Abstract
Squamous cell carcinoma (SCC) is a treatment‐refractory tumour which arises from the epithelium of diverse anatomical sites such as oesophagus, head and neck, lung and skin. Accumulating evidence has revealed a number of genomic, clinical and molecular features commonly observed in SCC of distinct origins. Some of these genetic events culminate in fostering the activity of ΔNp63, a potent oncogene which exerts its pro‐tumourigenic effects by regulating specific transcriptional programmes to sustain malignant cell proliferation and survival. In this review, we will describe the genetic and epigenetic determinants underlying ΔNp63 oncogenic activities in SCC, and discuss some relevant transcriptional effectors of ΔNp63, emphasizing their impact in modulating the crosstalk between tumour cells and tumour microenvironment (TME).
Keywords: p63, squamous cell carcinoma, transcription factor, tumour microenvironment
Abbreviations
- APC/C
anaphase‐promoting complex/cyclosome
- CSF2
colony stimulating factor 2
- DUSP6
dual specificity phosphatase 6
- DUSP7
dual specificity phosphatase 7
- ECM
extracellular matrix
- EGF‐R
epidermal growth factor receptor
- EMT
epithelial‐mesenchymal transition
- FGF7
fibroblast growth factor 7
- FGFR2
fibroblast growth factor receptor 2
- HA
hyaluronic acid
- HDAC1
histone deacetylases 1
- HDAC2
histone deacetylases 2
- HNSCC
squamous cell carcinoma of head and neck
- HPV
human papilloma viruses
- HβD
human beta defensins
- IGF‐1
insulin‐like growth factor 1
- IGFBP3
IGF binding protein 3
- IL
interleukin
- IRF6
interferon regulatory factor 6
- IRS1
Insulin Receptor Substrate 1
- KLF4
transcription factor Krüppel‐like factor 4
- MDSCs
myeloid‐derived immunosuppressor cells
- MMP9
matrix metalloproteinase 9
- NF‐κB
nuclear factor kappa‐light‐chain‐enhancer of activated B cells
- NUP62
nucleoporin 62
- SCC
squamous cell carcinoma
- SRCAP
Snf2‐related CREBBP activator protein
- TGFβ2
transforming growth factor‐beta 2
- TGFβR2
transforming growth factor beta receptor 2
- TGFβ
transforming growth factor beta
- TME
tumour microenvironment
- VEGF
vascular endothelial growth factor
1. Introduction
Squamous cell carcinoma (SCC) is a highly malignant cancer which arises from the squamous epithelium of oesophagus, head and neck, lung and skin. The incidence of SCC varies between different types ranging from 600 000 cases per year for head and neck SCC (HNSCC) to 450 000 cases for lung SCC (Ferlay et al., 2015). Although derived from distinct anatomical sites, SCC share a number of unified genomic, clinical and molecular features. The aetiology of SCC is strictly associated with the exposure to several environmental agents (Rothenberg and Ellisen, 2012). The most important risk factors are tobacco use, alcohol consumption and sun exposure. Infection with high‐risk types of human papilloma virus (HPV) has also been associated to the pathogenesis of a subset of HNSCC (Castellsague et al., 2016). Intriguingly, HPV‐positive oropharyngeal cancers are associated with a favourable prognosis, likely reflecting the low mutational rate of HPV‐positive tumours (Ang et al., 2010).
At clinical level, there is no effective therapy for HNSCC; the therapeutic options include surgery in combination with radiotherapy, chemotherapy or targeted approaches such as epidermal growth factor receptor (EGF‐R) inhibitor (Cetuximab). However, these strategies have not led to a significant increase in the overall 5‐year survival rate, which is approximately 50% for HNSCC patients and 18% for lung SCC (Gillison et al., 2015; Sacco and Cohen, 2015). Treatment failure, locoregional recurrence and comorbidities (especially for lung SCC) account for the majority of deaths (Langer et al., 2016; Leemans et al., 1994; Marur and Forastiere, 2016). In recent years, immune checkpoint inhibitors have been characterized as promising anti‐neoplastic agents in advanced HNSCC, although resistance to the immunotherapy has been observed in most patients ab initio (Mehra et al., 2018).
At molecular and genetic level, systematic sequencing studies have indicated that frequent genetic and molecular hits dysregulating the activity of two members of the p53 family, p53 itself and p63, are commonly observed in diverse SCC (Agrawal et al., 2011, 2012; Cancer Genome Atlas Research Network, 2012; Leemans et al., 2018; Pickering et al., 2013; Stransky et al., 2011). TP53 gene encodes a transcription factor able to prevent mutations in the genome by promoting cell cycle arrest, DNA repair, metabolic adaptation or apoptosis in response to different stresses (Aubrey et al., 2018; Charni et al., 2017; Kaiser and Attardi, 2018; Mello and Attardi, 2018; Sullivan et al., 2018). For these reasons, p53 is considered the guardian of the genome and its inactivation occurs in the majority of human cancers (Baugh et al., 2018; Muller and Vousden, 2013). In SCC, p53 mutation emerges as a dominant early genetic event with a mutational rate of 60–80% of cases and is associated with a more aggressive phenotype (Cancer Genome Atlas, 2015; Poeta et al., 2007). Several tumour‐derived p53 mutations are gain‐of‐function mutations and act as driver oncogenes promoting metastatic dissemination and drug resistance (Alexandrova et al., 2017; Amelio et al., 2018; Kim and Lozano, 2018; Morrison et al., 2017; Muller and Vousden, 2014; Parrales et al., 2018; Vaughan et al., 2017). In addition to gene mutation, others mechanisms may contribute to restraining p53 oncosuppressor activity in SCC, including overexpression/amplification of MDM2, infection with HPV E6 oncoprotein or transcriptional repression by EGF‐R signalling (Kolev et al., 2008; Scheffner et al., 1990; Wu and Prives, 2018).
In contrast to the high frequency of TP53 gene mutation, TP63 gene is rarely mutated in human cancers; rather it is subjected to different genetic and molecular events that either increase its expression or enhance its transcriptional activity (Stransky et al., 2011). The relevance of p63 during SCC initiation and progression is supported by studies performed in mouse models of squamous carcinogenesis. The skin‐specific genetic ablation of TP63 in a mouse model of chemical‐induced skin carcinogenesis induces a rapid and dramatic tumour regression, demonstrating the exquisite dependence of SCC on high levels of p63 (Ramsey et al., 2013). In the next paragraphs, we will try to provide a global picture of the oncogenic routes orchestrated by p63, emphasizing the molecular and genetic determinants enhancing its activity in various SCC.
2. p63 is a master regulator of epithelial development and homeostasis
p63 is a transcription factor belonging to the p53 family, which includes p53 itself, p63 and p73 (Candi et al., 2014; Nemajerova et al., 2018). TP63 gene is expressed as multiple isoforms arising by both alternative promoter usage and differential splicing events at the 3′ end of its RNA. The two main isoforms contain (TAp63) or lack (ΔNp63), the N‐terminal p53‐homologous transactivation domain (Dotsch et al., 2010; Yang et al., 1998). The ΔNp63α is the most abundant isoform expressed in the basal layer of stratified epithelia, where it maintains the proliferative potential and lineage specification of epithelial structures such as epidermis, lung, thymus and mammary gland (Candi et al., 2008; Mills et al., 1999; Shalom‐Feuerstein et al., 2011; Soares and Zhou, 2018). The critical importance of p63 during epithelial morphogenesis has been demonstrated in diverse animal models and by the functional association of TP63 mutations with human diseases. Genetic deletion of all p63 isoforms dramatically impairs the development of several epithelial tissues, such as thymus, breast and skin, resulting in premature death because of severe dehydration of the newborns (Mills et al., 1999; Yang et al., 1999). Notably, the selective genetic deletion of the ΔNp63 isoforms recapitulates the phenotype observed in the p63 global knock‐out mice, strongly indicating that ΔNp63 is the major p63 isoform governing epithelial morphogenesis (Romano et al., 2012). In humans, heterozygous mutations in TP63 cause several developmental disorders, which partially resemble the developmental defects observed in p63 null mice (Celli et al., 1999; Rinne et al., 2007).
Genome‐wide approaches convincingly established the critical role of p63 in regulating keratinocytes proliferation and differentiation (Kouwenhoven et al., 2010, 2015; Rivetti di Val Cervo et al., 2012; Truong et al., 2006). Recent reports unveiled the key role of ΔNp63 in controlling the epigenetic landscape of epithelial cells, as ΔNp63 recruits epigenetic modulators and chromatin remodelling factors in order to directly regulate numerous target genes involved in cell proliferation, differentiation and adhesion (Fessing et al., 2011; Mardaryev et al., 2014, 2016).
Based on its critical role in epithelial morphogenesis, it is not surprising that diverse transcriptional and post‐transcriptional mechanisms finely control ΔNp63 expression and activity during squamous differentiation (Lena et al., 2008; Rossi et al., 2006). Among them, Notch/ΔNp63 cross‐talk is particularly noteworthy, as it regulates the interplay between terminal differentiation and proliferation in squamous epithelia, as well being subjected to distinct oncogenic hits, as we will discuss later (Dotto, 2009; Nguyen et al., 2006). In mammals, four NOTCH receptors (NOTCH 1–4) are expressed and synthesized as precursors that undergo specific cleavages in response to ligand interaction. These cleavages release the NOTCH receptor intracellular domain, which translocates to the nucleus and modulates the expression of specific target genes involved in cell fate decisions (Bray, 2006). In the skin, Notch signalling and ΔNp63 activity are functionally interconnected in a finely regulated cross‐talk, which ensures activation of mitogenic and pro‐differentiative signals in distinct epithelial compartments (see Fig. 1, left panel) (Nguyen et al., 2006). In the basal layer of epidermis, ΔNp63 inhibits the expression of p21WAF1/Cip1 and HES1, a NOTCH1 target gene, thus sustaining cell cycle progression and repressing late differentiation stages (Nguyen et al., 2006; Westfall et al., 2003). ΔNp63 is also able directly to induce the expression of the Notch ligand JAG2, likely favouring the initial step of skin differentiation (Candi et al., 2007). In the suprabasal layer, NOTCH1 counteracts ΔNp63 activity in a negative feedback loop, directly or through the induction of the interferon regulatory factor 6 (IRF6), which may promote the proteasome‐dependent degradation of ΔNp63 (Moretti et al., 2010; Nguyen et al., 2006; Thomason et al., 2010). Interestingly, in human keratinocytes NOTCH1 expression might also be positively regulated by p53 (Kolev et al., 2008; Lefort et al., 2007).
Figure 1.
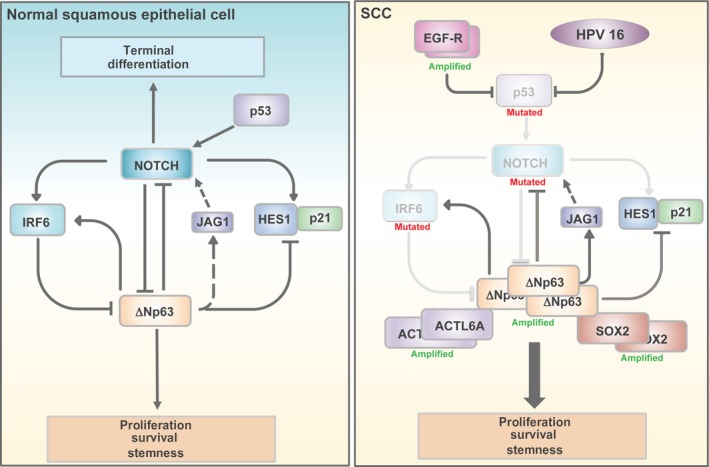
Schematic diagram of the molecular circuitry connecting squamous differentiation‐related genes in SCC. The genetic alterations of these genes are also reported.
3. Genetic determinants of ΔNp63 oncogenic activity in SCC
Over the past years, whole‐exome sequencing has allowed the identification of distinct genetic alterations associated with SCC development. Remarkably, a subset of functionally related genes (TP63, NOTCH, IRF6) controlling the squamous differentiation programme is dysregulated in 30–40% of SCC patients. In HNSCC and lung SCC, genomic amplification of the TP63 locus has been reported in up to 10% and 16% of the cases, respectively (Cancer Genome Atlas Network 2015; Cancer Genome Atlas Research Network, 2012; Pickering et al., 2013). In addition, overexpression of ΔNp63 itself is observed in the majority of invasive HNSCC as well as in lung, skin and oesophageal SCC (Agrawal et al., 2012; Reis‐Filho et al., 2003; Tonon et al., 2005). RNA sequencing data indicated that the ΔNp63 isoforms, mainly the α isoform, account for the high expression levels of p63. Indeed, ΔNp63 expression exceeds that of TAp63 more than 200‐fold in lung SCC. These data have been also confirmed in HNSCC cell lines, which express almost exclusively the ΔNp63α isoform (Compagnone et al., 2017; Rocco et al., 2006). Furthermore, some rare TP63 mutations reported in HNSCC samples are located in the TA domain, suggesting that ΔNp63 isoform expression is positively selected during tumour evolution (Stransky et al., 2011). Collectively, these data indicate that the major p63 isoform driving SCC tumourigenesis is ΔNp63, likely the ΔNp63α isoform (hereinafter referred as ΔNp63).
The relevance of ΔNp63 during SCC initiation and progression is also supported by studies performed in mouse models. The induction of moderate expression of ΔNp63 in the basal layer of stratified epithelia indeed causes the development of spontaneous epidermal cysts that resemble cutaneous premalignant lesions (Devos et al., 2017). Furthermore, in a model of chemical‐induced skin carcinogenesis, ΔNp63 expression accelerates the onset of papilloma occurrence, suggesting that ΔNp63 favours the early steps of SCC development (Devos et al., 2017).
In addition to gene amplification and/or overexpression, other genetic alterations foster ΔNp63 oncogenic activity in SCC. As stated before, Notch signalling is fundamental for the proper squamous epithelia homeostasis. In SCC, diverse genetic events dysregulate the Notch/ΔNp63 crosstalk. NOTCH1 point mutations occur in 11–15% of HNSCC, in 8% of lung SCC and in more than 40% of cutaneous SCC (Agrawal et al., 2011, 2012; Pickering et al., 2013). Point mutations in the NOTCH2 gene have been also reported in 11% of HNSCC samples; these mutations are mutually exclusive and exhibit minimal overlap with amplification of the TP63 gene (Stransky et al., 2011). These observations suggest that aberrations in squamous differentiation modulators, which act in the same functional axis, have overlapping functional consequences. Importantly, NOTCH mutations are loss‐of‐function, missense or nonsense mutations, a strong indication of a tumour‐suppressive function of Notch signalling in SCC. This conclusion is also supported by in vivo evidence showing that Notch1 inactivation in the mouse epidermis promotes skin tumourigenesis (Nicolas et al., 2003; Proweller et al., 2006). In addition to gene mutation, NOTCH activity is also restrained by the inhibitory action of ΔNp63 itself (see section ‘Genetic determinants of ΔNp63 oncogenic activity in SCC’). Furthermore, the high rate of TP53 mutation and the frequent amplification of EGF‐R locus in SCC may cooperate to maintain the low expression of NOTCH1, thus favouring tumour proliferation (Kolev et al., 2008).
As anticipated, another squamous differentiation‐related gene functionally connected with the Notch signalling and ΔNp63 is IRF6. IRF6 encodes for a transcription factor, acting as an important mediator of the Notch pro‐differentiation function. The Notch signalling sustains the expression of IRF6, which contributes to the activation of growth/differentiation‐related genes (Nguyen et al., 2006). At the same time IRF6 is able to restrain ΔNp63 protein levels by enhancing its protein degradation (Moretti et al., 2010). Diverse genetic events such as promoter methylation and gene mutation impair IRF6 activity in SCC (Rotondo et al., 2016). Mutations of the IRF6 gene have been reported in 7% of HNSCC patients and down‐regulation of IRF6 has been correlated with tumour invasive and differentiation status of SCC (Stransky et al., 2011).
Genomic amplification or overexpression of another squamous differentiation‐related gene, SOX2, also contribute to enhance and activate ΔNp63 oncogenic activity. The SOX2 gene lies approximately 10 Mb from TP63, and it is frequently amplified in lung, oesophageal and oral SCC (Ferone et al., 2016; Zhou et al., 2017). SOX2 and ΔNp63 proteins physically interact and exhibit overlapping genomic occupancy at a large number of loci in SCC, enhancing the expression of pro‐survival factors (Jiang et al., 2018; Watanabe et al., 2014).
Collectively, these data suggest that distinct genetic events, such as NOTCH mutations, TP63 genomic amplification/overexpression, TP53 mutation, IRF6 down‐modulation and SOX2 amplification (see Fig. 1, right panel), may promote an immature and more proliferative basal‐like phenotype by, at least in part, fostering ΔNp63 oncogenic activity.
4. Deregulation of factors controlling ΔNp63 levels and activity in SCC
In addition to these genetic lesions, SCC exhibits transcriptional alterations of factors involved in controlling ΔNp63 expression at both mRNA and protein level. One well‐established example is represented by ASPP2, a member of the ASPP family of proteins, which is able to repress ΔNp63 expression through a nuclear factor kappa‐light‐chain‐enhancer of activated B cells (NF‐κB)‐dependent mechanism (Tordella et al., 2013). Down‐regulation of ASPP2 is frequently observed in human HNSCC and is associated with increased p63 expression. Notably, Tp63 is required for the development of spontaneous SCC observed in ASPP2−/+ BALB/c heterozygous mice, implicating p63 as a critical mediator of ASPP2 tumour‐suppressive function in SCC (Tordella et al., 2013).
ΔNp63 levels can be also modulated by post‐transcriptional mechanisms, mainly by ubiquitin‐mediated proteolysis. Several E3 ubiquitin ligases targeting ΔNp63 have been identified so far, e.g. RACK1, NEDD4, ITCH, FBW7 and WWP1, each of them likely contributing to modulate ΔNp63 protein levels in tumours (Bakkers et al., 2005; Fomenkov et al., 2004; Galli et al., 2010; Li et al., 2008; Malatesta et al., 2013; Peschiaroli et al., 2010; Rossi et al., 2006). For instance, FBW7 is a tumour‐suppressor gene frequently mutated in a variety of solid tumours, including SCC of different origins (Xiao et al., 2018). Recently, Prives’s group has identified the anaphase‐promoting complex/cyclosome (APC/C) as a novel E3 ligase triggering ΔNp63 degradation in mitosis and during terminal differentiation (Rokudai et al., 2018). Although further studies are necessary to provide formal proof of the involvement of this degradative pathway in SCC development, some observations favour this conclusion. In detail, a ΔNp63 mutant refractory to APC/C activity increases keratinocyte proliferation, inhibits differentiation and has oncogenic activity in xenograft tumour transplantation assays. More importantly, high levels of STXBP4, a factor counteracting APC/C‐mediated ubiquitination of ΔNp63, significantly correlate with the accumulation of ΔNp63 in skin and lung SCC (Rokudai et al., 2018). However, this correlation might also result from the inhibitory effect exerted by STXBP4 on diverse ΔNp63 targeting E3 ligases, such as ITCH and RACK1 (Li et al., 2009).
Another recent report unveils a novel mechanism which can contribute to sustain ΔNp63‐mediated transcriptional activity in SCC. Hazawa et al. reported that nucleoporin 62 (NUP62) promotes ΔNp63 nuclear localization in SCC. NUP62 is highly expressed in stratified squamous epithelia and its abundance is further increased in SCC, thus maintaining elevated nuclear ΔNp63 levels (Hazawa et al., 2018).
In conclusion, these data clearly indicate that distinct genetic and molecular events foster ΔNp63 activity in SCC, strengthening the concept that ΔNp63 oncogenic activity is of critical importance for SCC initiation and progression.
5. Transcriptional effectors of ΔNp63 oncogenic function in SCC
Initially, it was hypothesized that the oncogenic role of ΔNp63 in SCC mainly relies on its ability to act as a dominant‐negative factor to p53/p73 proteins and repress their ability to activate the expression of target genes involved in apoptosis or cell cycle (DeYoung et al., 2006; Rocco et al., 2006; Westfall et al., 2003; Yang et al., 1998). In HNSCC cells, it has been showed that ΔNp63 is able to interact with p73, restraining the transcriptional activation of the pro‐apoptotic gene PUMA. Accordingly, down‐modulation of p63 relieves p73 and induces cell death, a phenomenon reversed by the overexpression of BCL‐2, an important anti‐apoptotic protein (Adams and Cory, 2018; Kale et al., 2018; Strasser and Vaux, 2018). Although this mechanism may be relevant for the pro‐survival effect exerted by ΔNp63, other observations suggested alternative molecular mechanisms by which ΔNp63 exerts its oncogenic function in SCC. First, ΔNp63 protein is able to transactivate specific target genes due to the presence of two transactivation domains, one located between the oligomerization domain and the SAM domain, and another at the first 26 amino acids (Dohn et al., 2001; Ghioni et al., 2002; Helton et al., 2006; Lena et al., 2015). Secondly, in lung SCC and HNSCC, ΔNp63 acts as a potent transcriptional repressor and diverse epigenetic factors have been implicated in ΔNp63‐mediated transcriptional repression. Thirdly, most SCC exhibits both overexpression of ΔNp63 and inactivating mutations in TP53, suggesting the existence of p53‐independent oncogenic functions of ΔNp63 (Neilsen et al., 2011; Nekulova et al., 2011). Last but not least, in p53 wild‐type lung SCC cells, depletion of p53 or p73 does not rescue the proliferation arrest caused by ΔNp63 knockdown (Gallant‐Behm et al., 2012). Collectively, these observations clearly indicate that the activation or repression of diverse transcriptional target genes is a critical oncogenic outcome of ΔNp63 activity in SCC, even though some transcriptional independent mechanisms have been described (Patturajan et al., 2002). Below, we will describe the transcriptional effectors of ΔNp63 oncogenic activity, emphasizing their role in controlling tumour proliferation, survival and dissemination.
5.1. ΔNp63‐repressed genes in SCC
The genesis and homeostasis of stratified epithelia, such as the epidermis, require the coordinated regulation of proliferation, adhesion, migration and differentiation. During the initiation and progression of SCC the fine balance between these processes is altered. Generally, SCC cells exhibit increased proliferative capabilities at the expense of a diminished ability to undergo the terminal differentiation programme. Probably one of the most important ΔNp63‐repressed genes involved in controlling the proliferation‐differentiation crosstalk is NOTCH1.
As we described before, ΔNp63 is able to inhibit Notch signalling mainly by transcriptional repression of the HES1 gene, a downstream target of NOTCH1. In addition to this indirect mechanism, a direct transcriptional effect of ΔNp63 on NOTCH1 gene expression has been also observed (Yugawa et al., 2010). In diverse normal and SCC cells, ΔNp63 may directly repress the transcription of NOTCH1 gene through a p53‐responsive element. Interestingly, the ability of ΔNp63 to affect NOTCH1 expression negatively has also been observed in the epidermis of ΔNp63 knock‐out mouse embryos (Romano et al., 2012), suggesting that the ΔNp63/NOTCH1 pathway might be indicative of a regenerative state of tumour cells. The ΔNp63‐mediated repression of NOTCH1 has two major consequences: it inhibits the NOTCH1‐mediated activation of terminal differentiation programme and sustains cell proliferation by suppressing the expression of the cyclin‐dependent kinase (CDK) inhibitor p21WAF1/Cip1. ΔNp63 may also repress p21WAF1/Cip1 expression directly (Westfall et al., 2003) or through an NF‐κB‐dependent mechanism (Nguyen et al., 2006) .
Another ΔNp63‐repressed gene involved in finely regulating the proliferation/differentiation crosstalk is the transcription factor Krüppel‐like factor 4 (KLF4). KLF4 is a key driver of squamous differentiation and is highly expressed in differentiating both oesophageal epithelial cells and keratinocytes, which, in cooperation with ZNF750, drives the commitment to terminal differentiation (Boxer et al., 2014; Sen et al., 2012). ΔNp63 is able directly to repress KLF4 transcription and enhance ZNF750 expression to activate an early step of squamous differentiation (Cordani et al., 2011; Sen et al., 2012). However, ZNF750 can also induce KLF4 expression (Sen et al., 2012), suggesting a complex interplay between ΔNp63 and effectors of early and terminal differentiation. Although it is not clear whether KLF4 and ZNF750 represent critical effectors of ΔNp63 oncogenic activity in SCC, several observations suggest their tumour‐suppressive function. Genetic ablation of KLF4 in the oesophagus results in delayed differentiation and development of precancerous squamous cell dysplasia (Tetreault et al., 2010). KLF4 is downregulated in human oesophageal squamous cell carcinoma (Luo et al., 2004; Wang et al., 2002) and KLF4 loss also promotes skin carcinogenesis in mice (Li et al., 2012). Down‐modulation of ZNF750 activity by gene mutation, deletion or under‐expression has been observed in human SCC, and low ZNF750 expression is associated with poor survival (Hazawa et al., 2017; Zhang et al., 2018). These data indicate that the ΔNp63/KLF4/ZNF750 axis might be important in the prevention of terminal differentiation and promotion of SCC tumourigenesis.
More recently, Ellisen's group has identified another ΔNp63 oncogenic pathway, which fine‐tunes the proliferation/differentiation crosstalk in SCC (Saladi et al., 2017). By comparing ChIP‐seq and RNA‐seq data in normal versus HNSCC cells, these authors have identified a tumour‐specific transcriptional profile directly regulated by ΔNp63. Notably, the expression of the majority of direct ΔNp63 target genes is upregulated in response to ΔNp63 silencing, indicating the dominant function of ΔNp63 as transcriptional repressor in SCC. WWC1 (KIBRA) is one of the top genes repressed by ΔNp63 in HNSCC. WWC1 encodes a cytosolic phosphoprotein that acts as a potent regulator of the Hippo pathway by favouring YAP cytoplasmic retention (Furth and Aylon, 2017; Yu et al., 2010; Zhao et al., 2007). By repressing the transcription of WWC1, ΔNp63 favours YAP nuclear activity, thereby promoting proliferation and inhibiting differentiation. As we will discuss later, HNSCC frequently displays amplification of ACTL6a, an epigenetic factor necessary for the ΔNp63‐mediated repression of WWC1.
The critical relevance of the ΔNp63‐mediated transcriptional repression in SCC evolution has also been highlighted by the identification of diverse ΔNp63‐negative target genes acting as key players of cell proliferation control in lung SCC. IGF binding protein 3, IGFBP3, is noteworthy, as its transcriptional control has been previously linked to the activity of p53 family members. The p53 activation is able to enhance the secretion of an active form of IGFBP3 which is capable of inhibiting mitogenic signalling by the insulin‐like growth factor IGF‐1 (Buckbinder et al., 1995). However, the p53‐dependent transactivation of IGFBP3 appears to be cell type‐specific, as in the H226 cell line, p53 activation does not lead to IGFBP3 induction (Gallant‐Behm et al., 2012). Conversely, in several SCC cells, IGFBP3 expression is under the negative control of ΔNp63, through an H2A.Z‐mediated transcriptional repression (Barbieri et al., 2005; Gallant‐Behm et al., 2012). Furthermore, diverse data have indicated that ΔNp63‐dependent regulation of IGFBP3 might be important for SCC progression. ΔNp63 and IGFBP‐3 expression are inversely correlated in human SCC samples and decreased IGFBP3 expression is correlated with unfavourable prognosis in human cancers (Barbieri et al., 2005; Chang et al., 2002; Katsaros et al., 2001), implying a potential relevance of IGFBP3 suppression in SCC development.
More recently, Espinosa's group has exploited a genome‐wide CRISPR‐Cas9 screen to identify the pathway required for the ΔNp63‐driven proliferation in lung SCC (Abraham et al., 2018). Upon ΔNp63 silencing, lung SCC cells undergo a block of proliferation which is dependent on the activation of the transforming growth factor beta (TGFβ) signalling. At molecular level, ΔNp63 directly represses the transcription of TGFB2 and TGFBR2, thus restraining the activation of the TGFβ signalling. Interestingly, the cell cycle arrest mediated by ΔNp63 silencing is also dependent on the expression of RHOA, a small GTPase activated by TGFβ signalling and commonly down‐regulated in lung SCC specimens. Although these data unveil a novel ΔNp63 oncogenic pathway controlling lung SCC proliferation, they do not address its effect in p53 mutated background. This is quite important, as the majority of SCC harbour mutations in the TP53 locus and TGFβ signalling can exert tumour‐prone or tumour‐suppressive effects depending on cell context as well as on the p53 status (Ikushima and Miyazono, 2010; Karlsson et al., 2017). Interestingly, the dependence on RHOA to sustain cell cycle arrest upon ΔNp63 depletion has not been observed in the immortalized keratinocyte cell line HaCaT, which harbours mutation of TP53 gene (Abraham et al., 2018). Furthermore, in HNSCC cells, ΔNp63 can enhance the pro‐tumourigenic function of TGFβ by repressing the expression of two microRNA (miRNA), miR‐527 and miR‐665, which target Smad4 and TGFβR2, respectively (Rodriguez Calleja et al., 2016). Therefore, the effect of ΔNp63 on TGFβ signalling might be cell context‐dependent and/or influenced by extrinsic factors, as we will discuss later.
In addition to miR‐527 and miR‐665, ΔNp63 can activate or repress the expression of diverse miRNA involved in a variety of tumour‐related processes (Lin et al., 2015; Ory et al., 2011; Ratovitski, 2013). However, the involvement of the ΔNp63‐miRNA pathway in SCC pathogenesis is not well characterized and our knowledge of its actions as critical effectors of ΔNp63 oncogenic activity is quite limited.
5.2. Epigenetic determinants of ΔNp63‐mediated transcriptional repression in SCC
In the past years, diverse molecular mechanisms underlying ΔNp63‐mediated transcriptional repression have been elucidated (see Fig. 2). The general picture is that ΔNp63 binds to and employs as co‐repressor several chromatin remodelling factors. In HNSCC, ΔNp63 can form a complex with the histone deacetylases HDAC1 and HDAC2 (Ramsey et al., 2011). HDACs mediate transcriptional regulation by increasing histone‐DNA affinity, thus promoting the formation of a chromatin‐condensed structure that prevents transcription factor binding (Li and Seto, 2016; Paluvai et al., 2018). ΔNp63 exploits this mechanism mainly for repressing pro‐apoptotic genes such as PUMA, then favouring cell survival. This anti‐apoptotic pathway can be disrupted by chemotherapeutic agents such as cisplatinum, which displace the HDAC/ΔNp63 complex from the PUMA promoter, thus favouring its expression and triggering, as a consequence, apoptosis. This pro‐survival mechanism may inhibit the treatment of p63 overexpressing cells with the HDAC inhibitors trichostatin A (TSA) and Vorinostat. Remarkably, HNSCC cell lines show a direct correlation between ΔNp63 protein levels and TSA sensitivity (Ramsey et al., 2011), and increased expression of HDAC is frequently observed in SCC where it predicts poor patient prognosis (Chang et al., 2009; Theocharis et al., 2011). This evidence supports the rationale of several ongoing clinical trials, which are coupling HDAC inhibitors with conventional chemotherapy for SCC treatment (Caponigro et al., 2016; Teknos et al., 2018).
Figure 2.
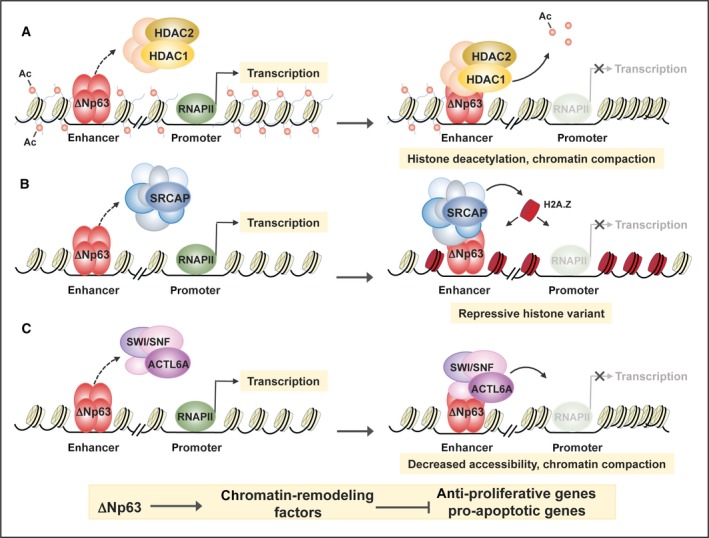
Model of the epigenetic mechanisms exploited by ΔNp63 to repress transcription in various SCC. (A) In HNSCC cells, ΔNp63 may recruit the histone deacetylates HDAC1 and HDAC2 to chromatin, preventing transcription factor binding to the promoters of pro‐apoptotic genes, such as PUMA. (B) In lung SCC cells, ΔNp63 is able to repress the transcription of anti‐proliferative genes by promoting H2A.Z incorporation. (C) In HNSCC cells, ΔNp63 interacts with the SWI/SNF subunit ACTL6A, inducing the repression of anti‐proliferative genes.
It is worth noting that the molecular mechanisms exploited by ΔNp63 to repress its target genes might be dependent on cell context (Gallant‐Behm et al., 2012). Indeed, in H226 cells, ΔNp63 represses the expression of its targets independently from HDAC action. In this lung SCC cell line, ΔNp63 takes part in a large chromatin remodelling protein complex called SRCAP that is able to exchange histone H2A with the histone variant H2A.Z and may act as a transcriptional repressor when arranged near the transcription start sites (Marques et al., 2010). H2A.Z deposition is involved in the repression of several ΔNp63 target genes such as ZHX2, NTN4, SAMD9L and IGFBP3, each of them likely contributing to the ΔNp63‐mediated control of cell proliferation (Gallant‐Behm et al., 2012).
More recently, the SWI/SNF complex has been involved in ΔNp63‐mediated transcriptional repression in HNSCC (Saladi et al., 2017). SWI/SNF is a multisubunit chromatin remodelling complex that catalyses nucleosome sliding or ejection, modulating DNA accessibility to the transcription machinery (Patel and Vanharanta, 2016; Wilson and Roberts, 2011). In HNSCC, ΔNp63 interacts with the SWI/SNF subunit ACTL6A, inducing the repression of WCC1 transcription, as we have described before. In contrast to what is observed in normal stratified epithelium, ACTL6A is frequently overexpressed in HNSCC and about 20% of HNSCC shows genomic co‐amplification of ACTL6A and TP63 loci (Saladi et al., 2017). Furthermore, elevated levels of ACTL6A expression are a negative prognostic factor of HNSCC patient survival, suggesting that ACTL6A has an oncogenic function in HNSCC (Saladi et al., 2017). Conversely, in different types of cancer, genomic analysis identified several loss‐of‐function mutations in various SWI/SNF subunits, suggesting a tumour‐suppressor role for this complex (Kadoch et al., 2013). This apparent contradiction can be mechanistically explained, taking into account that in epidermal progenitor cells, ACTL6A contributes to maintain the undifferentiated state preventing SWI/SNF complex, by binding and transactivating differentiation genes (Bao et al., 2013).
These molecular data together with the key importance of ΔNp63‐repressed pathways in mediating the mitogenic action of ΔNp63 in SCC clearly indicate that ΔNp63‐mediated transcriptional repression is a crucial oncogenic outcome of ΔNp63 in SCC as well as in other epithelial cancers (Regina et al., 2016a,2016b). On the other hand, as described in the next paragraph, ΔNp63 also exerts its oncogenic activity by enhancing the transcription of several pro‐tumourigenic factors, and epigenetic modifications are likely to be involved in this oncogenic function. Accordingly, a high mutation rate of the methyltransferase KMTD2, an ΔNp63 interacting epigenetic factor involved in the transcriptional activation of epithelial target genes, has been observed in SCC (Lin‐Shiao et al., 2018). Deciphering the global epigenetic landscape regulated by ΔNp63 and its involvement in SCC tumourigenesis needs to be fully explored and represents an important direction for future studies.
5.3. Oncogenic routes activated by ΔNp63 in SCC
Besides acting as a transcriptional repressor, ΔNp63 enhances the transcription of diverse pro‐tumourigenic genes playing a critical role in SCC tumourigenesis. Remarkably, a significant group of ΔNp63‐positive target genes codifies for proteins related to extracellular matrix (ECM) or involved in the growth factor‐mediated signalling (see Fig. 3). In the following sections, we will describe some of these ΔNp63‐activated genes, emphasizing their role in modulating the crosstalk of tumour cells, ECM and tumour microenvironment (TME).
Figure 3.
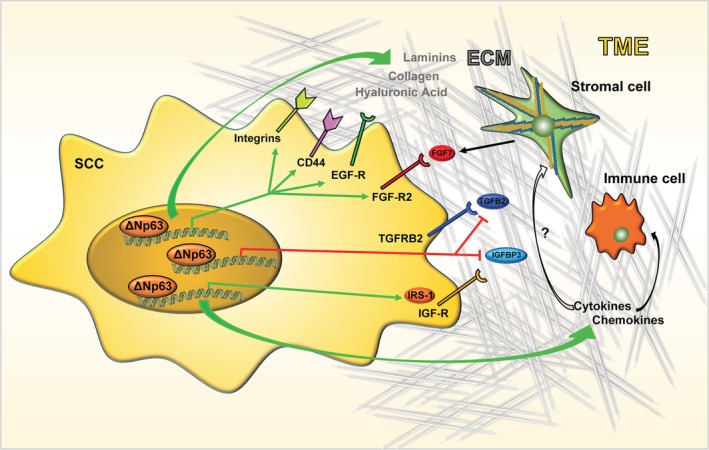
Schematic model of the ΔNp63 oncogenic routes in SCC. The green and red arrows indicate the pathways under positive and negative regulation by ΔNp63, respectively. See text for details.
5.3.1. ECM‐mediated signalling
The initial observations that ΔNp63 might actively participate in modulating the composition of ECM and activate ECM‐mediated signalling were reported in non‐transformed cells. In normal mammary epithelial cells, silencing of p63 induces cell detachment and anoikis, and the concomitant modulation of the expression of several adhesion molecules and ECM receptors. ΔNp63 enhances the expression of integrin receptors (e.g. ITGB1, ITGB4, ITGA6, ITGA3), ECM components such as laminin and collagen (e.g LAMC2 and COL17A1) and adhesion molecules (e.g. PERP and ZNF185) (Carroll et al., 2006; Ihrie et al., 2005; Kurata et al., 2004; Smirnov et al., 2019).
Although a formal demonstration that these ECM‐related factors are required for ΔNp63 oncogenic function in SCC is missing, several data indicate that they may significantly participate in SCC development. Tumour regression upon TP63 excision is associated with decrease of expression of ECM‐related proteins, such as collagen COL6A2 and COL17A1, LAMB3 and ITGB4 (Ramsey et al., 2013). In line with this evidence, the integrin‐mediated adhesion and signalling is one of the tumour‐specific programmes regulated by ΔNp63 in HNSCC cell lines (Saladi et al., 2017). Furthermore, increased suprabasilar expression of α6β4‐integrin occurs in more than 70% of HNSCC with higher invasive‐metastatic potential (Van Waes et al., 1995). Together with the pro‐tumourigenic pathway exerted by ITGB4/EGF‐R signalling crosstalk (Carroll et al., 2006), these data suggest that ECM‐mediated signalling might be functionally important during SCC development.
In agreement with this, our group has recently unveiled a novel ΔNp63‐dependent transcriptional oncogenic programme aimed at sustaining the synthesis of one important component of the ECM, hyaluronic acid (HA) (Compagnone et al., 2017). In human tumours, HA is pivotal for diverse tumour‐related processes, including cell migration, angiogenesis and metastasis (Chanmee et al., 2016; Jiang et al., 2011; Toole, 2004), and several types of cancers including SCC are characterized by relatively high amounts of tumour cell‐associated HA (Koyama et al., 2007, 2008; Kultti et al., 2014). We have demonstrated that ∆Np63 is able to drive the expression of the HA synthase gene HAS3 and the HA receptor CD44, thus favouring the HA/CD44‐mediated oncogenic signaling in HNSCC. In detail, ΔNp63 favours the activation of pro‐mitogenic and pro‐survival signals by EGF‐R in an HA‐dependent manner. Interestingly, EGF‐R expression has been positively correlated with that of HAS3 in human oesophageal SCC tumours (Twarock et al., 2011) and activation of EGF‐R enhances HAS3 expression in some tumour cells (Bourguignon et al., 2007; Chow et al., 2010), implying a positive feedback between EGF‐R signalling and ∆Np63‐HAS3 pathway in HNSCC. The functional importance of the ∆Np63‐mediated regulation of HA signalling in human tumours is highlighted by diverse observations. In HNSCC tumours, p63 expression is positively correlated with that of HAS3 and CD44, and the expression of three tumour‐related CD44 variants is associated with HNSCC progression in clinical samples (Maula et al., 2003; Misra et al., 2011). More importantly, high expression of TP63/HAS3 axis is a negative prognostic factor of HNSCC patient survival, indicating that the functional link between ΔNp63 and HA/CD44 signalling critically contributes to HNSCC progression and therapeutic response (Compagnone et al., 2017). Although these data reveal a novel ∆Np63 oncogenic route in HNSCC, further studies will be necessary to understand whether HA synthesis inhibition could be exploited as a novel therapeutically actionable pathway in HNSCC.
5.3.2. Growth factor‐mediated signalling
In addition to ECM‐related proteins, ∆Np63 controls the expression of ligands, receptors and intracellular mediators of diverse growth factors, thus favouring the activation of pro‐survival and mitogenic signals. One important example of such regulation is related to the activation of EGF‐R signalling. EGF‐R amplification has been observed in many SCC and its expression has been correlated with poor outcome (Chung et al., 2006; Ozawa et al., 1987; Sigismund et al., 2018; Temam et al., 2007; Wintergerst et al., 2018). The overall survival rate and time of relapse of HNSCC patients overexpressing EGF‐R is significantly shorter than those without EGF‐R overexpression. At the clinical level, anti‐EGFR monoclonal antibody (cetuximab) in combination with radiation or chemotherapy is currently approved as a targeted therapy for the treatment of selected HNSCC patients, even if gain of EGFR has not been clearly demonstrated to be predictive for the outcomes following cetuximab therapy (Bonner et al., 2006; De Pauw et al., 2018; Licitra et al., 2011).
∆Np63 oncogenic activity participates in sustaining EGF‐R‐mediated signalling by activating distinct pathways. In diverse epithelial tumours, ∆Np63 directly activates the transcription of EGF‐R, even though this mechanism seems to be tissue‐specific (Danilov et al., 2011). ∆Np63 is also able to induce the transcription of neuregulin (NRG1), a natural ligand of ErbB3 receptor (Forster et al., 2014). In HNSCC, full EGF‐R activation is dependent on the ErbB3‐neuregulin axis. (Redlich et al., 2018). Compared with other tumours, HNSCC have among the highest levels of NRG1, and tumours recurrence further increases neuregulin‐1 expression after curative treatments with radiation and chemotherapy (Wilson et al., 2011). In addition to transcriptional activation of these EGFR‐related factors, ∆Np63 might also sustain EGF‐R signalling by regulating the expression of ECM‐related factors, such as ITGB4, HA and CD44. Collectively, these data indicate that ∆Np63 exploits multiple oncogenic pathways to enhance EGF‐R‐mediated oncogenic signalling.
A recent report connected ∆Np63 transcriptional activity with the activation of another receptor tyrosine kinase, the insulin growth factor receptor 1 (IGFR‐1). In HNSCC cell lines ∆Np63 positively controls the transcription of the adaptor protein Insulin Receptor Substrate 1 (IRS1), an important mediator of the pro‐survival and mitogenic signalling of insulin and IGF‐1 (Frezza et al., 2018). The modulation of IRS1 levels by ∆Np63 is interesting in light of the role of ∆Np63 in preventing the expression of IGFBP3, which regulate the bioavailability and half‐life of circulating IGF‐1 (Barbieri et al., 2005). By repressing IGFBP3 and concomitantly activating IRS1 expression, ∆Np63 is able to enhance circulating IGF‐1 and increase its intracellular signalling, ultimately fostering the growth and survival of SCC cells. However, further studies are needed to clarify the clinical impact of the ∆Np63/IGF signaling, as contradictory findings on the prognostic impact of IGF signalling have been reported in HNSCC (Dale et al., 2015; Lara et al., 2011; Luo et al., 2014; Sun et al., 2011).
The regulation of the expression of FGF‐R2, a member of the fibroblast growth factor (FGF) receptor family, is another example of how ∆Np63 controls growth factor‐mediated signalling. In vitro and in vivo evidence demonstrated that FGF‐R2 is a relevant ∆Np63 transcriptional target gene exerting a pro‐tumourigenic signalling in SCC (Ramsey et al., 2013). Remarkably, activation of FGF‐R2 requires stromal‐dependent secretion of FGF7, the most specific ligand for FGF‐R2 (Wesche et al., 2011), resulting in a specific and spatial activation of FGF‐R2 signalling in tumour cells. Besides revealing a novel ∆Np63‐driven oncogenic pathway with a potential therapeutic impact, these data uncovered a functional link between ∆Np63 activity and tumour stroma interaction. In the next section, we will expand this concept by reporting a few examples of the interplay between ∆Np63 transcriptional activity and TME in SCC.
5.3.3. TME‐related signalling
Tumour development likely arises by the co‐evolution of tumour cells and cellular components of the stroma. The close relationship between tumour cells and the stroma is fundamental for adapting and modifying TME to sustain and enhance tumour growth and invasion (Bissell and Radisky, 2001; Junttila and de Sauvage, 2013; Schaaf et al., 2018). Stromal components include different types of cells, including cancer‐associated fibroblasts (CAF), endothelial cells and infiltrating immune cells. Tumour stroma interaction likely generates an intricate network of signals that significantly sustains the development of different types of cancer, and SCC is not an exception. HNSCC and other squamous carcinomas are characterized by a tight interaction of malignant cells with a dense fibrous stroma, and high levels of stromal infiltrate are associated with poor prognosis in SCC (Fujii et al., 2010; Takahashi et al., 2011).
Few data have established a functional link between ∆Np63 oncogenic activity and changes of the TME. By recruiting two NF‐κB family members, cRel and RelA, ∆Np63 co‐regulates a subset of pro‐inflammatory cytokines, such as interleukin (IL)‐1, IL‐6, IL‐8 and colony stimulating factor 2 (CSF2) (Yang et al., 2011). These pro‐inflammatory factors are known to attract infiltrating neutrophils and macrophages, and individually are known to promote the aggressive malignant behaviour that leads to poor prognosis of HNSCC (Allen et al., 2007; Chen et al., 1999). In human HNSCC samples and in ΔNp63 transgenic mice, ΔNp63 levels are correlated with the presence of infiltrating inflammatory cells (Yang et al., 2011). Interestingly, increased ΔNp63 expression and inflammatory cell infiltration in oral leukoplakias are associated with worse prognosis and higher rate of cancer progression (Saintigny et al., 2009).
By recruiting neutrophils and macrophages, ΔNp63 might also sustain tumour‐associated angiogenesis. In osteosarcoma cells, ectopic expression of ΔNp63 induces VEGF secretion by activating STAT3/HIF‐1α pathway in a cytokine‐dependent manner (Bid et al., 2014). Although this pro‐angiogenic pathway has not been formally proved to occur in SCC, another report suggested a link between ΔNp63 and lymphoangiogenesis in SCC. ΔNp63 enhances the transcription of three human beta defensins (HβD1, HβD2 and HβD4), a class of antimicrobial peptides secreted in inflammatory conditions by epithelial cells (Suarez‐Carmona et al., 2014). By doing this, ΔNp63 stimulates the migration of lymphatic endothelial cells in a CCR6‐dependent manner, which might explain the increased density of blood and lymphatic vessels observed in high ΔNp63‐expressing SCC.
Recent evidence in other epithelial tumours also confirmed a functional role of ΔNp63 in modelling TME. In triple negative breast cancer (TNBC) cells, ΔNp63 drives the recruitment of myeloid‐derived immunosuppressor cells (MDSCs) by direct activation of the chemokines CXCL2 and CCL22 (Kumar et al., 2018). Importantly, MDSCs secrete pro‐metastatic factors, such as MMP9 and chitinase 3‐like 1, which in turn promote TNBC tumour progression and metastasis.
These data indicate that ΔNp63‐activated pathways may modify TME to create a favourable niche for tumour progression. On the other hand, it is reasonable that TME could also influence ΔNp63 oncogenic activity. Although this relationship is far to be fully explored, there are a few indications that TME might be a critical determinant in controlling the biological outcome of ΔNp63 activity. The role of TGFβ in determining the pro‐migratory and pro‐invasive function of ΔNp63 is an example of the critical impact of TME on ΔNp63 oncogenic function. Several reports have ascribed opposite functions to ΔNp63 in controlling tumour dissemination in diverse tumour types, including SCC (Cho et al., 2010; Giacobbe et al., 2016; Wu et al., 2014). Generally, it has been established that the anti‐metastatic action of ΔNp63 in squamous tumours relies on its ability to maintain the epithelial identity and repress mesenchymal traits, thus inhibiting the EMT process. ΔNp63 deregulates the expression of several genes, such as POSTN, CDH2, L1CAM and WNT5A, which contributes to the anti‐migratory ability exerted by p63 in SCC (Barbieri et al., 2006). Conversely, other data have suggested a pro‐tumourigenic effect of ΔNp63 on tumour dissemination. Exogenous ΔNp63 can enhance the migration rate of oesophageal squamous carcinoma cells (Lee et al., 2014) and ΔNp63 is required for HNSCC cancer cell migration as well (Yang et al., 2011). Although these conflicting results might be ascribed to different experimental approaches or different cellular models, it is also possible that extrinsic factors could be determinant in regulating the outcome of ΔNp63 on cell migration and invasion. Accordingly, several reports have indicated in the TGFβ pathway a parameter controlling this ΔNp63 oncogenic function. In a TGFβ‐rich environment, osteosarcoma cells display increased motility and metastatic dissemination upon overexpression of ΔNp63 (Rodriguez Calleja et al., 2016). In SCC cells, TGFβ signalling activates ΔNp63 transcriptional activity, which in turn induces the transcription of two direct target genes, DUSP6 and DUSP7 (Vasilaki et al., 2016). Ablation of ΔNp63 or DUSP6/7 perturbs the TGFβ‐induced migration and invasion. Although the proposed molecular mechanism underlying the TGFβ‐dependent activation of ΔNp63 activity has not been fully clarified, these data suggest that TME could critically contribute to determine the oncogenic function of ΔNp63.
Other observations in different epithelial tumours further support this hypothesis. Basal‐like breast cancer cells that express ΔNp63 rely on mammary fibroblasts to initiate ECM reorganization, permiting collective invasion (Dang et al., 2011, 2015). Similarly, luminal B type breast cancer cells expressing ΔNp63 are limited to invading regions enriched in collagen I (Cheung et al., 2013). These data suggest that changes of TME during tumour evolution might be of extreme importance in the fine regulation of ΔNp63 oncogenic activity.
6. Concluding remarks
Squamous cell carcinoma is a highly malignant cancer and the therapeutic options have not led to a significant increase in the overall survival rate. A common feature of SCC of diverse epithelial origins is their dependence on the oncogenic function of the transcription factor ΔNp63. A variety of ΔNp63 transcriptional target genes involved in the regulation of cell adhesion, growth factor signalling, migration and invasion have been identified, and, presumably, each of them contributes to the positive regulation of SCC development. At the molecular level, ΔNp63 exerts its oncogenic function by recruiting distinct epigenetic factors, mainly transcriptional repressor complexes. ΔNp63 acts also as transcriptional activator and it is reasonable that these two opposite functions of ΔNp63 must be finely regulated during SCC initiation and progression. It would be intriguing to assess whether changes of the ΔNp63‐dependent epigenetic landscape are functionally linked to the different stages of SCC carcinogenesis and to investigate how ΔNp63 transcriptional activity as repressor or activator might be modulated in a stage‐dependent manner. These studies could unveil novel circuits linking ΔNp63 transcriptional activity and epigenetic alterations occurring in different phases of SCC development.
Another fascinating aspect of the ΔNp63‐mediated tumourigenesis is related to the functional relationship between ΔNp63 and TME. We dare to suggest that the primary oncogenic function of ΔNp63 is to activate pathways aimed to remodel ECM and TME in order to create a favourable niche for the growth and spread of malignant cells. This oncogenic function of ΔNp63 might be strictly related to the physiological function of ΔNp63 in processes such as wound healing and regeneration, in which TME modifications and ECM remodelling play a fundamental role. However, our knowledge on the functional interplay between TME and ΔNp63 in SCC as well as the role of epigenetic factors in this circuit is quite limited. Does TME modulate ΔNp63 transcriptional activity and expression? Does ΔNp63 play a role in regulating the immune landscape of SCC? Does ΔNp63 impact TME during metastatic spreading? ΔNp63/TME crosstalk could also be important in initial phases of SCC development. Genetic events altering p63 might cooperate with p53 mutations in order to trigger the initial clonal expansion of pre‐cancerous epithelial cells. These cancer‐primed cells could affect the surrounding stroma, contributing to the field cancerization, a phenomenon accounting for the frequent appearance of recurrent and multifocal tumours observed in HNSCC and lung SCC (Curtius et al., 2018; Dotto, 2014). How and whether ΔNp63 activity may be involved in promoting field cancerization remains an intriguing open question, which might reveal novel biomarker for cancer risk. Indeed, field cancerization can occur without any clinically detectable morphological changes, making its diagnosis difficult. In conclusion, we believe that further investigations on the oncogenic role of ΔNp63 in SCC would expand our knowledge on the pathogenesis of SCC, unveiling novel, therapeutically actionable pathways.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
AP, GM and VG wrote the paper. VG, CF and MAP prepared the figures.
Acknowledgements
This work has been supported by the MFAG AIRC #15523 grant awarded to A.P., AIRC #20473 grant, Regione Lazio through LazioInnova Progetto Gruppo di Ricerca n 85‐2017‐14986 to G.M., and Fondazione Luigi Maria Monti IDI‐IRCCS (Ministry of Health, RC#3.2). Partial support has been also provided by the Medical Research Council (UK) and MAECI Italy‐China cooperation grant (#PGR00774).
References
- Abraham CG, Ludwig MP, Andrysik Z, Pandey A, Joshi M, Galbraith MD, Sullivan KD and Espinosa JM (2018) ΔNp63α suppresses TGFB2 expression and RHOA activity to drive cell proliferation in squamous cell carcinomas. Cell Rep 24, 3224–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM and Cory S (2018) The BCL‐2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ 25, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J et al (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ et al (2012) Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Mirza SA, Xu S, Schulz‐Heddergott R, Marchenko ND and Moll UM (2017) p53 loss‐of‐heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain‐of‐function in vivo. Cell Death Dis 8, e2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CT, Ricker JL, Chen Z and Van Waes C (2007) Role of activated nuclear factor‐kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck 29, 959–971. [DOI] [PubMed] [Google Scholar]
- Amelio I, Mancini M, Petrova V, Cairns RA, Vikhreva P, Nicolai S, Marini A, Antonov AA, Le Quesne J, Baena Acevedo JD et al (2018) p53 mutants cooperate with HIF‐1 in transcriptional regulation of extracellular matrix components to promote tumour progression. Proc Natl Acad Sci U S A 115, E10869–E10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen‐Tan PF, Westra WH, Chung CH, Jordan RC, Lu C et al (2010) Human papillomavirus and survival of patients with oropharyngeal cancer. New Engl J Med 363, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey BJ, Kelly GL, Janic A, Herold MJ and Strasser A (2018) How does p53 induce apoptosis and how does this relate to p53‐mediated tumour suppression? Cell Death Differ 25, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J, Camacho‐Carvajal M, Nowak M, Kramer C, Danger B and Hammerschmidt M (2005) Destabilization of DeltaNp63alpha by Nedd4‐mediated ubiquitination and Ubc9‐mediated sumoylation, and its implications on dorsoventral patterning of the zebrafish embryo. Cell Cycle 4, 790–800. [DOI] [PubMed] [Google Scholar]
- Bao X, Tang J, Lopez‐Pajares V, Tao S, Qu K, Crabtree GR and Khavari PA (2013) ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF‐dependent induction of KLF4. Cell Stem Cell 12, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Perez CA, Johnson KN, Ely KA, Billheimer D and Pietenpol JA (2005) IGFBP‐3 is a direct target of transcriptional regulation by ΔNp63α in squamous epithelium. Can Res 65, 2314–2320. [DOI] [PubMed] [Google Scholar]
- Barbieri CE, Tang LJ, Brown KA and Pietenpol JA (2006) Loss of p63 leads to increased cell migration and up‐regulation of genes involved in invasion and metastasis. Can Res 66, 7589–7597. [DOI] [PubMed] [Google Scholar]
- Baugh EH, Ke H, Levine AJ, Bonneau RA and Chan CS (2018) Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ 25, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bid HK, Roberts RD, Cam M, Audino A, Kurmasheva RT, Lin J, Houghton PJ and Cam H (2014) ΔNp63 promotes pediatric neuroblastoma and osteosarcoma by regulating tumour angiogenesis. Can Res 74, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ and Radisky D (2001) Putting tumours in context. Nat Rev Cancer 1, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J et al (2006) Radiotherapy plus cetuximab for squamous‐cell carcinoma of the head and neck. New Engl J Med 354, 567–578. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Gilad E and Peyrollier K (2007) Heregulin‐mediated ErbB2‐ERK signaling activates hyaluronan synthases leading to CD44‐dependent ovarian tumour cell growth and migration. J Biol Chem 282, 19426–19441. [DOI] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J and Khavari PA (2014) ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev 28, 2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ (2006) Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7, 678–689. [DOI] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco‐Miguel S, Takenaka I, Faha B, Seizinger BR and Kley N (1995) Induction of the growth inhibitor IGF‐binding protein 3 by p53. Nature 377, 646–649. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Giamboi‐Miraglia A, Lena AM, Mantovani R, Knight R and Melino G (2007) ΔNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA 104, 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G and Knight R (2008) p63 in epithelial development. Cell Mol Life Sci 65, 3126–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Agostini M, Melino G and Bernassola F (2014) How the TP53 family proteins TP63 and TP73 contribute to tumourigenesis: regulators and effectors. Hum Mutat 35, 702–714. [DOI] [PubMed] [Google Scholar]
- Caponigro F, Di Gennaro E, Ionna F, Longo F, Aversa C, Pavone E, Maglione MG, Di Marzo M, Muto P, Cavalcanti E et al (2016) Phase II clinical study of valproic acid plus cisplatin and cetuximab in recurrent and/or metastatic squamous cell carcinoma of Head and Neck‐V‐CHANCE trial. BMC Cancer 16, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS and Ellisen LW (2006) p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 8, 551–561. [DOI] [PubMed] [Google Scholar]
- Castellsague X, Alemany L, Quer M, Halec G, Quiros B, Tous S, Clavero O, Alos L, Biegner T, Szafarowski T et al (2016) HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Inst 108, djv403. [DOI] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury‐Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A et al (1999) Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99, 143–153. [DOI] [PubMed] [Google Scholar]
- Chang HH, Chiang CP, Hung HC, Lin CY, Deng YT and Kuo MY (2009) Histone deacetylase 2 expression predicts poorer prognosis in oral cancer patients. Oral Oncol 45, 610–614. [DOI] [PubMed] [Google Scholar]
- Chang YS, Kong G, Sun S, Liu D, El‐Naggar AK, Khuri FR, Hong WK and Lee HY (2002) Clinical significance of insulin‐like growth factor‐binding protein‐3 expression in stage I non‐small cell lung cancer. Clin Cancer Res 8, 3796–3802. [PubMed] [Google Scholar]
- Chanmee T, Ontong P and Itano N (2016) Hyaluronan: a modulator of the tumour microenvironment. Cancer Lett 375, 20–30. [DOI] [PubMed] [Google Scholar]
- Charni M, Aloni‐Grinstein R, Molchadsky A and Rotter V (2017) p53 on the crossroad between regeneration and cancer. Cell Death Differ 24, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Malhotra PS, Thomas GR, Ondrey FG, Duffey DC, Smith CW, Enamorado I, Yeh NT, Kroog GS, Rudy S et al (1999) Expression of proinflammatory and proangiogenic cytokines in patients with head and neck cancer. Clin Cancer Res 5, 1369–1379. [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z and Ewald AJ (2013) Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155, 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MS, Chan IL and Flores ER (2010) ΔNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumourigenesis and metastasis. Cell Cycle 9, 2434–2441. [DOI] [PubMed] [Google Scholar]
- Chow G, Tauler J and Mulshine JL (2010) Cytokines and growth factors stimulate hyaluronan production: role of hyaluronan in epithelial to mesenchymal‐like transition in non‐small cell lung cancer. J Biomed Biotechnol 2010, 485468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Ely K, McGavran L, Varella‐Garcia M, Parker J, Parker N, Jarrett C, Carter J, Murphy BA, Netterville J et al (2006) Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol 24, 4170–4176. [DOI] [PubMed] [Google Scholar]
- Compagnone M, Gatti V, Presutti D, Ruberti G, Fierro C, Markert EK, Vousden KH, Zhou H, Mauriello A, Anemone L et al (2017) ΔNp63‐mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumourigenesis. Proc Natl Acad Sci U S A 114, 13254–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordani N, Pozzi S, Martynova E, Fanoni D, Borrelli S, Alotto D, Castagnoli C, Berti E, Vigano MA and Mantovani R (2011) Mutant p53 subverts p63 control over KLF4 expression in keratinocytes. Oncogene 30, 922–932. [DOI] [PubMed] [Google Scholar]
- Curtius K, Wright NA and Graham TA (2018) An evolutionary perspective on field cancerization. Nat Rev Cancer 18, 19–32. [DOI] [PubMed] [Google Scholar]
- Dale OT, Aleksic T, Shah KA, Han C, Mehanna H, Rapozo DC, Sheard JD, Goodyear P, Upile NS, Robinson M et al (2015) IGF‐1R expression is associated with HPV‐negative status and adverse survival in head and neck squamous cell cancer. Carcinogenesis 36, 648–655. [DOI] [PubMed] [Google Scholar]
- Dang TT, Prechtl AM and Pearson GW (2011) Breast cancer subtype‐specific interactions with the microenvironment dictate mechanisms of invasion. Can Res 71, 6857–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TT, Esparza MA, Maine EA, Westcott JM and Pearson GW (2015) ΔNp63α promotes breast cancer cell motility through the selective activation of components of the epithelial‐to‐mesenchymal transition program. Can Res 75, 3925–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilov AV, Neupane D, Nagaraja AS, Feofanova EV, Humphries LA, DiRenzo J and Korc M (2011) DeltaNp63alpha‐mediated induction of epidermal growth factor receptor promotes pancreatic cancer cell growth and chemoresistance. PLoS One 6, e26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pauw I, Lardon F, Van den Bossche J, Baysal H, Fransen E, Deschoolmeester V, Pauwels P, Peeters M, Vermorken JB and Wouters A (2018) Simultaneous targeting of EGFR, HER2, and HER4 by afatinib overcomes intrinsic and acquired cetuximab resistance in head and neck squamous cell carcinoma cell lines. Mol Oncol 12, 830–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos M, Gilbert B, Denecker G, Leurs K, Mc Guire C, Lemeire K, Hochepied T, Vuylsteke M, Lambert J, Van Den Broecke C et al (2017) Elevated ΔNp63α levels facilitate epidermal and biliary oncogenic transformation. J Invest Dermatol 137, 494–505. [DOI] [PubMed] [Google Scholar]
- DeYoung MP, Johannessen CM, Leong CO, Faquin W, Rocco JW and Ellisen LW (2006) Tumour‐specific p73 up‐regulation mediates p63 dependence in squamous cell carcinoma. Can Res 66, 9362–9368. [DOI] [PubMed] [Google Scholar]
- Dohn M, Zhang S and Chen X (2001) p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20, 3193–3205. [DOI] [PubMed] [Google Scholar]
- Dotsch V, Bernassola F, Coutandin D, Candi E and Melino G (2010) p63 and p73, the ancestors of p53. Cold Spring Harb Pers Biol 2, a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP (2009) Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer 9, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP (2014) Multifocal epithelial tumours and field cancerization: stroma as a primary determinant. J Clin Investig 124, 1446–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–E386. [DOI] [PubMed] [Google Scholar]
- Ferone G, Song JY, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij JP, Proost N, Gargiulo G and Berns A (2016) SOX2 Is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 30, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD, Poterlowicz K, Ferone G et al (2011) p63 regulates Satb1 to control tissue‐specific chromatin remodeling during development of the epidermis. J Cell Biol 194, 825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenkov A, Zangen R, Huang YP, Osada M, Guo Z, Fomenkov T, Trink B, Sidransky D and Ratovitski EA (2004) RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 3, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Forster N, Saladi SV, van Bragt M, Sfondouris ME, Jones FE, Li Z and Ellisen LW (2014) Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev Cell 28, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza V, Fierro C, Gatti E, Peschiaroli A, Lena AM, Petruzzelli MA, Candi E, Anemona L, Mauriello A, Pelicci PG et al (2018) ΔNp63 promotes IGF1 signalling through IRS1 in squamous cell carcinoma. Aging 10, 4224–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Yamazaki M, Muto M and Ochiai A (2010) Microvascular irregularities are associated with composition of squamous epithelial lesions and correlate with subepithelial invasion of superficial‐type pharyngeal squamous cell carcinoma. Histopathology 56, 510–522. [DOI] [PubMed] [Google Scholar]
- Furth N and Aylon Y (2017) The LATS1 and LATS2 tumour suppressors: beyond the Hippo pathway. Cell Death Differ 24, 1488–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant‐Behm CL, Ramsey MR, Bensard CL, Nojek I, Tran J, Liu M, Ellisen LW and Espinosa JM (2012) ΔNp63α represses anti‐proliferative genes via H2A.Z deposition. Genes Dev 26, 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli F, Rossi M, D'Alessandra Y, De Simone M, Lopardo T, Haupt Y, Alsheich‐Bartok O, Anzi S, Shaulian E, Calabro V et al (2010) MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J Cell Sci 123, 2423–2433. [DOI] [PubMed] [Google Scholar]
- Ghioni P, Bolognese F, Duijf PH, Van Bokhoven H, Mantovani R and Guerrini L (2002) Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol Cell Biol 22, 8659–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacobbe A, Compagnone M, Bongiorno‐Borbone L, Antonov A, Markert EK, Zhou JH, Annicchiarico‐Petruzzelli M, Melino G and Peschiaroli A (2016) p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 35, 1602–1608. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Chaturvedi AK, Anderson WF and Fakhry C (2015) Epidemiology of human papillomavirus‐positive head and neck squamous cell carcinoma. J Clin Oncol 33, 3235–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M, Lin DC, Handral H, Xu L, Chen Y, Jiang YY, Mayakonda A, Ding LW, Meng X, Sharma A et al (2017) ZNF750 is a lineage‐specific tumour suppressor in squamous cell carcinoma. Oncogene 36, 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M, Lin DC, Kobayashi A, Jiang YY, Xu L, Dewi FRP, Mohamed MS, Hartono Nakada M, Meguro‐Horike M, Horike SI et al (2018) ROCK‐dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep 19, 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton ES, Zhu J and Chen X (2006) The unique NH2‐terminally deleted (ΔN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the ΔN variant of p63. J Biol Chem 281, 2533–2542. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA and Attardi LD (2005) Perp is a p63‐regulated gene essential for epithelial integrity. Cell 120, 843–856. [DOI] [PubMed] [Google Scholar]
- Ikushima H and Miyazono K (2010) TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer 10, 415–424. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L, Xiao JF, Li CQ, Huang ML, Ding LW et al (2018) Co‐activation of super‐enhancer‐driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun 9, 3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Liang J and Noble PW (2011) Hyaluronan as an immune regulator in human diseases. Physiol Rev 91, 221–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR and de Sauvage FJ (2013) Influence of tumour micro‐environment heterogeneity on therapeutic response. Nature 501, 346–354. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J and Crabtree GR (2013) Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45, 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser AM and Attardi LD (2018) Deconstructing networks of p53‐mediated tumour suppression in vivo. Cell Death Differ 25, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale J, Osterlund EJ and Andrews DW (2018) BCL‐2 family proteins: changing partners in the dance towards death. Cell Death Differ 25, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson MC, Gonzalez SF, Welin J and Fuxe J (2017) Epithelial‐mesenchymal transition in cancer metastasis through the lymphatic system. Mol Oncol 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsaros D, Yu H, Levesque MA, Danese S, Genta F, Richiardi G, Fracchioli S, Khosravi MJ, Diamandi A, Gordini G et al (2001) IGFBP‐3 in epithelial ovarian carcinoma and its association with clinico‐pathological features and patient survival. Eur J Cancer 37, 478–485. [DOI] [PubMed] [Google Scholar]
- Kim MP and Lozano G (2018) Mutant p53 partners in crime. Cell Death Differ 25, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev V, Mandinova A, Guinea‐Viniegra J, Hu B, Lefort K, Lambertini C, Neel V, Dummer R, Wagner EF and Dotto GP (2008) EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol 10, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle‐Mustienes E, Smeenk L, Rinne T, Parsaulian L et al (2010) Genome‐wide profiling of p63 DNA‐binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet 6, e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, Oti M, Niehues H, van Heeringen SJ, Schalkwijk J, Stunnenberg HG, van Bokhoven H and Zhou H (2015) Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep 16, 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Hibi T, Isogai Z, Yoneda M, Fujimori M, Amano J, Kawakubo M, Kannagi R, Kimata K, Taniguchi S et al (2007) Hyperproduction of hyaluronan in neu‐induced mammary tumour accelerates angiogenesis through stromal cell recruitment: possible involvement of versican/PG‐M. Am J Pathol 170, 1086–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Kobayashi N, Harada M, Takeoka M, Kawai Y, Sano K, Fujimori M, Amano J, Ohhashi T, Kannagi R et al (2008) Significance of tumour‐associated stroma in promotion of intratumoural lymphangiogenesis: pivotal role of a hyaluronan‐rich tumour microenvironment. Am J Pathol 172, 179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultti A, Zhao C, Singha NC, Zimmerman S, Osgood RJ, Symons R, Jiang P, Li X, Thompson CB, Infante JR et al (2014) Accumulation of extracellular hyaluronan by hyaluronan synthase 3 promotes tumour growth and modulates the pancreatic cancer microenvironment. Biomed Res Int 2014, 817613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Wilkes DW, Samuel N, Blanco MA, Nayak A, Alicea‐Torres K, Gluck C, Sinha S, Gabrilovich D and Chakrabarti R (2018) DeltaNp63‐driven recruitment of myeloid‐derived suppressor cells promotes metastasis in triple‐negative breast cancer. J Clin Investig 128, 5095–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata S, Okuyama T, Osada M, Watanabe T, Tomimori Y, Sato S, Iwai A, Tsuji T, Ikawa Y and Katoh I (2004) p51/p63 Controls subunit alpha3 of the major epidermis integrin anchoring the stem cells to the niche. J Biol Chem 279, 50069–50077. [DOI] [PubMed] [Google Scholar]
- Langer CJ, Obasaju C, Bunn P, Bonomi P, Gandara D, Hirsch FR, Kim ES, Natale RB, Novello S, Paz‐Ares L et al (2016) Incremental innovation and progress in advanced squamous cell lung cancer: current status and future impact of treatment. J Thoracic Oncol 11, 2066–2081. [DOI] [PubMed] [Google Scholar]
- Lara PC, Bordon E, Rey A, Moreno M, Lloret M and Henriquez‐Hernandez LA (2011) IGF‐1R expression predicts clinical outcome in patients with locally advanced oral squamous cell carcinoma. Oral Oncol 47, 615–619. [DOI] [PubMed] [Google Scholar]
- Lee KB, Ye S, Park MH, Park BH, Lee JS and Kim SM (2014) p63‐Mediated activation of the beta‐catenin/c‐Myc signaling pathway stimulates esophageal squamous carcinoma cell invasion and metastasis. Cancer Lett 353, 124–132. [DOI] [PubMed] [Google Scholar]
- Leemans CR, Tiwari R, Nauta JJ, van der Waal I and Snow GB (1994) Recurrence at the primary site in head and neck cancer and the significance of neck lymph node metastases as a prognostic factor. Cancer 73, 187–190. [DOI] [PubMed] [Google Scholar]
- Leemans CR, Snijders PJF and Brakenhoff RH (2018) The molecular landscape of head and neck cancer. Nat Rev Cancer 18, 269–282. [DOI] [PubMed] [Google Scholar]
- Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D et al (2007) Notch1 is a p53 target gene involved in human keratinocyte tumour suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes Dev 21, 562–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom‐Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G and Candi E (2008) miR‐203 represses ‘stemness’ by repressing ΔNp63. Cell Death Differ 15, 1187–1195. [DOI] [PubMed] [Google Scholar]
- Lena AM, Duca S, Novelli F, Melino S, Annicchiarico‐Petruzzelli M, Melino G and Candi E (2015) Amino‐terminal residues of ΔNp63, mutated in ectodermal dysplasia, are required for its transcriptional activity. Biochem Biophys Res Comm 467, 434–440. [DOI] [PubMed] [Google Scholar]
- Li Y and Seto E (2016) HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med 6, a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou Z and Chen C (2008) WW domain‐containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin‐mediated proteasomal degradation and regulates apoptosis. Cell Death Differ 15, 1941–1951. [DOI] [PubMed] [Google Scholar]
- Li Y, Peart MJ and Prives C (2009) Stxbp4 regulates ΔNp63 stability by suppression of RACK1‐dependent degradation. Mol Cell Biol 29, 3953–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zheng H, Yu F, Yu T, Liu C, Huang S, Wang TC and Ai W (2012) Deficiency of the Kruppel‐like factor KLF4 correlates with increased cell proliferation and enhanced skin tumourigenesis. Carcinogenesis 33, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra L, Mesia R, Rivera F, Remenar E, Hitt R, Erfan J, Rottey S, Kawecki A, Zabolotnyy D, Benasso M et al (2011) Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first‐line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Oncol 22, 1078–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Li X, Zhang Y, Guo Y, Zhou J, Gao K, Dai J, Hu G, Lv L, Du J et al (2015) The microRNA feedback regulation of p63 in cancer progression. Oncotarget 6, 8434–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin‐Shiao E, Lan Y, Coradin M, Anderson A, Donahue G, Simpson CL, Sen P, Saffie R, Busino L, Garcia BA et al (2018) KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev 32, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, Ji J, Wang T, Zhi H, Wu M et al (2004) Discovery of Ca2+‐relevant and differentiation‐associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene 23, 1291–1299. [DOI] [PubMed] [Google Scholar]
- Luo J, Wen Q, Li J, Xu L, Chu S, Wang W, Shi L, Xie G, Huang D and Fan S (2014) Increased expression of IRS‐1 is associated with lymph node metastasis in nasopharyngeal carcinoma. Int J Clin Exp Pathol 7, 6117–6124. [PMC free article] [PubMed] [Google Scholar]
- Malatesta M, Peschiaroli A, Memmi EM, Zhang J, Antonov A, Green DR, Barlev NA, Garabadgiu AV, Zhou P, Melino G et al (2013) The Cul4A‐DDB1 E3 ubiquitin ligase complex represses p73 transcriptional activity. Oncogene 32, 4721–4726. [DOI] [PubMed] [Google Scholar]
- Mardaryev AN, Gdula MR, Yarker JL, Emelianov VU, Poterlowicz K, Sharov AA, Sharova TY, Scarpa JA, Joffe B, Solovei I et al (2014) p63 and Brg1 control developmentally regulated higher‐order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development 141, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev AN, Liu B, Rapisarda V, Poterlowicz K, Malashchuk I, Rudolf J, Sharov AA, Jahoda CA, Fessing MY, Benitah SA et al (2016) Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol 212, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M, Laflamme L, Gervais AL and Gaudreau L (2010) Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5, 267–272. [DOI] [PubMed] [Google Scholar]
- Marur S and Forastiere AA (2016) Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc 91, 386–396. [DOI] [PubMed] [Google Scholar]
- Maula SM, Luukkaa M, Grenman R, Jackson D, Jalkanen S and Ristamaki R (2003) Intratumoural lymphatics are essential for the metastatic spread and prognosis in squamous cell carcinomas of the head and neck region. Can Res 63, 1920–1926. [PubMed] [Google Scholar]
- Mehra R, Seiwert TY, Gupta S, Weiss J, Gluck I, Eder JP, Burtness B, Tahara M, Keam B, Kang H et al (2018) Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma: pooled analyses after long‐term follow‐up in KEYNOTE‐012. Br J Cancer 119, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello SS and Attardi LD (2018) Deciphering p53 signaling in tumour suppression. Curr Opin Cell Biol 51, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR and Bradley A (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713. [DOI] [PubMed] [Google Scholar]
- Misra S, Heldin P, Hascall VC, Karamanos NK, Skandalis SS, Markwald RR and Ghatak S (2011) Hyaluronan‐CD44 interactions as potential targets for cancer therapy. FEBS J 278, 1429–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson‐Lindahl E, Merlo G, Mills AA et al (2010) A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Investig 120, 1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison CD, Chang JC, Keri RA and Schiemann WP (2017) Mutant p53 dictates the oncogenic activity of c‐Abl in triple‐negative breast cancers. Cell Death Dis 8, e2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA and Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15, 2–8. [DOI] [PubMed] [Google Scholar]
- Muller PA and Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilsen PM, Noll JE, Suetani RJ, Schulz RB, Al‐Ejeh F, Evdokiou A, Lane DP and Callen DF (2011) Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro‐invasive secretome. Oncotarget 2, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekulova M, Holcakova J, Coates P and Vojtesek B (2011) The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett 16, 296–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemajerova A, Amelio I, Gebel J, Dotsch V, Melino G and Moll UM (2018) Non‐oncogenic roles of TAp73: from multiciliogenesis to metabolism. Cell Death Differ 25, 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, Tommasi di Vignano A et al (2006) Cross‐regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev 20, 1028–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP and Radtke F (2003) Notch1 functions as a tumour suppressor in mouse skin. Nat Genet 33, 416–421. [DOI] [PubMed] [Google Scholar]
- Ory B, Ramsey MR, Wilson C, Vadysirisack DD, Forster N, Rocco JW, Rothenberg SM and Ellisen LW (2011) A microRNA‐dependent program controls p53‐independent survival and chemosensitivity in human and murine squamous cell carcinoma. J Clin Investig 121, 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S, Ueda M, Ando N, Abe O and Shimizu N (1987) High incidence of EGF receptor hyperproduction in esophageal squamous‐cell carcinomas. Int J Cancer 39, 333–337. [DOI] [PubMed] [Google Scholar]
- Paluvai H, Di Giorgio E and Brancolini C (2018) Unscheduled HDAC4 repressive activity in human fibroblasts triggers TP53‐dependent senescence and favors cell transformation. Mol Oncol 12, 2165–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrales A, Thoenen E and Iwakuma T (2018) The interplay between mutant p53 and the mevalonate pathway. Cell Death Differ 25, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Vanharanta S (2016) Epigenetic determinants of metastasis. Mol Oncol 11, 79–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M, Nomoto S, Sommer M, Fomenkov A, Hibi K, Zangen R, Poliak N, Califano J, Trink B, Ratovitski E et al (2002) DeltaNp63 induces beta‐catenin nuclear accumulation and signaling. Cancer Cell 1, 369–379. [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Scialpi F, Bernassola F, El Sherbini el S and Melino G (2010) The E3 ubiquitin ligase WWP1 regulates DeltaNp63‐dependent transcription through Lys63 linkages. Biochem Biophys Res Commun 402, 425–430. [DOI] [PubMed] [Google Scholar]
- Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J et al (2013) Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov 3, 770–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J et al (2007) TP53 mutations and survival in squamous‐cell carcinoma of the head and neck. New Eng J Med 357, 2552–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, Millar SE, Pear WS and Parmacek MS (2006) Impaired notch signaling promotes de novo squamous cell carcinoma formation. Can Res 66, 7438–7444. [DOI] [PubMed] [Google Scholar]
- Ramsey MR, He L, Forster N, Ory B and Ellisen LW (2011) Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumour maintenance in squamous cell carcinoma. Can Res 71, 4373–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MR, Wilson C, Ory B, Rothenberg SM, Faquin W, Mills AA and Ellisen LW (2013) FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Investig 123, 3525–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski EA (2013) Phospho‐ΔNp63α‐dependent microRNAs modulate chemoresistance of squamous cell carcinoma cells to cisplatin: at the crossroads of cell life and death. FEBS Lett 587, 2536–2541. [DOI] [PubMed] [Google Scholar]
- Redlich N, Robinson AM, Nickel KP, Stein AP, Wheeler DL, Adkins DR, Uppaluri R, Kimple RJ, Van Tine BA and Michel LS (2018) Anti‐Trop2 blockade enhances the therapeutic efficacy of ErbB3 inhibition in head and neck squamous cell carcinoma. Cell Death Dis 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina C, Compagnone M, Peschiaroli A, Lena A, Annicchiarico‐Petruzzelli M, Piro MC, Melino G and Candi E (2016a) Setdb1, a novel interactor of ΔNp63, is involved in breast tumourigenesis. Oncotarget 7, 28836–28848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina C, Compagnone M, Peschiaroli A, Lena AM, Melino G and Candi E (2016b) ΔNp63α modulates histone methyl transferase SETDB1 to transcriptionally repress target genes in cancers. Cell Death Discovery 2, 16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis‐Filho JS, Simpson PT, Martins A, Preto A, Gartner F and Schmitt FC (2003) Distribution of p63, cytokeratins 5/6 and cytokeratin 14 in 51 normal and 400 neoplastic human tissue samples using TARP‐4 multi‐tumour tissue microarray. Virchows Arch Int J Pathol 443, 122–132. [DOI] [PubMed] [Google Scholar]
- Rinne T, Brunner HG and van Bokhoven H (2007) p63‐associated disorders. Cell Cycle 6, 262–268. [DOI] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahe C et al (2012) p63‐microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A 109, 1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco JW, Leong CO, Kuperwasser N, DeYoung MP and Ellisen LW (2006) p63 mediates survival in squamous cell carcinoma by suppression of p73‐dependent apoptosis. Cancer Cell 9, 45–56. [DOI] [PubMed] [Google Scholar]
- Rodriguez Calleja L, Jacques C, Lamoureux F, Baud'huin M, Tellez Gabriel M, Quillard T, Sahay D, Perrot P, Amiaud J, Charrier C et al (2016) ΔNp63α silences a miRNA program to aberrantly initiate a wound‐healing program that promotes TGFβ‐induced metastasis. Can Res 76, 3236–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokudai S, Li Y, Otaka Y, Fujieda M, Owens DM, Christiano AM, Nishiyama M and Prives C (2018) STXBP4 regulates APC/C‐mediated p63 turnover and drives squamous cell carcinogenesis. Proc Natl Acad Sci U S A 115, E4806–E4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S and Sinha S (2012) ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 139, 772–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM and Melino G (2006) The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci U S A 103, 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg SM and Ellisen LW (2012) The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Investig 122, 1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotondo JC, Borghi A, Selvatici R, Magri E, Bianchini E, Montinari E, Corazza M, Virgili A, Tognon M and Martini F (2016) Hypermethylation‐induced inactivation of the IRF6 gene as a possible early event in progression of vulvar squamous cell carcinoma associated with lichen sclerosus. JAMA Dermatol 152, 928–933. [DOI] [PubMed] [Google Scholar]
- Sacco AG and Cohen EE (2015) Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol 33, 3305–3313. [DOI] [PubMed] [Google Scholar]
- Saintigny P, El‐Naggar AK, Papadimitrakopoulou V, Ren H, Fan YH, Feng L, Lee JJ, Kim ES, Hong WK, Lippman SM et al (2009) ΔNp63 overexpression, alone and in combination with other biomarkers, predicts the development of oral cancer in patients with leukoplakia. Clin Cancer Res 15, 6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou H, Rajagopal J, Ramaswamy S and Ellisen LW (2017) ACTL6A is co‐amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell 31, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf MB, Garg AD and Agostinis P (2018) Defining the role of the tumour vasculature in antitumour immunity and immunotherapy. Cell Death Dis 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ and Howley PM (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129–1136. [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z and Khavari PA (2012) ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 22, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom‐Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, Melino G and Aberdam D (2011) ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ 18, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Avanzato D and Lanzetti L (2018) Emerging functions of the EGFR in cancer. Mol Oncol 12, 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Lena AM, Cappello A, Panatta E, Anemona L, Bischetti S, Annicchiarico‐Petruzzelli M, Mauriello A, Melino G, Candi E (2019) ZNF185 is a p63 target gene critical for epidermal differentiation and squamous cell carcinoma development. Oncogene 38, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares E and Zhou H (2018) Master regulatory role of p63 in epidermal development and disease. Cell Mol Life Sci 75, 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A et al (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A and Vaux DL (2018) Viewing BCL2 and cell death control from an evolutionary perspective. Cell Death Differ 25, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez‐Carmona M, Hubert P, Gonzalez A, Duray A, Roncarati P, Erpicum C, Boniver J, Castronovo V, Noel A, Saussez S et al (2014) ΔNp63 isoform‐mediated β‐defensin family up‐regulation is associated with (lymph)angiogenesis and poor prognosis in patients with squamous cell carcinoma. Oncotarget 5, 1856–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Galbraith MD, Andrysik Z and Espinosa JM (2018) Mechanisms of transcriptional regulation by p53. Cell Death Differ 25, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JM, Jun HJ, Ko YH, Park YH, Ahn YC, Son YI, Baek JH, Park K and Ahn MJ (2011) Insulin‐like growth factor binding protein‐3, in association with IGF‐1 receptor, can predict prognosis in squamous cell carcinoma of the head and neck. Oral Oncol 47, 714–719. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Ishii G, Taira T, Fujii S, Yanagi S, Hishida T, Yoshida J, Nishimura M, Nomori H, Nagai K et al (2011) Fibrous stroma is associated with poorer prognosis in lung squamous cell carcinoma patients. J Thoracic Oncol 6, 1460–1467. [DOI] [PubMed] [Google Scholar]
- Teknos TN, Grecula J, Agrawal A, Old MO, Ozer E, Carrau R, Kang S, Rocco J, Blakaj D, Diavolitsis V et al (2018) A phase 1 trial of Vorinostat in combination with concurrent chemoradiation therapy in the treatment of advanced staged head and neck squamous cell carcinoma. Invest New Drugs, 1–9. [DOI] [PubMed] [Google Scholar]
- Temam S, Kawaguchi H, El‐Naggar AK, Jelinek J, Tang H, Liu DD, Lang W, Issa JP, Lee JJ and Mao L (2007) Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol 25, 2164–2170. [DOI] [PubMed] [Google Scholar]
- Tetreault MP, Yang Y, Travis J, Yu QC, Klein‐Szanto A, Tobias JW and Katz JP (2010) Esophageal squamous cell dysplasia and delayed differentiation with deletion of kruppel‐like factor 4 in murine esophagus. Gastroenterology 139, 171–181e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis S, Klijanienko J, Giaginis C, Rodriguez J, Jouffroy T, Girod A, Alexandrou P and Sastre‐Garau X (2011) Histone deacetylase‐1 and ‐2 expression in mobile tongue squamous cell carcinoma: associations with clinicopathological parameters and patients survival. J Oral Pathol Med 40, 706–714. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H and Dixon J (2010) Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Investig 120, 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ et al (2005) High‐resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A 102, 9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole BP (2004) Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer 4, 528–539. [DOI] [PubMed] [Google Scholar]
- Tordella L, Koch S, Salter V, Pagotto A, Doondeea JB, Feller SM, Ratnayaka I, Zhong S, Goldin RD, Lozano G et al (2013) ASPP2 suppresses squamous cell carcinoma via RelA/p65‐mediated repression of p63. Proc Natl Acad Sci U S A 110, 17969–17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R and Khavari PA (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20, 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twarock S, Freudenberger T, Poscher E, Dai G, Jannasch K, Dullin C, Alves F, Prenzel K, Knoefel WT, Stoecklein NH et al (2011) Inhibition of oesophageal squamous cell carcinoma progression by in vivo targeting of hyaluronan synthesis. Mol Cancer 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Waes C, Surh DM, Chen Z, Kirby M, Rhim JS, Brager R, Sessions RB, Poore J, Wolf GT and Carey TE (1995) Increase in suprabasilar integrin adhesion molecule expression in human epidermal neoplasms accompanies increased proliferation occurring with immortalization and tumour progression. Can Res 55, 5434–5444. [PubMed] [Google Scholar]
- Vasilaki E, Morikawa M, Koinuma D, Mizutani A, Hirano Y, Ehata S, Sundqvist A, Kawasaki N, Cedervall J, Olsson AK et al (2016) Ras and TGF‐β signaling enhance cancer progression by promoting the ΔNp63 transcriptional program. Sci Signal 9, ra84. [DOI] [PubMed] [Google Scholar]
- Vaughan CA, Singh S, Grossman SR, Windle B, Deb SP and Deb S (2017) Gain‐of‐function p53 activates multiple signaling pathways to induce oncogenicity in lung cancer cells. Mol Oncol 11, 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN and Wu M (2002) Down‐regulation of gut‐enriched Kruppel‐like factor expression in esophageal cancer. World J Gastroenterol 8, 966–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, Fox C, Francis JM, Pedamallu CS, DeLuca DS et al (2014) SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Investig 124, 1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche J, Haglund K and Haugsten EM (2011) Fibroblast growth factors and their receptors in cancer. Biochem J 437, 199–213. [DOI] [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC and Pietenpol JA (2003) The ΔNp63α phosphoprotein binds the p21 and 14‐3‐3σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay‐Wells syndrome‐derived mutations. Mol Cell Biol 23, 2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG and Roberts CW (2011) SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11, 481–492. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Lee DY, Berry L, Shames DS and Settleman J (2011) Neuregulin‐1‐mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell 20, 158–172. [DOI] [PubMed] [Google Scholar]
- Wintergerst L, Selmansberger M, Maihoefer C, Schuttrumpf L, Walch A, Wilke C, Pitea A, Woischke C, Baumeister P, Kirchner T et al (2018) A prognostic mRNA expression signature of four 16q24.3 genes in radio(chemo)therapy‐treated head and neck squamous cell carcinoma (HNSCC). Mol Oncol 12, 2085–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D and Prives C (2018) Relevance of the p53‐MDM2 axis to aging. Cell Death Differ 25, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liang S, Bergholz J, He H, Walsh EM, Zhang Y and Xiao ZX (2014) ΔNp63α activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell Death Dis 5, e1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Yin C, Wang Y, Lv H, Wang W, Huang Y, Perez‐Losada J, Snijders AM, Mao JH and Zhang P (2018) FBXW7 deletion contributes to lung tumour development and confers resistance to gefitinib therapy. Mol Oncol 12, 883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D and McKeon F (1998) p63, a p53 homolog at 3q27‐29, encodes multiple products with transactivating, death‐inducing, and dominant‐negative activities. Mol Cell 2, 305–316. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C et al (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718. [DOI] [PubMed] [Google Scholar]
- Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, Duggal P, Allen C, Chuang R, Ehsanian R et al (2011) ΔNp63 versatilely regulates a Broad NF‐κB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Can Res 71, 3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM and Pan D (2010) Kibra functions as a tumour suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yugawa T, Narisawa‐Saito M, Yoshimatsu Y, Haga K, Ohno S, Egawa N, Fujita M and Kiyono T (2010) ΔNp63α repression of the Notch1 gene supports the proliferative capacity of normal human keratinocytes and cervical cancer cells. Can Res 70, 4034–4044. [DOI] [PubMed] [Google Scholar]
- Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M, Zhang J, Ren X, Wang Y, Yang X et al (2018) m(6)A‐mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis 9, 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L et al (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhao Z, Wang R, Han Y, Wang C, Yang F, Han Y, Liang H, Qi L, Wang C et al (2017) Identification of driver copy number alterations in diverse cancer types and application in drug repositioning. Mol Oncol 11, 1459–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]


