Significance
The p53 tumor suppressor protein is a critical stress response mediator. Polymorphisms in the human TP53 gene can alter protein function. We investigated an African-centric variant affecting codon 47 (P47S). Compared with those with the P47 variant, cells and mice expressing the serine 47 (S47) variant exhibit increased levels of coenzyme A (CoA) and glutathione (GSH). These molecules inhibit the induction of ferroptosis in S47 cells and mice. Elevated levels of CoA and GSH feedback on the p53 protein to negatively regulate the structure and function of this tumor suppressor protein. We identify CoA as a regulator of ferroptosis and show that the control of cell fate in response to certain ferroptosis inducers and p53 is reversible by altering the redox environment.
Keywords: p53, coenzyme A, ferroptosis, S-thiolation, polymorphism
Abstract
A population-restricted single-nucleotide coding region polymorphism (SNP) at codon 47 exists in the human TP53 gene (P47S, hereafter P47 and S47). In studies aimed at identifying functional differences between these variants, we found that the African-specific S47 variant associates with an impaired response to agents that induce the oxidative stress-dependent, nonapoptotic cell death process of ferroptosis. This phenotype is manifested as a greater resistance to glutamate-induced cytotoxicity in cultured cells as well as increased carbon tetrachloride-mediated liver damage in a mouse model. The differential ferroptotic responses associate with intracellular antioxidant differences between P47 and S47 cells, including elevated abundance of the low molecular weight thiols coenzyme A (CoA) and glutathione in S47 cells. Importantly, the disparate ferroptosis phenotypes related to the P47S polymorphism are reversible. Exogenous administration of CoA provides protection against ferroptosis in cultured mouse and human cells, as well as in a mouse model. The combined data support a positive role for p53 in ferroptosis and identify CoA as a regulator of this cell death process. Together, these findings provide mechanistic insight linking redox regulation of p53 to small molecule antioxidants and stress signaling pathways. They also identify potential therapeutic approaches to redox-related pathologies.
The p53 tumor suppressor protein is a critical stress signaling coordinator that plays a central role in defining the cellular response to many endogenous and exogenous stresses (1–3). As a sequence-specific transcription factor, p53 modulates the expression of a large and varied set of genes that promote the adaptation, survival, or elimination of targeted cells. The outcome is context dependent and influenced by cell type, nature, and degree of stress, as well as the local environment.
Human TP53 harbors naturally occurring SNPs that impact function (4, 5). A SNP affecting codon 47 of TP53 directs the insertion of either a proline (P47) or a serine (S47) in the protein (Pro47Ser, rs1800371, G/A). Interestingly, the presence of the S47 variant appears to be largely restricted to African-descent populations, at a minor allele frequency of ∼4% in Africans and 1% in African Americans (4, 6–8). Our prior studies showed that cells expressing the P47 and S47 isoforms generally exhibit a similar ability to induce apoptosis in response to most DNA-damaging agents and several chemotherapeutics. These isoforms also are comparable in the transactivation of the overwhelming majority of p53 target genes in response to such agents. Interestingly, however, we found that the S47 variant, in either heterozygous or homozygous form, associates with impaired induction of ferroptosis in mouse and human cells, as well as increased cancer risk, including increased susceptibility to breast cancer in premenopausal African Americans (7, 8).
The term ferroptosis describes the nonapoptotic, iron-dependent form of regulated cell death characterized by an accumulation of lethal lipid peroxides (9–11). Ferroptosis is triggered by a critical depletion of the antioxidant glutathione (GSH), or loss of lipid hydroperoxide repair systems. Oxytosis refers to nonreceptor-mediated oxidative glutamate toxicity, a process that also associates with GSH depletion. Because ferroptosis and oxytosis share key biochemical, genetic, and molecular features, they often are referred to together, and for simplicity both will be denoted herein as ferroptosis (9–12). The role of p53 in modulating the cellular response to ferroptosis-inducing agents has been controversial. On the one hand, several studies have found that p53 positively regulates ferroptosis (8, 13, 14). Conversely, others have reported that p53 inhibits, or delays the timing of, ferroptosis (15, 16). The underlying reason(s) for these conflicting results has been difficult to reconcile.
Here we find that S47 cells and mice exhibit an impaired response to several agents that induce ferroptosis, including exposure to excess glutamate or the hepatotoxin carbon tetrachloride (CCl4). These findings support a positive role for p53 in ferroptosis. We find that the different sensitivities of the P47 and S47 isoforms of p53 correlate with a greater intracellular abundance of coenzyme A (CoA) and GSH in the S47-containing cells. We find that redox changes feed back on p53 function by altering p53 oligomerization potential and transcriptional activity. The combined evidence supports CoA as a regulator of ferroptosis. It also shows that CoA and GSH act as mediators that set a threshold for ferroptotic sensitivity and for p53 activity. The regulation of CoA and GSH levels by p53, followed by the inhibitory feedback of these low molecular weight (LMW) thiols on p53 function, likely contribute to the conflicting results regarding the role of p53 in ferroptosis. These findings also suggest that these metabolites can reverse the cellular outcome to physiological induction of ferroptosis.
Results
Codon 47 Variants Differ in Ferroptosis Sensitivity.
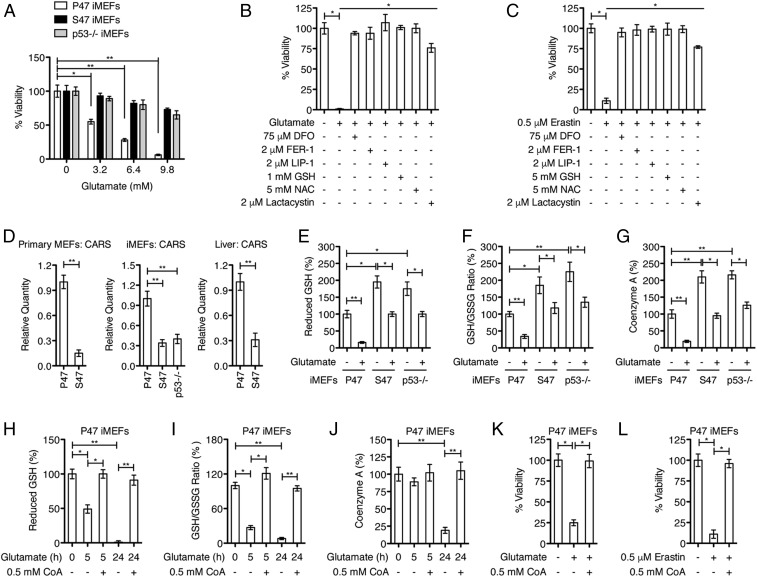
Exposure of cells to excess glutamate, as with the small molecule erastin, inhibits the uptake of cystine, leading to depletion of intracellular cysteine and GSH levels, and induction of ferroptosis (9–12). To study the sensitivity of S47 and P47 variants to ferroptosis, we analyzed the response of these cells to ferroptosis inducers in defined medium containing low serum and low glutamine. Under these conditions we found that, compared with cells with P47, those containing the S47 variant of p53 were resistant to cell death induced by increasing concentrations of glutamate (Fig. 1A). p53-null mouse embryonic fibroblasts (MEFs) (p53−/−) also were resistant (Fig. 1A). The greater resistance of S47 cells to glutamate was reproducible using multiple independent cultures of primary P47 and S47 MEFs, as well as several independently derived cultures of immortalized P47 and S47 MEFs (iMEFs), and human lymphoblastoid cells (LCLs) that are homozygous P47 or S47 (Fig. 1A and SI Appendix, Fig. S1 A and B). We next confirmed that the glutamate-induced death, like erastin killing, was mediated by ferroptosis (11, 12, 17), as they both were suppressed by cotreatment of the cells with recognized ferroptosis inhibitors deferoxamine (DFO), ferrostatin 1 (Fer-1), liproxstatin (Lip-1), and lactacystin, as well as by excess glutathione (GSH) and N-acetyl-l-cysteine (NAC) (Fig. 1 B and C).
Fig. 1.
Effect of codon 47 variants on sensitivity to glutamate toxicity and redox environment. (A) Cells, as indicated, were treated for 24 h with increasing concentrations of glutamate, and cell viability was assayed. Data represent the mean ± SD of multiple experiments (*P < 0.05, **P < 0.01). (B) P47 iMEFs were treated with PBS or 9.8 mM glutamate, with or without the indicated compounds, for 24 h, and analyzed for viability. Means and SD are shown (n = 4, *P < 0.05). (C) P47 iMEFs were treated with DMSO or 0.5 μM erastin, with or without the indicated compounds, for 24 h, and analyzed for viability. Means and SD are shown (n = 4, *P < 0.05). (D) qRT-PCR analyses of CARS in primary MEFs, iMEFs, and livers, as indicated. Relative expression data are plotted as the mean ± SD of multiple experiments (**P < 0.01). (E–G) iMEFs were treated with PBS or 6.4 mM glutamate for 24 h, as indicated. Cells were collected, washed with PBS, and analyzed for reduced GSH, GSH/GSSG ratio, and CoA abundance. Means and SD are shown (n = 4, *P < 0.05, **P < 0.01). (H–J) P47 iMEFs were treated with PBS, 9.8 mM glutamate, or 9.8 mM glutamate plus 0.5 mM CoA for 5 h or 24 h, as indicated. Intracellular levels of reduced GSH, GSH/GSSG ratio, and CoA abundance were determined. Means and SD are shown (n = 4, *P < 0.05, **P < 0.01). (K) P47 iMEFs were treated with PBS or 6.4 mM glutamate, with or without indicated 0.5 mM CoA, for 24 h, and analyzed for viability. Means and SD are shown (n = 4, *P < 0.05). (L) P47 iMEFs were treated with DMSO or 0.5 μM erastin, with or without indicated 0.5 mM CoA, for 24 h, and analyzed for viability. Means and SD are shown (n = 4, *P < 0.05).
Cellular Abundance of CoA and GSH Impacts Ferroptosis.
To investigate the underlying basis for the altered ferroptosis in S47 cells, we conducted metabolomics and transcriptional comparisons between P47 and S47 cells, including iMEFs and LCLs. An initial metabolomics profile revealed an increased abundance of several transsulfuration pathway metabolites in S47 versus P47 cells, including cystathionine, S-adenosylhomocysteine, and CoA (SI Appendix, Fig. S1C). The transsulfuration pathway promotes the de novo synthesis of cysteine (18, 19). These results likely are related to our transcriptional analyses showing that S47 cells exhibit much lower basal expression of cysteinyl-tRNA synthetase (CARS) and of the cystine/glutamate antiporter gene SLC7A11 (Fig. 1D and SI Appendix, Fig. S1D); genetic knockdown of CARS activates the transsulfuration pathway and confers resistance to erastin- and glutamate-induced ferroptosis (18, 19).
We followed this up with an analysis of the basal levels of the antioxidants GSH and CoA in P47 and S47 cells grown in the defined medium used for the glutamate toxicity assays. We found that S47 cells consistently displayed higher relative levels of GSH, a higher GSH/glutathione disulfide (GSSG) ratio, and increased CoA, compared with P47 cells (Fig. 1 E–G and SI Appendix, Fig. S1 E and F). While the differences in GSH and CoA between P47 and S47 cells were most consistent under the defined culture conditions, they also were present in cells cultured in 10% serum. Importantly, increased levels of CoA and GSH were evident in livers from P47 and S47 mice (SI Appendix, Fig. S1G). We next assessed the level of CoA and GSH in glutamate-treated cells. Not surprisingly we found that exposure of cells to excess glutamate, which is predicted to reduce cystine import, decreased GSH and CoA levels. Again, however, cells containing the S47 variant showed a dampened response and were more similar to p53-null cells, compared with P47 (Fig. 1 E–G). We reasoned that, if CoA abundance is an important factor in sensitivity to ferroptosis, then restoring CoA levels might be protective. We found that providing CoA in the culture medium, as reported (20), inhibited the glutamate-induced intracellular depletion of CoA, as well as of GSH (Fig. 1 H–J). Notably, CoA protected these nontransformed mouse cells against glutamate-induced cell death (Fig. 1K), cell death by erastin (Fig. 1L), as well as l-cystine starvation (SI Appendix, Fig. S2A). In extending these studies to human cells, we found that CoA also was protective against death induced by l-cystine depletion, exogenous glutamate, or erastin in a diverse group of nontransformed (IMR90 and WI38) and transformed cell lines (1205Lu, WM852, and HT-1080) (SI Appendix, Fig. S2 B–D). The combined data implicate CoA as a broad regulator of ferroptosis sensitivity.
Differential Activation of Ferroptosis Biomarkers.
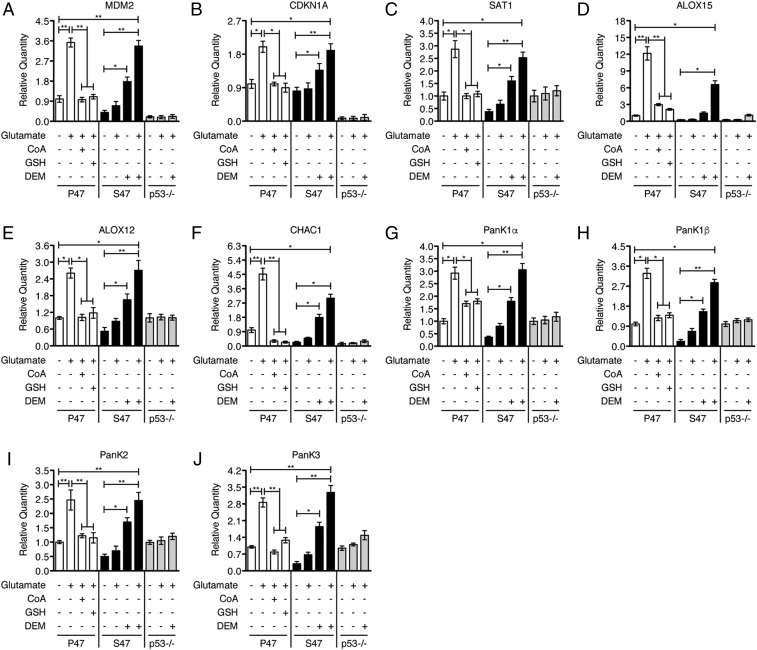
We next employed quantitative real-time PCR (qRT-PCR) analyses to examine gene expression changes in response to glutamate. This analysis included previously identified p53 target genes, including SAT1, CHAC1, three related PanK genes (PanK1, PanK2, and PanK3), MDM2 and CDKN1A (13, 14). SAT1 (spermidine/spermine N1-acetyltransferase 1) controls polyamine catabolism and oxidative stress responses (14). The three related PanK genes encode pantothenate kinases, which are enzymes that control the rate-limiting step in CoA biosynthesis (21–23). CHAC1 (cation transport regulator homolog 1) encodes a stress-response protein that digests glutathione, regulating redox potential and enhancing ferroptosis (13, 24). In addition to these p53 target genes, we examined the well-recognized ferroptosis biomarkers ALOX15 and ALOX12, which encode proteins that catalyze the peroxidation of polyunsaturated fatty acids to drive ferroptosis (11, 25, 26). Interestingly, ALOX15 has been identified as a downstream effector of p53-induced SAT1 in promoting ferroptosis (14). Transcripts from untreated controls, and from cells exposed to glutamate for 5 h, were tested. We found that these p53 target genes and ferroptosis biomarkers were transcriptionally up-regulated in glutamate-treated P47 cells. In contrast, this set of transcripts showed modest, if any, change in S47 and p53-null cells (Fig. 2). Of note, we found that the induction of these genes in glutamate-treated S47 cells could be restored by cotreating the cells with diethyl maleate (DEM) (Fig. 2), which is a small molecule that reduces the intracellular level of GSH and CoA (SI Appendix, Fig. S3). The results suggested that the increased GSH and CoA in S47 cells might be impeding the ability of the S47 variant of p53 to transactivate these genes. Conversely, as expected, providing DEM together with glutamate had no effect on the induction of these p53 target genes in p53-null cells (Fig. 2). Using Western blot analysis, we confirmed that the expression of PANK protein isoforms correlated well with the qRT-PCR results under these experimental conditions (SI Appendix, Fig. S4). The combined data support the premise that S47 cells are defective in ferroptosis induction, in part due to increased levels of GSH and CoA. These data also suggest that the ability of p53 to transactivate target genes was negatively impacted by excess LMW thiols, but that reducing the level of GSH and CoA in S47 cells could restore this activity.
Fig. 2.
Effect of GSH or CoA change on gene expression in response to glutamate. (A–J) qRT-PCR analyses of the indicated genes in P47, S47, and p53-null (p53−/−) iMEFs. P47 iMEFs were pretreated with 0.5 mM CoA or 1 mM GSH for 2 h followed by 6.4 mM glutamate for 5 h. S47 and p53-null (p53−/−) iMEFs were treated with 6.4 mM glutamate, 50 μM DEM, or 6.4 mM glutamate + 50 μM DEM, as indicated, for 5 h. P47, S47, and p53−/− iMEFs treated with PBS served as controls. Relative expression data are plotted as the mean ± SD (n = 3, *P < 0.05, **P < 0.01).
Redox Change and Reversible Regulation of p53.
It remained formally possible that S47 cells were defective in the transactivation of genes like PanK1-3 and SAT1 in response to all stresses, not simply ferroptotic stresses. To assess this, we next analyzed the response of P47 and S47 iMEFs to Nutlin-3, which stabilizes p53 by blocking MDM2 binding (27). Cells were treated with or without Nutlin-3 (10 μM) in our defined media for 24 h and gene expression changes were analyzed by qRT-PCR. We found that SAT1, ALOX12/15, PanK1-3, MDM2, and CDKN1A were transactivated comparably in Nutlin-treated P47 and S47 cells; as expected, these genes were not up-regulated in p53-null MEFs (SI Appendix, Fig. S5 A–I). Therefore, the S47 defect in transactivation of these p53 targets occurred only in response to certain ferroptosis inducers. Given these results, we asked if altering the redox state, by enhancing antioxidant concentration, would negatively impact the gene expression pattern in P47 cells. Consistent with this premise, we found that cotreatment of the P47 cells with glutamate and either CoA or GSH inhibited the transcriptional activation of these genes (Fig. 2). Thus, the transcriptional response of P47 and S47 variants to particular ferroptosis inducers can be reversed by altering the level of CoA and/or GSH.
Cellular Redox Milieu Alters p53 Oligomerization and Function.
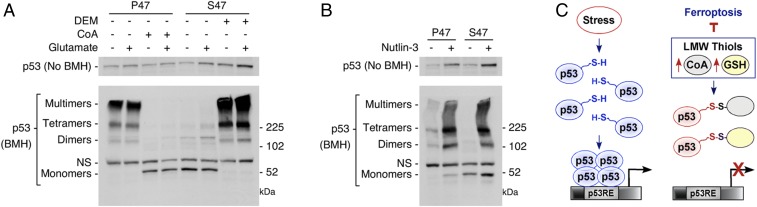
Previous in vitro and in vivo studies have shown that p53 is subject to redox regulation (28–31). Conserved cysteine residues at or near the DNA-binding domain are among critical redox targets that impact oligomerizaton status and function. The presence of reduced cysteine residues allows tetramerization and functional activation of the p53 protein. In contrast, oxidized cysteines can form mixed disulfides with GSH and other thiols, which interferes with tetramerization. Given these data, we tested the hypothesis that p53 oligomerization and/or transcriptional function could be impacted by the different redox environment of P47 and S47 cells. For these studies, p53 oligomerization and higher order structure were monitored using the chemical cross-linker bismaleimidohexane (BMH), which covalently conjugates free (reduced) sulfhydryl groups, linking cysteine residues within 13 Å. BMH treatment generated clear, consistent differences in the p53 cross-linking patterns. In untreated- or glutamate-treated P47 cells, the predominant p53 products migrated near or above the 225-kDa marker; this is most consistent with presence of tetramers and higher molecular weight oligomers (Fig. 3A). In contrast, in the untreated- or glutamate-exposed S47 cells, the p53 products appear to be predominantly monomeric, and there is little evidence for species of the molecular weight of dimers or tetramers (Fig. 3A).
Fig. 3.
Effect of GSH and CoA modulation on p53 oligomerization potential. (A) P47 and S47 iMEFs were treated with PBS, 6.4 mM glutamate, 0.5 mM CoA, 6.4 mM glutamate + 0.5 mM CoA, 50 μM DEM, or 6.4 mM glutamate + 50 μM DEM, as indicated, for 5 h. Protein lysates were analyzed by Western blot analysis for the proteins indicated (Top). p53 proteins were cross-linked with BMH, resolved by SDS/PAGE, and detected by Western blotting with a p53-specific antibody (Lower). (NS, nonspecific band). (B) P47 and S47 iMEFs were treated with DMSO or 10 μM Nutlin-3 for 24 h. Proteins were prepared and analyzed as described in A. (C) Proposed model for the influence of LMW thiols, such as GSH and CoA, on p53 tetramerization and function.
We next tested the potential to modify p53 oligomerization by changing the redox environment. P47 cells were incubated with CoA alone, or CoA together with glutamate. BMH cross-linking of lysates from the CoA-treated cells produced a pattern comparable to that of untreated S47 cells; namely, we observed what appeared to be primarily monomeric forms of p53 (Fig. 3A). In contrast, in S47 cells, treatment with DEM or DEM plus glutamate now allowed formation of higher molecular weight complexes, including what appeared to be dimeric, tetrameric, and oligomeric species (Fig. 3A). Notably, there was no difference in p53 higher order structure following treatment of the S47 and P47 cells with Nutlin-3 (Fig. 3B). The combined data support a model in which a reversible redox state in the S47 and P47 cells directly impacts p53 transcriptional function, likely by impacting protein conformation and structure (Fig. 3C).
Redox Status Modulates Outcomes in a Mouse Model of Liver Injury.
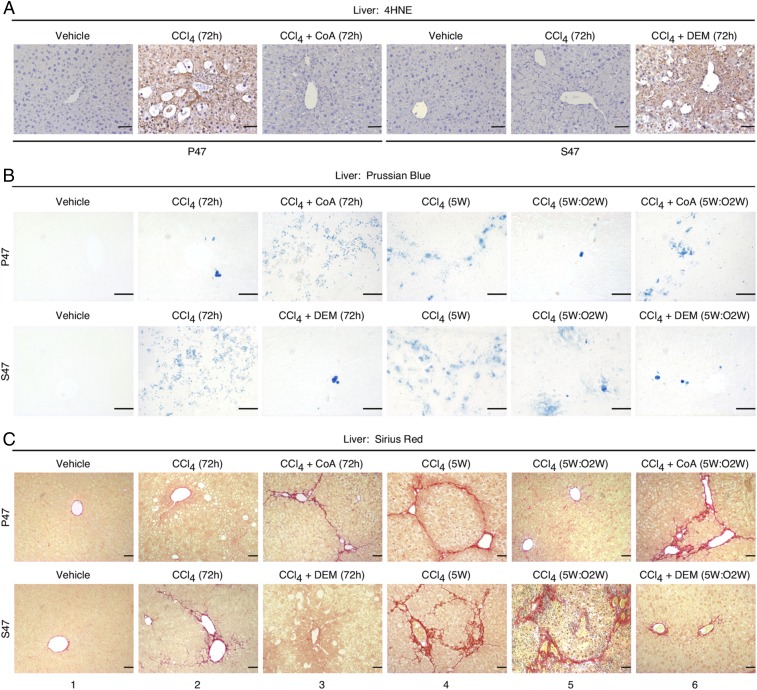
We next sought to extend this study by analyzing a more physiologically relevant setting of ferroptosis in mice. For these studies, we employed a well-established model of liver injury induced by the hepatotoxin CCl4. CCl4 induces liver damage that displays key features of ferroptosis, including increased lipid peroxidation, GSH depletion, and SAT1 induction (32); notably these features are inhibited by prior- or coadministration of the ferroptosis inhibitors DFO, GSH, or NAC (33, 34). We subjected P47 and S47 mice (8), 8 wk of age, to acute CCl4 treatment by i.p. injection of CCl4 (1 μL/g), once a day for 3 d. In P47 but not S47 livers, this treatment led to the transcriptional up-regulation of the ferroptosis biomarkers ALOX12/15 as well as the p53 target genes SAT1, PanK1-3, MDM2, CDKN1A, CHAC1, and PTGS2 (a marker induced downstream of ferroptosis-induced lipid peroxidation) (11, 13) (SI Appendix, Fig. S6 A–C). This transcriptional response was accompanied by an increase in lipid peroxidation in P47 livers, which was detected by immunostaining with antibody against the toxic reactive lipid intermediate, 4-hydroxynonenal (4HNE) (Fig. 4A). In contrast, the S47 livers exhibited decreased lipid peroxidation (Fig. 4A), consistent with impaired ferroptosis in these livers.
Fig. 4.
Effect of P47S polymorphism and redox environment on liver response to CCl4. (A) P47 and S47 Hupki mice were treated, as indicated. Liver sections from the proposed cohorts (n = 7 per genotype per treatment) were stained with 4HNE. The brown staining indicates positive 4HNE staining. (Scale bars, 50 μM.) (B and C) Representative Prussian Blue (B)- and Sirius Red (C)-stained liver sections from P47 and S47 mice that were treated as indicated with one of the following: Three injections of CCl4 (once a day for 3 d), with or without cotreatment with DEM or CoA, as indicated, and examined at 72 h; challenged with a chronic CCl4 treatment for 5 wk (5W); or chronic CCl4 treatment, with or without cotreatment with DEM or CoA, as indicated, for 5 wk followed by a 2-wk recovery (5W:O2W). (Scale bars, 50 μM.)
CCl4 is known to induce iron accumulation and fibrosis in the liver (35–37). Therefore, we next stained for iron in P47 and S47 livers treated with CCl4. CCl4 induction led to increased iron accumulation in S47 livers (Fig. 4B); this increased iron accumulation may be due to the defect in ferroptosis in these cells, which would normally remove cells with iron accumulation. We also stained livers with Sirius Red, which detects collagen and is commonly used to assess fibrosis. Following acute treatment with CCl4, P47 livers showed little evidence for fibrosis (Fig. 4C, Top, panel 2). In contrast, fibrosis was clearly evident in the S47 livers, especially around portal areas and central veins (Fig. 4C, Bottom, panel 2).
We next investigated the progression and resolution of fibrosis by subjecting the P47 and S47 mice to chronic CCl4 treatment for 5 wk, as previously described (35, 36). The livers were harvested either just after the last injection (CCl4:5W) or 2 wk after the last CCl4 treatment (CCl4:5W:O2W). In response to this chronic treatment protocol, the development of fibrosis was evident in the P47 mice (CCl4:5W) (Fig. 4C, Top, panel 4), but the fibrosis largely resolved following the 2-wk recovery period (CCl4:5W:O2W) (Fig. 4C, Top, panel 5). In contrast, livers of S47 mice displayed a greater degree of initial fibrosis (CCl4:5W) (Fig. 4C, Bottom, panel 4) and a notable impairment of fibrosis resolution (CCl4:5W:O2W) (Fig. 4C, Bottom, panel 5). This phenotype of the S47 livers resembles that reported for CCl4-treated p53-null mice (35, 36). These findings support the premise that S47 mice are defective for ferroptosis induction by CCl4 and suggest that this ferroptotic defect leads to iron accumulation and fibrosis.
Given these results, we then asked whether changing GSH and CoA abundance would act as response mediators following acute exposure to CCl4. The P47 mice were given CoA along with CCl4; this markedly reduced the induction of ferroptosis biomarkers and increased lipid peroxidation in livers (Fig. 4A and SI Appendix, Fig. S6 A–C). This altered phenotype in the P47 mice closely resembled that of the S47 mice treated with toxin alone (Fig. 4C, Bottom, panels 2 and 5). On the other hand, treating S47 mice with the toxin plus DEM, to reduce GSH and CoA, resulted in the expected decrease in tissue CoA, GSH, and GSH/GSSG abundance (SI Appendix, Fig. S6D). Concomitantly, this cotreatment led to a transcriptional up-regulation of ferroptosis/p53 marker genes (SI Appendix, Fig. S6 A–C), an increase in tissue 4HNE staining (Fig. 4A), and less fibrosis (Fig. 4C, Bottom, panels 3 and 6); overall, this phenotype closely resembled that of the CCl4-treated P47 mice (Fig. 4C, Top, panels 2 and 5). The data show that changing the tissue redox status and CoA/GSH abundance can effectively reverse the fate of cells and the ability of p53 to transactivate genes involved in ferroptosis.
Discussion
The role of p53 in ferroptosis has been controversial. Early findings indicated that an acetylation-defective variant of p53, denoted p53-3KR, was largely transcriptionally inactive and incapable of inducing growth arrest and apoptosis, yet was competent at ferroptosis induction and tumor suppression in an unstressed mouse model (13, 38). Some subsequent studies reported contradictory results related to p53 (15, 16). For example, p53-null tumor cells were found to be either more sensitive, or more resistant, to ferroptosis than the WT counterparts. Other studies suggested that p53 actions may delay the onset of ferroptosis, suggesting context dependence in regulating the role of p53 in ferroptosis.
Key differences exist in these studies that may help explain the disparate findings. First, some of these studies were performed in cells cultured in 10% serum, so the concentrations of thiols are nonphysiological. Most of these studies were performed exclusively in vitro and were not corroborated in animal models. In this work, we have tried to maintain more relevant concentrations of glutathione, and under these conditions the differences in ferroptosis resistance in S47 cells are most dramatic. Additionally, we show compelling data that CCl4, which has been reported to induce iron accumulation, cell death by lipid peroxidation, and GSH depletion (37), is a bona fide inducer of ferroptosis in the liver. Moreover, we confirm the defect in ferroptosis in S47 livers that was seen in cultured cells. Our combined data demonstrate the important role of p53 in regulating the ferroptotic response.
While its normal physiologic functions remain to be established, the regulated cell death pathway of ferroptosis has been implicated in a diverse array of human diseases, including tissue ischemia/reperfusion injury, neurodegenerative diseases, and cancers. Accumulating evidence supports the premise that ferroptosis is tumor suppressive under diverse metabolic stress conditions (13, 38–41). It is reasonable to hypothesize, therefore, that the p53-regulated response to glutamate-induced ferroptosis prevents the accumulation of damaged cells, contributing to its tumor suppressor actions. Previous studies, for example, have provided evidence for a functional contribution of p53 in the glutamate-induced cell death of neuronal cells. Terminally differentiated neurons, if forced by oncogenic mutations to reenter the cell cycle, may be predisposed to undergo programmed cell death, which is likely to be tumor suppressive for these normally noncycling cells (42). p53 activation promotes glutamate-mediated cytotoxicity in normal cells and transformed cells (43, 44), while p53 deficiency protects neurons from cell death (45). Addressing the physiologic functions and regulatory mechanisms of p53-mediated ferroptosis in diverse biological/pathologic settings will be important for ongoing efforts to develop therapeutic approaches to treating human disease.
We also report that the LMW thiol and essential cofactor, CoA, is a regulator of ferroptosis sensitivity. Interestingly, the first and rate-limiting step in CoA biosynthesis is carried out by pantothenate kinases, encoded by PanKs, which are p53 target genes (13, 21, 23). The p53-mediated up-regulation of PanK enzymes might be expected to help restore CoA levels and help maintain viability under conditions of moderate or temporary metabolic or oxidative stress. It is important to note, however, that in addition to transcriptional regulation by p53, PanK enzyme activity is subject to feedback inhibition by elevated levels of CoA and its thioesters, so several feedback loops are present in this process (22, 46).
The role of CoA in ferroptosis sensitivity remains to be further determined. This thiol can act as a cellular antioxidant (47, 48). Additionally, CoA is a key regulator of lipid metabolism and has been reported to protect against the injurious effects of reactive oxygen species to inhibit lipid peroxidation, to enhance GSH levels, and to stimulate the incorporation of fatty acids into membrane phospholipids, promoting repair (49, 50). Finally, it remains possible that CoA contributes to an increased abundance of acetyl-CoA, affecting epigenetic modulation and gene expression control (51). We are testing these possibilities, which are not mutually exclusive, at this time.
We find that excess LMW thiols feed back and inhibit p53 function, thus dampening the impact of p53 on the control of ferroptosis. The mechanism of feedback inhibition involves control of p53 oligomerization. Previous studies of p53 have implicated cysteine residues in protein conformation and site-specific DNA binding via redox regulation (28–31). The combined results are most consistent with a model where, under situations in which LMW thiols are limiting, p53 can be functionally activated, rendering cells sensitive to type I ferroptosis inducers. However, under conditions in which LMW thiols are present in high abundance, these can feed back to inhibit p53 transcriptional function, leading to reduced induction of p53 target genes and loss of the impact of p53 on ferroptosis sensitivity, such as occurs in S47 cells. It is important to note that there are several regulators of GSH and CoA abundance in cells, and these may dominate in certain tumors. For example, the activity of the master regulator of the antioxidant response, Nrf2, can be impacted by mutant forms of p53, thereby rendering some tumor cells with mutant p53 more sensitive to ferroptotic stresses (52). It will be important to determine the dominance and interplay of these other regulators as well and to continue to confirm results in animal models where possible.
Materials and Methods
All experimental procedures involving mice were conducted in accordance with protocols approved by The Institutional Animal Care & Use Committee, Office of Animal Welfare of the Perelman School of Medicine at the University of Pennsylvania, and conformed to the guidelines outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals (53). Materials and experimental procedures for cell culture, animal models, RNA isolation, qRT-PCR, cell viability assays, GSH/GSSG studies, CoA analyses, tissue staining and immunohistochemistry, protein isolation, Western blotting, and quantification and statistical analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by research grants from the National Institutes of Health (NIH R01 CA139319 to M.E.M., D.L.G., and J.I.-J.L.; R01 CA102184 to M.E.M.; and P01 CA114046 to M.E.M., D.L.G., and J.I.-J.L.). The Molecular Pathology & Imaging Core at the Perelman School of Medicine, University of Pennsylvania, is funded by research grants from the NIH (NIH P30 DK050306, P01 CA098101, and P01 DK049210). The Proteomics and Metabolomics Facility at The Wistar Institute is funded by Cancer Center Support Grant CA010815. We apologize to the many investigators whose work could not be cited because of space constraints.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821277116/-/DCSupplemental.
References
- 1.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 3.Pfister NT, Prives C. Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb Perspect Med. 2017;7:a026054. doi: 10.1101/cshperspect.a026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu S, Barnoud T, Kung CP, Reiss M, Murphy ME. The African-specific S47 polymorphism of p53 alters chemosensitivity. Cell Cycle. 2016;15:2557–2560. doi: 10.1080/15384101.2016.1215390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: Cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem. 2005;280:24245–24251. doi: 10.1074/jbc.M414637200. [DOI] [PubMed] [Google Scholar]
- 7.Murphy ME, et al. A functionally significant SNP in TP53 and breast cancer risk in African-American women. NPJ Breast Cancer. 2017;3:5. doi: 10.1038/s41523-017-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jennis M, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30:918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–143. doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stockwell BR, et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewerenz J, Ates G, Methner A, Conrad M, Maher P. Oxytosis/ferroptosis-(Re-) emerging roles for oxidative stress-dependent non-apoptotic cell death in diseases of the central nervous system. Front Neurosci. 2018;12:214. doi: 10.3389/fnins.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci USA. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 16.Tarangelo A, et al. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 2018;22:569–575. doi: 10.1016/j.celrep.2017.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maher P. Proteasome inhibitors prevent oxidative stress-induced nerve cell death by a novel mechanism. Biochem Pharmacol. 2008;75:1994–2006. doi: 10.1016/j.bcp.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23:270–278. doi: 10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimada K, Stockwell BR. tRNA synthase suppression activates de novo cysteine synthesis to compensate for cystine and glutathione deprivation during ferroptosis. Mol Cell Oncol. 2015;3:e1091059. doi: 10.1080/23723556.2015.1091059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan B, et al. Extracellular 4′-phosphopantetheine is a source for intracellular coenzyme A synthesis. Nat Chem Biol. 2015;11:784–792. doi: 10.1038/nchembio.1906. [DOI] [PubMed] [Google Scholar]
- 21.Chang GS, et al. A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep. 2014;8:514–527. doi: 10.1016/j.celrep.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leonardi R, et al. Modulation of pantothenate kinase 3 activity by small molecules that interact with the substrate/allosteric regulatory domain. Chem Biol. 2010;17:892–902. doi: 10.1016/j.chembiol.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SJ, et al. p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle. 2013;12:753–761. doi: 10.4161/cc.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen MS, et al. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2α-ATF4 pathway. Oncotarget. 2017;8:114588–114602. doi: 10.18632/oncotarget.23055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagan VE, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang WS, et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci USA. 2016;113:E4966–E4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 28.Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 29.Sun XZ, et al. Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxid Redox Signal. 2003;5:655–665. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- 30.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 32.Zahedi K, et al. Hepatocyte-specific ablation of spermine/spermidine-N1-acetyltransferase gene reduces the severity of CCl4-induced acute liver injury. Am J Physiol Gastrointest Liver Physiol. 2012;303:G546–G560. doi: 10.1152/ajpgi.00431.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Z, et al. N-acetylcysteine protects against liver injure induced by carbon tetrachloride via activation of the Nrf2/HO-1 pathway. Int J Clin Exp Pathol. 2015;8:8655–8662. [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammed A, Abd Al Haleem EN, El-Bakly WM, El-Demerdash E. Deferoxamine alleviates liver fibrosis induced by CCl4 in rats. Clin Exp Pharmacol Physiol. 2016;43:760–768. doi: 10.1111/1440-1681.12591. [DOI] [PubMed] [Google Scholar]
- 35.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagor AT, et al. Supplementation of fresh ucche (Momordica charantia L. var. muricata Willd) prevented oxidative stress, fibrosis and hepatic damage in CCl4 treated rats. BMC Complement Altern Med. 2015;15:115. doi: 10.1186/s12906-015-0636-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang SJ, et al. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao M, Jiang X. To eat or not to eat-the metabolic flavor of ferroptosis. Curr Opin Cell Biol. 2018;51:58–64. doi: 10.1016/j.ceb.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao M, et al. Role of mitochondria in ferroptosis. Mol Cell. 2019;73:354–363.e3. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heintz N. Cell death and the cell cycle: A relationship between transformation and neurodegeneration? Trends Biochem Sci. 1993;18:157–159. doi: 10.1016/0968-0004(93)90103-t. [DOI] [PubMed] [Google Scholar]
- 43.Choi HJ, Kang KS, Fukui M, Zhu BT. Critical role of the JNK-p53-GADD45α apoptotic cascade in mediating oxidative cytotoxicity in hippocampal neurons. Br J Pharmacol. 2011;162:175–192. doi: 10.1111/j.1476-5381.2010.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grilli M, Memo M. Possible role of NF-kappaB and p53 in the glutamate-induced pro-apoptotic neuronal pathway. Cell Death Differ. 1999;6:22–27. doi: 10.1038/sj.cdd.4400463. [DOI] [PubMed] [Google Scholar]
- 45.Morrison RS, et al. Loss of the p53 tumor suppressor gene protects neurons from kainate-induced cell death. J Neurosci. 1996;16:1337–1345. doi: 10.1523/JNEUROSCI.16-04-01337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackowski S, Leonardi R. Deregulated coenzyme A, loss of metabolic flexibility and diabetes. Biochem Soc Trans. 2014;42:1118–1122. doi: 10.1042/BST20140156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchiya Y, et al. Protein CoAlation: A redox-regulated protein modification by coenzyme A in mammalian cells. Biochem J. 2017;474:2489–2508. doi: 10.1042/BCJ20170129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuchiya Y, et al. Protein CoAlation and antioxidant function of coenzyme A in prokaryotic cells. Biochem J. 2018;475:1909–1937. doi: 10.1042/BCJ20180043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen YQ, Zhao SP, Zhao YH. Efficacy and tolerability of coenzyme A vs pantethine for the treatment of patients with hyperlipidemia: A randomized, double-blind, multicenter study. J Clin Lipidol. 2015;9:692–697. doi: 10.1016/j.jacl.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Evans M, et al. Pantethine, a derivative of vitamin B5, favorably alters total, LDL and non-HDL cholesterol in low to moderate cardiovascular risk subjects eligible for statin therapy: A triple-blinded placebo and diet-controlled investigation. Vasc Health Risk Manag. 2014;10:89–100. doi: 10.2147/VHRM.S57116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 52.Liu DS, et al. Inhibiting the system xC-/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun. 2017;8:14844. doi: 10.1038/ncomms14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Institutes of Health 2011. Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC), 8th Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.