Abstract
TP53 is the most frequently mutated gene across all cancer types. Our understanding of its functions has evolved since its discovery four decades ago. Initially thought to be an oncogene, it was later realized to be a critical tumour suppressor. A significant amount of our knowledge about p53 functions have come from the use of antibodies against its various forms. The early anti-p53 antibodies contributed to the recognition of p53 accumulation as a common feature of cancer cells and to our understanding of p53 DNA-binding and transcription activities. They led to the concept that conformational changes can facilitate p53’s activity as a growth inhibitory protein. The ensuing p53 conformational-specific antibodies further underlined p53’s conformational flexibility, collectively forming the basis for current efforts to generate therapeutic molecules capable of altering the conformation of mutant p53. A subsequent barrage of antibodies against post-translational modifications on p53 has clarified p53’s roles further, especially with respect to the mechanistic details and context-dependence of its activity. More recently, the generation of p53 mutation-specific antibodies have highlighted the possibility to go beyond the general framework of our comprehension of mutant p53—and promises to provide insights into the specific properties of individual p53 mutants. This review summarizes our current knowledge of p53 functions derived through the major classes of anti-p53 antibodies, which could be a paradigm for understanding other molecular events in health and disease.
Keywords: antibodies, conformation, mutant, p53, post-translational modifications
Introduction
DNA tumour viruses were actively studied as a paradigm for tumourigenesis in the 1970s, and the mechanistic basis of virus-induced cellular transformation was heavily sought after during that period. It was in such attempts to identify proteins interacting with the viral SV40 T antigen that p53 was first identified as a co-immuno-precipitating protein (Kress et al., 1979; Lane and Crawford, 1979; Linzer and Levine, 1979). Thereafter, the field has evolved exponentially. The cloning of Tp53 which encodes p53, and its ability to transform cells led to the initial concept that p53 was an oncogene (Eliyahu et al., 1984; Jenkins et al., 1984; Parada et al., 1984; Wolf et al., 1984). However, identification of mutations in sporadic cancer samples and in familial cancers, concomitant to the growth inhibitory properties of both wild-type (WT) p53 and temperature-sensitive mutant p53 cDNA clones demonstrated that p53 was indeed a tumour suppressor (Baker et al., 1989; Eliyahu et al., 1989; Finlay et al., 1989; Malkin et al., 1990; Michalovitz et al., 1990; Srivastava et al., 1990; Hollstein et al., 1991). A large number of laboratories around the world have contributed to our current knowledge of all aspects of p53 functions: from it being a labile transcription factor which works as a tetramer to regulate target genes essential for tumour suppression, to it being heavily mutated in a large number of cancer types (excellently reviewed in Menendez et al., 2009; Zilfou and Lowe, 2009; Rivlin et al., 2011; Bieging et al., 2014; Kruiswijk et al., 2015). Recently, significant knowledge has been uncovered from the standpoint of mutant p53. These include the understanding that mutant p53 is not only a long-lived inactive protein but that it also can have adverse effects on the remaining WT p53 protein in what is now known as the dominant-negative (DN) effect, and the realization that it can acquire novel gain-of-functions (GOF) to actively drive tumourigenesis as an oncogenic protein (Freed-Pastor and Prives, 2012; Muller and Vousden, 2014; Sabapathy, 2015; Sabapathy and Lane, 2018).
Many of the discoveries in the p53 field have been attributed to the availability of a large number of antibodies generated against the various domains, conformations, and modifications of p53. Initial data demonstrating the DNA-binding ability of p53 can be further enhanced by incubation with antibodies, and the role of conformational changes in regulating p53 functions came about due to the generation of a series of very intricate anti-p53 antibodies that were capable of recognizing the many modified states of p53, underscoring the incredible importance of these antibodies in advancing the p53 field. This review will therefore explore the journey of our understanding of p53 functions over the years through the various classes of the anti-p53 antibodies, and hopes to provide an overview for the p53 field and beyond, as these studies could serve as a paradigm for the in-depth study of protein functions using antibodies.
Our understanding of p53 functions through the initial anti-p53 antibodies
The discovery of p53 depended on the fact that animals bearing tumours made an auto-antibody response to p53. These polyclonal antibodies have been detected in the sera of tumour bearing mice, hamsters, and human patients (DeLeo et al., 1979; Kress et al., 1979; Linzer and Levine, 1979; Rotter et al., 1980; Chandrasekaran et al., 1981; Crawford et al., 1982; Soussi, 2000). With the advent of the monoclonal method, it became possible to make monoclonal antibodies to p53 by taking splenic B cells from mice that had been immunized by injection with autologous transformed cells. The antibodies from the first four laboratories to produce such reagents were made in this way within two years of the discovery of p53. These include the following antibodies: PAb122 from the Tucker Gurney laboratory (Gurney et al., 1980); PAb421 from Lionel Crawford’s laboratory (Harlow et al., 1981); 200.47 from Lloyd Old’s laboratory (Dippold et al., 1981); and RA3 2C2 from the Baltimore laboratory (Rotter et al., 1980). The use of these early antibodies established that p53 accumulation is a common feature of transformed cells (Crawford et al., 1981; Benchimol et al., 1982; Thomas et al., 1983). In a very important set of experiments for the field, PAb122 and 200.47 antibodies were used to investigate the role of p53 in cell cycle and serum-stimulated DNA synthesis. Microinjection of these antibodies into the nucleus of quiescent cells blocked DNA synthesis with great efficiency (Mercer et al., 1982, 1984). The results were highly specific as PAb122 and 200.47 have distinct species specificity (Mercer et al., 1984). Both antibodies were reactive and active in mouse cells while 200.47 worked in hamster cells and PAb122 did not. Conversely, PAb122 was reactive with human p53 and inhibited human cell escape from serum arrest, without having an impact on the ongoing cell cycle (Mercer et al., 1984). At that time, the data were interpreted on the assumption that the antibodies would be antagonistic—that is, p53 was essential for entry into the cycle. As discussed below, it later emerged that the antibodies were in fact agonists, activating p53’s transcriptional activity and inducing cell cycle arrest presumably by induction of p21 and other p53 response genes.
A few years later, a second set of anti-p53 antibodies—PAb242, PAb246, and PAb248 became available, and all of these had their binding sites mapped using Escherichia coli expression constructs and steric competition assays (Yewdell et al., 1986). These antibodies were then widely distributed in the community without charge. Subsequently, purified recombinant p53 proteins made in E.coli were used as immunogens and led to the production of the widely used DO-1, DO-7, and PAb240 antibodies from the Lane and Vojtesek laboratories (Gannon et al., 1990; Vojtesek et al., 1992), in addition to many other anti-p53 monoclonal antibodies that have enjoyed less widespread usage (Table 1 and http://p53.free.fr/p53_info/p53_monoclonal_antibodies/p53_MabH.html). These first generation antibodies have been in very wide usage. In particular, the DO-1 and DO-7 antibodies are widely used in pathology to examine p53 protein expression in human tissues as they are highly effective in staining standard Formalin Fixed Paraffin Embedded sections.
Table 1.
Commonly used anti-p53 monoclonal antibodies.
| Antibody clone (s) | Region recognizeda | Human p53 epitope recognized | Commercially available | Remarks |
|---|---|---|---|---|
| DO-1 and DO-7 | aa20–25 |
|
Yes (for both) | Widely used for formalin-fixed human tumour sample (FFPE) analysis; |
| Microinjection into cells rescues senescence and cell cycle arrest due to PAb421 microinjection | ||||
| DO-2 | aa10–16 |
|
Yes | |
| DO-13 | aa26–35 |
|
Yes | |
| DO-14 | aa56–65 |
|
Yes | |
| PAb1801 | aa46–55 |
|
Yes | Microinjection into cells rescues senescence and cell cycle arrest due to PAb421 microinjection; |
| Microinjection into cells leads to increased nuclear p53; | ||||
| Stimulates the ability of p53 to protect p53-binding site from cleavage by DNase I in vitro | ||||
| PAb421 and PAb122 | aa370–378 |
|
Yes | Microinjection into cells leads to cell cycle arrest and increased DNA binding; |
| Microinjection into cells bocks DNA synthesis in quiescent cells; | ||||
| Stimulates dramatically the quantity of p53–DNA complexes that can be detected by electromobility shift assays in vitro | ||||
| PAb200-47 | aa81–95 |
|
No | Microinjection into cells blocks DNA synthesis in quiescent cells |
| PAb242 | aa9–25 |
|
Yes | |
| PAb248 and RA3 2C2 | aa157–192 |
|
No (for both) | |
| Pab241/PAb243 | aa296–305 |
|
No (for both) | |
| ICA-9 | aa388–393 |
|
No | Inhibits p53’s ability to bind to DNA upon DNA damage and activation |
aAmino-acid numbering is based on human p53 sequence.
bNon-homologous amino acids in mouse (mo) p53 compared to human (hu) p53 are indicated in red.
Please refer to http://p53.free.fr/p53_info/p53_monoclonal_antibodies/p53_MabH.html for information of more antibodies.
Many of these antibodies have been carefully characterized with respect to the precise epitopes they recognize using both synthetic peptide libraries and random phage display peptide libraries (Stephen and Lane, 1992; Stephen et al., 1995). From these early studies, a number of key points emerged. Firstly, most antibodies recognized epitopes in the amino- and carboxyl-terminus of p53 and were directed to its unstructured regions. This seems to relate to the intrinsic structural features of these regions, as selection from a random ‘single pot’ antibody library also discovered antibodies to these major amino- and carboxyl-terminal epitopes (Nissim et al., 1994). Secondly, the antibodies could show exquisite species specificity. For instance, the commonly used DO-1 antibody is able to bind to human but not mouse p53 due to a single amino acid change of D in human to G in mouse at the 21st amino acid of the human sequence (Table 1). Thirdly, these antibodies fell into two distinct types: those that could recognize p53 in denatured form in immunoblots, and those that could only react with the folded native protein. This latter class is active in immunoprecipitation and in immunofluorescence using fixed cells, but not in western blots, as they bind complex epitopes in the folded structure of p53 DNA-binding domain (DBD). Finally, several of these antibodies bind to epitopes that containing amino acids that were subject to post-translational modifications (PTMs). For example, the PAb421 epitope is rendered less reactive when p53 is phosphorylated in cells exposed to DNA damaging agents (Hupp and Lane, 1995).
A major use of these antibodies in the study of p53 regulation was derived from the discovery that antibodies to the carboxyl-terminal 30 amino acids of p53 like PAb421 and PAb122 could massively enhance the specific DNA-binding ability of p53 in gel shift electromobility shift assays (Hupp et al., 1992; Hecker et al., 1996). The activity seems to act by neutralizing a negative regulatory effect of the p53 carboxyl-terminal region. It can be mimicked by deletion of the last 30 amino acids, by phosphorylation of sites in the region, and by the action of molecular chaperones (Hupp et al., 1992). The use of the monovalent Fab fragment of the PAb421 antibody combined with gel shifts incorporating the DO-1 antibody showed that the PAb421 monovalent Fab fragment was active in stimulation of p53 DNA-binding. These studies also showed that p53 bound to DNA as a tetramer and all four carboxyl-termini had to bind to the PAb421 antibody for the enhanced DNA-binding to take place (Hupp and Lane, 1994). In support of the idea that this regulation of p53 DNA-binding function was mediated by allosteric mechanisms, a new anti-p53 antibody, i.e. ICA-9, directed to the extreme carboxyl-terminus of p53 was found to inhibit the p53 DNA-binding function of p53 that had been activated by phosphorylation or the binding of PAb421 (Hupp and Lane, 1994). By contrast, an antibody against the amino terminus, PAb1801 (Banks et al., 1986), was later found to reduce the rate of p53 dissociation from DNA. It therefore increased p53’s ability to protect a cognate site from DNase I digestion and consequently stimulated the ability of p53 to protect the p21 promoter from DNase I cleavage (Cain et al., 2000). These latter data provided evidence for the amino-terminus of p53 to possess an auto-inhibitory function that is mechanistically different from the inhibitory region at the carboxyl-terminus.
These initial efforts led to antibody micro-injection studies that asked the question whether p53 transcription could be activated in living cells. Using human reporter cells that contained a bacterial β-galalacsidose gene under the control of a p53 response element, it was clearly demonstrated that the antibody PAb421 could induce p53’s transcriptional activity and cause a cell cycle arrest (Hupp et al., 1995). At the time, there were some concerns that the injection phenomena itself could be partly responsible for these observations by creating a stress signal. However, in subsequent work, the perfect control was realized when the DO-1 antibody, and another p53 antibody against the amino-terminal region, PAb1801, were shown to be able to reverse the effects of PAb421 and block p53-dependent transcription (Gire and Wynford-Thomas, 1998). In a remarkable and well-controlled set of experiments, it was further shown that injection of either DO-1 or PAb1801 could also block the p53 transcription response to DNA damage, allowing senescent human fibroblasts to efficiently resume a ‘young’ morphology and re-enter the cell cycle (Gire and Wynford-Thomas, 1998). The activity of these antibodies seemed to block the interaction of p53 with the transcription apparatus rather than its degradation, as the injection of PAb1801 increased rather than decreased the levels of nuclear p53. Further detailed biochemical work using WT p53 produced in Baculoviruses showed that both active and latent forms of p53 were produced and differed in their PTMs that included phosphorylation at the carboxyl-terminus of the protein (Hupp and Lane, 1995). Growing insect cells in low serum or treating them with UV irradiation led to clear changes in the modification and DNA-binding function of WT p53. In other studies, the binding of antibodies to the p53 binding domain of MDM2 was shown to activate p53 and antibodies to the amino-terminus of p53 were found able to protect it from thermal denaturation (Hansen et al., 1996; Böttger et al., 1997). Collectively, these early studies using microinjection of the anti-p53 monoclonal antibodies significantly enhanced our understanding of p53’s role in DNA-binding and cellular functions, which were further pursued in other biochemical assays.
Lessons from p53 conformation-specific antibodies
As alluded to above, most of the anti-p53 antibodies generated have been against the amino- or carboxyl-terminal regions. Only a few have been initially generated against the DNA-binding region. Of these, the first two were PAb240 and PAb246, which recognize regions between amino acids 211–217 and 201–212, respectively (Yewdell et al., 1986; Gannon et al., 1990; Stephen and Lane, 1992; Wang et al., 2001). PAb240 was shown to detect mutated p53, and PAb246 was shown to detect WT p53 (Gannon et al., 1990; Wang et al., 2001). In addition, PAb1620 which was earlier developed for SV40 T antigen (Ball et al., 1984), was shown to recognize the WT conformation of p53 (Milner et al., 1987). It recognizes the specific regions between amino acids 145–157 and 201–212 (Wang et al., 2001), the latter being similar to the PAb246 epitope and thus was capable of recognizing WT p53 (Milner et al., 1987; Cook and Milner, 1990). A few other conformation-specific antibodies were also subsequently generated (Legros et al., 1994; Vojtesek et al., 1995), of which some are highlighted in Table 2.
Table 2.
List of p53 conformation-specific monoclonal antibodies.
| Antibody clone | Region recognized | Human p53 epitope recognized | Conformation detected | Commercially available |
|---|---|---|---|---|
| PAb246 | aa201–212 | LRVEYLDDRNTF—hu |
WT (mouse) | Yes |
LYPEYLEDRQTF—moa,b | ||||
| PAb1620 | aa145–157 and aa201–212 (as above) |
LWVDSTPPPGTRV—hu
a
|
WT (human) | Yes |
LWVSATPPAGSRV—mo | ||||
| PAb240 | aa211–217 | TFRHSVV—hu |
Mutant | Yes |
TFRHSVV—mo | ||||
| DO-12 | aa256–270 | TLEDSSGNLLGRNSF—hu |
Mutant | Yes |
TLEDSSGNLLGRDSF—mo | ||||
| DO-11 | aa181–190 | SDSDGLAPPQ—hu |
Mutant | Yes |
SDGDGLAPPQ—mo |
aBold and underlined, specific amino acids recognized on epitope by antibody.
bNon-homologous amino acids in mouse (mo) p53 compared to human (hu) p53 are indicated in red.
In essence, based on a large number of studies on cell lines expressing WT or mutant p53, it has been well established that un-mutated p53 is recognized by the WT conformation-specific antibodies such as PAb246 or PAb1620, and mutated p53 is recognized by mutant conformation-specific PAb240 antibody (Bartek et al., 1990; Milner and Medcalf, 1991; Milner, 1995). Moreover, these studies correlated the p53 conformation to its biological functions. For instance, temperature-sensitive p53 mutants that adopt a WT conformation at the permissive temperature were found to be biologically active, whereas they lost their functions at a higher temperature when in a mutant conformation (Michalovitz et al., 1990; Martinez et al., 1991; Yonish-Rouach et al., 1991; Gaitonde et al., 2000). Furthermore, the binding affinities of heat shock proteins (HSP) (70 and 90) tended to be higher towards p53 in the mutant conformation (Gannon and Lane, 1991; Ory et al., 1994; Blagosklonny et al., 1996), correlating with the longer half-life of mutant p53 (Gronostajski et al., 1984; Reihsaus et al., 1990). On the other-hand, PAb246+ p53 had a much shorter half-life than the HSP70-bound PAb240+ p53 (Finlay et al., 1988). Treatment of cells with geldanamycin, a HSP inhibitor led to the destabilization of mutant p53 and to the loss of the PAb240+ epitope (Blagosklonny et al., 1995; Blagosklonny, 2002), directly linking the HSP-binding and mutant p53 conformation.
However, exceptions have been noted to the general principle of mutation status of p53 to its conformation, and these can be broadly classified into two categories. In the first case, while mutations in p53 often lead to a conformational change, not all p53 mutants exclusively adopt a mutant conformation recognizable by PAb240 (Schmieg and Simmons, 1993; Webley et al., 2000). This finding highlighted that the degree of conformational change depends on the amino acids affected, which may have a differential effects on the overall conformation. Moreover, mutated p53 in mutant conformation can be induced to adopt a WT conformation localized in the nucleus, as demonstrated by the treatment of mutant p53-expressing cells with DMSO (Ryan and Clarke, 1994). This observation suggested that conformational change of mutated p53 is not an irreversible feature, and together with other observations described in the previous section that PAb421 enhanced the DNA-binding of mutant p53, formed the basis for current therapeutic strategies to reverse the conformation of mutant p53 to a WT conformation capable of tumour suppression. Lead drugs such as APR-246 (Prima-Met) that are capable of enabling the refolding of mutant p53 to a conformation detectable by PAb1620 but not by PAb240, currently are in clinical trials (Bykov and Wiman, 2014). In addition, a recent study has emphasized this point further using mutant p53 expressed in embryonic stem cells. In the study, mutant p53 was shown to exhibit a WT conformation due to binding of several proteins such as the CCT complex, Trim24, Nedd4, USP7, and Aurora A (Rivlin et al., 2014), highlighting that mutant p53 can indeed be made to adopt a WT conformation and be functionally activated. Following this up, the authors performed a screen to search for peptides capable of restoring the WT conformation to mutant p53, and successfully identified a series of peptides capable of restoring the WT conformation that led to the regression of mutant p53-expressing tumours in vivo (Tal et al., 2016).
In the second category, un-mutated WT p53 has been shown to lose its WT conformation and adopt a mutant conformation in multiple instances. Such conformational changes of WT p53 appear to occur during cell cycle progression, as a decrease in reactivity to PAb246 and a concomitant increase in reactivity to PAb240 was noted when fresh medium was added to cells (Milner and Watson, 1990). Similarly, stimulation of normal lymphocytes with the mitogen concanavalin A or treatment of acute myelogenous leukaemia cells with growth factors (such as granulocyte-macrophage colony-stimulating factor and interleukin 3) induced a mutant-like conformation recognizable by PAb240 of WT p53 in cells (Zhang et al., 1992; Wu et al., 1993; Zhang and Deisseroth, 1994), though this change was not observed by another group (Mosner and Deppert, 1992). Furthermore, WTp53 tended to lose the mutant-specific PAb240 epitope in G1-arrested cells (Ullrich et al., 1992). These data collectively indicate a positive correlation between the mutant conformation of WT p53 with cellular proliferation. The general consensus therefore appears to be that stimulation of cells to enter into the cell cycle leads to p53 adopting a mutant conformation transiently, to allow for the relief of inhibitory effects on proliferation to enable the cells to divide (Milner, 1991). These data also indicate that the conformational change of p53 is indeed flexible and can be modulated, depending on the cellular context.
In another context, treatment of cells with metal chelators also led to an increase in PAb240 reactivity of p53 concomitant to a decrease in PAb1620 reactivity (Coffer and Knowles, 1994). This was accompanied by decreased DNA-binding and p21 target gene expression (Verhaegh et al., 1998). Temperature shift also affected p53 conformation, which appeared to be associated with properties of chelation (Hainaut et al., 1995). Exposure to high temperatures led to mutant conformation and enhanced chelation, and on the contrary, lower temperatures resulted in a WT conformation and p53 was protected from chelation (Hainaut et al., 1995). As p53 activity is dependent on its binding to metals such as Zinc (Rainwater et al., 1995), it is not unexpected that chelation results in a mutant conformation which would disrupt its functionality (Hainaut and Milner, 1993a), again indicating the flexibility of p53 to adopt a mutant conformation in adverse conditions not congruent with tumour suppression. Similarly, oxidation of p53 also led to a decrease in reactivity to PAb246 and a concomitant decrease in DNA binding (Hainaut and Milner, 1993b). Interestingly, fibroblasts isolated from Alzheimer’s patients experiencing elevated oxidative stress express p53 with reduced activity that is in the mutant conformation recognizable by PAb240 (Uberti et al., 2006), supporting the paradigm that inactive WT p53 could be found in a mutant conformation. Furthermore, WT p53 in mouse embryonic stem cells which is expressed at very high levels adopted the mutant conformation upon differentiation when its levels and activity decreased significantly (Sabapathy et al., 1997). However, one report has suggested that in vitro translated WT p53 bound to target gene promoters adopted the mutant conformation (Halazonetis et al., 1993). While these latter report do not support and in fact appear to be diametrically opposite to the general idea of WT p53 adopting a mutant conformation when inactive, most studies have established a strong positive correlation between the loss of functionality and the mutant conformation of p53.
Mapping of the PAb240 epitope by phage display to the RHSVV sequence allowed the detailed examination of why the epitope is hidden in the WT conformation and becomes exposed in the mutant state (Stephen and Lane, 1992). This sequence is buried in the crystal structure of the DBD and hidden from the surface of p53 (Cho et al., 1994). Thus, p53 must be unfolded or ‘melted’ from its normal 3D structure, as in most p53 mutants, to react with PAb240. This in turn also explains why PAb240 is able to detect WT p53 in immunoblotting methods but not in immunoprecipitation or other methods that preserved protein conformation. Another two antibodies recognizing the DBD also exhibited the same behaviour as PAb240 (Vojtesek et al., 1995), and epitope mapping indicated that their binding sites on mutant p53 is substantially unfolded in its ‘mutant’ conformation. All these studies together provide an explanation for the flexibility observed in p53 conformation during different cellular states.
Although many of the studies described in this section were performed over two decades ago, the findings are still relevant and consistent with our current understanding of p53 functions, underscoring the value of these earlier experiments using the p53 conformation-specific antibodies.
Role of p53 PTMs—a view from the p53 PTM-specific antibodies
Phosphorylation
The last decade of the 20th century has brought with it a major revolution in biology—the possibility to generate antibodies against specific PTM on individual amino acid residues of various proteins. This has transformed the way we comprehend the dynamic nature of protein regulation in situ. The coordinated changes that a protein undergoes as a function of time to regulate the appropriate outcomes can now be followed. The p53 field has benefitted tremendously from this technology as many antibodies against a multitude of modified residues have been generated. Among these, the phosphorylation-specific and acetylation-specific antibodies have been key in our understanding of p53 functions, mainly due to the prior information that p53 is a phospho-protein and is often acetylated (Jay et al., 1981; Meek and Eckhart, 1988; Gu and Roeder, 1997). Early studies have used in vitro kinase or acetylation assays to establish that these modifications occur on p53, and have even identified the potential residues that are modified. However, our current knowledge of the cellular context in which such modifications operate, the synergy or the competition among them in regulating p53 functions, could only have been possible with the advent of these p53 PTM-specific antibodies. The roles of PTMs in the regulation of p53 functions have been excellently reviewed elsewhere (Meek and Anderson, 2009; Brooks and Gu, 2011), and herein, we will specifically present the role of the PTM-specific antibodies in shaping our understanding of p53 functions.
The first series of p53 phosphorylation-specific antibodies were reported around 1997/1998. The first confirmation of a previously suspected phosphorylation event using these antibodies was that serine 15 (S15) phosphorylation is robustly induced by DNA-damaging agents such as UV and γ-irradiation (IR) (Shieh et al., 1997; Siliciano et al., 1997; Banin et al., 1998; Canman et al., 1998). These antibodies also established that S15 phosphorylation attenuated the binding of p53 to its negative regulator MDM2, likely due to a conformational change (Shieh et al., 1997). Concurrently, S392 phosphorylation was also reported to be induced by DNA damage and crucial for p53 tetramerization (Sakaguchi et al., 1997). However, the phospho-S392-specific antibodies revealed that this site was phosphorylated only upon UV irradiation, and not upon IR (Blayden and Hupp, 1998; Kapoor and Lozano, 1998; Lu et al., 1998), highlighting the cellular contexts in which this phosphorylation event occurs. In addition, a flurry of other reports led to other significant findings. For instance, the significance of the amino-terminal phosphorylation events was deciphered through the phospho-S6, S9, threonine 18 (T18), S20, S33, S37, S46, T55, and T81-specific antibodies, as well as the carboxyl-terminal phospho-S315, S378, and S392-specific antibodies. All of the afore-mentioned sites were phosphorylated upon DNA damage (Ko et al., 1997; Sakaguchi et al., 1998, 2000; Waterman et al., 1998; Bulavin et al., 1999; Shieh et al., 1999, 2000; Chao et al., 2000; Higashimoto et al., 2000; Blaydes et al., 2001; Buschmann et al., 2001; Saito et al., 2002, 2003; Wu et al., 2002; Li et al., 2004, 2007), except T55, which was found phosphorylated in unstimulated cells and was dephosphorylated upon DNA damage (Gatti et al., 2000; Li et al., 2004) (Figure 1).
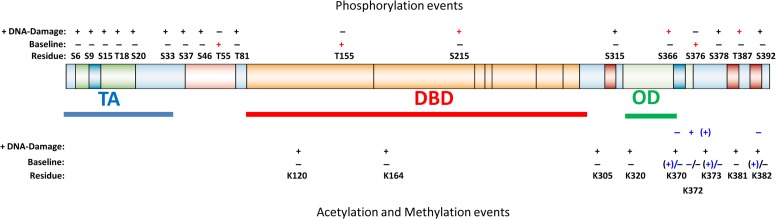
Figure 1.
Schematic of the major PTMs on p53 evaluated with PTM-specific antibodies. Phosphorylation status on the specific residues at baseline and upon DNA damage, as evaluated using the respective phosphorylation-specific antibodies, are highlighted in the upper panel above the p53 cartoon. ‘+’ indicates phosphorylation. ‘+’ in black indicates activation of p53 stability/function upon phosphorylation, whereas in red indicates degradation of p53 after phosphorylation. Lower panel shows the status of acetylation (black font) and methylation (blue font) at baseline and upon DNA damage, determined using the respective PTM-specific antibodies. All acetylation events reported so far lead to p53 activation (indicated by ‘+’). However, only methylation at K372 leads to p53 activation. Methylation at the other three sites lead to inactivation of p53, represented by ‘(+)’. Amino acid residues are reflected (not to scale), together with the major p53 domains. TA, transactivation; DBD, DNA-binding domain; OD, oligomerization domain. Other PTMs on p53, such as ubiquitination and SUMOylation, are not included in this review.
In the former case of phosphorylation upon DNA damage, several interesting points have been collectively noted. Firstly, almost all tested pleiotropic DNA-damage signals, such as IR, UV irradiation, camptothecin (CPT), and adriamycin invariably led to the phosphorylation of the amino-terminal sites, unlike the signal specificity seen with S392 phosphorylation at the carboxyl-terminus. In addition, genotoxic stress signals that activate the JNK pathway, such as UV irradiation, hydrogen peroxide, sorbitol, and heat shock, also resulted in phosphorylation at T81 (Buschmann et al., 2001). Although all these sites are generally phosphorylated by cellular stimulation, the kinetics of phosphorylation differed in some cases (Shieh et al., 2000; Saito et al., 2003). For instance, while S20 phosphorylation was rapid upon IR, it was slower upon UV irradiation (Shieh et al., 2000). Secondly, phophorylation of these PTM sites, especially S6, S9, S15, S20, and S46, were somewhat dependent on ATM (Saito et al., 2002), indicating a coordinating role of specific residues in bringing about consequent effects. In the case of S15 that serves as a nuclear event to enable subsequent phosphorylation on adjacent sites, its phosphorylation led to the subsequent phosphorylation on T18 by CK1, and its binding to MDM2 (Sakaguchi et al., 2000), arguing for a critical role of phosphorylation events in mediating p53 stabilization. Similarly, UV irradiation-induced, p38 kinase-dependent phosphorylation at S33 and S47 was found to be required for the phosphorylation at S15 and S37 (Bulavin et al., 1999). Likewise, S33 and S37 phosphorylation was critical for p53 acetylation at the carboxyl terminal (K382) (Lambert et al., 1998; Sakaguchi et al., 1998). These phosphorylation events therefore appear to be critical for p53 stabilization and augmentation of its functional activity, beyond the primary mechanism of p53 stabilization upon DNA damage through phosphorylation and inactivation of MDM2 (Gannon et al., 2012). However, treatment of cells with ALLN could stabilize p53 without phosphorylation on any of these amino-terminal sites (Sakaguchi et al., 2000). Thus, though important in the physiological context, the phosphorylation events on the amino-terminal of p53 may not be absolutely necessary for p53 stabilization, but are likely to play an auxiliary role. Yet, these events would likely be critical in promoting p53 activity, noted with S392 phosphorylation which was critical for p53 oligomerization and functioning (Sakaguchi et al., 1997). Consistently, proteasomal inhibition-mediated p53 stabilization is often found not to activate p53 functions (Ashcroft et al., 2000), though this is subject to some controversy (Olsson et al., 2007).
The other case is where DNA damage leads to dephosphorylation. T55 was found to be basally phosphorylated in a manner-dependent on TAF1 in unstimulated cells, resulting in its degradation (Gatti et al., 2000). T55 phosphorylated p53 was bound to the nuclear export protein CRM1, which led to its cytoplasmic localization and consequent degradation (Li et al., 2004). DNA damage resulted in a decrease in this phosphorylation event, contributing to p53 stabilization. A similar phenomenon was also observed whereby p53 is phosphorylated at T155 in the DBD by the COP9-signalasome complex (CSN) and results in its nuclear export (Lee et al., 2017). In this case, glycosylation on S149 reversed T155 phosphorylation and stabilized p53 (Yang et al., 2006). Moreover, phosphorylation at S215 by the PAK4 kinase, which is overexpressed in cancers, decreased p53 transactivation potential and promoted the metastasis of hepatocellular carcinoma (Xu et al., 2016). Likewise, carboxyl-terminal phosphorylation of p53 also led to the deactivation or degradation of p53. For example, phosphorylation at S376 was noted in unstimulated cells, but its level was reduced upon DNA damage, resulting in the stabilization of p53 (Waterman et al., 1998). These examples together highlight that basal p53 phosphorylation on specific sites in unstimulated contexts is required to keep p53 labile and inactive. Hence, coordinated dephosphorylation/phosphorylation at these sites upon DNA damage, together with additional stimulatory phosphorylation at other sites lead to p53 activation. A specific example of coordinated dephosphorylation/phosphorylation regulating p53 activity was demonstrated by using the phospho-S376 and S378-specific antibodies. Loss of phosphorylation at S376 led to the binding of p53 to 14-3-3 and increased DNA-binding ability and the consequent phosphorylation at the adjacent S378 site (Waterman et al., 1998).
Finally, phosphorylation upon stress also controlled the deactivation and degradation of p53. S366 and T387 were phosphorylated upon DNA damage by the Chk1/2 kinases, and S366 phosphorylation by IKK led to p53 degradation (Ou et al., 2005; Xia et al., 2009). Notably, S366 phosphorylation facilitated β-TrcP1 binding which contributed to the ubiquitination and degradation of p53 in a manner independent of MDM2. Expectedly, the phosphorylation kinetics at S366/T387 and S378 were different, with the phosphorylation of the former two sites occurring later than S378 (Ou et al., 2005), indicating that these sites are phosphorylated to attenuate the initiating signal upon completion of the intended response. Consistently, studies using the S366 phosphorylation-specific antibody demonstrated that the GAS41–PP2CP complex which is amplified in cancers, dephosphorylated S366 on p53 upon UV irradiation, thereby attenuating the signal (Park et al., 2011).
Taken together, the availability of these phosphorylation-specific antibodies has revealed three important principles of the regulation of p53, as depicted in Figure 1. Most of the DNA damage-induced phosphorylation occurs in the amino-terminal sites and leads to p53 activation; several sites are critical for keeping p53 phosphorylated at the basal state through its degradation; and finally, selected sites are phosphorylated upon DNA damage to attenuate the activating signal and hence destabilize p53.
Acetylation
While phosphorylation appears to be an important regulator of p53 stability, acetylation of p53 (primarily on the carboxyl-terminal sites) upon DNA damage is critical to regulate p53 transcriptional activity. The first set of p53 acetylation-specific antibodies generated was against the K320, K373, and K382 residues (Sakaguchi et al., 1998; Liu et al., 1999). K320 and K373 sites were found to be acetylated upon DNA damage, though by different acetyl-transferases. Whereas K320 was acetylated by PCAF, K373 was acetylated by p300, similar to K382 (Sakaguchi et al., 1998; Liu et al., 1999). However, acetylation of both K320 and K382 upon DNA damage was dependent on KAISO, a transcription factor that is induced by DNA damage and bound to the p53/p300 complex (Koh et al., 2014). These acetylation events increased the p53 DNA-binding and transcriptional activation. Similarly, K305 was acetylated by IR and UV irradiation, and associated with increased p53 transcriptional activity (Wang et al., 2003). Relatively recent analyses using other acetylation-specific antibodies have demonstrated that sites such as K164 and K381 of p53 are also acetylated upon DNA damage (Tang et al., 2008; Ryu et al., 2017).
However, unlike S33/S37-facilitated K382 acetylation, phosphorylation at S15, a critical event in p53 activation, was not strictly required for K320 and K373 acetylation in mouse cells (Chao et al., 2000), indicating that only certain phosphorylation events are associated with enabling the acetylation of p53. Nevertheless, while acetylation at the carboxyl-terminal sites appeared to be critical for p53 transcriptional activity, specificity of target gene activation was found to be associated with its acetylation on K120 in the DBD (Sykes et al., 2006; Tang et al., 2006). Tip60 was identified to acetylate K120 upon DNA damage, leading to selective transactivation of apoptotic target genes such as Bax and Puma (Sykes et al., 2006). Furthermore, K120 is mutated in cancers, underscoring the importance of this site in apoptosis induction and the target gene specificity afforded by its acetylation.
While treatment of cancer cells with DNA-damaging agents such as CPT was shown to induce K382 acetylation and the consequent activation of p53, this was not the case upon treatment of neuronal cells with HDAC inhibitors. It was demonstrated that HDAC inhibitors-mediated K381 and K382 acetylation was associated with reduced p53 activity due to reduced p53 association with the pro-apoptotic PUMA promoter (Brochier et al., 2013), highlighting context-dependent consequences of K382 acetylation. Moreover, specificity among HDAC inhibitors also appear to exist in the regulation of K373/K382 acetylation. While treatment with depsipeptide, a HDAC inhibitor, leads to K373/K382 acetylation, treatment with TSA, another HDAC inhibitor, does not (Zhao et al., 2006). These data suggest two significant points. Firstly, the signalling and cellular contexts appear to dictate the p53 acetylation profile (be it the inducing signal or the responding cell type). Secondly, the consequence of K382 acetylation under different contexts could be different. While K382 acetylation is often associated with p53-mediated transactivation of target genes upon DNA damage, this is not the case upon HDAC inhibitor treatment. These findings imply that the mode of activation is critical in determining the eventual effects on p53 functions, which is likely influenced by other associated events yet to be identified. These studies together indicate that detection of the acetylation at such sites by acetylation-specific antibodies may not be evidence for positive p53 transcriptional activity, and analysis of other related parameters such as target gene expression is required for affirmation.
Methylation
p53 has been shown to be methylated on at least four lysine residues: K370, K372, K373, and K382 (Chuikov et al., 2004; Huang et al., 2006, 2010; Shi et al., 2007), of which methylation only at K372 appears to correlate functionally with activated p53. Chuikov et al. found that K372 could be mono-methylated by Set7/9, using a mono-methyl-K372-specific antibody (Chuikov et al., 2004), and this methylation was shown to occur upon DNA damage (Ivanov et al., 2007). K372 methylation led to the consequent acetylation at K373 and an increase in p53 transcriptional activity and stability. Furthermore, mono-methylation at K372 was shown to inhibit the methylation of p53 on K370, using another methyl-K370-specific antibody (Huang et al., 2006). Interestingly, methylation of K370 by Smyd2 prevented p53 from binding to DNA, and this methylation was inhibited by DNA damage (Huang et al., 2006). Thus, the interplay of methylation alternately at K370 initially and thereafter at K372 upon DNA damage orchestrates the regulation of p53 transcriptional activity.
Similar to K370 methylation, K382 methylation by Set8 was found to decrease p53 activity (Shi et al., 2007). This is a site that is also acetylated upon DNA damage. Concomitant to a reduction in methylation at K382, acetylation was induced by DNA damage and resulted in p53 activation (Shi et al., 2007). By contrast, di-methylation at K373 by the histone methyl-transferases G9a and Glp did not lead to any appreciable changes in the overall levels of methylated K373 upon DNA damage, suggesting that this PTM event correlates with inactive p53 (Huang et al., 2010). Unlike the acetylated and phosphorylated residues on p53 against which several monoclonal antibodies are available, most of the initial work on p53 methylation has been done with rabbit polyclonal antibodies. Hence, there is a need to follow-up on these findings in future to explore the relevance of these events in various cellular signalling contexts.
Collectively, the available p53 PTM-specific antibodies have provided the field with a good description of the specific p53 PTM events that occur during the various cellular states, despite these being a subset of all possible PTMs on p53 (DeHart et al., 2014). Nevertheless, several caveats need to be kept in mind while recognizing the importance of these PTMs on p53. Firstly, these p53 PTM-specific antibodies can only measure the relative presence or absence of changes in these individual modifications, but not the absolute values as to which sites are highly or only marginally modified. Secondly, what we interpret is based on what the p53 PTM-specific antibodies are capable of recognizing, especially in the context of regions that contain multiples sites with potentially differing PTM residues. For instance, the region encompassing amino acids 370–386 is extensively modified by acetylation, ubiquitination, phosphorylation, and methylation. Hence, whether the ability of a PTM antibody to recognize its intended site is influenced by other adjacent PTM events should be kept in mind while interpreting the data. Thirdly, as alluded to above, some PTM events have been reported by occasional reports and require further in-depth investigation using monoclonal antibodies in multiple systems, to solidify our understanding of these events. Finally, whereas modification-specific antibodies can provide valuable information regarding the occurrence of modifications, establishing their function can be significantly more challenging. Whether these events are critical and absolutely necessary for p53 functions in vivo can only be clarified with the generation of mice and cell lines that contain a modified residue mimicking the activated or the inactivated state. Mice and cell lines with individual alterations on these residues have both confirmed their proposed roles, as well have failed to fully recapitulate all the expected roles in some cases (Wu et al., 2002; Bruins et al., 2004; Sluss et al., 2004; Lee et al., 2011), indicating that a single PTM event alone may not be sufficient to regulate all p53 functions. A case in point is the strain of mice lacking four acetylation sites on p53, which are prone to tumour development, compared to mice in which only three of the acetylation residues were altered and resistant to tumour development (Li et al., 2012; Wang et al., 2016). Thus, the functional roles of the p53 PTM events unravelled by the PTM-specific antibodies need to be verified with the generation of new and improved in vivo cell lines and mice strains.
New kid in the block—p53 mutation-specific antibodies
We recently reported the generation and characterization of antibodies to individual p53 mutants (Hwang et al., 2018). Essentially, our efforts were aimed at generating antibodies that are capable of recognizing a single amino acid change as seen in p53 missense mutations identified in cancers. This was inspired by the early observations that the DO-1 antibody recognizing the amino-terminus of human p53 was unable to detect mouse p53, though both differ by only one amino acid in the epitope (Table 1), indicating that highly specific antibodies can be generated to discern a single amino acid substitution. Hence, we had embarked on generating antibodies against the three hot-spot p53 mutants: R175H, R248Q, and R273H in our initial study. By tweaking the antigen design and monoclonal antibody generation protocols, we were able to successfully generate mutation-specific antibodies for the three p53 mutations. These antibodies were effective in a variety of biochemical assays as well as in immunohistochemical assays, capable of detecting the presence of mutant p53 in clinical samples. This work highlights the potential utility of these mutation-specific antibodies in evaluating large panels of archival clinical material to assess the clinical features of the individual mutations, which could provide significant information for personalized therapy. The utility of these latest generation of p53 antibodies in tumour imaging and therapy are currently being explored, based on previous studies that have shown the ability of other antibodies to penetrate cellular membranes to target their intended antigens (Hong and Zeng, 2012). Thus, antibody engineering methods are currently being employed to improve the bioavailability of these mutation-specific antibodies within the cells.
Conclusions and future outlook
The availability of various classes of antibodies has been critical in our understanding of p53 functions over the years, and will contribute further to the expansion of our knowledge of p53. While a substantial amount of work described in this review was performed over two decades ago, most of the results and their interpretations still remain relevant and very instrumental in our knowledge of p53 functions. Notably, many of the data have been key to our current efforts to therapeutically target mutant p53. Despite their limitations, the antibodies to the various forms of p53 have served their purpose well. With the advent of advanced proteomics approaches, future work can be built on from the basics provided by these earlier studies, in terms of quantifying the changes and evaluating multiple changes as a whole using a PTM signature, rather than at an individual level. Moreover, the recent demonstration of the possibility to generate mutation-specific antibodies will likely allow us to clarify the roles of individual mutations strewn across the entire length of p53, differentiating the strongest ones with GOFs that would lead to severe phenotypes, and thus, poor prognosis, from the others. The possibility to generate antibodies against a single amino acid change could also mean that antibodies against the common single nucleotide polymorphisms (SNP) in p53, such as the R72P or the R47P, could be generated to clarify their biological functions. Interestingly, the enhanced expression of one allele over the other in the heterozygote state was shown to differ between the normal vs. cancer tissues with respect to the R72P SNP (Siddique et al., 2005). Thus, the molecular basis of allele-dominance and the consequent biological outcomes can be teased out with such antibodies. Finally, the possibility of generating antibodies against a single amino acid substitution can be expanded beyond p53 to essentially mutations or SNPs in any other proteins, and could transform the way we understand their functions in both health and disease.
Acknowledgements
We thank NMRC and ASTAR for funding support in the authors’ laboratories. The authors thank the reviewers for their valuable suggestions. We apologize for not being able to include all possible references due to space limits.
Conflict of interest
none declared.
References
- Ashcroft M., Taya Y., and Vousden K.H. (2000). Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20, 3224–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.J., Fearon E.R., Nigro J.M., et al. (1989). Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244, 217–221. [DOI] [PubMed] [Google Scholar]
- Ball R.K., Siegl B., Quellhorst S., et al. (1984). Monoclonal antibodies against simian virus 40 nuclear large T tumour antigen: epitope mapping, papova virus cross-reaction and cell surface staining. EMBO J. 3, 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S., Moyal L., Shieh S.Y., et al. (1998). Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- Banks L., Matlasheswski G., and Crawford L. (1986). Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Euro. J. Bio. 159, 529–534. [DOI] [PubMed] [Google Scholar]
- Bartek J., Iggo R., Gannon J., et al. (1990). Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene 5, 893–899. [PubMed] [Google Scholar]
- Benchimol S., Pim D., and Crawford L. (1982). Radioimmunoassay of the cellular protein p53 in mouse and human cell lines. EMBO J. 1, 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging K.T., Mello S.S., and Attardi L.D. (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M. (2002). Hsp-90-associated oncoproteins: multiple targets of geldanamycin and its analogs. Leukemia 16, 455–462. [DOI] [PubMed] [Google Scholar]
- Blagosklonny M.V., Toretsky J., and Neckers L. (1995). Geldanamycin selectively destabilizes and conformationally alters mutated p53. Oncogene 11, 933–939. [PubMed] [Google Scholar]
- Blagosklonny M.V., Toretsky J., Bohen S., et al. (1996). Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc. Natl Acad. Sci. USA 93, 8379–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydes J.P., and Hupp T.R. (1998). DNA damage triggers DRB-resistant phosphorylation of human p53 at the CK2 site. Oncogene 17, 1045–1052. [DOI] [PubMed] [Google Scholar]
- Blaydes J.P., Luciani M.G., Pospisilova S., et al. (2001). Stoichiometric Phosphorylation of Human p53 at Ser315 stimulates p53-dependent Transcription. J. Bio. Chem. 276, 4699–4708. [DOI] [PubMed] [Google Scholar]
- Brochier C., Dennis G., Rivieccio M.A., et al. (2013). Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J. Neurosci. 33, 8621–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C.L., and Gu W. (2011). The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2, 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins W., Zwart E., Attardi L.D., et al. (2004). Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol. Cell. Biol. 24, 8884–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin D.V., Saito S., Hollander M.C., et al. (1999). Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18, 6845–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann T., Potapova O., Bar-Shira A., et al. (2001). Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol. Cell. Biol. 21, 2743–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov V.J., and Wiman K.G. (2014). Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 588, 2622–2627. [DOI] [PubMed] [Google Scholar]
- Böttger A., Böttger V., Sparks A., et al. (1997). Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol. 7, 860–869. [DOI] [PubMed] [Google Scholar]
- Cain C., Miller S., Ahn J., et al. (2000). The N terminus of p53 regulates its dissociation from DNA. J. Biol. Chem. 275, 39944–39953. [DOI] [PubMed] [Google Scholar]
- Canman C.E., Lim D.-S., Cimprich K.A., et al. (1998). Activation of the ATM kinase by ionizing radiation and phosphorylation ofp53. Science 281, 1677–1679. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran K., McFarland V.W., Simmons D.T., et al. (1981). Quantitation and characterization of a species-specific and embryo stage-dependent 55-kilodalton phosphoprotein also present in cells transformed by simian virus 40. Proc. Natl Acad. Sci. USA 78, 6953–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C., Saito S.I., Anderson C.W., et al. (2000). Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl Acad. Sci. USA 97, 11936–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P.D., et al. (1994). Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265, 346–355. [DOI] [PubMed] [Google Scholar]
- Chuikov S., Kurash J.K., Wilson J.R., et al. (2004). Regulation of p53 activity through lysine methylation. Nature 432, 353–360. [DOI] [PubMed] [Google Scholar]
- Coffer A.I., and Knowles P.P. (1994). Divalent metal ions induce conformational change in pure, human wild-type p53 tumor suppressor protein. Biochi. Biophys. Acta 1209, 279–285. [DOI] [PubMed] [Google Scholar]
- Cook A., and Milner J. (1990). Evidence for allosteric variants of wild-type p53, a tumour suppressor protein. British J. Cancer 61, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L., Pim D., and Bulbrook R. (1982). Detection of antibodies against the cellular protein p53 in sera from patients with breast cancer. Int. J. Cancer 30, 403–408. [DOI] [PubMed] [Google Scholar]
- Crawford L.V., Pim D.C., Gurney E.G., et al. (1981). Detection of a common feature in several human tumor cell lines--a 53,000-dalton protein. Proc. Natl Acad. Sci. USA 78, 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHart C.J., Chahal J.S., Flint S.J., et al. (2014). Extensive post-translational modifications on active and inactivated forms of endogenous p53. Mol. Cell. Proteomics 13, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo A.B., Jay G., Appella E., et al. (1979). Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc. Natl Acad. Sci. USA 76, 2420–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold W.G., Jay G., DeLeo A.B., et al. (1981). p53 transformation-related protein: detection by monoclonal antibody in mouse and human cells. Proc. Natl Acad. Sci. USA 78, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu D., Michalovitz D., Eliyahu S., et al. (1989). Wild-type p53 can inhibit oncogene-mediated focus formation. Proc. Natl Acad. Sci. USA 86, 8763–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu D., Raz A., Gruss P., et al. (1984). Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 312, 646. [DOI] [PubMed] [Google Scholar]
- Finlay C.A., Hinds P.W., and Levine A.J. (1989). The p53 proto-oncogene can act as a suppressor of transformation. Cell 57, 1083–1093. [DOI] [PubMed] [Google Scholar]
- Finlay C., Hinds P., Tan T., et al. (1988). Activating mutations for transformation by p53 produce a gene product that forms an hsc70-p53 complex with an altered half-life. Mol. Cell. Biol. 8, 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor W.A., and Prives C. (2012). Mutantp53: one name, many proteins. Genes Dev. 26, 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitonde S.V., Riley J.R., Qiao D., et al. (2000). Conformational phenotype of p53 is linked to nuclear translocation. Oncogene 19, 4042–4049. [DOI] [PubMed] [Google Scholar]
- Gannon J., Greaves R., Iggo R., et al. (1990). Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 9, 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon J.V., and Lane D.P. (1991). Protein synthesis required to anchor a mutant p53 protein which is temperature-sensitive for nuclear transport. Nature 349, 802–806. [DOI] [PubMed] [Google Scholar]
- Gannon H.S., Woda B.A., and Jones S.N. (2012). ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 21, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti A., Li H.H., Traugh J.A., et al. (2000). Phosphorylation of human p53 on Thr-55. Biochemistry 39, 9837–9842. [DOI] [PubMed] [Google Scholar]
- Gire V., and Wynford-Thomas D. (1998). Reinitiation of DNA synthesis and cell division in senescent human fibroblasts by microinjection of anti-p53 antibodies. Mol. Cell. Biol. 18, 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R.M., Goldberg A., and Pardee A. (1984). Energy requirement for degradation of tumor-associated protein p53. Mol. Cell. Biol. 4, 442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., and Roeder R.G. (1997). Activation of p53 sequence-specific DNA-binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Gurney E., Harrison R., and Fenno J. (1980). Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct sublcasses of large T antigen and for similarities among nonviral T antigens. J. Virol. 34, 752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P., Butcher S., and Milner J. (1995). Temperature sensitivity for conformation is an intrinsic property of wild-typep53. British J. Cancer 71, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P., and Milner J. (1993. a). A structural role for metal ions in the ‘wild-type’ conformation of the tumor suppressor protein p53. Cancer Res. 53, 1739–1742. [PubMed] [Google Scholar]
- Hainaut P., and Milner J. (1993. b). Redox modulation of p53 conformation and sequence-specific DNA-binding in vitro. Cancer Res. 53, 4469–4473. [PubMed] [Google Scholar]
- Halazonetis T.D., Davis L.J., and Kandil A. (1993). Wild-type p53 adopts a ‘mutant’-like conformation when bound to DNA. EMBO J. 12, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S., Hupp T.R., and Lane D.P. (1996). Allosteric regulation of the thermostability and DNA-binding activity of human p53 by specific interacting proteins. J. Bio. Chem. 271, 3917–3924. [DOI] [PubMed] [Google Scholar]
- Harlow E., Crawford L.V., Pim D.C., et al. (1981). Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39, 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker D., Page G., Lohrum M., et al. (1996). Complex regulation of the DNA-binding activity of p53 by phosphorylation: differential effects of individual phosphorylation sites on the interaction with different binding motifs. Oncogene 12, 953–961. [PubMed] [Google Scholar]
- Higashimoto Y., Saito S.i., Tong X.H., et al. (2000). Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J. Bio. Chem. 275, 23199–23203. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., et al. (1991). p53 mutations in human cancers. Science 253, 49–53. [DOI] [PubMed] [Google Scholar]
- Hong C.W., and Zeng Q. (2012). Awaiting a new era of cancer immunotherapy. Cancer Res. 72, 3715–3719. [DOI] [PubMed] [Google Scholar]
- Huang J., Dorsey J., Chuikov S., et al. (2010). G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 285, 9636–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Perez-Burgos L., Placek B.J., et al. (2006). Repression of p53 activity by Smyd2-mediated methylation. Nature 444, 629. [DOI] [PubMed] [Google Scholar]
- Hupp T.R., and Lane D.P. (1994). Allosteric activation of latent p53 tetramers. Curr. Biol. 4, 865–875. [DOI] [PubMed] [Google Scholar]
- Hupp T.R., and Lane D.P. (1995). Two distinct signaling pathways activate the latent DNA-binding function of p53 in a casein kinase II-independent manner. J. Biol. Chem. 270, 18165–18174. [DOI] [PubMed] [Google Scholar]
- Hupp T.R., Meek D.W., Midgley C.A., et al. (1992). Regulation of the specific DNA-binding function of p53. Cell 71, 875–886. [DOI] [PubMed] [Google Scholar]
- Hupp T.R., Sparks A., and Lane D.P. (1995). Small peptides activate the latent sequence-specific DNA-binding function of p53. Cell 83, 237–245. [DOI] [PubMed] [Google Scholar]
- Hwang L.A., Phang B.H., Liew O.W., et al. (2018). Monoclonal antibodies against specific p53 hotspot mutants as potential tools for precision medicine. Cell Rep. 22, 299–312. [DOI] [PubMed] [Google Scholar]
- Ivanov G.S., Ivanova T., Kurash J., et al. (2007). Methylation-acetylation interplay activates p53 in response to DNA damage. Mol. Cell. Biol. 27, 6756–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay G., Khoury G., DeLeo A.B., et al. (1981). p53 transformation-related protein: detection of an associated phosphotransferase activity. Proc. Natl Acad. Sci. USA 78, 2932–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J., Rudge K., and Currie G. (1984). Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature 312, 651. [DOI] [PubMed] [Google Scholar]
- Kapoor M., and Lozano G. (1998). Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc. Natl Acad. Sci. USA 95, 2834–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L.J., Shieh S.Y., Chen X., et al. (1997). p53 is phosphorylated by CDK7-cyclin H in a p36MAT1-dependent manner. Mol. Cell. Biol. 17, 7220–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh D.I., Han D., Ryu H., et al. (2014). KAISO, a critical regulator of p53-mediated transcription of CDKN1A and apoptotic genes. Proc. Natl Acad. Sci. USA 111, 15078–15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress M., May E., Cassingena R., et al. (1979). Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 31, 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C.F., and Vousden K.H. (2015). p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393. [DOI] [PubMed] [Google Scholar]
- Lambert P.F., Kashanchi F., Radonovich M.F., et al. (1998). Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048–33053. [DOI] [PubMed] [Google Scholar]
- Lane D.P., and Crawford L.V. (1979). T antigen is bound to a host protein in SY40-transformed cells. Nature 278, 261–263. [DOI] [PubMed] [Google Scholar]
- Lee E.W., Oh W., Song H.P., et al. (2017). Phosphorylation of p53 at threonine 155 is required for Jab1-mediated nuclear export of p53. BMB Rep. 50, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.K., Tong W.M., Wang Z.Q., et al. (2011). Serine 312 phosphorylation is dispensable for wild-type p53 functions in vivo. Cell Death Differ. 18, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros Y., Meyer A., Ory K., et al. (1994). Mutations in p53 produce a common conformational effect that can be detected with a panel of monoclonal antibodies directed toward the central part of the p53 protein. Oncogene 9, 3689–3694. [PubMed] [Google Scholar]
- Li H.H., Cai X., Shouse G.P., et al. (2007). A specific PP2A regulatory subunit, B56γ, mediates DNA damage-induced dephosphorylation of p53 at Thr55. EMBO J. 26, 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Kon N., Jiang L., et al. (2012). Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.H., Li A.G., Sheppard H.M., et al. (2004). Phosphorylation on Thr-55 by TAF1 mediates degradation ofp53: a role for TAF1 in cell G1 progression. Mol. Cell 13, 867–878. [DOI] [PubMed] [Google Scholar]
- Linzer D.I., and Levine A.J. (1979). Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17, 43–52. [DOI] [PubMed] [Google Scholar]
- Liu L., Scolnick D.M., Trievel R.C., et al. (1999). p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Taya Y., Ikeda M., et al. (1998). Ultraviolet radiation, but not γ radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc. Natl Acad. Sci. USA 95, 6399–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D., Li F.P., Strong L.C., et al. (1990). Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250, 1233–1238. [DOI] [PubMed] [Google Scholar]
- Martinez J., Georgoff I., and Levine A.J. (1991). Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 5, 151–159. [DOI] [PubMed] [Google Scholar]
- Meek D.W., and Anderson C.W. (2009). Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 1, a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W., and Eckhart W. (1988). Phosphorylation of p53 in normal and simian virus 40-transformed NIH 3T3 cells. Mol. Cell. Biol. 8, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D., Inga A., and Resnick M.A. (2009). The expanding universe of p53 targets. Nat. Rev. Cancer 9, 724. [DOI] [PubMed] [Google Scholar]
- Mercer W.E., Avignolo C., and Baserga R. (1984). Role of the p53 protein in cell proliferation as studied by microinjection of monoclonal antibodies. Mol. Cell. Biol. 4, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer W.E., Nelson D., DeLeo A.B., et al. (1982). Microinjection of monoclonal antibody to protein p53 inhibits serum-induced DNA synthesis in 3T3 cells. Proc. Natl Acad. Sci. USA 79, 6309–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalovitz D., Halevy O., and Oren M. (1990). Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell 62, 671–680. [DOI] [PubMed] [Google Scholar]
- Milner J. (1991). A conformation hypothesis for the suppressor and promoter functions of p53 in cell growth control and in cancer. Proc. Biol. Sci. 245, 139–145. [DOI] [PubMed] [Google Scholar]
- Milner J. (1995). Flexibility: the key to p53 function? Trends Biol. Sci. 20, 49–51. [DOI] [PubMed] [Google Scholar]
- Milner J., Cook A., and Sheldon M. (1987). A new anti-p53 monoclonal antibody, previously reported to be directed against the large T antigen of simian virus 40. Oncogene 1, 453–455. [PubMed] [Google Scholar]
- Milner J., and Medcalf E. (1991). Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65, 765–774. [DOI] [PubMed] [Google Scholar]
- Milner J., and Watson J. (1990). Addition of fresh medium induces cell cycle and conformation changes inp53, a tumour suppressor protein. Oncogene 5, 1683–1690. [PubMed] [Google Scholar]
- Mosner J., and Deppert W. (1992). Conformational analysis of p53 in resting and concanavalin A-stimulated mouse lymphocytes. Oncogene 7, 661–666. [PubMed] [Google Scholar]
- Muller P.A., and Vousden K.H. (2014). Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim A., Hoogenboom H.R., Tomlinson I., et al. (1994). Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 13, 692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A., Manzl C., Strasser A., et al. (2007). How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 14, 1561–1575. [DOI] [PubMed] [Google Scholar]
- Ory K., Legros Y., Auguin C., et al. (1994). Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO J. 13, 3496–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y.H., Chung P.H., Sun T.P., et al. (2005). p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol. Biol. Cell 16, 1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L.F., Land H., Weinberg R.A., et al. (1984). Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature 312, 649. [DOI] [PubMed] [Google Scholar]
- Park J.H., Smith R.J., Shieh S.Y., et al. (2011). The GAS4-PP2Cβ complex dephosphorylates p53 at serine 366 and regulates its stability. J. Biol. Chem. 286, 10911–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater R., Parks D., Anderson M.E., et al. (1995). Role of cysteine residues in regulation of p53 function. Mol. Cell. Biol. 15, 3892–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reihsaus E., Kohler M., Kraiss S., et al. (1990). Regulation of the level of the oncoprotein p53 in non-transformed and transformed cells. Oncogene 5, 137–145. [PubMed] [Google Scholar]
- Rivlin N., Brosh R., Oren M., et al. (2011). Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2, 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin N., Katz S., Doody M., et al. (2014). Rescue of embryonic stem cells from cellular transformation by proteomic stabilization of mutant p53 and conversion into WT conformation. Proc. Natl Acad. Sci. USA 111, 7006–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter V., Witte O.N., Coffman R., et al. (1980). Abelson murine leukemia virus-induced tumors elicit antibodies against a host cell protein, P50. J. Virol. 36, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J.J., and Clarke M.F. (1994). Alteration of p53 conformation and induction of apoptosis in a murine erythroleukemia cell line by dimethylsulfoxide. Leuk. Res. 18, 617–621. [DOI] [PubMed] [Google Scholar]
- Ryu H.W., Shin D.H., Lee D.H., et al. (2017). HDAC6 deacetylates p53 at lysines 381/382 and differentially coordinates p53-induced apoptosis. Cancer Lett. 391, 162–171. [DOI] [PubMed] [Google Scholar]
- Sabapathy K. (2015). The contrived mutant p53 oncogene–beyond loss of functions. Front. Oncol. 5, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabapathy K., Klemm M., Jaenisch R., et al. (1997). Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 16, 6217–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabapathy K., and Lane D.P. (2018). Therapeutic targeting ofp53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 15, 13. [DOI] [PubMed] [Google Scholar]
- Saito S., Goodarzi A.A., Higashimoto Y., et al. (2002). ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J. Biol. Chem. 277, 12491–12494. [DOI] [PubMed] [Google Scholar]
- Saito S.i., Yamaguchi H., Higashimoto Y., et al. (2003). Phosphorylation site interdependence of human p53 posttranslational modifications in response to stress. J. Biol. Chem. 278, 37536–37544. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera J.E., Saito S.i., et al. (1998). DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Saito S.i., Higashimoto Y., et al. (2000). Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 275, 9278–9283. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Sakamoto H., Lewis M.S., et al. (1997). Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry 36, 10117–10124. [DOI] [PubMed] [Google Scholar]
- Schmieg F., and Simmons D. (1993). p53 mutants with changes in conserved region II: three classes with differing antibody reactivity, SV40 T antigen binding and ability to inhibit transformation of rat cells. Oncogene 8, 2043–2050. [PubMed] [Google Scholar]
- Shi X., Kachirskaia I., Yamaguchi H., et al. (2007). Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 27, 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S.Y., Ahn J., Tamai K., et al. (2000). The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14, 289–300. [PMC free article] [PubMed] [Google Scholar]
- Shieh S.Y., Ikeda M., Taya Y., et al. (1997). DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91, 325–334. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Taya Y., and Prives C. (1999). DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M.M., Balram C., Fiszer-Maliszewska L., et al. (2005). Evidence for selective expression of the p53 codon 72 polymorphs: implications in cancer development. Cancer Epidemiol. Biomarkers Prev. 14, 2245–2252. [DOI] [PubMed] [Google Scholar]
- Siliciano J.D., Canman C.E., Taya Y., et al. (1997). DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11, 3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluss H.K., Armata H., Gallant J., et al. (2004). Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell. Biol. 24, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T. (2000). p53 antibodies in the sera of patients with various types of cancer: a review. Cancer Res. 60, 1777–1788. [PubMed] [Google Scholar]
- Srivastava S., Zou Z., Pirollo K., et al. (1990). Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li–Fraumeni syndrome. Nature 348, 747. [DOI] [PubMed] [Google Scholar]
- Stephen C.W., Helminen P., and Lane D.P. (1995). Characterisation of epitopes on human p53 using phage-displayed peptide libraries: insights into antibody-peptide interactions. J. Mol. Biol. 248, 58–78. [DOI] [PubMed] [Google Scholar]
- Stephen C.W., and Lane D.P. (1992). Mutant conformation of p53: precise epitope mapping using a filamentous phage epitope library. J. Mol. Biol. 225, 577–583. [DOI] [PubMed] [Google Scholar]
- Sykes S.M., Mellert H.S., Holbert M.A., et al. (2006). Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal P., Eizenberger S., Cohen E., et al. (2016). Cancer therapeutic approach based on conformational stabilization of mutant p53 protein by small peptides. Oncotarget 7, 11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Luo J., Zhang W., et al. (2006). Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839. [DOI] [PubMed] [Google Scholar]
- Tang Y., Zhao W., Chen Y., et al. (2008). Acetylation is indispensable for p53 activation. Cell 133, 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R., Kaplan L., Reich N., et al. (1983). Characterization of human p53 antigens employing primate specific monoclonal antibodies. Virology 131, 502–517. [DOI] [PubMed] [Google Scholar]
- Uberti D., Lanni C., Carsana T., et al. (2006). Identification of a mutant-like conformation of p53 in fibroblasts from sporadic Alzheimer’s disease patients. Neurobiol. Aging 27, 1193–1201. [DOI] [PubMed] [Google Scholar]
- Ullrich S.J., Mercer W., and Appella E. (1992). Human wild-type p53 adopts a unique conformational and phosphorylation state in vivo during growth arrest of glioblastoma cells. Oncogene 7, 1635–1643. [PubMed] [Google Scholar]
- Verhaegh G.W., Parat M.O., Richard M.J., et al. (1998). Modulation of p53 protein conformation and DNA-binding activity by intracellular chelation of zinc. Mol. Carcinog. 21, 205–214. [DOI] [PubMed] [Google Scholar]
- Vojtesek B., Bartek J., Midgley C., et al. (1992). An immunochemical analysis of the human nuclear phosphoprotein p53: new monoclonal antibodies and epitope mapping using recombinant p53. J. Immunol. Methods 151, 237–244. [DOI] [PubMed] [Google Scholar]
- Vojtesek B., Dolezalova H., Lauerova L., et al. (1995). Conformational changes in p53 analysed using new antibodies to the core DNA-binding domain of the protein. Oncogene 10, 389–393. [PubMed] [Google Scholar]
- Wang S.J., Li D., Ou Y., et al. (2016). Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 17, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.L., Sait F., and Winter G. (2001). The ‘wildtype’ conformation of p53: epitope mapping using hybrid proteins. Oncogene 20, 2318. [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Tsay Y.G., Tan B.C., et al. (2003). Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J. Biol. Chem. 278, 25568–25576. [DOI] [PubMed] [Google Scholar]
- Waterman M.J., Starvidi E.S., Waterman J.L., et al. (1998). ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19, 175–178. [DOI] [PubMed] [Google Scholar]
- Webley K.M., Shorthouse A.J., and Royds J.A. (2000). Effect of mutation and conformation on the function of p53 in colorectal cancer. J. Pathol. 191, 361–367. [DOI] [PubMed] [Google Scholar]
- Wolf D., Harris N., and Rotter V. (1984). Reconstitution of p53 expression in a nonproducer Ab-MuLV-transformed cell line by transfection of a functional p53 gene. Cell 38, 119–126. [DOI] [PubMed] [Google Scholar]
- Wu Z., Earle J., Saito S.I., et al. (2002). Mutation of mouse p53 Ser23 and the response to DNA damage. Mol. Cell. Biol. 22, 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang M., Li X., et al. (1993). Conformation changes of p53 proteins in regulation of murine T lymphocyte proliferation. Cell. Mol. Biol. Res. 39, 27–31. [PubMed] [Google Scholar]
- Xia Y., Padre R.C., De Mendoza T.H., et al. (2009). Phosphorylation of p53 by IκB kinase 2 promotes its degradation by β-TrCP. Proc. Natl Acad. Sci. USA 106, 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.T., Lai W.L., Liu H.F., et al. (2016). PAK4 phosphorylates p53 at serine 215 to promote liver cancer metastasis. Cancer Res. 76, 5732–5742. [DOI] [PubMed] [Google Scholar]
- Yang W.H., Kim J.E., Nam H.W., et al. (2006). Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nature Cell Biol. 8, 1074. [DOI] [PubMed] [Google Scholar]
- Yewdell J., Gannon J., and Lane D.P. (1986). Monoclonal antibody analysis of p53 expression in normal and transformed cells. J. Virol. 59, 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonish-Rouach E., Resnftzky D., Lotem J., et al. (1991). Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352, 345. [DOI] [PubMed] [Google Scholar]
- Zhang W., and Deisseroth A.B. (1994). Conformational change of p53 protein in growth factor-stimulated human myelogenous leukemia cells. Leuk. Lymphoma 14, 251–255. [DOI] [PubMed] [Google Scholar]
- Zhang W., Hu G., Estey E., et al. (1992). Altered conformation of the p53 protein in myeloid leukemia cells and mitogen-stimulated normal blood cells. Oncogene 7, 1645–1647. [PubMed] [Google Scholar]
- Zhao Y., Lu S., Wu L., et al. (2006). Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21Waf1/Cip1. Mol. Cell. Biol. 26, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilfou J.T., and Lowe S.W. (2009). Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 1, a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]