Abstract
Parkinson’s disease (PD) is a progressive, neurodegenerative disease that presents with significant motor symptoms, for which there is no diagnostic chemical test. We have serendipitously identified a hyperosmic individual, a “Super Smeller” who can detect PD by odor alone, and our early pilot studies have indicated that the odor was present in the sebum from the skin of PD subjects. Here, we have employed an unbiased approach to investigate the volatile metabolites of sebum samples obtained noninvasively from the upper back of 64 participants in total (21 controls and 43 PD subjects). Our results, validated by an independent cohort (n=31), identified a distinct volatiles-associated signature of PD, including altered levels of perillic aldehyde and eicosane, the smell of which was then described as being highly similar to the scent of PD by our “Super Smeller”.
Short abstract
Sebum from the upper back, analyzed using a mass spectrometer hyphenated to an odor port reveals a unique volatilome associated with Parkinson’s disease (PD) smell, useful for diagnosing PD noninvasively.
Background
Physicians in ancient times, including Hippocrates, Galenus, and Avicenna, used odor as a diagnostic tool. Although the olfactory skills of physicians are not routinely used in modern medicine, it is well documented that a number of conditions, predominantly metabolic and infectious diseases, are associated with a unique odor,1 but there is scant evidence for odors as symptoms of neurodegenerative disorders. To the best of our knowledge, this is the first study that demonstrates the use of sebum as biofluid to screen for Parkinson’s disease (PD). There have been a small number of metabolomics studies of PD using various biofluids such as blood, feces, saliva, urine, and cerebrospinal fluid, as well as insect and mouse models of PD as described in this recent review by Shao and Le;2 there is no mention of a “PD odor”. Joy Milne, a Super Smeller whose husband Les was diagnosed with PD in 1986, has demonstrated a unique ability to detect PD by odor.3 Joy has an extremely sensitive sense of smell, and this enables her to detect and discriminate odors not normally detected by those of average olfactory ability. Preliminary tests with T-shirts and medical gauze indicated the odor was present in areas of high sebum production, namely, the upper back and forehead, and not present in armpits.3 Sebum is a waxy, lipid-rich biofluid excreted by the sebaceous glands in the skin, overproduction of which known as seborrhea, is a known non-motor symptom of PD.4,5 PD skin has recently been shown to contain phosphorylated α-synuclein, a molecular hallmark of PD.6,7 Identification and quantification of the compounds that are associated with this distinctive PD odor could enable rapid, early screening of PD as well as provide insights into molecular changes that occur as the disease progresses and enable stratification of the disease in the future.
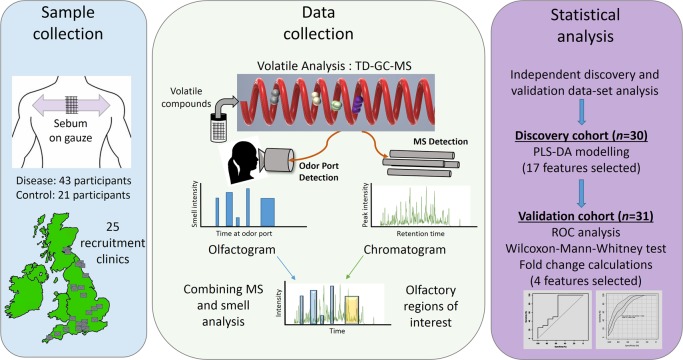
Volatile organic compounds (VOCs) are often associated with characteristic odors, although some volatiles may also be odorless. The term “volatilome” describes the entirety of the volatile organic and inorganic compounds that may originate from any organism, or object, which may be analytically characterized. For any given sample under ambient conditions in a confined environment, collecting, identifying, and measuring molecules in its headspace will then define its volatilome. Such measurements can be performed with thermal desorption–gas chromatography–mass spectrometry (TD–GC–MS), where a sample is placed in a closed vessel. The sample is then heated to encourage the production of volatiles, and the headspace is captured for analysis by GC–MS. Investigation of volatile metabolites using mass spectrometry has proven to be extremely useful in clinical studies8−11 as well as in the analysis of the consistency and provenance of edible items.12−14 Recently, TD-GC-MS has been used as a volatilome analysis platform for the detection of compounds from bacteria implicated in ventilator associated pneumonia,10 for differentiation between odors due to human and animal decomposition,15 as well as aerosol detection of the fumes from e-cigarettes.16 This versatility of TD-GC-MS for samples from many sources renders it highly suitable for use in identifying the metabolites that give rise to the distinct scent of PD. We have established a workflow that starts in clinics with the collections of sebum samples from the upper backs of PD patients along with matched control subjects and progresses to the discovery of disease specific volatile metabolites, the odor of which is confirmed by our Super Smeller (Figure 1, Supporting Information and Table S1A).
Figure 1.
Schematic outline of the workflow described in this study - from sample collection to biomarker discovery. Parkinson’s disease patient samples and control participant samples were collected from 25 sites across the UK using gauze swabs to sample the sebum from the top back region from 64 people. Thermal desorption–gas chromatography–mass spectrometry (TD–GC–MS) analysis was performed alongside olfactory analysis, results of which were then combined. Statistical analysis was performed on two independent cohorts. Data from discovery cohort consisting of 30 participants were used to create a partial least-squares-discriminant analysis (PLS-DA) model, and differential features found as a result were then targeted for the presence in a separate validation cohort consisting of 31 participants. The significance of these biomarkers was tested using receiver operating characteristic (ROC) analyses and the Wilcoxon-Mann–Whitney test. Finally, four features that showed similar statistical significance and expression in both cohorts were selected for biological interpretation of data.
In the current study, VOCs from the sample headspace were measured in two cohorts: a “discovery” cohort (n = 30) and a “validation” cohort (n = 31), to validate discovered biomarkers17 (for demographics of each cohort see Tables S1B and S5). A third cohort consisting of three drug-naïve PD participants was used for GC-MS analysis in conjunction with a human Super Smeller via an odor port (Figure 1). This proof of principal study provides the first description of the skin volatilome in PD compared to control subjects.
Study Participants
The participants for this study were part of a nationwide recruitment process taking place at 25 different NHS clinics. The participants for this study were selected at random from these sites. The study was performed in three stages. The first two stages (discovery and validation) consisted of 61 samples (a mixture of control, PD participants on medication, and drug naïve PD subjects as shown in Table S1B). The first cohort was used for volatilome discovery, and the second cohort was used to validate the significant features discovered in first cohort. A third cohort consisting of three drug naïve PD participants was used for smell analysis from the Super Smeller. Ethical approval for this project (IRAS project ID 191917) was obtained by the NHS Health Research Authority (REC reference: 15/SW/0354). The metadata analysis for these participants is reported in Table S1B. The study design was as outlined in Figure 1.
Sample Collection
The sampling involved each subject being swabbed on the upper back with a medical gauze. The gauze with sebum sample from the participant’s upper back was sealed in background-inert plastic bags and transported to the central facility at the University of Manchester, where they were stored at −80 °C until the date of analysis.
Analytical Method: TD–GC–MS Analysis
Description of the Technique
A dynamic headspace (DHS) GC–MS method was developed for the analysis of gauze swabs which contained sampled participant sebum. DHS is a sample preparation capability for subsequent GC application using the GERSTEL MultiPurpose sampler (MPS) that concentrates VOCs from liquid or solid samples. The sample is incubated while the headspace is purged with a controlled flow of inert gas through an adsorbent tube. Once extraction and preconcentration are completed, the adsorbent tube is automatically desorbed using the GERSTEL thermal desorption unit (TDU). Analytes are then cryofocused on the GERSTEL cool injection system (CIS) programmed temperature vaporizer (PTV) injector before being transferred to the GC for analysis.
In order to correlate the PD molecular signature to the PD smell, the same setup was used in combination with the GERSTEL olfactory detection port (ODP). The ODP allows detection of odorous compounds as they elute from the GC, by smell. In fact, the gas flow is split as it leaves the column between the detector of choice (in our case MS) and the ODP to allow simultaneous detection on the two analytical tools. The additional smell profile information can then be acquired as an olfactogram. Voice recognition software and intensity registration allow direct annotation of the chromatogram.
Method Details
Gauze swabs were transferred into 20 mL headspace vials and then analyzed by DHS–TD–GC–MS. For the DHS preconcentration step, samples were incubated for 5 min at 60 °C before proceeding with the trapping step. Trapping was performed by purging 500 mL of the sample headspace at 50 mL·min–1 through a Tenax TA adsorbent tube kept at 40 °C (GERSTEL, Germany). Dry nitrogen was used as the purge gas. To release the analytes, the adsorbent trap was desorbed in the TDU in splitless mode. The TDU was kept at 30 °C for 1 min then ramped at 12 °C·s–1 to 250 °C and held for 5 min. Desorbed analytes were cryofocused in the CIS injector. The CIS was operated in solvent vent mode, using a vent flow of 80 mL·min–1 and applying a split ratio of 10. The initial temperature was kept at 10 °C for 2 min, then ramped at 12 °C·s–1 to 250 °C and held for 10 min. The GC analysis was performed on an Agilent GC 7890B coupled to an Agilent MSD 5977B equipped with high efficiency source (HES) operating in EI mode. Separation was achieved on an Agilent HP-5MS Ultra inert 30 m × 0.25 mm × 0.25 μm column. The column flow was kept at 1 mL·min–1. The oven ramp was programmed as follows: 40 °C held for 5 min, 10 °C·min–1 to 170 °C, 8 °C·min–1 to 250 °C, 10 °C·min–1 to 260 °C held for 2 min for a total run time of 31 min. The transfer line to the MS was kept at 300 °C. The HES source was kept at 230 °C and the Quadrupole at 150 °C. The MSD was operated in scan mode for the mass range between 30 and 800 m/z. For the olfactometry approach, the chromatographic flow was split between the mass spectrometer and the GERSTEL olfactory detection port (ODP3) using Agilent Technologies Capillary Flow Technology (three-way splitter plate equipped with makeup gas). The ODP3 transfer line was kept at 100 °C, and humidity of the nose cone was maintained constant.
Data Preprocessing and Deconvolution
TD–GC–MS data were converted to open source mzXML format using ProteoWizard. Each cohort data set was deconvolved separately using eRah package for R.18 Upon deconvolution, in discovery cohort 207 features and in validation cohort 210 features were assigned to detected peaks. The deconvolved analytes were assigned putative identifications by matching fragment spectra with compound spectra present in the Golm database, NIST library, and Fiehn GCMS library. In discovery cohort 163 features were assigned a putative identification, and in validation cohort 156 features were assigned a putative identification. The resulting matrices for each cohort consisted of variables and their respective area under the peak for each sample. All data were normalized for age and total ion count to account for confounding variables (Table S1B).
Statistical Analyses
The discovery cohort data analysis included a global analysis of all the detected compounds. PLS-DA modeling was carried out using all the measured features. We have not included PCA results because, using this unsupervised clustering method, we were unable to see any clustering of data. We attribute this to the complex nature of metabolomics data especially for volatile metabolites. This results in high dimensionality of the data, and it is unrealistic to expect that the separation between PD and controls is the most dominating variance in the data and thus results in poorer display on PCA/MDS plots. Often supervised modeling is required to train the models to find defined differences by overcoming noise.
The data were log10-scaled, and Pareto scaled prior to Wilcoxon-Mann–Whitney analysis, PLS-DA, and the production of ROC curves. The PLS-DA modeling was performed and executed using MATLAB (2018a),19,20 and the MATLAB functions are freely available from our in-house cluster toolbox hosted at https://github.com/biospec. ROC curves were generated using the R package called pROC.21 The samples from both cohorts were also analysed together combined as a single dataset, thus increasing sample size and providing better statistical power while evaluating the performance of this panel of biomarkers (Figure 2c, Figure S1). ROC curves were generated by Monte Carlo cross validations (MCCV) using balanced subsampling. In each of the MCCV, two-thirds of the samples were used to evaluate the feature importance. The top two, three, five, seven, and nine important features were then used to build classification models, which were validated using the remaining one-third of the samples. The process was repeated 500 times to calculate the average performance and confidence interval of each model. Classification and feature ranking were performed using a PLS-DA algorithm using two latent variables (Figure 2c).
Figure 2.
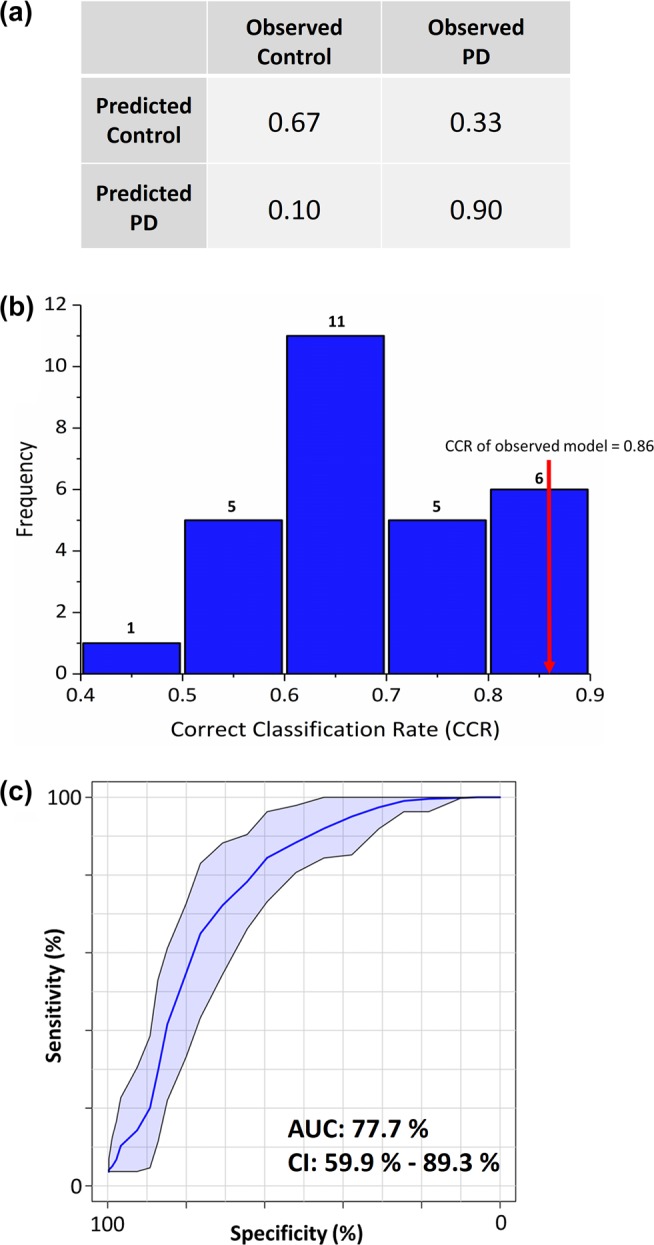
PLS–DA classification model. (a) Classification matrix of PLS–DA model validated using 5-fold cross validation showing 90% correct prediction of Parkinson’s disease samples. (b) PLS–DA modeling was further tested using permutation tests (where the output classification was randomized; n = 28), and results were plotted as a histogram which showed frequency distribution of correct classification rate (CCR) which yielded CCRs ranging between 0.4 and 0.9 for permutated models. The observed model was significantly better than most of the permuted models (p < 0.1), shown by the red arrow. (c) ROC plot generated using combined samples from both cohorts and the panel of four metabolites that was common and differential between control and PD. The shaded blue area indicates 95% confidence intervals calculated by Monte Carlo cross validation (MCCV) using balanced subsampling with multiple repeats.
When performing k-nearest neighbors analysis, k was chosen to be 5 given the small sample size, and the distance parameter used was Euclidean distance which was used as weights such that closer neighbors of a query point have a greater influence than the neighbors farther away. During random forest analysis of the same data, 10 decision trees were grown, and the growth control was achieved by not splitting into subsets smaller than five. The SVM model was built using LIBSVM, implemented in e1071 package of R,22 with a linear kernel. The cost (C) and regression loss epsilon (∈) were determined by performing a grid search and were set at C = 10 and ∈ was set at 0.10.
Safety Statement
No unexpected or unusually high safety hazards were encountered in the course of this work.
Results and Discussion
A partial least-squares discriminant analysis (PLS–DA) model was built using the discovery cohort data (Figure 2). The classification accuracy of this model was validated by a bootstrapping approach (n = 1000). The variables contributing to classification (n = 17) were selected using variable importance in projections (VIP) scores where VIP > 1. We note at this stage that one of the 17 metabolites found is 3,4-dihydroxy mandelic acid, a metabolite of norepinephrine in humans. This catechol is also a metabolite of L-dopa, one of the most commonly prescribed medication for Parkinson’s. In this study, 3,4-dihydroxy mandelic acid is observed in both drug naïve participants and control participants, indicating its presence may originate from endogenous mandelic acid instead of PD drugs. Norepinephrines including 3,4-dihydroxy mandelic acid are key molecules in the anabolism of brain neurotransmitters. Changes in neurons and neurotransmitters are an extremely well-known characterization of PD;23 for instance, the decrease of dopamines, a precursor to 3,4-dihydroxy mandelic acid, is a known characterization of PD. It could, therefore, be hypothesized that the presence of endogenous 3,4-dihydroxy mandelic acid could be indicative of altered levels of neurotransmitters in PD.
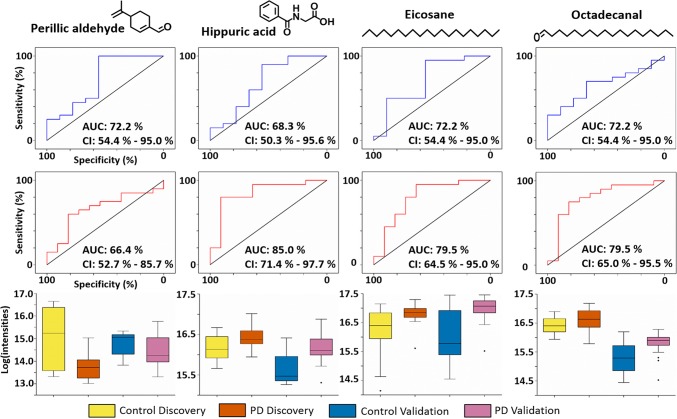
The measured volatilome in the validation cohort data (from a different population than the discovery cohort) was targeted for the presence or absence of these discovered biomarkers. Out of these 17 metabolites, 13 were also found in the validation cohort data, and nine of these had retention times that allowed us to confidently assign them as identical (Table S2). These nine biomarkers found in both cohorts were selected for further analysis and statistical testing. To evaluate the performance of these biomarkers, we conducted receiver operating characteristic (ROC) analyses with data from both the discovery cohort and the validation cohort (Figure S1). ROC curves and Wilcoxon-Mann–Whitney tests as well as fold-change calculations on individual metabolites shows four out of these nine metabolites had a similar trend in regulation between the discovery and validation cohorts, and their performance was also similar as measured by AUC (Table 1, Figure 3). The results from the combined analysis using both cohorts as a single experiment indicate increased confidence in the data (p-values in Table 1, confidence intervals in Figure S1).
Table 1. Panel of Four Volatile Metabolites That Were Found to Be Differential between Parkinson’s and Control Samples, with Similar Trends Observed in Expression and AUC Curves Measured by ROC Analysesa.
| FDR
corrected p-value (Mann–Whitney test) |
expression
(PD/control) |
||||||
|---|---|---|---|---|---|---|---|
| putative identification | parent mass | ΔRT (min) | discovery | validation | combined | discovery | validation |
| perillic aldehyde | 150.22 | 0.15 | 0.0279 | 0.0403 | <0.0001 | down | down |
| hippuric acid | 179.17 | 0.09 | 0.1908 | 0.0403 | 0.1833 | up | up |
| eicosane | 282.56 | 0.03 | 0.0279 | 0.0403 | 0.0013 | up | up |
| octadecanal | 170.34 | 0.12 | 0.2605 | 0.0604 | 0.3040 | up | up |
Perillic aldehyde and Eicosane were significantly down-regulated and up-regulated in PD, respectively (FDR corrected p < 0.05) in both cohorts.
Figure 3.
ROC curves and box plots for analytes of interest: In each panel from top to bottom: ROC curves for both discovery (blue) and validation (red) cohort for four analytes common to both experiments. Confidence intervals were computed with 2000 stratified bootstrap replicates, and diagonal black line represents random guess. Box plots show comparison of means of log scaled peak intensities of these analytes, where black dots were outliers.
We adhered to the Metabolomics Standards Initiative (MSI) guidelines for data analysis and for assignment of identity to features of interest,17 and all identified features are at MSI level two, which means these are putatively annotated compounds (i.e. without chemical reference standards, based upon physicochemical properties and/or spectral similarity with public/commercial spectral libraries). The compounds perillic aldehyde and eicosane are significantly different between PD and control in both the cohorts (p < 0.05): perillic aldehyde was observed to be lower in PD samples, whereas eicosane was observed at significantly higher levels. Although hippuric acid and octadecanal were not significantly different (p > 0.05), the AUC and box plots (Figure 3) between the two cohorts were comparable and showed similar trends of being increased in PD. Previous studies have reported varying abundances of these compounds in other biofluids (Table 2).
Table 2. Known Normal Abundances of Molecules of Interest (listed in Table 1) Measured Using Mass Spectrometry Approaches, As Reported in the Literature.
| molecule | biospecimen | abundance | reference |
|---|---|---|---|
| eicosane | feces | not quantified | Garner et al.26 |
| saliva | not quantified | Soini et al.27 | |
| saliva | not quantified | Costello et al.28 | |
| hippuric acid | cerebrospinal fluid | 3.0 (0.0–0.5) μM | Hoffman et al.29 |
| urine | not quantified | Hanhineva et al.30 | |
| blood | 16.74 ± 11.16 μM | Duraton et al.31 | |
| octadecanal | feces | not quantified | Dixon et al.32 |
| perillic aldehyde | saliva | not quantified | Costello et al.28 |
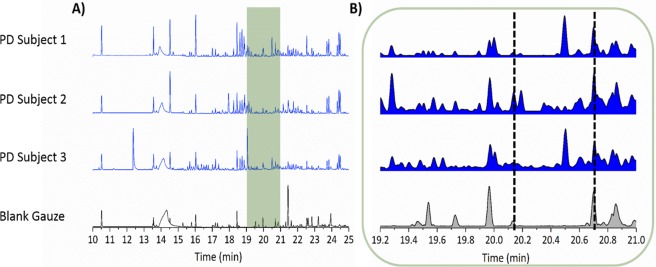
Using an odor port attached to the GC–MS instrument, the Super Smeller identified times at which any smell was present and also more importantly the times at which a specific “musky” smell of PD was detected. Data were presented in the form of an olfactogram, where the presence and relative intensity of each smell were recorded at its corresponding chromatographic retention time. Olfactogram results obtained from the odor port were overlaid on the respective total ion chromatogram from GC–MS (Figure 4A). There was significant overlap between regions that contained up-regulated compounds and regions in which a smell similar or identical to that of PD scent was present. In the chromatographic trace the region between 19 and 21 min is of particular interest (Figure 4B) since the smell associated with the mixture of analytes in that window was described as “very strong” and “musky”. This is the same region where three compounds, viz. hippuric acid, eicosane, and octadecanal, have been detected in both cohorts, and all three were found to be up-regulated in PD subjects.
Figure 4.
Comparison of GC–MS chromatogram to description of olfactory data described by the Super Smeller: GC–MS chromatogram from three drug naïve Parkinson’s subjects and a blank gauze. (A) The 10–25 min retention time range of the chromatographic analysis in which the Super Smeller described various odors associated with different GC–MS peaks. The overlaid green shaded area shows the overlap between real time GC–MS analysis and the Super Smeller describing a “strong PD smell” via the odor port. (B) A zoom of the green highlighted area from A. This region is of particular interest as three out of four identified compounds are found here (Tables 1 and S2); it encompasses the time during which the Super Smeller described a musky PD-like scent as being “very strong” (between the time lines at 19 and 21 min) for the PD samples and not for the blank. It can be noted that none of these compounds are found in blank gauze (bottom chromatogram) within the same retention time window as shown by normalized relative peak intensities to the highest peak in each chromatogram. The area between black dotted lines highlights the presence of compounds in PD samples but complete absence in the blank gauze.
In order to validate mass spectrometry led biomarkers and to verify the resultant scent, the candidate compounds listed in Table S2 (n = 17) were purchased and spiked onto gauze swabs (Table S3). An exploratory study with our Super Smeller was performed in which multiple mixtures of compounds (n = 5) were spiked onto both blank gauze swabs and swabs that contained control sebum. Two final dispensed volumes of the mixtures were tested (40 and 100 μL), and all compounds used were at a single concentration (10 μM). In these blinded tests, the Super Smeller grouped the samples in order of PD-like odor. She was able to isolate the swabs with a sebum background matrix and described them as more familiar to the PD-like smell than without control sebum. Further tests utilized control sebum as a background matrix for spiking candidate compounds, and a range of concentrations was then selected for testing. Mixtures of the candidate compounds (n = 17) were prepared at a range of concentrations (10 μM, 5 μM, 0.5 μM, 0.05 μM, 0.005 μM) and presented to the Super Smeller in a second blinded test; she was again asked to rank in order of PD-like smell. These results demonstrated she could detect (although not in any systematic order) the whole range of concentrations offered, and a concentration between 0.05 μM and 0.5 μM gave her the best response. A validation study consisting of three compound mixtures with significance from the MS analysis aimed to distinguish the combination that best gave rise to the most PD-like smell. Three mixture combinations were chosen at a single concentration (0.5 μM): all candidate compounds (n = 17), all compounds identified in both the discovery and validation cohorts (n = 9), and the panel of compounds expressed in same direction and differential between PD and control (n = 4). The mixture of nine compounds was consistently described as being most akin to the PD-like odor and was slightly overlapped by description and rank with the mixture of four compounds. The mixture of 17 compounds was grouped as the same “smell” as the other two combinations; however they were described as significantly weaker. We hypothesize this is due to a lower concentration of each compound in the mixture and thus higher interference from background solvent smell. The results from these tests are depicted in Figure S2 whereby the intensity and correlation to the PD-like smell partition the groups of samples tested. We do not conclude that these chemicals alone constitute the unique smell associated with PD; rather, we demonstrate that they contribute to it.
From results obtained from three independent sets of data, from different people with one underlying factor (i.e., PD) separating them, it was clear that several volatile features were found to be significantly different between control and PD participants. There were no significant differences observed between PD participants on medication and drug naïve PD participants (p > 0.05 for all measured volatiles), indicating that the majority of the analyzed volatilome and by inference sebum are unlikely to contain drug metabolites associated with PD medication. In addition, applying machine learning approaches such as k-nearest neighbors, random forest, and support vector machines (SVM) did not lead to a classification between drug naïve PD participants and PD participants on medication (results in Table S4).
Perillic aldehyde and octadecanal are ordinarily observed as plant metabolites or food additives. It can be hypothesized that with increased and altered sebum secretion such lipid-like hydrophobic metabolites may be better captured or retained on the sebum-rich skin of PD subjects. Skin disorders in PD have been observed previously, and seborrheic dermatitis (SD) in particular has been flagged as a premotor feature of PD.23 It has been reported by Arsenijevic and co-workers5 that PD patients who suffer from SD have increased Malassezia spp. density on their skin and commensurate higher lipase activity required metabolically by yeast. This increased lipase activity could correlate with the enhanced production of eicosane, perillic aldehyde, and octadecanal as highly lipophilic molecules since Malassezia spp. requires specific exogenous lipids for growth. Eicosane is reported as being produced by Streptomyces spp. as an antifungal agent24 which also supports its increased presence on the skin of PD sufferers. The effects observed in our study could also signal altered microbial activity on the skin of PD patients that may affect the skin microflora resulting inchanges in the production of metabolites such as hippuric acid.25 These potential explanations for the change in odor in PD patients suggest a change in skin microflora and skin physiology that is highly specific to PD.
Conclusion
In conclusion, our study highlights the potential of comprehensive analysis of sebum from PD patients and raises the possibility that individuals can be screened noninvasively based on targeted analysis for these volatile biomarkers. We do acknowledge that the current study is limited with smaller sample size, but the power of this study is a different validation cohort that consisted of completely different participants. This validation cohort was able to verify the findings and classification model built using data from our discovery cohort. A larger study with extended olfactory data from human smellers as well as canine smellers in addition to headspace analyses is the next step in further characterizing the PD sebum volatilome. This will enable the establishment of a panel of volatile biomarkers associated with PD and will open new avenues for stratification as well as facilitate earlier detection of PD and further the understanding of disease mechanisms.
Acknowledgments
We thank Michael J Fox Foundation and Parkinson’s UK for funding this study. We also thank our nationwide NIHR recruitment centers for their enthusiasm and rigor during the recruitment process. We are very grateful to all the participants who took part in this study as well as PIs and nurses across all the recruiting centers who helped us with the recruitment of participants.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00879.
Description of participant metadata, list of candidate biomarkers, additional statistical testing, qualitative smell analysis, and list of all the PIs from recruitment centers that participated in this study (PDF)
Author Contributions
D.K.T., E.S., C.L., J.M., and P.B. performed or contributed to the sample preparation and data collection. D.K.T. and Y.X. performed statistical analysis and validation of data. D.K.T., E.S., J.M., R.G., and P.B. designed the experiments. P.B., R.G., M.S., and T.K. conceived and supervised the study. D.K.T. wrote the manuscript, and all authors commented and contributed to the final version of manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Shirasu M.; Touhara K. The scent of disease: volatile organic compounds of the human body related to disease and disorder. J. Biochem. 2011, 150 (3), 257–266. 10.1093/jb/mvr090. [DOI] [PubMed] [Google Scholar]
- Shao Y.; Le W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol. Neurodegener. 2019, 14 (1), 3. 10.1186/s13024-018-0304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J. Joy of super smeller: sebum clues for PD diagnostics. Lancet Neurol. 2016, 15, 138–139. 10.1016/S1474-4422(15)00396-8. [DOI] [PubMed] [Google Scholar]
- Krestin D. The Seborrhoeic Facies as a Manifestation of Post-Encephalitic Parkinsonism and Allied Disorders. QJM 1927, os-21 (81), 177–186. 10.1093/qjmed/os-21.81.177. [DOI] [Google Scholar]
- Ravn Jørgensen A.-H.; Thyssen J. P.; Egeberg A. Skin disorders in Parkinson’s disease: potential biomarkers and risk factors. Clin., Cosmet. Invest. Dermatol. 2017, 10, 87–92. 10.2147/CCID.S130319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zange L.; Noack C.; Hahn K.; Stenzel W.; Lipp A. Phosphorylated alpha-synuclein in skin nerve fibres differentiates Parkinson’s disease from multiple system atrophy. Brain 2015, 138 (Pt 8), 2310–2321. 10.1093/brain/awv138. [DOI] [PubMed] [Google Scholar]
- Donadio V.; Incensi A.; Leta V.; Giannoccaro M. P.; Scaglione C.; Martinelli P.; Capellari S.; Avoni P.; Baruzzi A.; Liguori R. Skin nerve alpha-synuclein deposits: a biomarker for idiopathic Parkinson disease. Neurology 2014, 82 (15), 1362–9. 10.1212/WNL.0000000000000316. [DOI] [PubMed] [Google Scholar]
- Bellmann-Weiler, A. P. a. W. F. a. J. W. a. C. A. a. H. W. a. R. K. a. J. B. a. C. T. a. C. A. M. a. G.;Pizzini A.; Filipiak W.; Wille J.; Ager C.; Wiesenhofer H.; Kubinec R.; Blaško J.; Tschurtschenthaler C.; Mayhew C. A.; Weiss G.; Bellmann-Weiler R. Analysis of volatile organic compounds in the breath of patients with stable or acute exacerbation of chronic obstructive pulmonary disease. Journal of Breath Research 2018, 12, 036002. 10.1088/1752-7163/aaa4c5. [DOI] [PubMed] [Google Scholar]
- Chang J.-E.; Lee D.-S.; Ban S.-W.; Oh J.; Jung M. Y.; Kim S.-H.; Park S.; Persaud K.; Jheon S. Analysis of volatile organic compounds in exhaled breath for lung cancer diagnosis using a sensor system. Sens. Actuators, B 2018, 255, 800–807. 10.1016/j.snb.2017.08.057. [DOI] [Google Scholar]
- Lawal O.; Muhamadali H.; Ahmed W. M.; White I. R.; Nijsen T. M. E. E.; Goodacre R.; Fowler S. J. consortium, o. b. o. t. B., Headspace volatile organic compounds from bacteria implicated in ventilator-associated pneumonia analysed by TD-GC/MS. Journal of Breath Research 2018, 12, 026002. 10.1088/1752-7163/aa8efc. [DOI] [PubMed] [Google Scholar]
- Rattray N. J. W.; Hamrang Z.; Trivedi D. K.; Goodacre R.; Fowler S. J. Taking your breath away: metabolomics breathes life in to personalized medicine. Trends Biotechnol. 2014, 32 (10), 538–548. 10.1016/j.tibtech.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Xie J.; Deng L.; Zhou Y.; Yao S.; Zeng K. Analysis of changes in volatile constituents and expression of genes involved in terpenoid metabolism in oleocellosis peel. Food Chem. 2018, 243, 269–276. 10.1016/j.foodchem.2017.09.142. [DOI] [PubMed] [Google Scholar]
- Gatzias I. S.; Karabagias I. K.; Kontakos S. P.; Kontominas M. G.; Badeka A. V. Characterization and differentiation of sheep’s milk from Greek breeds based on physicochemical parameters, fatty acid composition and volatile profile. J. Sci. Food Agric. 2018, 98, 3935. 10.1002/jsfa.8914. [DOI] [PubMed] [Google Scholar]
- Gerhardt N.; Birkenmeier M.; Schwolow S.; Rohn S.; Weller P. Volatile-Compound Fingerprinting by Headspace-Gas-Chromatography Ion-Mobility Spectrometry (HS-GC-IMS) as a Benchtop Alternative to 1H NMR Profiling for Assessment of the Authenticity of Honey. Anal. Chem. 2018, 90, 1777–1785. 10.1021/acs.analchem.7b03748. [DOI] [PubMed] [Google Scholar]
- Rosier E.; Cuypers E.; Dekens M.; Verplaetse R.; Develter W.; Van de Voorde W.; Maes D.; Tytgat J. Development and validation of a new TD-GC/MS method and its applicability in the search for human and animal decomposition products. Anal. Bioanal. Chem. 2014, 406, 3611–3619. 10.1007/s00216-014-7741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J. S.; Myers C. Electronic cigarette solutions and resultant aerosol profiles. Journal of chromatography. A 2015, 1418, 192–199. 10.1016/j.chroma.2015.09.034. [DOI] [PubMed] [Google Scholar]
- Goodacre R.; Broadhurst D.; Smilde A. K.; Kristal B. S.; Baker J. D.; Beger R.; Bessant C.; Connor S.; Capuani G.; Craig A.; Ebbels T.; Kell D. B.; Manetti C.; Newton J.; Paternostro G.; Somorjai R.; Sjöström M.; Trygg J.; Wulfert F. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 2007, 3 (3), 231–241. 10.1007/s11306-007-0081-3. [DOI] [Google Scholar]
- Domingo-Almenara X.; Brezmes J.; Vinaixa M.; Samino S.; Ramirez N.; Ramon-Krauel M.; Lerin C.; Díaz M.; Ibáñez L.; Correig X.; Perera-Lluna A.; Yanes O. eRah: A Computational Tool Integrating Spectral Deconvolution and Alignment with Quantification and Identification of Metabolites in GC/MS-Based Metabolomics. Anal. Chem. 2016, 88 (19), 9821–9829. 10.1021/acs.analchem.6b02927. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Muhamadali H.; Sayqal A.; Dixon N.; Goodacre R. Partial Least Squares with Structured Output for Modelling the Metabolomics Data Obtained from Complex Experimental Designs: A Study into the Y-Block Coding. Metabolites 2016, 6 (4), 38. 10.3390/metabo6040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Goodacre R. On Splitting Training and Validation Set: A Comparative Study of Cross-Validation, Bootstrap and Systematic Sampling for Estimating the Generalization Performance of Supervised Learning. Journal of Analysis and Testing 2018, 2 (3), 249–262. 10.1007/s41664-018-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X.; Turck N.; Hainard A.; Tiberti N.; Lisacek F.; Sanchez J.-C.; Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011, 12 (1), 77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-C.; Lin C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2 (3), 1–27. 10.1145/1961189.1961199. [DOI] [Google Scholar]
- Werner F.-M.; Coveñas R. Classical Neurotransmitters and Neuropeptides involved in Parkinson’s disease: Focus on anti-Parkinsonian Drugs. Curr. Drug Ther. 2015, 10 (2), 66–81. 10.2174/157488551002151222154335. [DOI] [Google Scholar]
- Ahsan T.; Chen J.; Zhao X.; Irfan M.; Wu Y. Extraction and identification of bioactive compounds (eicosane and dibutyl phthalate) produced by Streptomyces strain KX852460 for the biological control of Rhizoctonia solani AG-3 strain KX852461 to control target spot disease in tobacco leaf. AMB Express 2017, 7, 54. 10.1186/s13568-017-0351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W. R.; Anfora A. T.; Liu J.; Schultz P. G.; Lesley S. A.; Peters E. C.; Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (10), 3698–3703. 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C. E.; Smith S.; de Lacy Costello B. d. L.; White P.; Spencer R.; Probert C. S. J.; Ratcliffe N. M. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007, 21 (8), 1675–1688. 10.1096/fj.06-6927com. [DOI] [PubMed] [Google Scholar]
- Soini H. A.; Klouckova I.; Wiesler D.; Oberzaucher E.; Grammer K.; Dixon S. J.; Xu Y.; Brereton R. G.; Penn D. J.; Novotny M. V. Analysis of Volatile Organic Compounds in Human Saliva by a Static Sorptive Extraction Method and Gas Chromatography-Mass Spectrometry. J. Chem. Ecol. 2010, 36 (9), 1035–1042. 10.1007/s10886-010-9846-7. [DOI] [PubMed] [Google Scholar]
- de Lacy Costello B.; Amann A.; Al-Kateb H.; Flynn C.; Filipiak W.; Khalid T.; Osborne D.; Ratcliffe N. M. A review of the volatiles from the healthy human body. J. Breath Res. 2014, 8 (1), 014001 10.1088/1752-7155/8/1/014001. [DOI] [PubMed] [Google Scholar]
- Hoffmann G. F.; Meier-Augenstein W.; Stöckler S.; Surtees R.; Rating D.; Nyhan W. L. Physiology and pathophysiology of organic acids in cerebrospinal fluid. J. Inherited Metab. Dis. 1993, 16 (4), 648–669. 10.1007/BF00711898. [DOI] [PubMed] [Google Scholar]
- Hanhineva K.; Brunius C.; Andersson A.; Marklund M.; Juvonen R.; Keski-Rahkonen P.; Auriola S.; Landberg R. Discovery of urinary biomarkers of whole grain rye intake in free-living subjects using nontargeted LC-MS metabolite profiling. Mol. Nutr. Food Res. 2015, 59 (11), 2315–2325. 10.1002/mnfr.201500423. [DOI] [PubMed] [Google Scholar]
- Duranton F.; Cohen G.; De Smet R.; Rodriguez M.; Jankowski J.; Vanholder R.; Argiles A. European Uremic Toxin Work, G., Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 2012, 23 (7), 1258–1270. 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon E.; Clubb C.; Pittman S.; Ammann L.; Rasheed Z.; Kazmi N.; Keshavarzian A.; Gillevet P.; Rangwala H.; Couch R. D. Solid-phase microextraction and the human fecal VOC metabolome. PLoS One 2011, 6 (4), e18471–e18471. 10.1371/journal.pone.0018471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.