83) that safely eradicate deleterious conformers with minimal disruption of natively folded proteins and complexes.
Figure 1.
AAA+ protein translocases transform misfolded proteins. AAA+ proteins like Hsp104 or PAN recognize a range of misfolded conformers, including amyloid fibers, amorphous aggregates, oligomers, or unstable monomers, and unfold and remodel these substrates to their native fold and function.
Finally, and perhaps most intriguingly, it may be possible to unlock protein-disaggregase activity of native human AAA+ proteins via pharmacologic modulation, thus obviating the complex challenges associated with genetic engineering. For instance, pharmacologic upregulation of the cyclic adenosine monophosphate/protein kinase A pathway leads to proteasome phosphorylation that stimulates proteasome activity (84). Remarkably, the same treatment administered to transgenic mice expressing pathogenic tau led to decreased tau deposition and improved cognitive performance (85). A recent study demonstrated that proteasomes directly fragment tau and α-synuclein fibrils, although these fragments are more cytotoxic than untreated fibrils, somewhat confounding a beneficial role for the proteasome in eliminating misfolded protein conformers (86). However, proteasome stimulation through phosphorylation may enable the complete eradication of toxic protein conformers. Similarly, although Hsp104 lacks a direct human homolog, emerging human genetics data implicates several human AAA+ proteins in neurodegenerative diseases, including p97/valosin-containing protein (87, 88) and torsin A (89, 90, 91), and suggests that these proteins may have some level of protein-disaggregase activity that may become overwhelmed in neurodegeneration. Likewise, another AAA+ protein, RuvBL, might disassemble protein aggregates (92). It will be exciting to delineate how these proteins interface with the human proteostasis network and to explore the potentiation of these endogenous human AAA+ proteins via small-molecule modulation to enhance human proteostasis (30, 83). Clearly, a wide range of literature now points to a critical utility of AAA+ proteins to counteract protein misfolding in neurodegenerative disease, and it will be interesting to explore the biochemical basis of protein unfolding by endogenous human AAA+ proteins in the future.
Acknowledgments
This work was supported by National Science Foundation Graduate Research Fellowship DGE-1321851 (K.L.M) and National Institutes of Health grants T32GM071399 (Z.M.M.), F31NS101807 (Z.M.M.), and R01GM099836 (J.S.).
Editor: Brian Salzberg.
Footnotes
Zachary M. March and Korrie L. Mack contributed equally to this work.
References
- 1.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Balch W.E., Morimoto R.I., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 3.Lindquist S.L., Kelly J.W. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb. Perspect. Biol. 2011;3:a004507. doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thibaudeau T.A., Anderson R.T., Smith D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018;9:1097. doi: 10.1038/s41467-018-03509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindberg I., Shorter J., McLean P.J. Chaperones in neurodegeneration. J. Neurosci. 2015;35:13853–13859. doi: 10.1523/JNEUROSCI.2600-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forman M.S., Trojanowski J.Q., Lee V.M. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 7.Chuang E., Hori A.M., Shorter J. Amyloid assembly and disassembly. J. Cell Sci. 2018;131:jcs189928. doi: 10.1242/jcs.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos S., Pacheco C., Aguayo L.G. Features of alpha-synuclein that could explain the progression and irreversibility of Parkinson’s disease. Front. Neurosci. 2015;9:59. doi: 10.3389/fnins.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim W.S., Kågedal K., Halliday G.M. Alpha-synuclein biology in Lewy body diseases. Alzheimers Res. Ther. 2014;6:73. doi: 10.1186/s13195-014-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recasens A., Dehay B. Alpha-synuclein spreading in Parkinson’s disease. Front. Neuroanat. 2014;8:159. doi: 10.3389/fnana.2014.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snead D., Eliezer D. Alpha-synuclein function and dysfunction on cellular membranes. Exp. Neurobiol. 2014;23:292–313. doi: 10.5607/en.2014.23.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitler A.D., Shorter J. Prime time for alpha-synuclein. J. Neurosci. 2007;27:2433–2434. doi: 10.1523/JNEUROSCI.0094-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.March Z.M., King O.D., Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitler A.D., Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–187. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson B.S., Snead D., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Z., Diaz Z., Gitler A.D. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann M., Sampathu D.M., Lee V.M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 19.Gasset-Rosa F., Lu S., Cleveland D.W. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 2019 doi: 10.1016/j.neuron.2019.02.038. Published online March 7, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann J.R., Gleixner A.M., Donnelly C.J. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019 doi: 10.1016/j.neuron.2019.01.048. Published online February 27, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 22.Hanson P.I., Whiteheart S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 23.Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 24.Shorter J., Houry W.A. Editorial: the role of AAA+ proteins in protein repair and degradation. Front. Mol. Biosci. 2018;5:85. doi: 10.3389/fmolb.2018.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snider J., Thibault G., Houry W.A. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 2008;9:216. doi: 10.1186/gb-2008-9-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shorter J., Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSantis M.E., Leung E.H., Shorter J. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackrel M.E., DeSantis M.E., Shorter J. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cushman-Nick M., Bonini N.M., Shorter J. Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet. 2013;9:e1003781. doi: 10.1371/journal.pgen.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shorter J. Designer protein disaggregases to counter neurodegenerative disease. Curr. Opin. Genet. Dev. 2017;44:1–8. doi: 10.1016/j.gde.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vashist S., Cushman M., Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem. Cell Biol. 2010;88:1–13. doi: 10.1139/o09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gates S.N., Yokom A.L., Southworth D.R. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science. 2017;357:273–279. doi: 10.1126/science.aan1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shorter J., Southworth D.R. Spiraling in control: structures and mechanisms of the Hsp104 disaggregase. Cold Spring Harb. Perspect. Biol. 2019 doi: 10.1101/cshperspect.a034033. Published online February 11, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweeny E.A., Shorter J. Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J. Mol. Biol. 2016;428:1870–1885. doi: 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez Y., Lindquist S.L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 36.Chernoff Y.O., Lindquist S.L., Liebman S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 37.Halfmann R., Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 38.Shorter J., Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 39.Klaips C.L., Hochstrasser M.L., Serio T.R. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. ELife. 2014;3 doi: 10.7554/eLife.04288. Published online December 9, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweeny E.A., Shorter J. Prion proteostasis: hsp104 meets its supporting cast. Prion. 2008;2:135–140. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glover J.R., Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 42.Parsell D.A., Kowal A.S., Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 43.Castellano L.M., Bart S.M., Shorter J. Repurposing Hsp104 to antagonize seminal amyloid and counter HIV infection. Chem. Biol. 2015;22:1074–1086. doi: 10.1016/j.chembiol.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shorter J., Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace E.W., Kear-Scott J.L., Drummond D.A. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsell D.A., Taulien J., Lindquist S. The role of heat-shock proteins in thermotolerance. Philos. Trans. R Soc. Lond. B Biol. Sci. 1993;339:279–285. doi: 10.1098/rstb.1993.0026. discussion 285–286. [DOI] [PubMed] [Google Scholar]
- 47.Tessarz P., Mogk A., Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 48.Lee D.H., Goldberg A.L. Hsp104 is essential for the selective degradation in yeast of polyglutamine expanded ataxin-1 but not most misfolded proteins generally. Biochem. Biophys. Res. Commun. 2010;391:1056–1061. doi: 10.1016/j.bbrc.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preston G.M., Guerriero C.J., Brodsky J.L. Substrate insolubility dictates Hsp104-dependent endoplasmic-reticulum-associated degradation. Mol. Cell. 2018;70:242–253.e6. doi: 10.1016/j.molcel.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erives A.J., Fassler J.S. Metabolic and chaperone gene loss marks the origin of animals: evidence for Hsp104 and Hsp78 chaperones sharing mitochondrial enzymes as clients. PLoS One. 2015;10:e0117192. doi: 10.1371/journal.pone.0117192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mosser D.D., Ho S., Glover J.R. Saccharomyces cerevisiae Hsp104 enhances the chaperone capacity of human cells and inhibits heat stress-induced proapoptotic signaling. Biochemistry. 2004;43:8107–8115. doi: 10.1021/bi0493766. [DOI] [PubMed] [Google Scholar]
- 52.Bao Y.P., Cook L.J., Rubinsztein D.C. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. J. Biol. Chem. 2002;277:12263–12269. doi: 10.1074/jbc.M109633200. [DOI] [PubMed] [Google Scholar]
- 53.Carmichael J., Chatellier J., Rubinsztein D.C. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2000;97:9701–9705. doi: 10.1073/pnas.170280697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duennwald M.L., Echeverria A., Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo Bianco C., Shorter J., Aebischer P. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J. Clin. Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shorter J. The mammalian disaggregase machinery: hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y.H., Han Y.L., Dong X.P. Heat shock protein 104 inhibited the fibrillization of prion peptide 106-126 and disassembled prion peptide 106-126 fibrils in vitro. Int. J. Biochem. Cell Biol. 2011;43:768–774. doi: 10.1016/j.biocel.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 58.Vacher C., Garcia-Oroz L., Rubinsztein D.C. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum. Mol. Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 59.Satyal S.H., Schmidt E., Morimoto R.I. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrin V., Régulier E., Déglon N. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington’s disease. Mol. Ther. 2007;15:903–911. doi: 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- 61.Mack K.L., Shorter J. Engineering and evolution of molecular chaperones and protein disaggregases with enhanced activity. Front. Mol. Biosci. 2016;3:8. doi: 10.3389/fmolb.2016.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackrel M.E., Shorter J. Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis. Model. Mech. 2014;7:1175–1184. doi: 10.1242/dmm.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackrel M.E., Yee K., Shorter J. Disparate mutations confer therapeutic gain of Hsp104 function. ACS Chem. Biol. 2015;10:2672–2679. doi: 10.1021/acschembio.5b00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sweeny E.A., Jackrel M.E., Shorter J. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell. 2015;57:836–849. doi: 10.1016/j.molcel.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tariq A., Lin J., Shorter J. Potentiating Hsp104 activity via phosphomimetic mutations in the middle domain. FEMS Yeast Res. 2018;18 doi: 10.1093/femsyr/foy042. Published online August 1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackrel M.E., Shorter J. Engineering enhanced protein disaggregases for neurodegenerative disease. Prion. 2015;9:90–109. doi: 10.1080/19336896.2015.1020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackrel M.E., Shorter J. Protein-remodeling factors as potential therapeutics for neurodegenerative disease. Front. Neurosci. 2017;11:99. doi: 10.3389/fnins.2017.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Torrente M.P., Chuang E., Shorter J. Mechanistic insights into Hsp104 potentiation. J. Biol. Chem. 2016;291:5101–5115. doi: 10.1074/jbc.M115.707976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yasuda K., Clatterbuck-Soper S.F., Mili S. FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J. Cell Biol. 2017;216:1015–1034. doi: 10.1083/jcb.201608022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michalska K., Zhang K., Joachimiak A. Structure of Calcarisporiella thermophila Hsp104 disaggregase that antagonizes diverse proteotoxic misfolding events. Structure. 2018;27:449–463.e7. doi: 10.1016/j.str.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith D.M., Benaroudj N., Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 72.Joachimiak L.A., Walzthoeni T., Frydman J. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell. 2014;159:1042–1055. doi: 10.1016/j.cell.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Humbard M.A., Miranda H.V., Maupin-Furlow J.A. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zwickl P., Ng D., Goldberg A.L. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J. Biol. Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
- 75.Smith D.M., Kafri G., Goldberg A.L. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 76.Benaroudj N., Goldberg A.L. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat. Cell Biol. 2000;2:833–839. doi: 10.1038/35041081. [DOI] [PubMed] [Google Scholar]
- 77.Brooks C., Snoberger A., Sokolov M. Archaeal unfoldase counteracts protein misfolding retinopathy in mice. J. Neurosci. 2018;38:7248–7254. doi: 10.1523/JNEUROSCI.0905-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kolesnikov A.V., Rikimaru L., Kisselev O.G. G-protein betagamma-complex is crucial for efficient signal amplification in vision. J. Neurosci. 2011;31:8067–8077. doi: 10.1523/JNEUROSCI.0174-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lobanova E.S., Finkelstein S., Arshavsky V.Y. Transducin gamma-subunit sets expression levels of alpha- and beta-subunits and is crucial for rod viability. J. Neurosci. 2008;28:3510–3520. doi: 10.1523/JNEUROSCI.0338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lobanova E.S., Finkelstein S., Arshavsky V.Y. Proteasome overload is a common stress factor in multiple forms of inherited retinal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:9986–9991. doi: 10.1073/pnas.1305521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dexter P.M., Lobanova E.S., Arshavsky V.Y. Transducin β-subunit can interact with multiple G-protein γ-subunits to enable light detection by rod photoreceptors. eNeuro. 2018;5 doi: 10.1523/ENEURO.0144-18.2018. Published online June 11, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Juszkiewicz S., Hegde R.S. Quality control of orphaned proteins. Mol. Cell. 2018;71:443–457. doi: 10.1016/j.molcel.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shorter J. Engineering therapeutic protein disaggregases. Mol. Biol. Cell. 2016;27:1556–1560. doi: 10.1091/mbc.E15-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lokireddy S., Kukushkin N.V., Goldberg A.L. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc. Natl. Acad. Sci. USA. 2015;112:E7176–E7185. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Myeku N., Clelland C.L., Duff K.E. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 2016;22:46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cliffe R., Sang J.C., Ye Y. Filamentous aggregates are fragmented by the proteasome holoenzyme. Cell Rep. 2019;26:2140–2149.e3. doi: 10.1016/j.celrep.2019.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson J.O., Mandrioli J., Traynor B.J., ITALSGEN Consortium Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buchan J.R., Kolaitis R.M., Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ozelius L.J., Hewett J.W., Breakefield X.O. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 90.Pappas S.S., Darr K., Dauer W.T. Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. ELife. 2015;4:e08352. doi: 10.7554/eLife.08352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pappas S.S., Li J., Dauer W.T. A cell autonomous torsinA requirement for cholinergic neuron survival and motor control. ELife. 2018;7:e36691. doi: 10.7554/eLife.36691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Narayanan A., Meriin A., Cisse I.I. A first order phase transition mechanism underlies protein aggregation in mammalian cells. ELife. 2019;8:e39695. doi: 10.7554/eLife.39695. [DOI] [PMC free article] [PubMed] [Google Scholar]