Abstract
Introduction:
Multiple genetic mechanisms have been identified in EGFR-mutant lung cancers as mediators of acquired resistance (AR) to EGFR tyrosine kinase inhibitors (TKI) but many cases still lack a known mechanism.
Methods:
To identify novel mechanisms of AR, we performed targeted large panel sequencing on 374 consecutive patients with metastatic EGFR-mutant lung cancer, including 174 post-TKI samples of which 38 patients also had a matched pre-TKI sample. Alterations hypothesized to confer AR were introduced into drug-sensitive EGFR-mutant lung cancer cell lines (H1975, HCC827, PC9) using CRISPR-Cas9 genome editing. MSK-LX138cl, a cell line with EGFR ex19del and PJA2/BRAF fusion, was generated from an EGFR TKI-resistant patient sample.
Results:
We identified 4 patients (2.3%) with a BRAF fusion (3 AGK/BRAF, 1 PJA2/BRAF) in samples obtained at AR to EGFR TKI (2 post-erlotinib; 2 post-erlotinib and post-osimertinib). Pre-TKI samples were available in 2/4 patients and both were negative for BRAF fusion. Induction of AGK/BRAF fusion in H1975 (L858R+T790M), PC9 (ex19del) and HCC827 (ex19del) cells increased phosphorylation of BRAF, MEK1/2, ERK1/2 and STAT3, and conferred resistance to growth inhibition by osimertinib. MEK inhibition with trametinib synergized with osimertinib to block growth. Alternately, a pan-RAF inhibitor as a single agent blocked growth of all cell lines with mutant EGFR and BRAF fusion.
Conclusion:
BRAF fusion is a mechanism of AR to EGFR TKI in approximately 2% of patients. Combined inhibition of EGFR and MEK (with osimertinib and trametinib) or BRAF (with a pan-RAF inhibitor) are potential therapeutic strategies that should be explored.
Keywords: Lung adenocarcinoma, EGFR, BRAF fusion, acquired resistance, osimertinib, CRISPR-Cas9
INTRODUCTION
Long term clinical benefits of EGFR tyrosine kinase inhibitors (EGFR TKI) in EGFR mutant non-small cell lung cancers remain limited by the development of drug resistance [1]. The most common mechanism of resistance to first and second generation EGFR TKIs, the EGFR T790M mutation (60%), can be targeted successfully with third generation drugs such as osimertinib [2]. Recently, osimertinib has been approved as first line therapy for EGFR mutant lung cancer [3]. As expected, disease progression due to acquired resistance to osimertinib has emerged, with the most common alteration described to date being the EGFR C797S mutation [4]. Activation of parallel signaling pathways (amplification of HER2 and MET) or of downstream signaling (mutations in KRAS, PIK3CA, and BRAF) has also been shown to induce resistance to osimertinib and other EGFR TKIs [5]. Several acquired gene rearrangements, including fusions involving ALK, BRAF, FGFR3, NTRK1 and RET have recently been reported as possible mechanisms of resistance to EGFR TKI [6–11]. Clinically, a combination of an EGFR TKI with a drug targeting either ALK or RET, has been shown to be effective in lung cancer patients with an EGFR mutation and the respective gene fusion. However, only RET or FGFR3 fusions have been experimentally proven to confer resistance to EGFR inhibitors in EGFR-mutant lung cancers [7, 12, 13].
Activating BRAF fusions typically occur at a very low frequency across a wide variety of cancers with the exception of pilocytic astrocytomas [9, 14]. BRAF rearrangements are divided into N-terminal deletions (NTD), kinase domain duplications (KDD) and BRAF fusions. Overall, BRAF alterations are present in 4.4 % NSCLC, and rearrangements represent 4.3 % of all alterations [10]. Rearrangements retain the BRAF kinase domain at the 3’ end, while lacking the N-terminal inhibitory domain. Due to the lack of the N-terminal BRAF inhibitory domain, rearrangements result in constitutive dimerization of RAF proteins independently of RAS activation [15], thereby activating downstream MAP kinase signaling.
Here, we report 4 cases of EGFR mutant NSCLC with concurrent expression of a BRAF fusion and provide functional data supporting BRAF fusions as a mechanism of acquired resistance to EGFR therapy in EGFR mutant lung adenocarcinomas. In addition, we present data supporting treatment strategies focusing on targeting RAF with pan-RAF inhibitors.
METHODS
Generation of AGK/BRAF fusion using CRISPR/Cas9 in EGFR mutant lung cancer cell lines
gRNA design and cloning.
In order to faithfully model a BRAF fusion, gRNAs were designed to generate a fusion linking AGK exons 1–2 with BRAF exons 8 – 18. Specifically, four guide RNAs (gRNAs) were designed (Supplemental table 1) to target the intron between AGK exons 2 and 3 or BRAF exons 7 and 8 and selected using http://crispr.mit.edu/. The placement of these gRNAs aimed to minimize splicing interference by being positioned at least 50 bp-250 bp from the splice site. gRNA cloning was performed according to previously published protocols [16]. 1 μg px458 (containing Cas9 and GFP cDNAs) was digested with 1 μl FastDigest Bpil, 1 μl fast AP, 2 μl 10× FastDigest buffer diluted in 6 μl water at a final volume of 20 μl at 37°C for 45 min. The resulting DNA was cleaned up with PCR clean up kit to a final concentration of 15–30 ng/μl. 1 μl of each pair of gRNA was then phosphorylated and annealed with 1 μl 10× T4 ligation buffer, 0.5 μl T4 polynucleotide kinase and 6.5 μl RNAse/DNAse-free water to a final volume of 10 μl, in a Biorad C1000 Touch thermocycler for 30 min at 37°C, then decrements in temperature from 95°C to 25°C in 5 min intervals. Annealed gRNAs were ligated into px458 using 50 ng of vector, 1.5 μl of annealed gRNAs (1:100 dilution), 5 μl of quick ligase buffer, 1 μl quick ligase with addition of RNAse/DNAse-free water to a final volume of 10 μl, at room temperature for 30 min. The annealed and ligated plasmids were then transformed using TOP10 chemically competent E. coli. The plasmid DNA was extracted and insertion of gRNAs in px458 was confirmed by Sanger sequencing.
Validation of gRNAs.
To test the efficacy of fusion generation, human bronchoepithelial cells (HBEC) (plated at a density of 500,000 cells/well in 6-well plates) were transfected with each possible pair of gRNAs with 1 μg of each CRISPR/Ca9 construct. After 48 h, GFP was readily visible, and after 72 h each pool of cells was harvested for RNA extraction, cDNA synthesis and RT-PCR to detect the fusion mRNA. The pool of cells containing BRAF gRNA #4 and AGK gRNA #4 generated detectable fusion mRNA. This pairing (BRAF gRNA #4 and AGK gRNA #4) was transfected into H1975, HCC827 and PC9 cells plated at a density of 1,500,000 cells/10 cm dish with 7.5 μg of each plasmid and fusion mRNA detected as described above.
Clonal selection.
Transfected cells were maintained in growth media supplemented with 0.5 μM osimertinib to select for a drug-resistant population. The resistant cells were then replated at a density of 500 cells/10 cm dish to isolate single-cell clones. Colonies were then picked with cloning discs and replated in a 96-well plate for expansion in RPMI containing 10% FBS, 1% antibiotic/antimycotic and 0.5 μM osimertinib. Once clones began to proliferate readily, cells were serially moved to larger containers and then subjected to RT-PCR for fusion detection.
Generation of BRAF fusion constructs.
The cDNA encoding PJA2/BRAF fusion was amplified from MSK-LX138cl cells by PCR and cloned into pENTR/TOPO vector. The cDNA encoding AGK/BRAF fusion was generated by first amplifying the AGK and BRAF fragments from HEK-293T cells and then assembling the two pieces into Kpn I -EcoRV-digested pENTR1A vector by Gibson assembly. These BRAF fusion cDNAs were then subcloned into pLX303 lentiviral transfer vector. The primer sequences used for plasmid construction are shown in Supplemental Table 2. All constructs were confirmed by DNA sequencing. Lentiviruses harboring the two fusions were generated using HEK-293T cells and then used to infect cells the lung cancer lines. Cells stably expressing the cDNAs were selected with blasticidin (20 μg/mL) for 10 days.
shRNA infection and BRAF knockdown.
shRNAs (TRCN0000195066 – Sh2, TRCN0000195609 – Sh3 and TRCN0000196844 – Sh1) were obtained from Sigma-Aldrich (St. Louis, MO). Lentiviruses were generated using HEK-293T cells and were used to infect cells. Cells were plated in 6-well plates at a density of 150,000–500,000 cells/well and then infected 24 h later with viral supernatant (MOI=7) expressing BRAF shRNAs or non-targeting sequence, mixed with 10 μg/mL polybrene. Infected cells were selected with 5–10 μg/mL puromycin then replated for viability testing with osimertinib or used to detect BRAF by RT-qPCR.
RESULTS
Co-expression of EGFR mutation and BRAF fusions in lung cancer samples refractory to EGFR TKI.
We performed targeted large panel sequencing using the MSK-IMPACT assay [14, 17] on 374 consecutive patients with metastatic EGFR mutant lung cancers, including 200 tested prior to receiving an EGFR TKI, 136 tested after progression on an EGFR TKI, and 38 patients with both types of samples. We identified four patients (4/174; 2.3%) with disease progression on EGFR TKI (two on erlotinib; two on erlotinib, followed by osimertinib) with an EGFR mutation and a BRAF fusion (3 AGK/BRAF and 1 PJA2/BRAF). In one patient for which paired pre- and post TKI samples were available, an AGK/BRAF fusion was present only after progression on erlotinib supporting the hypothesis that BRAF fusions are possible acquired mechanisms of drug resistance. The other 3 EGFR TKI-resistant patient samples lacked pre-EGFR TKI data from the same site sample, however one patient had pre-EGFR TKI MSK-IMPACT testing on a lung biopsy and a BRAF fusion on a pleural effusion (Supplemental table 5). Importantly, none of 200 patients whose tumor sample was obtained prior to EGFR TKI treatment showed a concurrent BRAF fusion.
Clinical course of two patients with matched pre- and post-TKI samples
Case 1.
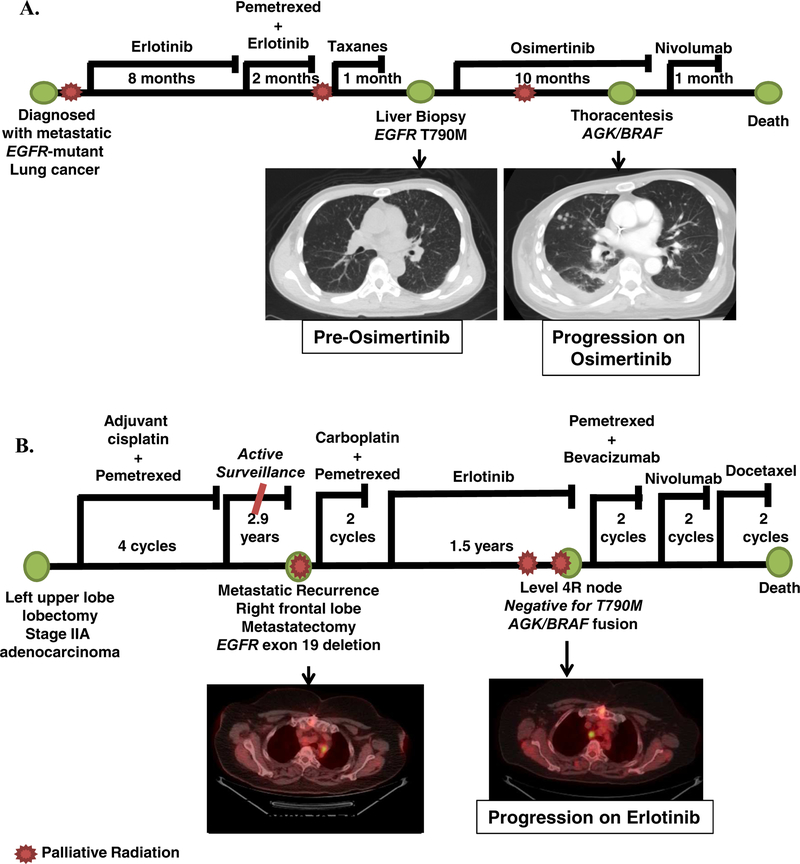
A 48-year-old man, never smoker, presented with a right cranial nerve 12 palsy and was found to have metastatic lung adenocarcinoma to the brain, liver, bone, and lymphangitic spread in the lungs (patient 2, Supplemental table 4) (Figure 1). Evaluation of the right upper lobe biopsy sample by PCR revealed a 15-base pair EGFR exon 19 deletion. The patient received whole brain radiation therapy followed by single agent erlotinib 150 mg daily. A complete depiction of the treatment course and diagnosis is shown in Figure 1A. The patient initially had a robust clinical and radiologic response but after 7.7 months of treatment was noted to have progressive disease in the bone and subsequently liver. A biopsy was performed on the liver and MSK-IMPACT [17] testing revealed a newly acquired EGFR T790M mutation. The patient was subsequently given osimertinib 80 mg daily with initial response. After 9.5 months the patient had radiologic and clinical progression with right-sided chest pain and a pleural effusion. A thoracentesis was done with MSK-IMPACT testing revealing a BRAF structural rearrangement. Targeted RNAseq using MSK-Fusion Solid, a panel assay based on anchored multiplex PCR [18], was performed and showed the AGK/BRAF fusion (Supplemental Table 5). The patient was changed to nivolumab but showed further clinical deterioration and died 2 weeks later (Figure 1).
FIGURE 1. Clinical information for patients with resistance to EGFR TKI and BRAF fusions.
A. Disease course of patient 2, found to have acquired resistance to osimertinib and AGK/BRAF fusion. B. Disease course of patient 4, found to have acquired resistance to erlotinib and progressive mediastinal disease with AGK/BRAF fusion.
Case 2.
A 69-year-old woman, former smoker (22 pack-years) was diagnosed initially with stage IIa (pT2aN1M0) lung adenocarcinoma status post a left upper lobe lobectomy and mediastinal node dissection followed by adjuvant cisplatin and pemetrexed for 4 cycles (patient 4, Supplemental table 4) (Figure 1). She was maintained on active surveillance for 2.9 years at which time she was noted to have a metastatic recurrence to the brain. She underwent a metastasectomy with pathology of a right frontal lobe metastasis showing a 1.8 cm tumor with predictive immunohistochemistry negative for EGFR L858R and ALK. MSK-IMPACT analysis revealed an EGFR exon 19 deletion. A timeline of the clinical course is shown in Figure 1B. An interval CT scan found enlarging mediastinal lymph nodes and the patient was given erlotinib 150 mg daily. The patient had a clinical and radiologic response to erlotinib, remaining on it for 1.5 years at which point she was noted to have progression in her mediastinal lymph nodes. MSK-IMPACT analysis of the post-erlotinib treatment sample identified the original EGFR exon 19 deletion as well as a new AGK/BRAF fusion (Supplemental Table 5). She was placed on multiple lines of cytotoxic chemotherapy, however, she had continued disease progression and died.
Generation of isogenic EGFR mutant cell lines with BRAF fusions.
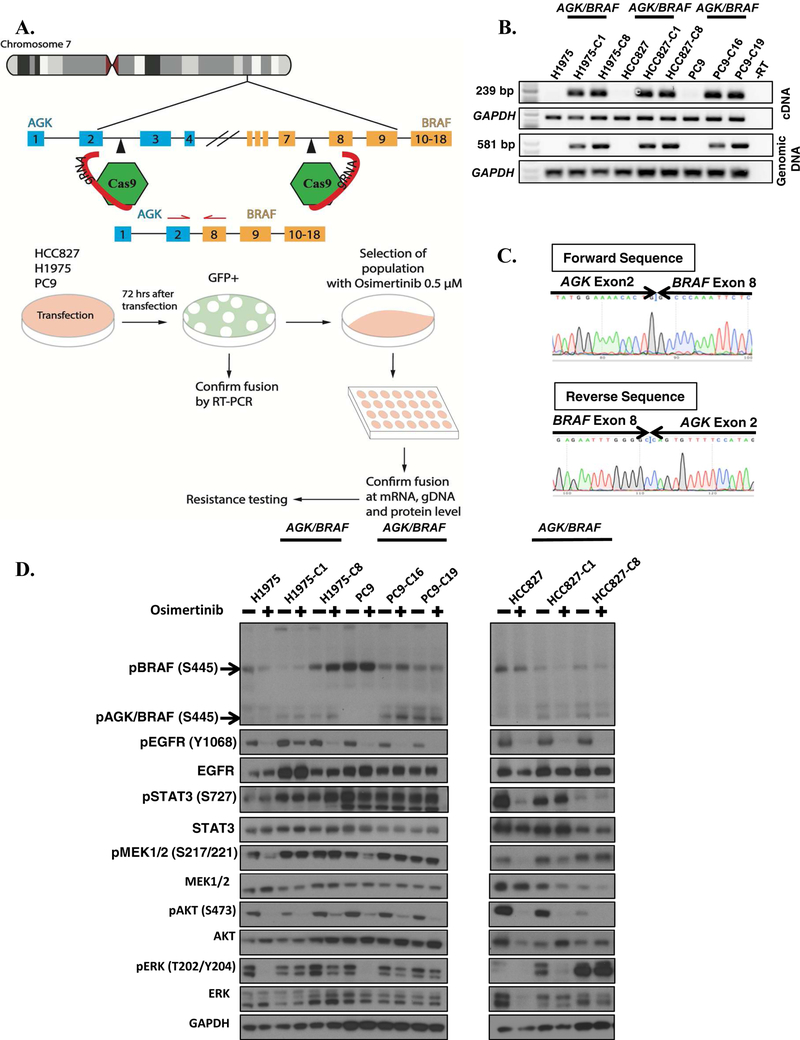
To examine the influence that acquired BRAF rearrangements have on sensitivity to EGFR TKIs, we inserted an AGK/BRAF fusion into EGFR mutant lung cancer cell lines H1975 (T790M, L858R), HCC827 (EGFR Ex19del) and PC9 (EGFR Ex19del) using CRISPR/Cas9 genome editing [16, 19]. An outline of the experimental scheme is illustrated in Figure 2A. Guides were designed to target intron 1 of AGK and intron 7 of BRAF in order to generate a fusion of AGK exon 2 with exon 8 of BRAF (Figure 2A) and cloned into pX458 expression plasmid. Plasmids were introduced and cells selected with osimertinib as described in Methods. All osimertinib-resistant clones were positive for the AGK/BRAF fusion by PCR (22 H1975, 16 HCC827 and 20 PC9). Two clones of each cell line that was positive for AGK/BRAF fusion at the mRNA and genomic DNA level were selected for further analysis (Figure 2B). The fusion junction of the two genes was confirmed by Sanger sequencing (Figure 2C).
FIGURE 2. Generation and characterization of CRISPR/Cas9 AGK/BRAF cell lines.
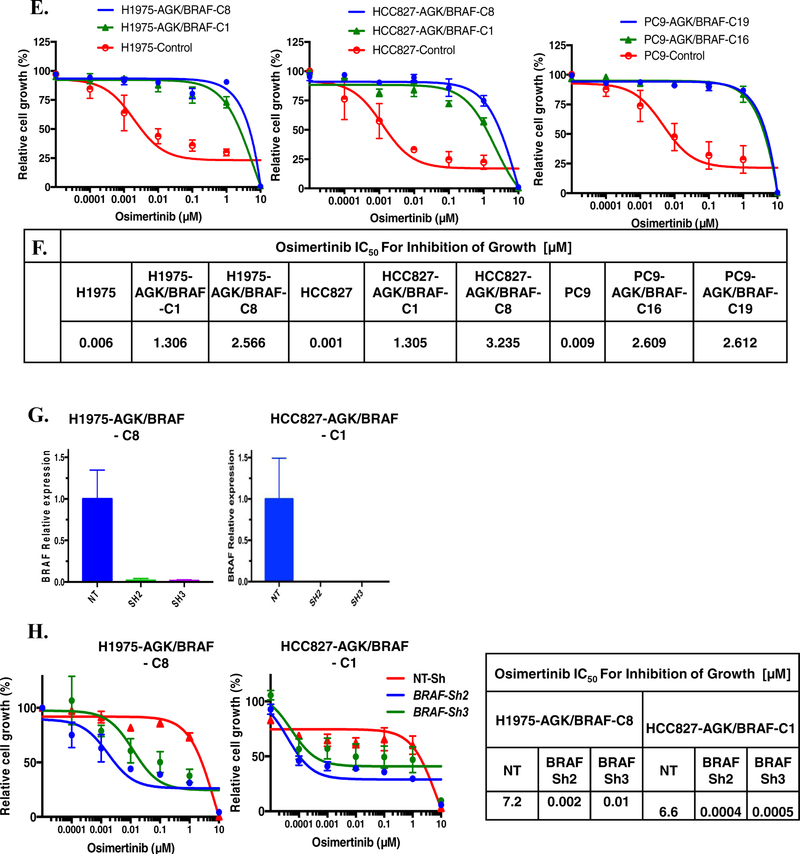
A. CRISPR/Cas9 genome engineering protocol scheme. B. Expression of AGK/BRAF. C. Sanger sequencing of PCR product confirming fusion in cell line of AGK exon 2 with exon 8 of BRAF. D. H1975 and PC9 cells expressing AGK/BRAF were treated with 0.5 osimertinib for 1 h, whole-cell extracts prepared and then lysates were subjected to immunoblotting. Results represent 3 independent experiments. E. The indicated cell lines were plated at a density 7,500/well in 96-well plates, treated with osimertinib for 96 h and the IC50 values for growth inhibition are shown in the Table in panel F. G-H. Cells were plated at a density of 150,000 cells/well in 6-well plates then infected 24 h later with lentiviruses harboring the indicated shRNAs and then selected with puromycin 48 h later. BRAF mRNA levels was determined by qRT-PCR (G) or cells were replated in osimertinib and growth assessed 96 later (H). IC50 values for growth inhibition were determined by nonlinear regression analysis using Graphpad Prism software. Results represent the mean ± SD of 2 experiments.
Expression of AGK/BRAF fusion in EGFR mutant cell lines induces resistance to EGFR TKIs.
Western blot analysis with phospho-specific BRAF anti-sera confirmed the presence of the AGK/BRAF chimeric protein at the predicted molecular mass of 50 kDa. All cells expressing the AGK/BRAF fusion showed enhanced phosphorylation of MEK1/2 and ERK1/2 (Figure 2D) compared to the parental control cells. Importantly, treatment with osimertinib (0.5 μM) blocked activation of MEK1/2, ERK1/2 and STAT3 in parental cells but not in the AGK/BRAF-expressing cells, probably due to EGFR-independent activation of the pathways by the BRAF fusion (Figure 2D).
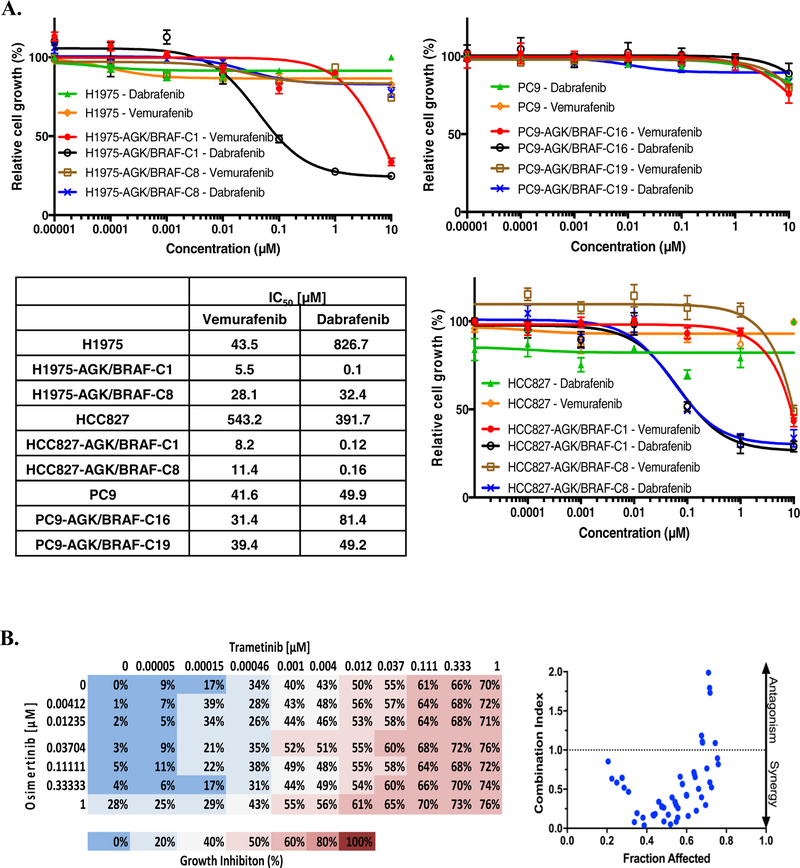
To confirm that expression of BRAF fusions can impede osimertinib efficacy, we assessed viability of EGFR mutant cell lines expressing AGK/BRAF fusion in dose-response studies. Osimertinib inhibited growth of parental HCC827, H1975 and PC9 with IC50 value of 0.001, 0.006 and 0.009 μM, respectively. In contrast, clones expressing AGK/BRAF were approximately 1000-fold less sensitive to osimertinib than the parental lines (Figures 2E and 2F). To provide orthogonal validation of these results obtained with the CRISPR-generated AGK/BRAF isogenic lines, we also expressed AGK/BRAF and PJA2/BRAF fusions using lentiviral plasmids harboring the respective fusion cDNAs in H1975 and PC9 cells (Supplementary figure 1A). As shown in Supplementary Figure 1B, expression of the two BRAF fusions in H1975 and PC9 cells resulted in lower sensitivity to osimertinib. To ensure that the resistance to EGFR TKIs was due to expression of BRAF fusions, we examined viability in two independent clones (H1975-AGK/BRAF-C8 and HCC827-AGK/BRAF-C1) in which BRAF was knocked down with two independent shRNAs targeting kinase domain of BRAF. Expression of BRAF mRNA was reduced by 98.2–100% by the two shRNAs, compared to cells expressing a non-targeting control shRNA (Figure 2G). HCC827 and H1975 clones expressing the AGK/BRAF fusion were resensitized to osimertinib upon treatment with these shRNAs targeting the portion of BRAF included in the fusion, confirming that the resistance we observed was due to expression of the fusion (>100 fold decrease in IC50 compared to non-targeting control) (Figure 2H). Similar results were obtained in cells expressing AGK/BRAF and PJA2/BRAF cDNAs (Supplementary figure 1C and 1D). Taken together, these results confirm BRAF fusions as resistance mechanism to EGFR TKI.
An EGFR TKI-resistant patient derived cell line with EGFR Ex19 del and PJA2/BRAF.
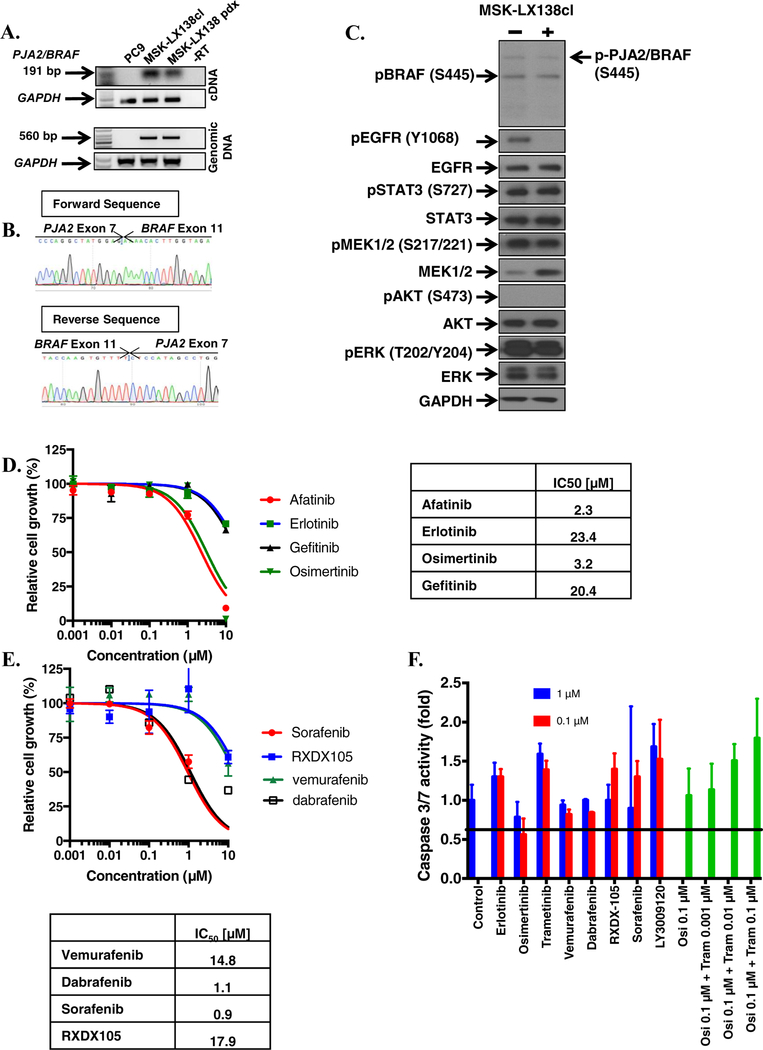
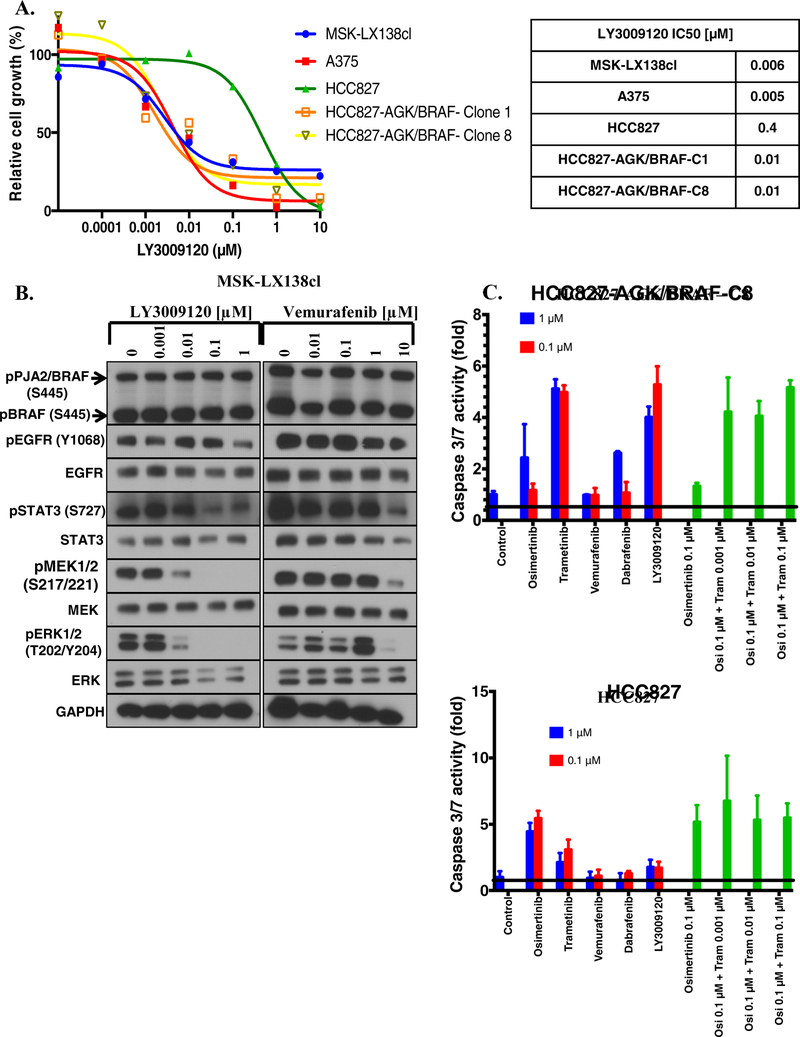
We generated a patient derived xenograft (MSK-LX138) and paired cell line (referred to as MSKLX138cl) from tissue isolated from patient 1 after resistance to EGFR TKI emerged. MSK-IMPACT analysis of this tumor sample showed a PJA2-BRAF fusion and we confirmed the presence and expression of the fusion at genomic DNA, mRNA and protein level (Figure 3A, 3B, 3C). In agreement with the MSK-IMPACT data, Sanger sequencing of the PCR amplicon indicated a fusion between exon 7 of PJA2 and exon 11 of BRAF (Figure 3B), and EGFR exon 19 deletion (Supplementary figure 2A). Similar to the observations in the isogenic lung cancer cell lines with AGK/BRAF fusion that we generated, MSK-LX138cl cells exhibited high levels of MEK and ERK phosphorylation, which persisted despite osimertinib treatment. Interestingly, this cell line exhibited no AKT phosphorylation (Figure 3C), suggesting that BRAF fusions can drive growth and survival signals independently of the PI3K-AKT pathway. Growth of MSK-LX138cl cells was insensitive to EGFR TKIs (afatinib, erlotinib, osimertinib, gefitinib) and several BRAF inhibitors (vemurafenib, dabrafenib, sorafenib, RXDX-105) (Figure 3D–E). Similar results with BRAF inhibitors were obtained in the established CRISPR fusion clones (Figure 4A). As BRAF fusions function as dimers, the lack of activity of vemurafenib and dabrafenib was not unexpected. In agreement with the reduced sensitivity of these cells to EGFR and BRAF inhibitors, there was no apoptosis-related induction in caspase 3/7 activity in response to treatment with several EGFR and RAS-MAPK pathway inhibitors as single agents (osimertinib, erlotinib, trametinib, RXDX-105, sorafenib, dabrafenib) further supporting the resistant nature of this cell line (Figure 3F).
FIGURE 3. Characterization of an EGFR mutant cell line with PJA2/BRAF fusion.
A. Expression of PJA2/BRAF in a patient-derived cell line, MSK-LX138cl, and xenograft tissue. B. Sanger sequencing of PCR product confirming a fusion between PJA2 exon 7 and BRAF exon 11. C. MSK-LX138cl cells were treated with 0.5 osimertinib for 1 h, whole-cell extracts prepared and then lysates were subjected to immunoblotting. Results represent 3 independent experiments. D-E. MSK-LX138cl cells were plated at a density 7,500/well in a 96-well plates, treated with EGFR (D) or BRAF (E) inhibitors. IC50 values for growth inhibition were determined by nonlinear regression analysis using Graphpad Prism software. F. 50,000 MSK-LX138cl cells/well were plated in 96-well plates, the following day they were treated with 0.1 or 1 μM of the indicated agents or a combination of osimertinib, and trametinib. Caspase 3/7 activity was determined 48 h later. Results represent the mean ± SD of 2 experiments.
FIGURE 4. Combined inhibition of MEK and EGFR is effective and reducing growth of cells with EGFR mutation.
A. Cells were plated at a density of 7,500 cells/well in 96-well plate and treated with vemurafenib or dabrafenib for 96 h then growth was determined. IC50 values for growth inhibition are shown (Right). B. MSK-LX138cl cells were plated at a density of 7,500 cells/well in 96-well plates and treated with trametinib (0–1μM) and osimertinib (0–1μM) for 96 h. Growth inhibition was then determined and combination index calculated using CompuSyn software. Data represent the mean value of growth inhibition ratio at each concentration of the drugs in two independent experiments. Dot plot indicates the combination index and fraction affected (inhibition ratio) of various drug concentrations.
Combined inhibition of MEK and EGFR as potential therapy.
Given the presence of two actionable oncogenes in our cell lines and patients, we tested the hypothesis that co-inhibition of MEK and EGFR could be an efficient therapeutic strategy. Treatment with trametinib alone decreased growth of all cell lines with EGFR mutation alone, or with a BRAF fusion; cell lines with the BRAF fusion were more sensitive, consistent with published data showing only limited activity of MEK inhibitors in EGFR-mutated cell lines [20]. Indeed, we found that only a maximum inhibition of 50% cell growth was achieved by trametinib (results not shown). This may indicate that a subpopulation of cells from lines expressing a BRAF fusion can still survive after MEK inhibition. Taken together, these support the notion that MEK inhibition alone is not sufficient to fully inhibit growth of BRAF-fusion positive cell lines. To determine the combinatorial effect of osimertinib and trametinib we employed the method of Chou-Talalay that was developed to identify if two or more agents act in a synergistic manner [21]. We calculated the combination index (CI) of multiple combinations of different drug concentrations, where CI<1 represents synergy, CI=1 represents an additive effect and CI>1 represents an antagonistic effect, to determine if the two agents can act synergistically. The experiment was performed in MSK-LX138cl (Figure 4B) as well as two of our isogenic EGFR mutant cell lines with AGK/BRAF fusion (H1975-C1, HCC827-C1). Our results show that trametinib and osimertinib acted in a synergistic manner to inhibit growth up to a concentration of 100 nM.
LY3009120, a pan-RAF inhibitor is effective as a single agent in BRAF-fusion-positive cell lines.
BRAF rearrangements are grossly insensitive to BRAF inhibitors (eg vemurafenib, dabrafenib) due to homo- and heterodimerization with other RAF proteins [22, 23]. We also show above, as previously described [24], that BRAF inhibitors are not effective in cell lines with BRAF fusions. However, RAF inhibitors such as BGB659 and LY3009120 are expected to be effective against the fusions because they can bind and inhibit all RAF isoforms [25–29]. Treatment of MSK-LX138cl with LY3009120 resulted in complete inhibition of growth with IC50 = 0.006 μM (Figure 5A). The melanoma cell line A375, which harbors a BRAF V600E mutation [30], was used as a positive control in these experiments. We next examined the effect of LY3009120 on caspase 3/7 activity. LY3009120 caused a small but significant increase in caspase 3/7 activity, similar to that observed with trametinib+osimertinib in MSK-LX138cl (Figure 4F) and HCC827-AGK/BRAF-C1 cells, but not in the parental HCC827 cell line (Figure 5C). Finally, to better understand how LY3009120 inhibits growth, we examined its effect on phosphorylation of BRAF fusion and other downstream signaling proteins. LY3009120 at 0.1 μM caused a profound reduction in phosphorylation of MEK, ERK and STAT3 (Figure 5B).
FIGURE 5. The pan-RAF inhibitor LY3009120 effectively inhibits growth of EGFR mutant cell lines with BRAF fusions.
A. Cells were plated in 96-well plates, treated with LY3009120 and then growth determined (Left). IC50 values for growth inhibition were determined by nonlinear regression analysis using GraphPad Prism software (Right). B. MSK-LX138cl cells were treated with the indicated concentration of LY3009120 or vemurafenib for 1 h then whole-cell extracts prepared and immunoblotted for the indicated proteins. Results are representative of two independent experiments. C. Cells were treated with 0.1 or 1 μM of the indicated compounds or osimertinib/trametinib combinations and caspase 3/7 activity determined 48 h later.
DISCUSSION
The present study establishes BRAF fusions as a mechanism of resistance to EGFR TKI in EGFR-mutant NSCLC and offers two potential therapeutic strategies to circumvent the drug resistance, namely combined inhibition of MEK and EGFR, or inhibition of BRAF fusion. Of seven patients with BRAF fusion present on MSK-IMPACT (n = 374), four had concomitant EGFR driver mutations suggesting that the co-occurrence of EGFR mutations and BRAF fusion is a rare but recurrent event. Importantly, all 4 patients were resistant to EGFR TKIs (erlotinib or osimertinib), and in two of four cases the BRAF fusions were confirmed to be acquired because pre-TKI samples were available and negative for BRAF fusion. AGK, which, like BRAF, resides at 7q34, encodes a mitochondrial membrane protein involved in lipid and glycerolipid metabolism, whereas PJA2 protein (encoded by a gene at 5q21) has E2-dependent E3 ubiquitin-protein ligase activity. AGK/BRAF fusions have been described in melanomas, gliomas, and cancers of the lung, thyroid, and breast [14]. To the best of our knowledge, this is the first report of a PJA2/BRAF fusion. Multiple cellular models including one patient-derived cell line and isogenic EGFR-mutated lung cancer cell line models demonstrated that expression of BRAF fusion mediated resistance to EGFR TKIs. In these models, we observed activation of the MEK-ERK pathway independently of EGFR. We further found that loss of BRAF fusion expression restores sensitivity to EGFR TKI. Taken together these results indicate that BRAF fusions drive resistance to EGFR TKIs in a subset of EGFR mutant NSCLC and provides a pathway for developing a viable therapeutic strategy.
It is customary to express a cDNA for a given gene either by lipid-based transfection or viral transduction to determine if expression of the gene alters biological properties such as response to small molecules. These methods of gene expression typically result in supraphysiologic levels of the protein because multiple copies of the cDNA are usually introduced, and expression is driven from a strong, constitutively active viral promoter. Therefore, there is often a concern that the observed results may be an artifact of overexpression of the protein. To avoid this pitfall, we engineered the AGK/BRAF fusion using CRISPR-Cas9-mediated genome editing in lung cancer cells harboring drug-sensitive EGFR mutations. The advantage of this method over conventional exogenous cDNA expression is that the BRAF fusion is expressed from the endogenous, non-amplified allele, resulting in a physiological level of protein expression. Using this approach, we demonstrated that a physiological level of AGK/BRAF fusion was able to induce resistance to EGFR inhibitors in EGFR-mutant lung cancer cell lines. These results were concordant with data obtained by overexpression of BRAF fusions using lentiviral vectors harboring the respective cDNAs. Our approach represents a methodological advance that can be used to model other fusion genes as mechanisms of resistance.
Although BRAF fusion may seem to be an obvious therapeutic target, FDA-approved BRAF inhibitors have not been effective against BRAF fusions [24, 31–33] and have not always been effective against BRAF mutants. As previously described, BRAF V600E-positive colorectal cancers have notoriously been insensitive to selective BRAF inhibitors (vemurafenib or dabrafenib), due to feedback activation of EGFR and subsequent RAF dimerization [34]. A similar resistance mechanism has been noted in BRAF V600E melanomas, where RAF inhibition abrogates ERK activation, which then terminates the high ERK negative feedback on RTK-dependent activation of RAS, leading to consequent RAF dimerization and ERK reactivation [35]. This ineffectiveness of FDA-approved BRAF drugs against BRAF fusions is highlighted by the fact that an AGAP3/BRAF fusion has been proposed as a mechanism of acquired resistance to vemurafenib in BRAF V600E melanoma [24]. When compounds like vemurafenib and dabrafenib bind to the first site within the BRAF dimer, affinity for the second dimer site is significantly lower, explaining why traditional BRAF inhibitors do not abrogate ERK phosphorylation in BRAF fusion-positive tumors [36]. BRAF V600E mutants can become resistant to BRAF inhibitors via formation of BRAF dimers by the mechanism described above [23, 37], which prompted development of pan-RAF inhibitors [26, 38]. Yao et al have reported superiority of a dual RAF inhibitor BGB659 over BRAF inhibitors [36]. Similarly, Peng et al reported the development of LY3009120, a pan-RAF inhibitor that binds simultaneously to two sites of BRAF dimers and equally inhibits all RAF isoforms (ARAF, BRAF, CRAF) with similar affinity [25–29]. We found LY3009120 to be very active against cell lines with BRAF fusions as a single agent. Taken together, this indicates that pan-RAF inhibition is a superior single agent strategy for targeting these acquired BRAF fusions. Notably, we (N.X.R.) have recently shown that another promising RAF inhibitor, PLX8394, selectively disrupts BRAF dimers, sparing CRAF homodimer-dependent RAF function in normal cells, which may lead to a better safety profile clinically [39]. Clinical studies will be needed to determine if RAF inhibitors are effective against tumors with BRAF fusions as a single agent or should be combined with an EGFR TKI to maximize the effectiveness.
To identify potential therapeutic strategies for patients with co-expression of EGFR mutant and BRAF fusion that can be utilized immediately, we investigated the effectiveness of a combination of trametinib and osimertinib. We found that combined inhibition of MEK and EGFR with these drugs inhibited growth of isogenic cell line models as well as a patient-derived cell line with a BRAF fusion and EGFR mutation (established from a patient who did not respond to EGFR drugs) in a synergistic manner. The maximum inhibition of growth that we observed was 76%, which may have been due to reactivation of HER2 and ERK following the prolonged treatment of cells with osimertinib and trametinib, respectively (data not shown). Nevertheless, these results suggest that combined MEK and EGFR inhibition is a possible treatment strategy in the absence of more effective FDA-approved RAF inhibitors that are effective against BRAF fusions.
Supplementary Material
ACKNOWLEDGEMENTS:
This work was supported by NIH grants P01 CA129243, P30 CA008748, and U54 OD020355. The authors are grateful to Igor Odintsov and Dr. Zebing Liu for critical reading of the manuscript and helpful suggestions during the course of this study, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST STATEMENTS
Morana Vojnic, Daisuke Kubota, Christopher Kurzatkowski, Ken Suzawa, Michael Offin, Adam J. Schoenfeld, Andrew J. Plodkowski and J. T. Poirier have no relevant conflicts of interest to disclose.
Maria E. Arcila is a consultant for AstraZeneca and received support from Astrazeneca, Invivoscribe, and Raindance Technologies.
Alexander Drilon is a consultant for Ignyta, LOXO Oncology, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Genentech Roche, Takeda, and has received research funding from Foundation Medicine.
Helena A Yu is a consultant for AstraZeneca and received research funding from Astellas Pharma, Incyte, Lilly Oncology, Novartis, Daiichi, and AstraZeneca.
Charles M. Rudin is a consultant for Bristol-Myers Squibb, Abbvie, Seattle Genetics, Harpoon Therapeutics, Genentech Roche, and AstraZeneca.
Mark G. Kris is a consultant for Pfizer, Astrazeneca and Regeneron.
Neal X. Rosen has received research funding from Chugai, is on the Scientific Advisory Boards of Chugai, Astra-Zeneca, Beigene, Daichi-Sankyo and received consulting fees from Novartis, Boeringher-Ingelheim, Array, Revolutionary Medicines, and Tarveda. He has equity in Beigene, Kura, Z-Lab, Araxes and Fortress.
Gregory J. Riely received research funding from Novartis, Roche, Genentech, Millenium, GlaxoSmithKline, Pfizer, Infinity Pharmaceuticals and, ARIAD. He has received expense coverage from Merck Sharp & Dohme. He has a patent application submitted for pulsatile use of erlotinib to treat or prevent brain metastases.
Romel Somwar has received research support from Helsinn Healthcare.
Marc Ladanyi has received advisory board compensation from Boehringer Ingelheim, AstraZeneca, Bristol-Myers Squibb, Takeda, and Bayer, and research support from LOXO Oncology and Helsinn Healthcare.
REFERENCES:
- 1.Stewart EL, et al. , Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res, 2015. 4(1): p. 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goss G, et al. , Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol, 2016. 17(12): p. 1643–1652. [DOI] [PubMed] [Google Scholar]
- 3.Soria JC, et al. , Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med, 2018. 378(2): p. 113–125. [DOI] [PubMed] [Google Scholar]
- 4.Thress KS, et al. , Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med, 2015. 21(6): p. 560–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho C-C, et al. , Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. Journal of Thoracic Oncology, 2017. 12(3): p. 567–572. [DOI] [PubMed] [Google Scholar]
- 6.Ou S, et al. , MA 12.03 Kinase Fusions as Recurrent Mechanisms of Acquired Resistance in EGFR-Mutated Non-Small Cell Lung Cancer (NSCLC). Journal of Thoracic Oncology, 2017. 12(11): p. S1848. [Google Scholar]
- 7.Offin M, et al. , Acquired ALK and RET Gene Fusions as Mechanisms of Resistance to Osimertinib in EGFR-Mutant Lung Cancers. JCO Precision Oncology, 2018(2): p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klempner SJ, et al. , Emergence of RET rearrangement co-existing with activated EGFR mutation in EGFR-mutated NSCLC patients who had progressed on first- or second-generation EGFR TKI. Lung Cancer, 2015. 89(3): p. 357–359. [DOI] [PubMed] [Google Scholar]
- 9.Ross JS, et al. , The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer, 2016. 138(4): p. 881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheikine Y, et al. , BRAF in Lung Cancers: Analysis of Patient Cases Reveals Recurrent BRAF Mutations, Fusions, Kinase Duplications, and Concurrent Alterations. JCO Precision Oncology, 2018(2): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrock AB, et al. , Receptor Tyrosine Kinase Fusions and BRAF Kinase Fusions are Rare but Actionable Resistance Mechanisms to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol, 2018. 13(9): p. 1312–1323. [DOI] [PubMed] [Google Scholar]
- 12.Piotrowska Z, et al. , Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou SI, et al. , Emergence of FGFR3-TACC3 fusions as a potential by-pass resistance mechanism to EGFR tyrosine kinase inhibitors in EGFR mutated NSCLC patients. Lung Cancer, 2017. 111: p. 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zehir A, et al. , Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 2017. 23(6): p. 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HS, et al. , Oncogenic BRAF fusions in mucosal melanomas activate the MAPK pathway and are sensitive to MEK/PI3K inhibition or MEK/CDK4/6 inhibition. Oncogene, 2017. 36(23): p. 3334–3345. [DOI] [PubMed] [Google Scholar]
- 16.Ran FA, et al. , Genome engineering using the CRISPR-Cas9 system. Nat Protoc, 2013. 8(11): p. 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng DT, et al. , Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn, 2015. 17(3): p. 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Z, et al. , Anchored multiplex PCR for targeted next-generation sequencing. Nat Med, 2014. 20(12): p. 1479–84. [DOI] [PubMed] [Google Scholar]
- 19.Spraggon L, et al. , Generation of conditional oncogenic chromosomal translocations using CRISPR-Cas9 genomic editing and homology-directed repair. J Pathol, 2017. 242(1): p. 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiba M, et al. , MEK inhibitors against MET-amplified non-small cell lung cancer. Int J Oncol, 2016. 49(6): p. 2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou TC and Talalay P, Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul, 1984. 22: p. 27–55. [DOI] [PubMed] [Google Scholar]
- 22.Sievert AJ, et al. , Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A, 2013. 110(15): p. 5957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulikakos PI, et al. , RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature, 2011. 480(7377): p. 387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulkarni A, et al. , BRAF Fusion as a Novel Mechanism of Acquired Resistance to Vemurafenib in BRAF(V600E) Mutant Melanoma. Clin Cancer Res, 2017. 23(18): p. 5631–5638. [DOI] [PubMed] [Google Scholar]
- 25.Peng SB, et al. , Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell, 2015. 28(3): p. 384–98. [DOI] [PubMed] [Google Scholar]
- 26.Vakana E, et al. , LY3009120, a panRAF inhibitor, has significant anti-tumor activity in BRAF and KRAS mutant preclinical models of colorectal cancer. Oncotarget, 2017. 8(6): p. 9251–9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao YM, et al. , Mouse PDX Trial Suggests Synergy of Concurrent Inhibition of RAF and EGFR in Colorectal Cancer with BRAF or KRAS Mutations. Clin Cancer Res, 2017. 23(18): p. 5547–5560. [DOI] [PubMed] [Google Scholar]
- 28.Wei WJ, et al. , Obatoclax and LY3009120 Efficiently Overcome Vemurafenib Resistance in Differentiated Thyroid Cancer. Theranostics, 2017. 7(4): p. 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain P, et al. , CRAF gene fusions in pediatric low-grade gliomas define a distinct drug response based on dimerization profiles. Oncogene, 2017. 36(45): p. 6348–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giard DJ, et al. , In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst, 1973. 51(5): p. 1417–23. [DOI] [PubMed] [Google Scholar]
- 31.Corcoran RB, et al. , EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov, 2012. 2(3): p. 227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montagut C and Settleman J, Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett, 2009. 283(2): p. 125–34. [DOI] [PubMed] [Google Scholar]
- 33.Holderfield M, et al. , Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer, 2014. 14(7): p. 455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prahallad A, et al. , Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature, 2012. 483(7387): p. 100–3. [DOI] [PubMed] [Google Scholar]
- 35.Lito P, et al. , Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell, 2012. 22(5): p. 668–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao Z, et al. , BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell, 2015. 28(3): p. 370–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulikakos PI, et al. , RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature, 2010. 464(7287): p. 427–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henry JR, et al. , Discovery of 1-(3,3-dimethylbutyl)-3-(2-fluoro-4-methyl-5-(7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl)phenyl)urea (LY3009120) as a pan-RAF inhibitor with minimal paradoxical activation and activity against BRAF or RAS mutant tumor cells. J Med Chem, 2015. 58(10): p. 4165–79. [DOI] [PubMed] [Google Scholar]
- 39.Yao Z, et al. , RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat Med, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, et al. , A public genome-scale lentiviral expression library of human ORFs. Nat Methods, 2011. 8(8): p. 659–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somwar R, et al. , Identification and preliminary characterization of novel small molecules that inhibit growth of human lung adenocarcinoma cells. J Biomol Screen, 2009. 14(10): p. 1176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.