Abstract
We present Rsubread, a Bioconductor software package that provides high-performance alignment and read counting functions for RNA-seq reads. Rsubread is based on the successful Subread suite with the added ease-of-use of the R programming environment, creating a matrix of read counts directly as an R object ready for downstream analysis. It integrates read mapping and quantification in a single package and has no software dependencies other than R itself. We demonstrate Rsubread’s ability to detect exon–exon junctions de novo and to quantify expression at the level of either genes, exons or exon junctions. The resulting read counts can be input directly into a wide range of downstream statistical analyses using other Bioconductor packages. Using SEQC data and simulations, we compare Rsubread to TopHat2, STAR and HTSeq as well as to counting functions in the Bioconductor infrastructure packages. We consider the performance of these tools on the combined quantification task starting from raw sequence reads through to summary counts, and in particular evaluate the performance of different combinations of alignment and counting algorithms. We show that Rsubread is faster and uses less memory than competitor tools and produces read count summaries that more accurately correlate with true values.
INTRODUCTION
RNA sequencing (RNA-seq) is currently the method of choice for performing genome-wide expression profiling. One of the most popular strategies for measuring expression levels is to align RNA-seq reads to a reference genome and to count the number of aligned reads that overlap each annotated gene (1–3). Alternatively, reads might be counted by exon or by exon–exon junction (4). Read mapping and read counting thus constitute a common workflow by which raw reads are summarized into a count matrix that can be used for downstream analyses. These two steps often represent the most computationally expensive part of an RNA-seq analysis, with mapping and counting both contributing substantially to the total cost.
The last decade has seen rapid development of splice-aware read alignment software. TopHat was the first successful and popular RNA-seq aligner (5). Later aligners such as STAR (6), Subread, Subjunc (7) and HISAT (8) were dramatically faster while maintaining or improving on accuracy. RNA-seq read counting algorithms have developed at almost the same pace, including BEDTools (9), featureCounts (1), htseq-count (3) and Rcount (10). Some of these tools are under continuous development and this article particularly highlights recent improvements in the Subread algorithms.
R is one of the world’s most popular programming languages (11). The TIOBE Programming Community index places it 14th overall at the time of writing and first amongst languages designed specifically for statistical analysis (https://www.tiobe.com/tiobe-index). Building on R, Bioconductor is arguably the world’s most prominent software development project in statistical bioinformatics (12). Bioconductor contains many highly cited packages for the analysis of RNA-seq read counts, including limma (13,14), edgeR (15) and DESeq2 (16) for differential expression analyses and DEXSeq (4) for analysis of differential splicing. Key attractions of Bioconductor include the ease-of-use of the R programming environment, the well organized package management system, the wealth of statistical and annotation resources, the interoperability of different packages and the ability to document reproducible analysis pipelines.
All the Bioconductor RNA-seq data analysis packages rely, however, on read alignment and summarization, which typically have to be performed outside of R. The aligners and quantification tools mentioned above, for example, are written in C, C++, Python or a mixture of those languages. More than one programming language might be used even within a single tool with, for example, Python scripts often found in read mapping tools otherwise written in C or C++. This complicates the analysis pipeline, introducing additional software dependencies and creating substantial obstacles for non-expert uses.
QuasR is a Bioconductor package that attempts to fill the gap, providing RNA-seq read alignment and read counting in the form of R functions (17). QuasR is however an interface to C programs from 2010 or earlier, specifically to Bowtie version 1.1.1 (18), SpliceMap 3.3.5.2 (19) and SeqAn 1.1 (20). These older tools do not reflect the considerable improvements in algorithms achieved during the last 8 years.
This article presents Rsubread, a Bioconductor package that implements current high-performance RNA-seq read alignment and read counting algorithms in the form of R functions. The Rsubread algorithms build on the ‘seed-and-vote’ mapping paradigm (7) and earlier featureCount routines (1) with important new developments that substantially improve performance. The Rsubread user interface provides added functionality and ease-of-use associated with the R Programming environment.
Rsubread integrates read mapping and quantification in a single package and has no software dependencies other than R itself. It has the ability to detect exon–exon junctions de novo and to quantify expression at the level of either genes, exons or exon junctions. Except for read alignment itself, all Rsubread functions produce standard R data objects, allowing seamless integration with downstream analysis packages. The resulting read counts can be input directly into a wide range of downstream statistical analyses using other Bioconductor packages. Rsubread allows RNA-seq data analyses, from raw sequence reads to scientific results, to be conducted entirely in R (21,22).
We take the opportunity in this article to compare Rsubread with the current versions of the popular non-R tools TopHat2, STAR and HTSeq as well as to counting functions in the Bioconductor infrastructure packages. Previous studies have almost always evaluated the performance of read aligners or quantifiers separately (1,7,23,24), an approach that does not reflect all aspects of the user experience. Here, we instead consider the performance of the tools on the combined quantification task starting from raw sequence reads through to summary counts. In particular this allows us to evaluate the performance of different combinations of alignment and counting algorithms. Using SEQC data and simulations, tools are compared on their ability to return read counts that correctly represent the expression levels of genes and exons and on their ability to detect exon–exon junctions. The accuracy of the read counts is assessed by correlating with known expression values. We also measure the time taken and memory used to achieve the results. We note that counting can take longer than alignment for some tools.
This article reports the following results. First we describe the Rsubread package workflow. We summarize the index building, alignment and read counting functionalities of the package in turn, with emphasis on recently added features. Then Rsubread is compared with the most popular competing tools for alignment and quantification. The comparisons are carried out successively at the gene level, then the exon level and finally for detection of exon–exon junctions. All comparisons report accuracy, time taken and memory used. The results show that Rsubread outperforms other tools on all three metrics. It is faster and uses less memory than competitor tools and produces read count summaries that more accurately correlate with true values.
MATERIALS AND METHODS
Software tools
This study compares Rsubread 1.30.9 with aligners STAR 2.6.0c and TopHat 2.1.1 and with quantifiers HTSeq 0.10.0, IRanges 2.14.10, GenomicRanges 1.32.3 and DEXSeq 1.26.0. Rsubread, IRanges, GenomicRanges and DEXSeq are R packages available from http://www.bioconductor.org. Due to version number bumping with each Bioconductor release, Rsubread version 1.30.9 is the same as Rsubread 1.32.0. STAR and TopHat2 are Unix command-line tools written in C++ and Python available from https://github.com/alexdobin/STAR and https://ccb.jhu.edu/software/tophat respectively. HTSeq is a Python library available from https://pypi.org/project/HTSeq.
To make a fair comparison across different workflows, aligners and quantifiers were run with similar settings as far as possible. All aligners were instructed to output no more than one alignment per read. STAR was run in 2-pass mode. Rsubread and STAR were set to output name-sorted reads for paired end data, but TopHat2 supports only location-sorted reads. All aligners were permitted to use gene annotation as an added resource to help detect exon–exon junctions. All aligners were run with 10 threads. featureCounts is the only quantifier that supports multithreading and was run with 4 threads in the evaluation.
All timings and comparisons reported in this article were undertaken on a CentOS 6 Linux server with 24 Intel Xeon 2.60 GHz CPU cores and 512GB of memory.
SEQC data
As an example of real RNA-seq data with known expression profiles, data generated by the SEQC Project (2) was used. Two particular FASTQ files were used, one generated from sequencing of Human Brain Reference RNA (HBRR) and one from Universal Human Reference RNA (UHRR). Each file contains 15 million 100 bp read-pairs and was generated from an Illumina HiSeq sequencer.
The SEQC Project includes expression values measured by TaqMan RT-PCR for slightly over 1000 genes for both HBRR and UHRR. 958 of these TaqMan validated genes were found to have matched symbols with genes in the RNA-seq data. The TaqMan RT-PCR expression values are available from the seqc Bioconductor package.
Simulations
FASTQ files containing simulated paired-end reads were generated using the same GRCh38/hg38 genome and gene annotation as for the SEQC data. Germline variants including SNPs and short indels were introduced to the reference genome at the rates of 0.0009 and 0.0001 respectively, before sequence reads were extracted from the genome. Base substitution errors in sequencing were simulated according to Phred scores at the corresponding positions in randomly selected RNA-seq reads from an actual RNA-seq library (GEO accession GSM1819901), ensuring that the error profile of simulated reads is similar to that of real RNA-seq reads.
FPKM values were generated from an exponential distribution and randomly assigned to genes. The FPKM values were then mapped back to genewise read counts according to known gene lengths in order to achieve a library size of 15 million read pairs. Fragment lengths were randomly generated according to a normal distribution with mean 200 and variance 30. Fragment lengths <110 or >300 were reset to 110 or 300 respectively. Given the fragment length, a pair of 100 bp sequences was randomly selected from exonic base positions of a gene assuming that all exons for that gene are equally expressed and sequentially spliced.
Annotation
All evaluations and simulations used Rsubread’s built-in RefSeq gene annotation for human genome GRCh38/hg38 (build 38.2). This is identical to NCBI annotation except that overlapping exons from the same gene are merged to produce a non-overlapping set of exons for each gene. This simplification reduces ambiguity and somewhat improves the performance of all the read quantification tools. This annotation contains 28 395 genes and 261 752 exons.
Access to data and code
Rsubread can be installed by typing install (‘Rsubread’) at the R prompt, where install is a function in the BiocManager package. Complete data and code necessary to reproduce the figures and results presented in this article is available from http://bioinf.wehi.edu.au/Rsubread/.
RESULTS
The Rsubread workflow
The Rsubread pipeline for read mapping and quantification consists of five R functions (Table 1). buildIndex builds an index of the reference genome. This is a once-off operation for each reference genome, as the same index file can be used for multiple projects. Either align or subjunc is used to align sequence reads to the reference genome and featureCounts produces a matrix of counts. propmapped is optional and produces a table of mapping statistics. All functions return R objects. buildIndex, align and subjunc also write files to disk.
Table 1.
The main Rsubread functions for read alignment and quantification
| Function | Description |
|---|---|
| buildIndex | Create hash table of target genome |
| align | Basic alignment with soft-clipping, for gene-level analyses |
| subjunc | Alignment with identification of exon–exon junctions |
| propmapped | Compute mapping statistics |
| featureCounts | Compute count matrix for specified genomic features |
Other Rsubread functions are briefly discussed in the Discussion section.
All functions operate on an entire set of RNA samples at once rather than one at a time. A typical user-workflow is to start with an R dataframe associating the FastQ filenames with genotypic or phenotypic variables representing experimental conditions. The dataframe column containing the filenames is passed to Rsubread and an annotated R matrix of counts is returned, with samples in the original order, after calls to align (or subjunc) and featureCounts.
A variety of new functionality has been added to the Rsubread functions in recent Bioconductor releases. Particularly large speed improvements were achieved with the Bioconductor 3.8 release in October 2018, corresponding to Rsubread versions 1.30.9 and later.
Rsubread also installs with curated RefSeq gene annotation for human and mouse and includes a 55-page User’s Guide.
Building the index
buildIndex creates a hash table of the target genome from a FASTA file. The index can be built at either single-base or 3-base resolution. Building the full index at single-base resolution takes slightly longer than the gapped index (about 40 minutes vs 15 minutes for the human or mouse genomes) and produces a larger file (about 15Gb versus 5Gb), but allows subsequent alignment to proceed more quickly. The index needs to be built only once for each genome so single-based resolution has been set as the default since Rsubread version 1.30.4. Improvements to buildindex’s hashing algorithms, using a more aggressive divide-and-conquer strategy, reduced the time needed to build a full index more than two-fold in October 2018. Building a gapped index might still however be an efficient choice for smaller projects and was the default in older versions of the software.
Alignment
Alignment itself is performed by either the align or subjunc functions. Both functions accept raw reads, in the form of Fastq, SAM or BAM files, and output read alignments in either SAM or BAM format. align and subjunc also write VCF files containing detected indels and subjunc outputs BED files of exon–exon junctions. At the R level, both functions return an R data.frame containing the total number of reads, the number of uniquely mapped reads, the number of multi-mapping reads and other mapping statistics.
The align function is exceptionally flexible. It performs local read alignment and reports the largest mappable region for each read, with unmapped read bases being soft-clipped. Its unique seed-and-vote design makes it suitable for RNA-seq as well as for genomic DNA sequencing experiments. It automatically detects insertions and deletions. align is recommended for gene-level expression analyses of RNA-seq or for any type of DNA sequencing.
The subjunc function is similar to align but provides comprehensive detection of exon–exon junctions and reports full alignments of junction-spanning reads. subjunc is recommended for any RNA-seq analysis requiring intra-gene resolution.
Both align and subjunc achieve high accuracy via a two-pass process. The first pass is the seed-and-vote step, by which a large number of 16mer subreads from each read are mapped to the genome using the hash table. This step detects indels and exon–exon junctions and determines the major mapping location of the read. The second pass undertakes a detailed local re-alignment of each read with the aid of collected indels and junctions. Subread and Subjunc were the first aligners to implement such a two-pass strategy (7), although the use of indels in the second pass is new.
align and subjunc support reads from any of the major Next Generation Sequencing (NGS) technologies. Users can specify the amount of computer memory and the number of threads to be used, enabling the aligners to run efficiently on a variety of computer hardwares from super-computers to personal computers.
The propmapped function calculates the proportion of reads or fragments that are successfully mapped, a useful quality assessment metric.
Improvements to the alignment algorithms
Recent modifications to the alignment algorithms have improved performance in a variety of ways. Aligned reads are, by default, written to disk without reordering so that paired reads are in successive positions in the file, but reads can now optionally be sorted by genomic location with little increase in execution time. align and subjunc can now optionally use gene annotation in order to refine the search for exon–exon junctions. The required annotation object can be generated conveniently by using Bioconductor organism packages created by the Bioconductor core team or, for human or mouse, Rsubread’s built-in annotation can be used. align and subjunc can now detect multiple short indels even within the same read. Both functions also handle long indels up to 200 bp.
More candidate locations are now examined for multi-mapping reads. Previously paired-end reads were restricted to stipulated minimum and maximum possible fragment lengths, but now align and subjunc can align read pairs arbitrarily far apart if the alignment is sufficiently good and no more canonical alignment is available. A weighting strategy is used to give preference to alignments within the expected fragment length bounds. Gene fusions are now supported by allowing different subreads from the same read to map to different chromosomes.
In a major development, improved support for multithreaded input and output halved the execution time for align and subjunc in the Bioconductor 3.8 release.
Counting reads
The featureCounts function counts the number of reads or read-pairs that overlap any specified set of genomic features. It can assign reads to any type of genomic region. Regions may be specified as simple genomic intervals, such as promoter regions, or can be collections of genomic intervals, such as genes comprising multiple exons. Any set of genomic features can be specified in GTF, GFF or SAF format, either as a file or as an R data.frame. SAF is a Simplified Annotation Format with columns GeneID, Chr, Start, End and Strand.
featureCounts produces a matrix of genewise counts suitable for input to gene expression analysis packages such as limma (13), edgeR (15) or DESeq2 (16). Alternatively, a matrix of exon-level counts can be produced suitable for differential exon usage analyses using limma, edgeR or DEXSeq (4).
featureCounts outputs the genomic length and position of each feature as well as the read count, making it straightforward to calculate summary measures such as RPKM (reads per kilobase per million reads).
featureCounts includes a large number of powerful options that allow it to be optimized for different applications. Reads that overlap more than one feature can be ignored, multi-counted or counted fractionally. Reads can be extended before counting to allow for probable fragment length. Minimum overlap or minimum quality score metrics can be specified. Reads can be counted in a strand specific or non-specific manner.
Counting reads for genes, exons and junctions
A common use of featureCounts is to count reads by gene, and this forms the basis of most gene expression analyses. In this mode, each exon is considered by Rsubread to be a ‘feature’ and each gene is a ‘meta-feature’ comprising a collection of exons. To count reads at the gene level, each read is counted once for a gene if it overlaps one or more of the exons that make up that gene.
Another common procedure is to count reads by exon, a practice that permits alternative exon usage or alternative splicing to be investigated. In this mode, a read is typically counted once for every exon that it overlaps. Of particular importance is the mapping and counting of reads that span two or more exons in the same gene, i.e., that span one or more exon-junctions. Junction-spanning reads typically account for ∼20–30% of reads in an RNA-seq dataset.
featureCounts also provides a third option. In this new approach, subjunc is first used to detect all exon-junctions in an RNA sample. Then featureCounts is used to count (i) the number of reads spanning each junction and (ii) the number of reads entirely internal to each exon. This junction+internal approach provides complete information and has the advantage of counting each read exactly once.
Gene-level counting can be performed using alignments from either align or subjunc, but align is recommended because it provides the most mapped reads. For exon-level counting, subjunc aligner should be used for read mapping as it comprehensively detects exon-junctions and performs a complete alignment for each read.
New functionality for the counting algorithms
As with alignment, recent modifications to featureCounts have either improved performance or increased functionality. Earlier versions of featureCounts required the input BAM files to be name-sorted, i.e. for paired reads to immediately follow one another in the file, but featureCounts now works with any ordering. This avoids any unnecessary overheads when processing BAM files produced by STAR or TopHat, for which location-sorting is the default. featureCounts now handles reads of arbitrary length, allowing it to count reads from long-read sequencers such as those from Oxford Nanopore Technologies.
The original behavior of featureCounts was to count any read that overlaps a feature, even by a single nucleotide. That behavior is still the default, but featureCounts now offers unrivaled flexibility in terms of being able to specify how a read should overlap with a feature before it is counted. Users can specify that reads should overlap a feature by a minimum number of bases, or alternatively overlap a minimum percentage of the feature. Reads can be reduced to a single base so that users can count, for example, the number of read-start positions than overlap a set of genomic features. Alternatively reads can be extended in a directional manner to represent the putative DNA or RNA fragment from which the read originated. Reads can even be shifted by a specified number of bases in a 3’ or 5’ direction before counting is done. Other new functionality includes the junction+internal counting described above.
Built-in annotation
Rsubread comes with annotation for human and mouse genes already installed, so that GTF or SAF files do not need to be specified for these species. The built-in annotation follows NCBI RefSeq gene annotation with the simplification that overlapping exons from the same gene are merged. This simplification reduces annotation ambiguity and proves beneficial for most RNA-seq expression analyses. Built-in annotation is provided for the mm9, mm10, hg19 and hg38 genome builds. Rsubread’s built-in hg38 annotation was used for the simulations and comparisons reported in this article.
Quantification at the gene-level: speed and memory
We now compare the Rsubread gene-level workflow, which consists of align and featureCounts, to other workflows that generate read counts for genes. Rsubread is compared to aligners TopHat2 (25) and STAR (26) combined with quantification tools htseq-count (3), summarizeOverlaps and featureCounts. Google Scholar searches suggest that these are currently the most popular tools for generating gene-level counts. htseq-count is part of the HTSeq Python library. summarizeOverlaps is a function in the Bioconductor package GenomicRanges.
First we assessed the running time of the read aligners on the SEQC UHRR sample (Figure 1). align was faster when run with the full genome index (align-F) as opposed to the gapped index (align-G), with both options being faster than STAR or TopHat2. align-F was more than three times as fast as STAR and 25 times as fast as TopHat2.
Figure 1.
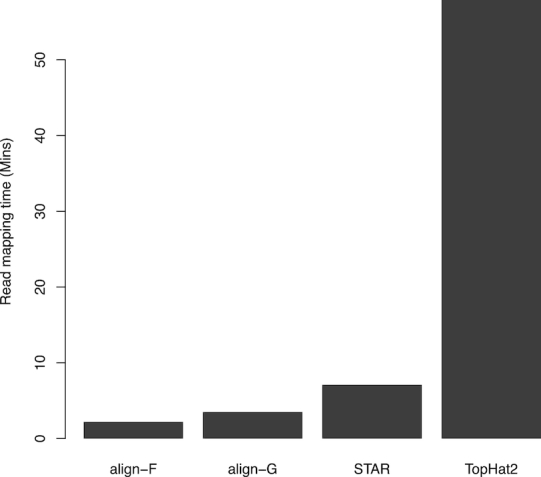
Run times of read aligners. Each aligner used ten threads to map 15 million 100 bp read-pairs from the SEQC UHRR sample to the human reference genome GRCh38. Rsubread::align is faster than STAR or TopHat2 regardless of whether the full index (align-F) or a gapped index (align-G) is used.
TopHat2 and align-G had the smallest memory footprints for the same operation (supplementary Figure S1). align-F used twice as much memory and STAR over four times as much.
Next we ran the quantification tools to assign the mapped UHRR reads to RefSeq human genes. This showed featureCounts to be 16–175 times faster than the other tools (Figure 2). featureCounts was equally as fast regardless of the alignment used. summarizeOverlaps and htseq-count were slower when working on the TopHat2 alignment than the STAR alignment. In this evaluation, align and STAR output name-sorted aligned reads whereas TopHat2 output location-sorted reads.
Figure 2.
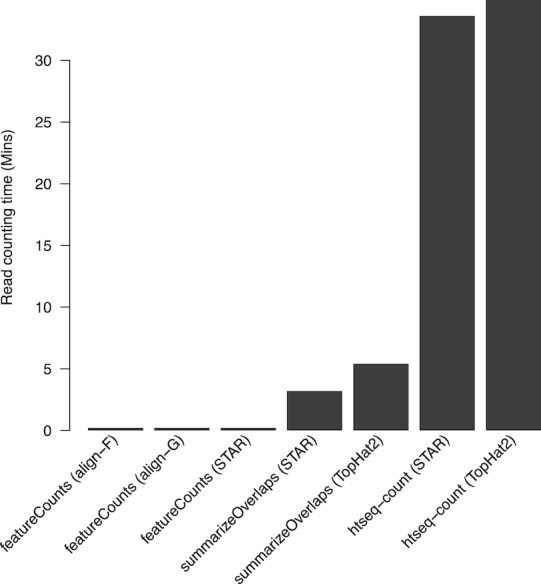
Running time of different quantification tools. Labels under each bar indicate the quantification method and the aligner (in parenthesis) that produced the mapped reads used for counting. Mapped reads were assigned to NCBI RefSeq human genes. featureCounts is the only tool that supports multi-threaded read counting and it was run with four threads.
featureCounts used easily the least memory for the quantification step (supplementary Figure S2). htseq-count used only slightly more memory than featureCounts when read pairs were name-sorted, but it used >40 times more memory when the read pairs were location-sorted. summarizeOverlaps had high memory usage for both name-sorted and location-sorted reads because it loads all the reads into memory at once.
In summary, Rsubread outperformed the STAR-based workflows in both speed and memory use. While TopHat2 has a slightly smaller memory footprint than Rsubread for alignment, it was far too slow to be competitive.
Quantification at the gene-level: accuracy
Next we compared workflows for accuracy in quantifying gene expression levels. First we ran the workflows on the UHRR and HBRR samples from the SEQC Project. Read counts generated from each workflow were then compared to the expression levels of 958 genes as measured by TaqMan RT-PCR, a high-throughput quantitative PCR technique. RNA-seq counts were converted to log2-FPKM (fragments per kilobases per million) values and TaqMan RT-PCR data were also converted to log2 scale.
Rsubread workflows are found to yield the highest correlation with TaqMan RT-PCR data for both UHRR and HBRR samples (Table 2). All workflows produce higher correlation for HBRR sample than for UHRR sample, which is expected because the UHRR sample is made up from multiple cancer cell lines.
Table 2.
Gene-level accuracy comparison. The table gives Pearson correlations between true log2-expression levels and log2-FPKM values produced by each workflow. The align-F + featureCounts workflow gives the best correlation in each case
| Workflow | UHRR | HBRR | Simulation |
|---|---|---|---|
| align-F + featureCounts | 0.851 | 0.870 | 0.955 |
| align-G + featureCounts | 0.850 | 0.869 | 0.955 |
| STAR + featureCounts | 0.848 | 0.867 | 0.901 |
| STAR + htseq-count | 0.845 | 0.864 | 0.877 |
| STAR + summarizeOverlaps | 0.845 | 0.864 | 0.877 |
| TopHat2 + htseq-count | 0.843 | 0.863 | 0.921 |
| TopHat2 + summarizeOverlaps | 0.843 | 0.863 | 0.921 |
Columns ‘UHRR’ and ‘HBRR’ are for the SEQC UHRR and SEQC HBRR samples respectively. For the SEQC columns, the log2-expression values of 958 genes measured by TaqMan RT-PCR are taken as true values. Column ‘Simulation’ shows simulation results for 28 395 genes. For all columns, an offset of 1 was added to raw gene counts to avoid taking logarithms of zeros.
Next we compared the workflows on simulated data. All the workflows were run on a simulated FASTQ file of 15 million read pairs. Read counts were converted to log2-FPKM and compared to the known log2-FPKM values from which the simulated sequence reads were generated. The Rsubread workflows are again found to achieve the best correlation with the true expression values (Table 2). Both Rsubread workflows yield correlation >0.95, much higher than those for other workflows.
In general, align was more accurate than STAR and featureCounts was more accurate than either htseq-count or summarizeOverlaps. summarizeOverlaps uses the same counting strategy as that developed by htseq-count and therefore gives the same results.
Quantification at the exon level: speed and memory
Next we compared workflows to obtain exon-level read counts. Rsubread workflows for exon-level analysis comprise the subjunc program, which was run with a full genome index (subjunc-F) or a gapped index (subjunc-G), and the featureCounts programs. Rsubread was compared to TopHat2 + dexseq_count, TopHat2 + countOverlaps, STAR + dexseq_count, STAR + countOverlaps and STAR + featureCounts. dexseq_count.py is a Python script that comes with the DEXSeq package for counting RNA-seq reads by exon. countOverlaps is a function in IRanges package. For all pipelines, reads spanning multiple exons were counted for all the relevant exons.
All workflows were run on the SEQC UHRR data. The results were qualitatively similar to those observed at the gene-level. subjunc-F was faster than subjunc-G and both were faster than STAR and TopHat2 (supplementary Figure S3). subjunc-F was more than twice as fast as STAR and 20 times as fast as TopHat2.
For read counting, featureCounts is more than an order of magnitude faster than countOverlaps or dexseq_count, regardless of which aligner output was used (supplementary Figure S4). Dexseq_count was the slowest counting tool.
subjunc uses much less memory than STAR (supplementary Figure S5) and featureCounts uses less memory than dexseq_count or countOverlaps (supplementary Figure S6). As for the gene-level results, TopHat2 used slightly less memory than subjunc-G but at too high a price in terms of running time.
In summary, subjunc-F and featureCounts constitute the fastest workflow for exon-level analysis of RNA-seq data. featureCounts uses the least memory of any quantification tool and subjunc uses less memory than STAR.
Quantification at the exon level: accuracy
We used the same simulated data as for the gene-level comparison to assess the accuracy of the exon-level workflows. Overlapping exons found between genes were removed from analysis to avoid counting ambiguity. Exons from genes appearing in more than one chromosome, or appearing in both strands of the same chromosome, were also removed because dexseq_count cannot process such exons. 5000 exons were excluded from this analysis in total (out of 261 752 exons). Read counts from remaining exons were then converted to log2-FRKMs for comparison.
As was seen in the gene-level comparison, the two Rsubread workflows both outperformed the other workflows (Table 3). subjunc and TopHat2 were more accurate than STAR. featureCounts was more accurate than countOverlaps and countOverlaps was more accurate than dexseq_count. The accuracy of the workflows was affected by both read mapping and counting. The STAR + dexseq_count workflow had the worst correlation of the workflows. Replacing dexseq_count with featureCounts improved the accuracy, but it remained lower than that for the two pure Rsubread workflows.
Table 3.
Exon-level accuracy comparison. The table shows the Pearson correlation between the true log2-FPKM expression values of exons and log2-FPKM values produced by each workflow. The Rsubread workflows give the best correlation with the true values
| Workflow | Correlation |
|---|---|
| subjunc-F + featureCounts | 0.982 |
| subjunc-G + featureCounts | 0.982 |
| STAR + featureCounts | 0.950 |
| STAR + dexseq_count | 0.927 |
| STAR + countOverlaps | 0.950 |
| TopHat2 + dexseq_count | 0.942 |
| TopHat2 + countOverlaps | 0.980 |
An offset of 1 was added to raw exon counts to avoid taking logarithms of zero values.
Detection and quantification of exon–exon junctions
Exon-exon junctions can be discovered directly by the correct mapping of junction-spanning reads (junction reads). Most RNA-seq aligners report the locations of exon splice sites (donor and receptor sites). The number of reads supporting the splice sites detected are often reported as well, providing a quantitive measurement for the junctions events. Analysis of discovered junctions and their abundance is an important step in the discovery of alternative splicing events. Junction data can be further combined with exon-level and gene-level expression data in order to detect differentially spliced genes.
Nevertheless, junction reads can be difficult to map correctly because they may span an intron tens of thousands of bases long and indeed might span more than one intron. Here we assess the performance of the aligners for mapping junction reads and calling junctions. We used the same simulated data as above. There are 233 021 exon–exon junctions in the simulated data and 25% of the simulation reads are junction reads.
subjunc-F and subjunc-G had better overall performance than STAR or TopHat2 as measured by the F1 summary of precision and recall (Table 4). In particular, subjunc-F and subjunc-G outperformed STAR and TopHat2 in mapping of junction reads by a clear margin. STAR was slightly less sensitive than the other aligners in mapping junctions reads or detecting junctions.
Table 4.
Aligner performance in mapping junction reads and reporting exon–exon junctions. Results are based on simulated data
| Junctions | Junction reads | |||||
|---|---|---|---|---|---|---|
| Workflow | Recall | Prec | F1 | Recall | Prec | F1 |
| subjunc-F | 99.80 | 99.53 | 99.66 | 95.51 | 98.18 | 96.82 |
| subjunc-G | 99.82 | 99.43 | 99.63 | 95.50 | 98.16 | 96.81 |
| STAR | 98.48 | 99.87 | 99.17 | 89.87 | 98.06 | 93.78 |
| TopHat2 | 99.12 | 99.91 | 99.51 | 90.15 | 98.57 | 94.17 |
Column ‘Recall’ gives the percentage of correctly called junctions (or junction reads) out of all junctions (or junction reads) generated in the simulated dataset. Column ‘Precision’ gives the percentage of correctly called junctions (or junction reads) out of all reported junctions (or junction reads). Column ‘F1’ gives the F1 score that is the harmonic mean of precision and recall.
DISCUSSION
Read mapping and quantification are computationally-intensive operations that lay the foundation for most analyses of NGS data. The time-consuming and resource-hungry nature of these operations is a major bottleneck for larger projects. Meanwhile, R is an easy-to-learn scripting language that is widely used for statistical analyses of NGS data once the processing of the raw reads is completed. Rsubread provides functions for read mapping and quantification within the R programming environment, allowing an entire NGS analysis, from reads to results, to be completed in a single R session. Rsubread can work with Bioconductor packages limma, edgeR and DESeq2 to complete an entire RNA-seq analysis in R from read mapping through to the discovery of genes that exhibit significant expression changes (21,22). It has proved a valuable resource for NGS analyses in biomedical research (27).
The use of a pure R environment means that users do not need to install additional software tools, learn user-interfaces or assimilate documentation outside of the R environment. The R functions provide careful checking for input parameters and user-friendly error messages. At a more advanced level, an entire analysis workflow can be documented using reproducible research packages such as knitr or Sweave (28).
Rsubread consists of over 60 000 lines of source code, but the user interface is concise and easy to navigate (Table 1). Each of the Rsubread functions offers optional arguments, but these are accessed only as needed and in most cases the default arguments perform well. All functions and options are documented via the R help system and are summarized in the Rsubread User’s Guide.
As well as ease-of-use, this study has shown that Rsubread outperforms the most popular competing alignment and quantification tools regardless of programming language. Rsubread was found to be faster, to use less memory and to provide more accurate expression quantification than competitor tools. The speed difference was substantial, with Rsubread more than three times as fast as its nearest competitor for gene-level alignment and an order of magniture faster for quantification. Rsubread was consistently the best performer on all metrics considered except that TopHat2 used slightly less memory for alignment. TopHat2 however was an order of magnitude slower and hence is not considered competitive overall. The improved accuracy of the Rsubread workflows should translate into more accurate downstream analyses such as discovery of differentially expressed genes.
The performance of the Rsubread aligners ultimately derives from the efficiency of the seed-and-vote mapping strategy implemented in Subread (7). Seed-and-vote tiles each sequence reads with a large number of short seeds, each of which can be mapped to the genome using an extremely efficient hashing operation. This enables align and subjunc to very quickly detect all possible candidate mapping locations for a read. The top candidate locations with a high number of votes received from the seeds can be quickly followed up. Gaps between mapped seeds are filled by using a banded Smith–Waterman dynamic programming procedure. Local alignment proceeds very quickly because the gaps are small and gap length is pre-determined by the flanking seeds. For the mapping of paired-end reads, the end with most votes serves as the anchor read and the other end is re-mapped by taking into account the expected distance between the two ends. Rsubread builds on this fundamental framework to provide general purpose RNA-seq functionality. The package is under continuous development and substantial performance improvements continue to be achieved.
Although the main focus of this study is on the analysis of RNA-seq data, Rsubread can be used also for the analysis of other types of sequencing data such as histone ChIP-seq and ATAC-seq. The align function can be used for read mapping and featureCounts can be used to produce read counts for promoter regions, gene bodies, windows or regions to provide a measurement of peak abundance (29,30).
The Rsubread package includes other functions beyond the scope of this article including sublong (for long read mapping), exactSNP (SNP identification), atgcContent (compute nucleotide frequencies), detectionCall, and promoterRegions.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jenny Dai and Timothy Triche, Jr, for code contributions to Rsubread.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian National Health and Medical Research Council [Program grant 1054618 and Fellowship 1058892 to G.K.S.; Project grant 1023454 to G.K.S. and W.S.; Project grant 1128609 to W.S.]; Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIIS; Walter and Eliza Hall Institute Centenary Fellowship funded by a donation from CSL Ltd. (to W.S.) Funding for open access charge: Australian National Health and Medical Research Council Project grant 1128609 to W.S..
Conflict of interest statement. None declared.
REFERENCES
- 1. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general-purpose read summarization program. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 2. Su Z., Labaj P.P., Li S., Thierry-Mieg J., Thierry-Mieg D., Shi W., Wang C., Schroth G.P., Setterquist R.A., Thompson J.F. et al.. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014; 32:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anders S., Reyes A., Huber W.. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012; 22:2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liao Y., Smyth G.K., Shi W.. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quinlan A., Hall I.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmid M.W., Grossniklaus U.. Rcount: simple and flexible RNA-Seq read counting. Bioinformatics. 2015; 31:436–437. [DOI] [PubMed] [Google Scholar]
- 11. R Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2018; Vienna. [Google Scholar]
- 12. Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. et al.. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015; 12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Law C.W., Chen Y., Shi W., Smyth G.K.. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014; 15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson M., McCarthy D., Smyth G.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaidatzis D., Lerch A., Hahne F., Stadler M.B.. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015; 31:1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Au K.F., Jiang H., Lin L., Xing Y., Wong W.H.. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010; 38:4570–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Döring A., Weese D., Rausch T., Reinert K.. SeqAn an efficient, generic C++ library for sequence analysis. BMC Bioinformatics. 2008; 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., Lun A.T.L., Smyth G.K.. From reads to genes to pathways: differential expression analysis of RNA-seq experiments using Rsubread and the edgeR quasi-likelihood pipeline [version 2; referees: 5 approved]. F1000Research. 2016; 5:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmid M.W. RNA-Seq data analysis protocol: Combining in-house and publicly available data. Methods Mol. Biol. 2017; 1669:309–335. [DOI] [PubMed] [Google Scholar]
- 23. Engstrom P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Ratsch G., Goldman N., Hubbard T.J., Harrow J., Guigo R. et al.. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013; 10:1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baruzzo G., Hayer K.E., Kim E.J., Camillo1 B.D., FitzGerald G.A., Grant G.R.. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat. Methods. 2017; 14:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fong C.Y., Gilan O., Lam E.Y., Rubin A.F., Ftouni S., Tyler D., Stanley K., Sinha D., Yeh P., Morison J. et al.. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015; 525:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie Y. Dynamic Documents with R and knitr. 2013; Boca Raton: CRC Press. [Google Scholar]
- 29. Pal B., Bouras T., Shi W., Vaillant F., Sheridan J., Fu N., Breslin K., Jiang K., Ritchie M., Young M. et al.. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep. 2013; 3:411–426. [DOI] [PubMed] [Google Scholar]
- 30. de Santiago I., Carroll T.. Analysis of ChIP-seq data in R/Bioconductor. Methods Mol. Biol. 2018; 1689:195–226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


