Using electrophysiology, North et al. show that ectopic signalling in dorsal root ganglion neuron somata is directly associated with neuropathic pain. Transcriptome analysis reveals gene modules and signalling pathways in neuronal plasticity and immune function that may underlie this spontaneous activity, as well as evidence of sex differences.
Keywords: spontaneous activity, neuropathy, DRG transcriptomics, machine learning in healthcare
Abstract
Neuropathic pain encompasses a diverse array of clinical entities affecting 7–10% of the population, which is challenging to adequately treat. Several promising therapeutics derived from molecular discoveries in animal models of neuropathic pain have failed to translate following unsuccessful clinical trials suggesting the possibility of important cellular-level and molecular differences between animals and humans. Establishing the extent of potential differences between laboratory animals and humans, through direct study of human tissues and/or cells, is likely important in facilitating translation of preclinical discoveries to meaningful treatments. Patch-clamp electrophysiology and RNA-sequencing was performed on dorsal root ganglia taken from patients with variable presence of radicular/neuropathic pain. Findings establish that spontaneous action potential generation in dorsal root ganglion neurons is associated with radicular/neuropathic pain and radiographic nerve root compression. Transcriptome analysis suggests presence of sex-specific differences and reveals gene modules and signalling pathways in immune response and neuronal plasticity related to radicular/neuropathic pain that may suggest therapeutic avenues and that has the potential to predict neuropathic pain in future cohorts.
Introduction
Significant effort has been placed on development of molecularly targeted therapies for neuropathic pain given the tremendous unmet need and consequent expanding chronic pain epidemic (van Hecke et al., 2014). Yet, numerous promising therapeutics derived from discoveries in animal models have failed in clinical trials (Hill, 2000; Gavva et al., 2008). A variety of factors have been proposed as possible causes for these failures with basic cellular-level and molecular differences between animals and humans commonly implicated (Borsook et al., 2014; Gereau et al., 2014). More efficient translation may be facilitated through direct study of human tissues and/or cells. Prior laboratory studies with human dorsal root ganglion (DRG) neurons from foetal tissue, post-mortem organ donation, and patients undergoing surgical treatments for chronic pain have attempted to make such confirmations by probing a wide array of basic histochemical and electrophysiological parameters (Baumann et al., 1996; Borsook et al., 2014; Davidson et al., 2014; Li et al., 2015, 2017). However, a key gap in knowledge is direct comparison of DRG neuron electrophysiology and paired gene expression profiling from patients with and without chronic neuropathic pain. Using a unique cohort of patients, here we provide detailed electrophysiological characterization and RNA sequencing (RNA-seq) of DRG neurons and tissue, respectively, from people with neuropathic pain. Our results provide clear evidence of spontaneous activity in sensory neurons as a driver of neuropathic pain; and our RNA-seq data suggest key pathways for targeted therapeutics and reveal potential biomarkers for neuropathic pain.
Materials and methods
Study approval
Written informed consent for participation, including use of tissue samples, was obtained from each patient prior to inclusion. The protocol was reviewed and approved by the M.D. Anderson and The University of Texas at Dallas Institutional Review Boards and all experiments conform to relevant guidelines and regulations.
Clinical data collection
Clinical data were obtained from patients undergoing treatment at MD Anderson Cancer Center for malignant tumours involving the spine through a combination of retrospective review of medical records and prospective data collection at the time of study enrolment. These data included basic patient demographics, medical history, and clinical symptoms. Preoperative MRI was evaluated for radiographic evidence of spinal cord or nerve root compression. Spinal cord compression was evaluated according to the epidural spinal cord compression scale (Bilsky et al., 2010). Presence of nerve root compression was determined based on a documented report from a neuroradiologist or review by a neurosurgeon. Axial spine pain was defined as present if there was a documented history of pain complaint in the midline in the neck or back, or if physical exam findings indicative of the axial spine as a pain generator was present. Axial spine pain was determined as absent if there was no documentation of a history of midline pain in the neck/back and a documented denial of axial pain, nor any physical exam findings indicative of axial spine as a pain generator. Determination of presence or absence of radicular/neuropathic pain was performed for each dermatome associated with a harvested dorsal root ganglion and consistent with the guidelines for probable or definite neuropathic pain from the Assessment Committee of the Neuropathic Pain Special Interest Group of the International Association for the Study of Pain (IASP) (Haanpaa et al., 2011). Specifically, pain was deemed present if the patient had documented symptoms of spontaneous pain, sensory loss, paraesthesia, dysaesthesia, hyperalgesia, or allodynia in a distribution at or within two classically defined dermatomes of the harvested ganglion. Neuropathic pain was considered absent if the patient had no history of any symptoms defined in part 1 or if the ganglion was harvested from the side contralateral to reported pain in a patient with only unilateral symptoms. Any remaining scenario was categorized as indeterminate and neurons from these ganglia excluded from analysis of associations with clinical data. Of note, although some patients had a history of chemotherapy treatment, the DRG collected here were outside the dermatomes affected by length-dependent neuropathy. Detailed clinical characteristics for the entire cohort are found in Supplementary Table 1.
Human dorsal root ganglion neuron preparation
Human DRG neurons were prepared as described previously (Li et al., 2015, 2017) and based largely on additional prior work (Davidson et al., 2014). Briefly, each donor was undergoing surgical treatment that necessitated ligation of spinal nerve roots to facilitate tumour resection or spinal reconstruction. Spinal roots were ligated proximal to the DRG, spinal root sharply cut both proximal and distal to the DRG, and excised DRG transferred immediately into cold (∼4°C) and sterile balanced salt solution containing nutrients. DRG were transported to the laboratory on ice in a sterile, sealed 50-ml centrifuge tube. Upon arrival to the laboratory, each ganglion was carefully dissected from the surrounding connective tissues and sectioned into three to four parts. One section was immediately frozen in RNAlater (Ambion) and saved for subsequent RNA sequencing. One or two sections of DRG were further cut into several ∼1–2-mm pieces and cells dissociated for electrophysiology recording. Further details on the DRG cell dissociation, recording procedures can be found in the Supplementary material.
RNA sequencing
Total RNA from 21 quartered DRG samples from 15 patients were purified using TRIzolTM (ThermoFisher) and subjected to ribosomal RNA depletion and total RNA Tru-seq library preparation according to the manufacturer’s instructions (Illumina). Tru-seq total RNA library kit with ribosomal RNA depletion (Illumina) was used to generate sequencing libraries. Fifty cycle, single-end sequencing of these RNA-seq libraries was performed on the Illumina Hi-Seq sequencing platform. Obtained sequencing reads were mapped to the reference genome in a strand-aware fashion, retaining only uniquely mapped reads, based on the reference transcriptome annotations and the reference human genome hg19 in the NCBI Entrez/RefSeq database (Maglott et al., 2005). The bowtie2 tool (with maximum allowed alignment mismatch ≤2) (Langmead and Salzberg, 2012) was used for mapping reads and the Subread package was used for counting mapped reads (Liao et al., 2013). Read counts were normalized to transcripts per million for downstream analysis.
Random Forest-based prediction of cohort membership
We performed a proof-of-principle analysis for predicting the pain categorization of each sample based solely on the RNA abundance profile using the predictive classification model Random Forest, which uses an ensemble of decision trees to classify samples, and which has been used successfully in whole genome assay studies (Chen and Ishwaran, 2012). We built separate classifiers to discriminate between male-pain and male-no pain samples; and between male-pain and female-pain samples, solely based on the RNA profile of the autosomal gene expression profile of the corresponding sample. We performed leave-one-out cross validation analysis, by training our Random Forest model on all but one of the samples. We then blinded ourselves to the cohort membership of the held out sample (referred to as the test sample), and then predicted the label of the test sample using its RNA profile. This analysis was performed on every sample in turn to generate a cohort membership prediction for every sample based on their individual RNA profiles. Our leave-one-out cross validation approach provides an alternative to an independent validation cohort for confirming whether conclusions drawn from our present cohort about discriminative gene sets can be successfully applied to new datasets.
Statistics and computation: clinical, electrophysiological and RNA-seq data analysis
Clinical and electrophysiological data were analysed with GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). Unless otherwise specified, data are expressed as mean ± standard error of mean (SEM). Continuous variables were analysed with Mann-Whitney U-test. Fisher’s exact test was used for analysis of contingency tables. Details of the computational analysis of the RNA-seq data (including the random forest classification algorithm) can be found in the Supplementary material.
Data availability
The neurophysiological data can be shared on request. The full transciptomic dataset and code for analysis is available at: https://www.utdallas.edu/bbs/painneurosciencelab/sensoryomics/hdrgclinical/. Raw sequencing datasets are available from the dbGaP repository as single-end read libraries (phs001158.v2.p1).
Results
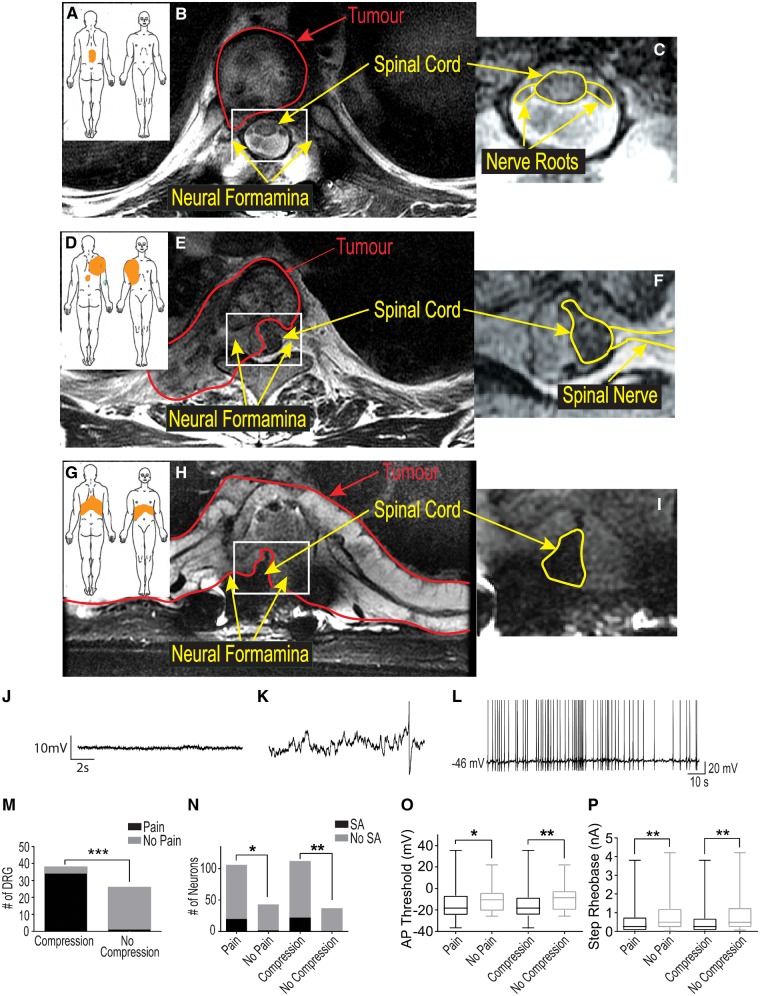
Sixty-six DRG were collected from 26 (eight female and 18 male) patients whose clinical data, including opioid consumption, are summarized in Supplementary Table 1. The donor cohort in this study is unique in that all donors had a complete medical history available for review allowing us to make clear distinctions between pain and no pain samples for electrophysiological and RNA-seq analyses. The majority of patients (n = 17) were afflicted with metastatic carcinoma to the spine versus primary malignancies of bone (n = 7) or local extension of a primary carcinoma (n = 2); and most (n = 25) had a history consistent with axial spine pain. Presence versus absence of associated dermatomal radicular/neuropathic pain was determined for all donated ganglia. These criteria defined three patient groups. The first was composed of six patients with isolated axial spine pain, but without any radicular/neuropathic pain (Fig. 1A–C). Group 2 included 15 patients with unilateral radicular/neuropathic pain (Fig. 1D and E) and Group 3 included five patients with bilateral radicular/neuropathic pain (Fig. 1G–I). Radicular/neuropathic pain was strongly associated with radiographic evidence of nerve root compression (Fig. 1K, 35/39 compressed ganglia with pain, P < 0.001). At maximal intensity, median visual analogue pain scale (VAS) was 7.72 for the entire cohort and no statistically significant difference between VAS score for patients with versus without radicular/neuropathic symptoms (7.72 versus 7.53, P = 0.85). The majority of patients’ radicular/neuropathic symptoms were present for more than 6 months (12/20) and there were no patients without symptoms dating back at least 1 month.
Figure 1.
DRG neurons from dermatomes with radicular/neuropathic pain show ectopic spontaneous activity and hyperexcitability. Pain diagrams and MRI spinal images for three categories of patients are shown in A–I. The orange shaded area in A, D and G indicate where patients marked the location of their pain. This was either localized to the spine without signs of radicular/neuropathic pain (axial pain only, A); showed radiation only to one side (unilateral radicular/neuropathic pain, D); or pain that radiated to both sides of the body (bilateral radicular/neuropathic pain). The large MRI scan in B shows that patients with axial pain often only had tumours (outlined in red) that did not compress the nerve roots or spinal cord. Patients with unilateral neuropathic pain (E) typically had tumours that compressed one or more nerve roots on one side and part of the spinal cord. Patients with bilateral neuropathic pain typically had compression of one or more roots on both sides and the spinal cord (H). The area in B, E and H outlined in white are magnified in C, F, and I to show the spinal cord and nerve roots better (outlined in yellow). A representative recording of the resting membrane potential with an expanded time base for a cell without spontaneous activity is shown in J while a similar recording for a cell with spontaneous activity is shown in K to illustrate the spontaneous depolarizing fluctuations (DSFs) in membrane potential that occurred in these cells. A single action potential is shown at the right of this trace occurring atop one of the larger of these DSFs. The representative trace shown in L illustrates the irregular pattern of action potentials typically seen in cells with spontaneous activity. The bar graphs in M show that radiological evidence of nerve compression was strongly associated with signs of radicular/neuropathic pain; while in N the bar graphs show the relationship of radicular/neuropathic pain and nerve compression with spontaneous activity (SA). The box and whisker plots in O and P show that DRG neurons from a dermatome with pain and/or nerve compression had a more depolarized spike threshold potential and lower rheobase, respectively.
Whole-cell patch clamp recordings were performed on samples from 17 patients from a total of 28 DRG after dissociation and >24 h in culture. The median patched cells per patient was nine (range 2–26). Spontaneous activity was recorded in 13% of neurons (20/149), from 39% of donated DRG (11/28), and in 59% of patients (10/17). Representative analogue traces show the baseline membrane potential in a non-spontaneous activity neuron was stable (Fig. 1J); whereas the exploded view of the baseline membrane potential (Fig. 1K) and compressed time base (Fig. 1L) for a neuron with spontaneous activity show the occurrence of spontaneous depolarizations of membrane potential was only observed in cells with spontaneous activity (Fig. 1K) and these cells typically showed an irregular pattern of action potentials (Fig. 1L). Statistical analysis relating the clinical parameters to electrophysiology revealed significant associations of spontaneous activity and neuronal hyperexcitability [hyperpolarization of action potential threshold (Fig. 1M) and decrease in step rheobase (Fig. 1N)] with both radicular/neuropathic pain and radiographic nerve root compression (Fig. 1L and M, spontaneous activity: P < 0.05, spike threshold P < 0.05, rheobase: P < 0.05). Spontaneous activity was noted in 19% (20/106) of neurons from DRG with corresponding dermatomal pain and in 20% (22/112) of neurons with associated radiographic nerve root compression. spontaneous activity was noted in only 4.6% (2/43) of neurons from DRG without associated dermatomal pain and in none (0/37) of the neurons from DRG without radiographic nerve root compression. Differences in resting membrane potential, neuron size, capacitance, action potential profile and kinetics were not significantly correlated with either radicular/neuropathic pain or nerve root compression (Table 1). No significant relationships for these same parameters were found for age, sex, axial spine pain, radiographic spinal cord compression, prior chemotherapy, prior radiation treatment, or a history of length-dependent peripheral neuropathy (this latter symptom affected dermatomes that were not sampled).
Table 1.
Detailed neurophysiological parameters for DRG neurons organized by groups
| Group | Diameter, µm | RMP, mV | C, pF | Rheobase, nA | Spike threshold, mV | AP peak, mV | AP overshoot, mV | AP rise time, ms | AP fall time, ms | AHP amplitude, mV | Tau, ms |
|---|---|---|---|---|---|---|---|---|---|---|---|
| All neurons | 43.7 ± 0.8 | −57.9 ± 0.9 | 190 ± 10 | 0.7 ± 0.08 | −14.1 ± 1.3 | 77.0 ± 1.4 | 62.4 ± 1.7 | 1.9 ± 0.1 | 6.0 ± 0.3 | 15.7 ± 0.5 | 41.1 ± 3.0 |
| With pain | 42.1 ± 0.8 | −57.8 ± 1.0 | 200 ± 20 | 0.6 ± 0.09* | −14.8 ± 1.7* | 78.0 ±1.5 | 63.4 ± 1.8 | 1.8 ± 0.1 | 6.1 ± 0.4 | 15.9 ± 0.6 | 40.2 ± 3.5 |
| No pain | 45.1 ± 1.3 | −58.2 ± 1.8 | 180 ± 20 | 0.9 ± 0.17 | −11.3 ± 2.1 | 75.3 ± 3.5 | 60.9 ± 4.2 | 2.0 ± 0.3 | 5.5 ± 0.7 | 15.2 ± 1.2 | 45.3 ± 6.9 |
AHP = after-hyperpolarization; AP = action potential; C = capacitance; RMP = resting membrane potential.
*P < 0.05.
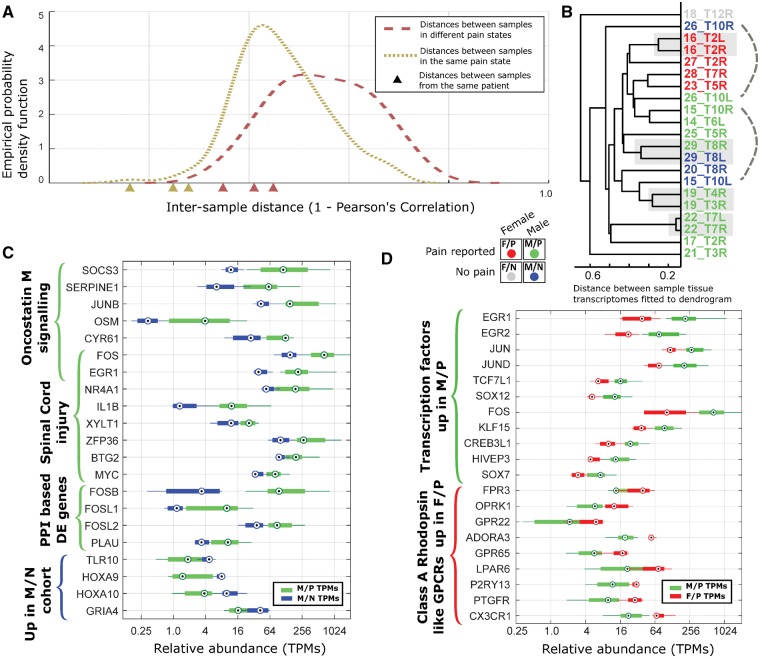
Pairwise distances between 21 sample transcriptomes were calculated from RNA-seq data (Supplementary Table 2). Samples were separated into two groups: those with associated dermatomal radicular/neuropathic pain and those without. Distribution of distances between pain and non-pain samples was higher on average (Fig. 2A). Twelve of 21 samples were from six donors with two sequenced DRGs each. The pairwise distance between donor-controlled pairs was smaller for their respective groups (Fig. 2A). Hierarchical clustering of pain and non-pain groups revealed that only a small number of genes are consistently differentially expressed between the groups. However, female pain samples were well correlated with each other (Fig. 2B) suggesting that sex of the sample is also influential in shaping the DRG transcriptome. Based on these insights, our 21 samples were partitioned into four cohorts by sex and pain state (male, female, pain, and no-pain).
Figure 2.
Differential expression analysis for human DRG transcriptomes. (A) Empirically estimated density function for pairwise transcriptome distances between samples with the same pain state and between samples with different pain states show overlap but a clear increase overall. Inter-sample distances for samples from the same patient (shown by triangles on the x-axis) are comparatively low with respect to the set of all distances. (B) Hierarchical clustering of RNA profiles for all samples, showing close distances between female pain samples. Standard hierarchical clustering was performed for all RNA-seq samples using expression patterns of high variability (entropy <3.5, see Fig. 3), expressed [transcripts per million (TPM) > 1.5 in at least one sample] genes with distance metric = 1 − Pearsons’ correlation coefficient, and average linkage. Four cohorts [male-pain (M/P), male-no pain (M/N), female-pain (F/P), and female-no pain (F/N)] are colour-coded. (C and D) Several representative differentially-expressed gene sets for the male-pain versus male-no pain (C) and for the male-pain versus female-pain (D) comparison.
The three male donor DRG pairs with pain in one dermatome, but not the other, were each analysed for differentially-expressed genes (Table 2 and Supplementary Tables 3 and 4). Several signalling pathways were enriched in the gene set upregulated in pain samples, including the TNF-alpha, TGF-beta, MAPK and TLR pathways (Letterio and Roberts, 1998; Morikawa et al., 2004; Wei et al., 2013; Cevikbas et al., 2014). Transcription factors linked to neuropathic pain in preclinical models, including FOS, FOSB and ATF3, and a number of well-known cytokine ligands including TNF, IL6 and CCL3 were also upregulated in at least two of the three pairs.
Table 2.
Fold change in transcripts per million in paired single patient samples (pain:no pain)
| Patient 15 | Patient 29 | Patient 26 | MAPK signalling | TNF-α signalling | TGF-β signalling | TLR signalling | OSM signalling | AHR signalling | SCI | |
|---|---|---|---|---|---|---|---|---|---|---|
| Transcription factors and co-factors | ||||||||||
| JUN | 2.62 | 1.24 | 1.83 | ✓ | ✓ | - | ✓ | - | ✓ | - |
| FOS | 2.60 | 2.00 | 10.40 | ✓ | - | - | ✓ | ✓ | - | ✓ |
| NFKB2 | 2.00 | 1.06 | 0.92 | - | ✓ | - | ✓ | - | - | - |
| RUNX2 | 2.75 | 1.22 | 1.93 | - | - | ✓ | - | - | - | - |
| FOSB | 13.55 | 13.53 | 283.90 | - | - | ✓ | - | - | - | - |
| ATF3 | 7.58 | 1.99 | 1.76 | - | - | ✓ | - | - | - | - |
| JUNB | 3.39 | 1.38 | 4.90 | - | - | ✓ | - | ✓ | ✓ | - |
| EGR1 | 6.28 | 2.47 | 10.19 | - | - | - | - | ✓ | - | ✓ |
| KLF10 | 2.35 | 1.28 | 1.47 | - | - | ✓ | - | - | - | - |
| NR4A1 | 3.21 | 1.01 | 1.76 | ✓ | - | - | - | - | - | ✓ |
| HES1 | 2.41 | 1.17 | 1.36 | - | - | - | - | - | ✓ | - |
| AHR | 2.02 | 1.26 | 1.04 | - | - | - | - | - | ✓ | - |
| BTG2 | 1.23 | 1.11 | 2.79 | - | - | - | - | - | - | ✓ |
| RNA binding proteins | ||||||||||
| ZFP36 | 2.29 | 1.31 | 4.12 | - | - | - | - | - | - | ✓ |
| Cytokine ligands | ||||||||||
| TNF | 3.71 | LE | 19.66 | ✓ | ✓ | - | ✓ | - | ✓ | ✓ |
| IL1B | 8.72 | LE | 9.09 | ✓ | - | - | ✓ | - | ✓ | ✓ |
| IL6 | 9.13 | LE | 4.68 | - | ✓ | - | ✓ | - | - | ✓ |
| IL12A | 3.01 | 1.40 | 1.31 | - | - | - | ✓ | - | ✓ | - |
| CCL2 | 3.04 | 1.00 | 0.93 | - | - | - | - | ✓ | - | ✓ |
| CCL3 | 12.52 | 3.45 | 75.40 | - | - | - | ✓ | - | - | - |
| CCL4 | 23.60 | 1.60 | 3035.20 | - | - | - | ✓ | - | - | - |
| CXCL2 | 1.92 | LE | 2.99 | - | - | - | - | - | - | ✓ |
| OSM | 10.39 | LE | 37.07 | - | - | - | - | ✓ | - | - |
| TGFB1 | 2.13 | 1.09 | 1.81 | ✓ | - | ✓ | - | - | ✓ | ✓ |
| TGFB3 | 4.77 | 1.35 | 1.81 | ✓ | - | ✓ | - | - | - | - |
| GDNF | 8.47 | LE | LE | - | - | - | - | - | - | ✓ |
| Cytokine regulators | ||||||||||
| SOCS3 | 14.41 | 2.84 | 4.64 | - | - | - | ✓ | ✓ | - | - |
| Neurotrophins | ||||||||||
| NGF | 2.30 | 1.04 | 1.39 | ✓ | - | - | - | - | - | - |
Genes involved in important signalling pathways or spinal cord injury, and their fold-change in the three pairs of samples from the same patients with differing pain states show several key transcription factors and cytokines to be upregulated in the pain state. LE = low expression; SCI = spinal cord injury.
✓ = gene set membership.
The analysis was broadened further to contrast the male-pain and male-no pain cohorts. The comparison yielded 70 genes that were upregulated and 52 genes that were downregulated in the male-pain cohort (Fig. 2C and Supplementary Table 5). Gene set enrichment analysis (Supplementary Table 6) showed an upregulated signature of genes related to spinal cord injury, and enrichment of several important signalling pathways (MAPK, TGFB, OSM and corticotrophin hormone pathways) that were similar to observations in the paired samples. Genes upregulated in pain samples include well known neuro-immune genes (CD93, CCL4, SOCS3), Schwann cell genes involved in rodent models of nerve injury (NR4A1, EGR1, EGR3), and genes known to be expressed in the human DRG and mouse sensory neurons (ARC, OMP, CHST1), suggesting crosstalk between immune cells and neurons/glia (Usoskin et al., 2015; Ray et al., 2018).
Interestingly, comparison of the male-pain and female-pain cohorts (Fig. 2D and Supplementary Table 7) yielded a more extensive set of differentially-expressed genes (426 autosomal genes upregulated in male-pain and 149 upregulated in female-pain cohorts). This could occur because some of the detected genes have sex-differential expression in baseline DRG while others could potentially underlie a sex-specific neuropathic pain pathology. It is interesting to note that based on gene set enrichment analysis (Supplementary Table 6), a different set of spinal cord injury-associated genes were upregulated in the female-pain cohort (TLR4, AIF1, OMG, C1QB) as compared to the male-enriched genes (EGR1, NR4A1, ZFP36, BTG2, MYC and others). Overlap with known lineage-specific gene modules in human macrophage lineages (Xue et al., 2014) suggests that some of the sex-differential gene expression in pain samples may be driven by macrophages (Supplementary Table 7). Human macrophage lineage-enriched genes up in the male-pain cohort (136 out of 426 autosomal genes) include CXCL2, TNF, and several transcription factors of the FOS-JUN family (FOS, FOSB, JUNB, JUND), while genes up in the female-pain cohort (75 of 149 autosomal genes) include several class A rhodopsin like G-protein coupled receptors (CX3CR1, ADORA3, P2RY13 and GPR65). While DRG-specific ion channels have been shown to be differentially expressed in mouse and rat models of neuropathic pain (Lacroix-Fralish et al., 2011; Zhang and Dougherty, 2014) and in human neuropathic pain (Li et al., 2018), we do not find statistically significant differences in abundances for ion channels expressed in human DRG (Supplementary Table 8), possibly due to regulation in translation or post-translation phases that are not reflected in RNA-seq data. Based on our cohort analysis, we found a set of ion channels (ANO8, GRIK5, GRIN1, HCN2, KCNAB2, KCNC1, KCNG1, KCNH2, KCNK3, PANX2) that have higher expression in the male-pain cohort compared to the female-pain cohort, again suggesting sexually dimorphic mechanisms (Supplementary Table 8).
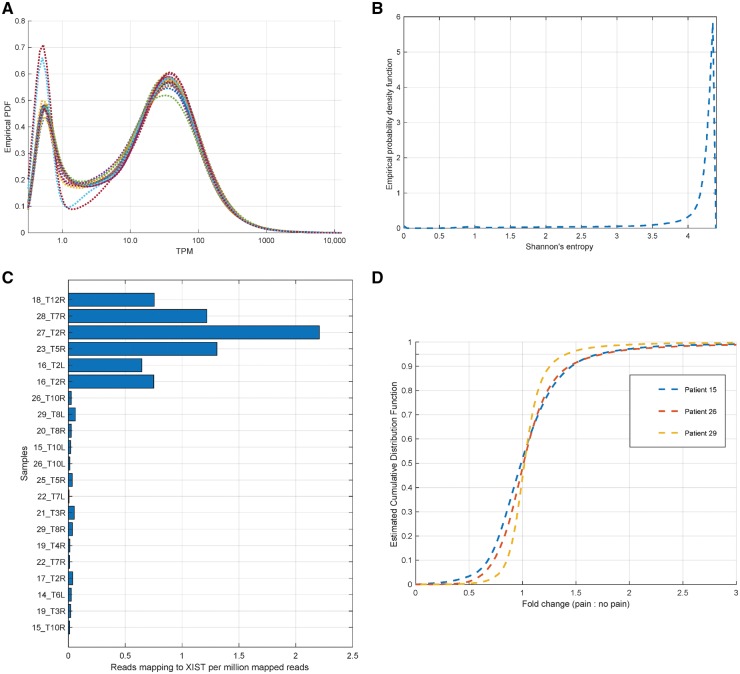
Multiple control analyses were performed on the data. The distribution of gene relative abundances (in transcripts per million) were plotted to ensure a similar distribution and comparable inflexion points in the distribution across samples (Fig. 3A). Genes with higher variability across samples in our dataset were identified in a cohort-agnostic manner using the notion of Shannon’s entropy (Fig. 3B). The sex of each sample was validated based on reads mapping to the XIST locus (Fig. 3C). For pain and non-pain samples derived from the same patient, the distribution of fold change in gene expression was quantified to identify the genes with the biggest change in abundance (Fig 3D).
Figure 3.
In silico controls for RNA-sequencing analysis. (A) The estimated probability density function for transcripts per million (TPMs) (smoothed by adding 0.5 to each value) show that all samples have approximately similar distributions over coding gene TPMs, along with a consistent number of genes expressed at 1.5 TPM or higher in each sample (between 13 850 and 14 715). (B) Genes with high variability in TPM across our datasets were identified in a fashion agnostic to clinical information by calculating Shannon’s entropy for each gene’s TPMs across RNA-seq samples, identifying genes with high variability (based on low entropy values in the left tail of the estimated distribution, value <3.5). Higher values correspond to more generic expression patterns. (C) Some well-known marker genes were checked in RNA-seq samples. The reported sex for each sample was independently verified using reads mapping to the XIST non-coding gene. (D) Estimated density function for the gene expression (for genes with transcripts per million >3.0 in either sample) fold change between pain and non-pain samples derived from the same patient, showing that a 2-fold change corresponds to the top 5th percentile.
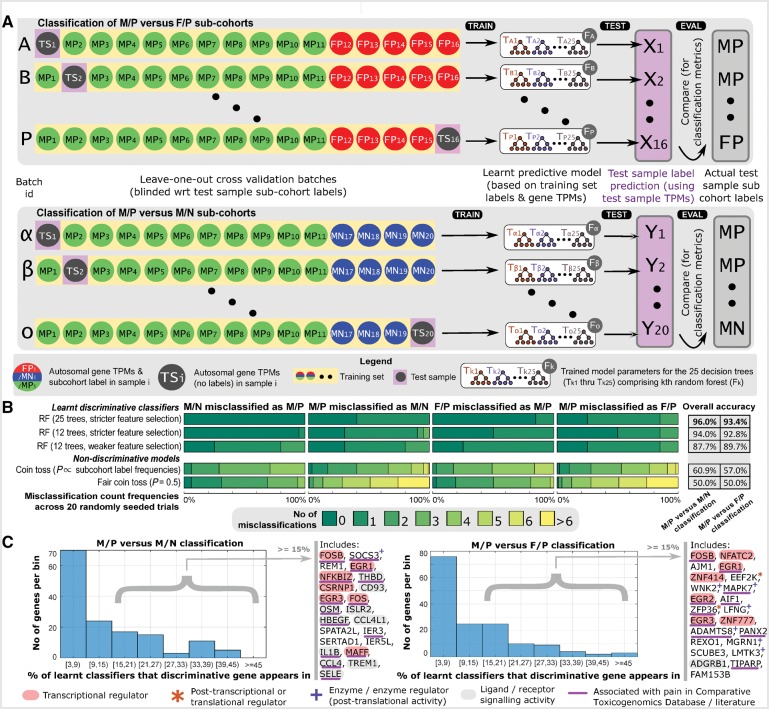
We predicted cohort membership for each sample (with the exception of the sole female-no pain sample) based on trained Random Forest classifiers. The cross validation training and testing batches we used are shown in Fig. 4A. We classified 11 male-pain and five female-pain samples using the male-pain versus female-pain classifier, and classified 11 male-pain and four male-no pain samples using the male-pain versus male-no pain classifier. This process was repeated 20 times (using random seeds to initialize the classifier training) to evaluate our classification algorithm. Of the 620 (31 classifications over 20 trials) predictions, we obtained a high (94.7%) accuracy in cohort membership prediction, suggesting that the gene expression changes we see are consistent and correlated and our classifier is able to harness this signal to perform classification (Fig. 4B). Random Forests are trained by identifying a set of discriminative features (in this case, genes) used to construct decision trees. We identified the genes that were most frequently chosen by the algorithm to construct Random Forests, since these were putatively the most reliable genes for discriminating across cohorts. For genes used in >15% of the trained Random Forests, we find that a majority of these genes overlap with the genes we identified in our cohort analysis in the previous section. They include genes coding for transcriptional regulators (like members of the FOS/JUN and EGR family), post transcriptional and translational regulators (ZFP36, EEF2K), transferases (WNK2, SOCS3, MAPK7), and signalling molecules (ISLR2, OSM, CD93, IL1B) (Fig 4C). Regulatory and signalling molecules in the discriminative gene set clearly suggests consistent usage of specific regulatory programs and signalling pathways, which could yield molecular signatures underlying human pain states in the future.
Figure 4.
Sample cohort prediction using Random Forests. (A) Leave one out cross validation schema is shown, with one sample (test sample) held out from training in each batch. The RNA profile of the test sample is then used by the trained classifier to predict its cohort membership, and the predicted cohort label is compared to actual cohort membership to evaluate the quality of classification. (B) Classification metrics for our optimal Random Forest model, using 25 decision trees, with no more than five decisions per tree, and using an input set of discriminative candidate genes is shown on top. Metrics from random forests built using 12 trees; as well as from random forests using 12 trees and a larger input set of candidate genes are also shown. Our classifier achieves discriminative results across a range of training parameters. We also show expected classification metrics for classifiers with no discriminative ability: based on models of biased and unbiased coin tosses. (C) A small set of genes are chosen for many of the random forests that we trained, suggesting a high predictive ability of these genes. Histograms show the number of genes that are chosen most frequently (in 3% to 45% of trained random forests) for both male-pain (M/P) versus female-pain (F/P) and male-pain versus male-no pain (M/N) classification. Genes chosen in >15% of the random forests include transcriptional/post transcriptional regulators, enzymes, and signalling molecules, many of which are associated with pain.
Finally, we compiled a list of studies that identified gene-neuropathic pain associations in humans or model species for the list of differentially expressed genes that we identified (Supplementary Table 9). Of ∼750 differently expressed genes across our analysis, 220 were identified in existing databases of pain-associated genes in humans. Therefore, while our dataset has substantial overlap with an existing knowledgebase in the field, we have identified a large cohort of new potential targets to investigate for neuropathic pain mechanisms based entirely on molecular investigation on patient samples.
Discussion
A key aspect of this study is the pairing of electrophysiology with RNA-seq for discovery of transcriptomic signatures of neuropathic pain. Though limited by a relatively small cohort and the multifactorial nature of each patient’s dermatomal pain (with potential contributions from local effects such as direct neural compression, peritumoral inflammation, tumour-derived soluble factors, and systemic conditions such as diabetes mellitus and/or prior treatments of patient’s malignancies), our findings allow several important conclusions.
First, there is a strong correlation between both radicular/neuropathic pain and radiographic nerve root compression to the presence of spontaneous activity and electrophysiological measures of hyperexcitability. Our results are similar to incidence of spontaneous activity reported in the literature for animal experiments with 10.3–20.5% for injured nerves versus 1.6–2.8% in controls (Liu et al., 2002; Ma and LaMotte, 2007; Li et al., 2017). Three physiological maladaptations were noted in recent work on the mechanisms underlying spontaneous activity in a model of spinal cord injury neuropathic pain. These included the development of a more positive resting membrane potential; a more hyperpolarized action potential threshold; and the occurrence of depolarizing spontaneous fluctuations in membrane potential (Odem et al., 2018). We found two of these occurring in human neurons with spontaneous activity, a more hyperpolarized action potential threshold (Table 1) and depolarizing spontaneous fluctuations (Fig. 1K). Therefore, we establish that the emergence of DRG neuron spontaneous activity and hyperexcitability are fundamental shared features between animal models of radicular/neuropathic pain and humans with clinically defined radicular/neuropathic pain.
It is perhaps surprising that significant changes in specific ion channels were only observed for the paired samples but not in the overall population analysis. There are a number of potential reasons for this. RNA-seq data measures the steady-state abundance of RNA species. This means that only changes at the transcriptional and post-transcriptional levels will be reflected in the data. There is clear evidence that specific ion channels contribute to ectopic spontaneous activity in human DRG neurons as shown by increased protein abundance changes and suppression of spontaneous activity using specific ion channel inhibitors (Li et al., 2017, 2018). But this can occur because of changes in translational regulation. Additionally, post translational regulation can also affect ion channel function. These changes would not be apparent in our datasets. Moreover, we performed bulk RNA-seq, with input coming from neuronal and non-neuronal cells, thus the signal for changes in a single sensory neuronal subpopulation (as would be the case for an ion channel such as Nav1.7) would be diluted in the bulk RNA-seq data. Future single cell assays (like imaging studies for in situ hybridization, or single cell RNA-seq) may be sufficiently sensitive to adequately capture such changes. Alternatively, changes in ion channel abundance may be temporally transient during the development of neuropathic pain. Electrophysiological recordings and RNA-sequencing are performed on the same donor DRG, but patients are at different times in the disease pathology. This cannot be controlled for in a clinical cohort like ours, but is always controlled for in animal studies, where much of the evidence for such changes originates. Finally, a combination of these points is likely.
The second broad conclusion that can be drawn here is that in male DRGs from painful dermatomes a transcriptional signature associated with spinal cord injury and enriched in signalling factors that converge on gp-130 receptors can be clearly identified. Given the known role of gp-130 expression in the DRG in preclinical pain models (Andratsch et al., 2009), our findings validate this pathway but unexpectedly implicate OSM and its receptor, OSMR, which forms a signalling complex with gp-130, in human neuropathic pain. This would not be predicted based on the preclinical literature which has predominately focused on IL6 as the primary mechanism for activating gp-130 in chronic pain. This finding has obvious implications for biological (e.g. antibody) development targeting this signalling system. We also uncover preliminary evidence of differences in transcriptomic signatures in the DRGs of males and females with neuropathic pain. While our cohort sizes are relatively modest and require further validation, this is consistent with emerging lines of evidence for sex differential neuroimmune response in preclinical models (Sorge et al., 2015; Lopes et al., 2017) and suggests the potential of sex-specific mechanisms for the development of neuropathic pain and spontaneous activity in DRG neurons.
Importantly, our study identifies sets of genes that are differentially expressed in the male-pain, male-no pain and female-pain cohorts. Our machine learning approach, which used a Random Forest model, finds that these genes have good predictive ability for identifying these cohorts, suggesting consistent changes in gene expression. We propose that this experimental framework will be useful in new datasets that are generated from independent projects to test if pain phenotypes can be reliably predicted from RNA-seq data. A limitation of this approach is that DRGs are not readily available from most clinical cohorts. However, some previous experiments in animal models have shown that certain immune cells can be predictive of transcriptomic changes in other nervous system areas in neuropathic pain (Massart et al., 2016). If this is also true in humans, it may eventually be possible to use a specific immune cell population as a proxy for transcriptomic changes in the DRG. This idea can be tested in ongoing studies with the clinical cohort described here.
Finally, while many of the genes we identified are known from previous human or (mostly) rodent studies, the majority of these have been understudied or not been studied in the context of neuropathic pain (e.g. OSM, discussed in the ‘Results’ section). Another excellent example is ISLR2. This mRNA encodes a protein called Linx that is known to play a role in the development of nociceptors (Mandai et al., 2009). Linx interacts with two well-known tyrosine receptor kinases, TrkA and TrkC, and previous work has shown a clear effect of this gene product in regulating how NGF signals through the TrkA receptor (Mandai et al., 2009). No previous studies have investigated the role of this gene in neuropathic pain but our machine learning approach identifies this gene as predictive of neuropathic pain phenotypes. Given the well-known role of NGF and TrkA signalling in pain, and the expanding clinical literature based on anti-NGF therapeutics with mixed results in neuropathic pain trials (Bannwarth and Kostine, 2014), we propose that this is an excellent example of a high-quality target for further exploration as a therapeutic intervention point.
In conclusion, our work provides the first evidence that neuropathic pain in humans is associated with spontaneous activity in the soma of DRG neurons. Combining this electrophysiological approach with bulk RNA-seq gives extensive new insight into mechanisms of neuropathic pain based entirely on clinical samples. Two important features of neuropathic pain emerging from this approach are marked sexual dimorphisms and nuances in known mechanisms that have important implications for therapeutic development. A caveat in consideration of these results is that the possibility exists that some of the results seen here could also be due to the influence of tumour-derived factors in addition to nerve injury.
Funding
This work was supported by grants from the National Institutes of Health: AI107067 (T.H.K.), NS 065926 (T.J.P.), CA200263 (P.M.D.), the Thompson Family Foundation Initiative (P.M.D.) and the H.E.B. Professorship in Cancer Research (P.M.D.).
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
Abbreviation
- DRG
dorsal root ganglion
References
- Andratsch M, Mair N, Constantin CE, Scherbakov N, Benetti C, Quarta S et al. A key role for gp130 expressed on peripheral sensory nerves in pathological pain. J Neurosci 2009; 29: 13473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannwarth B, Kostine M. Targeting nerve growth factor (NGF) for pain management: what does the future hold for NGF antagonists? Drugs 2014; 74: 619–26. [DOI] [PubMed] [Google Scholar]
- Baumann TK, Burchiel KJ, Ingram SL, Martenson ME. Responses of adult human dorsal root ganglion neurons in culture to capsaicin and low ph. Pain 1996; 65: 31–8. [DOI] [PubMed] [Google Scholar]
- Bilsky MH, Laufer I, Fourney DR, Groff M, Schmidt MH, Varga PP et al. Reliability analysis of the epidural spinal cord compression scale. J Neurosurg Spine 2010; 13: 324–8. [DOI] [PubMed] [Google Scholar]
- Borsook D, Hargreaves R, Bountra C, Porreca F. Lost but making progress–Where will new analgesic drugs come from? Sci Transl Med 2014; 6: 249sr3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevikbas F, Kempkes C, Buhl T, Mess C, Buddenkotte J, Steinhoff M. Role of interleukin-31 and oncostatin M in itch and neuroimmune communication. In: Carstens E, Akiyama T, editors. Itch: Mechanisms and Treatment. Boca Raton, FL: CRC Press/Taylor & Francis Publishers; 2014. [PubMed] [Google Scholar]
- Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics 2012; 99: 323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S, Copits BA, Zhang J, Page G, Ghetti A, Gereau RW. Human sensory neurons: membrane properties and sensitization by inflammatory mediators. Pain 2014; 155: 1861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavva NR, Treanor JJ, Garami A, Fang L, Surapaneni S, Akrami A et al. Pharmacological blockade of the vanilloid receptor TRPV1 elicits marked hyperthermia in humans. Pain 2008; 136: 202–10. [DOI] [PubMed] [Google Scholar]
- Gereau RW, Sluka KA, Maixner W, Savage SR, Price TJ, Murinson BB et al. A pain research agenda for the 21st century. J Pain 2014; 15: 1203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanpaa M, Attal N, Backonja M, Baron R, Bennett M, Bouhassira D et al. NeuPSIG guidelines on neuropathic pain assessment. Pain 2011; 152: 14–27. [DOI] [PubMed] [Google Scholar]
- Hill R. NK1 (substance P) receptor antagonists–why are they not analgesic in humans? Trends Pharmacol Sci 2000; 21: 244–6. [DOI] [PubMed] [Google Scholar]
- Lacroix-Fralish ML, Austin JS, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain 2011; 152: 1888–98. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annu Rev Immunol 1998; 16: 137–61. [DOI] [PubMed] [Google Scholar]
- Li Y, Adamek P, Zhang H, Tatsui CE, Rhines LD, Mrozkova P et al. The cancer chemotherapeutic paclitaxel increases human and rodent sensory neuron responses to TRPV1 by activation of TLR4. J Neurosci 2015; 35: 13487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, North RY, Rhines LD, Tatsui CE, Rao G, Edwards DD et al. DRG voltage-gated sodium channel 1.7 is upregulated in paclitaxel-induced neuropathy in rats and in humans with neuropathic pain. J Neurosci 2018; 38: 1124–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tatsui CE, Rhines LD, North RY, Harrison DS, Cassidy RM et al. Dorsal root ganglion neurons become hyperexcitable and increase expression of voltage-gated T-type calcium channels (Cav3.2) in paclitaxel-induced peripheral neuropathy. Pain 2017; 158: 417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013; 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CN, Devor M, Waxman SG, Kocsis JD. Subthreshold oscillations induced by spinal nerve injury in dissociated muscle and cutaneous afferents of mouse DRG. J Neurophysiol 2002; 87: 2009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes DM, Melek N, Edye M, Jager SB, McMurray S, McMahon SB et al. Sex differences in peripheral not central immune responses to pain-inducing injury. Sci Rep 2017; 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, LaMotte RH. Multiple sites for generation of ectopic spontaneous activity in neurons of the chronically compressed dorsal root ganglion. J Neurosci 2007; 27: 14059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2005; 33(Database issue): D54–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K, Guo T, St Hillaire C, Meabon JS, Kanning KC, Bothwell M et al. LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development. Neuron 2009; 63: 614–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R, Dymov S, Millecamps M, Suderman M, Gregoire S, Koenigs K et al. Overlapping signatures of chronic pain in the DNA methylation landscape of prefrontal cortex and peripheral T cells. Sci Rep 2016; 6: 19615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Tamura S, Minehata K, Donovan PJ, Miyajima A, Senba E. Essential function of oncostatin m in nociceptive neurons of dorsal root ganglia. J Neurosci 2004; 24: 1941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odem MA, Bavencoffe AG, Cassidy RM, Lopez ER, Tian J, Dessauer CW et al. Isolated nociceptors reveal multiple specializations for generating irregular ongoing activity associated with ongoing pain. Pain 2018; 159: 2347–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018; 159: 1325–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge RE, Mapplebeck JC, Rosen S, Beggs S, Taves S, Alexander JK et al. Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci 2015; 18: 1081–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015; 18: 145–53. [DOI] [PubMed] [Google Scholar]
- van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain 2014; 155: 654–62. [DOI] [PubMed] [Google Scholar]
- Wei XH, Na XD, Liao GJ, Chen QY, Cui Y, Chen FY et al. The up-regulation of IL-6 in DRG and spinal dorsal horn contributes to neuropathic pain following L5 ventral root transection. Exp Neurol 2013; 241: 159–68. [DOI] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014; 40: 274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dougherty PM. Enhanced excitability of primary sensory neurons and altered gene expression of neuronal ion channels in dorsal root ganglion in paclitaxel-induced peripheral neuropathy. Anesthesiology 2014; 120: 1463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The neurophysiological data can be shared on request. The full transciptomic dataset and code for analysis is available at: https://www.utdallas.edu/bbs/painneurosciencelab/sensoryomics/hdrgclinical/. Raw sequencing datasets are available from the dbGaP repository as single-end read libraries (phs001158.v2.p1).