Abstract
LDL, VLDL and other members of the low-density lipoparticles (LLPs) enter cells through a large family of receptors. The actual receptor ligand(s) in apolipoprotein B100, one of the main proteins of LLP, remain(s) unknown. The objective of this study was to identify true receptor ligand(s) in apo B100, a molecule of 4,563 residues. Apo B100 contains 33 analogues of Cardin-Weintraub Arginine/Lysine-based receptor ligand motifs and shares key Lysine motifs and sequence similarity with the LDL receptor-associated protein, RAP, MESD, and heat shock proteins. Eleven FITC-labeled synthetic peptides of 21 – 42 residues, with at least one ligand, were tested for binding and internalization using HeLa cells. All peptides bind but display different binding capacities and patterns. Peptides B0013, B0582, B2366, and B2932 mediate endocytosis and appear in distinct sites in the cytoplasm. B0708 and B3181 bind and remain on the cell surface as aggregates/clusters. Peptides B3119 (Site A) and B3347 (Site B), the putative ligands showed low binding and no cell entry capacity. Apo B100 regions in this study share similarities with related proteins of known function including chaperone proteins and Apo BEC Stimulating Protein, and not directly related proteins, e.g., the DNA-binding domain of Interferon Regulatory Factors, MSX2-Interacting Protein, and snake venom Zinc metalloproteinase-disintegrin-like proteins.
Keywords: Apo B100 – apolipoprotein B100, Apo E – apolipoprotein E, RAP – LDL receptor associated protein, LDL – Low Density Lipoprotein
INTRODUCTION
It is generally accepted that the main physiological role of Apo B100 in low density lipoprotein particles (LLPs) - very low-density lipoproteins (VLDL), intermediate density lipoproteins (IDL) and low density lipoprotein (LDL) - is transport and intracellular delivery of cholesterol, cholesteryl ester, triglycerides, phospholipids, and other low molecular weight compounds, such as vitamins, hormones (sterols, retinol and tocopherols) (1, 2). LLPs are also involved in the transport of a great variety of smaller proteins including multiple copies of apolipoprotein E (apo E) and other apolipoproteins such as apo C-I, -II, -III, A-I, -II, -IV, and M (3–5). Accrued evidence supports a model in which apo B100 and apo E achieve endocytosis via membrane receptors (6–8).
Apo B100 is a member of the large lipid transfer protein (LLTP) superfamily that includes microsomal triglyceride transfer protein (MTTP), vigilin, vitellogenins, and insect lipophorins (9–11). Non-apo B100 LLTPs are known to have multiple roles, including protein-protein interactions in immune response (12–14) and mRNA binding (15–17). Apo B100 is a massive, semi-hydrophobic molecule of 4,536 amino acids and, like other LLTPs, contains a variety of short sequence motifs indicative of potential function (7, 8, 18, 19).
Other functions of Apo B100 are supported by several reports implying a role for LDL and VLDL in cell signaling pathways (20–24). The strongest evidence that LLPs have functions beyond lipid transport is based on LDL’s ability to bind nucleic acids and efficiently transfect cells in culture and the rat in vivo (25, 26). LDL’s capacity to binding RNA is likely based on 11 Gly-X-X-Gly motifs in the apo B100 sequence. These motifs are characteristic of RNA-binding proteins (27, 28). In addition, the N-terminal 100-residue sequence of apo B100 contains several motifs also present in the DNA-binding domains of the Interferon Regulatory Factors, IRFs (21, 25). Two synthetic peptides covering residues 13 thru 94 were shown to bind plasmid DNA using electrophoresis mobility shift assays (EMSA) and to deliver it to the nucleus of HeLa cells in culture (25, 26). Two other synthetic peptides spanning the sequence between 3313Asp –Thr3401 were shown to bind DNA but did not bind or enter the cell. The latter sequence shares some motifs with known dengue helicases. The region between 3804Ser – Lys4006 contains several motifs also found in the signal transducer kinases, e.g. JAK1 and JAK2 (20). Kinase activity was shown experimentally using highly purified preparation of LDL (20).
Endocytosis of LLPs is mediated by a large gene family of receptors, Low-Density Lipoprotein Receptor – related Proteins (LRPs), with quite extensive set of ligand-binding modules that recognize a variety of ligand sequences (29–31). The diversity of the potential ligands provokes the question - what is/are the real LDL receptor ligand(s) on apo B100? The LDL receptor and Glycosaminoglycan (GAG) ligand motifs occur in Lysine/Arginine-rich clusters, XBBBXXBX (Type I), XBBXBX (Type II), and ΨBΨXB (Type III), where X is any amino acid and Ψ is a non-polar residue (32–34). These are known as Cardin-Weintraub motifs (34). Other elements that may be involved in forming clathrin-coated vesicles, i.e. endocytic motifs, have not been confirmed for either Apo E or Apo B100. Apo E contains four potential receptor ligand motifs; 144LRKRLLRD151, Type I; 141LRKLRKR147, Type II; and 071LKAYKS076 and 091ARLSKE096, Type III motifs, the critical residues for high affinity binding to LDL B/E receptor are Lysine residues in positions 143 and 146 of the Type II ligand motif (35, 36). Motif LRK that appears twice in tandem in Apo E is an analogue of motifs VKK and WKK in RAP (alpha-2-Macrogolublin Receptor-Associated Protein; AMRP_HUMAN; P3–533), an intracellular chaperone for the LDL receptor and related receptor proteins with 3D structure established using X-ray crystallography (37). There are other motifs in apo B100 and Apo E that are similar to the motifs in heat shock proteins, known chaperones for the LDLR family of proteins (29–31). Apo E also contains two potential endocytic motifs, 0059ELRALM0064 and 0151DADDL0155, analogues of E/D-x-x-x-L [L, I, M] and acidic motifs (38, 39). Apo E has been reported to bind several members of the LDL super-gene family of receptors (35, 40) and highly sulfated GAGs, such as heparin and heparan sulfate (34). LLPs, including LDL, are removed from plasma using GAGs affinity chromatography in a clinical application (41).
Published reports suggest that the apo B100 molecule contains several potential receptor ligand analogs. Two sites in apo B100 sequence were identified as potential receptor ligands based on their similarity to the apo E ligand (7, 8,19). Site A contains Type II ligand motif, 3145VKAQYKKNKHRHS3157 while Site B, 3351YKLEGTTRLTRKRGLKL3367, possesses an Arginine-rich, Type I motif. Hospattankar, et al. (42), proposed that multiple regions rich in positive charged residues in apo B100 have potential for binding to LDL receptors. Seven heparin-binding sites in Apo B100 were identified using proteolytic digestion methods (43). Sites encompassing 3134Leu-Lys3209 (overlaps with Site A), and 3356Thr-Ala3489 (partially overlaps with Site B), were found to have high affinity heparin and were also considered to possess LDL receptor binding capacity. In a separate report (44), four murine monoclonal antibodies, Mabs 2A, 5A, 7A, and 9A, to epitopes in three different regions of apo B100 were shown to reduce LDL interaction with the receptor by “up to 95%”. These apo B100 regions are defined as follows: 5A, 1480Asp-Val1693; 2A and 7A, 2152Gln-Asn2377; and 9A, 2657Gln-Lys3248.
The goal of the present study was to identify regions in the apo B100 sequence with Lysine-based motifs that may be involved in binding HeLa cells. Presence of membrane LDL receptors by HeLa cells is well known and their expression can be manipulated in various ways including nutrient reduction and/or including statins in the medium (45). Analyzing apo B100 sequence for content of Type I-III LDL receptor ligands and short Lysine-based receptor ligand motifs of RAP and heat shock proteins (46), we identified approximately sixty potential receptor binding sites and a plethora of endocytic motifs. Fluorescence-labeled (FITC) synthetic peptides of eleven regions of apo B100 with at least one potential receptor ligand and some with candidate internalization motifs were used to assess their interaction with HeLa cells, i.e. binding, internalization and possible nuclear translocation. In this report, we qualitatively describe cell-binding properties of these peptides, along with our insights into other possible functions of the selected regions of Apo B100.
MATERIALS AND METHODS
Sequences included in this study were obtained from NIH NCBI PUBMED Protein and Structure databases. Synthetic peptides used in these studies, shown in Table 1, are based on regions in the apo B100 which contain a candidate LDL receptor ligand motif and share sequence similarity with known functional regions in other proteins.
Table 1.
Carboxyl-terminus FITC-labeled Apolipoprotein B100 Synthetic Peptides Containing LDL Receptor Ligand Analogues.
| Peptide | Sequence | Residues | Mass |
|---|---|---|---|
| B0013 | 0013KDATRFKHLRKYTYNYEAESSSGVPGTADSRSATRINCKVEL0054 | 41 | 4751.2 |
| B0055 | 0055EVPQLCSFILKTSQCTLKEVYGFNPEGKALLKKTKNSEEFAA0096 | 41 | 4691.4 |
| B0575 | 0575SEELDIQDLKKLVKEVLKESQ0595 | 21 | 2471.8 |
| B0582 | 0582DLKKLVKEVLKESQLPTVMDFRKFSRNYQ0610 | 29 | 3541.1 |
| B0708 | 0708DHFGYTKDDKHEQDMVNGIMLSVEKLIKDLKSKEVPE0744 | 37 | 4317.9 |
| B2054 | 2054ERNRQTIIWLENVQRNLKHINIDQFVRKYRAALGKLPQQAN2095 | 41 | 4985.7 |
| B2366 | 2366VGFIDDAVKKLNELSFKTFIEDVNKFLDMLIKKLKSFDYHQF2407 | 41 | 5025.8 |
| B2932 | 2932SNKINSKHLRVNQNLVYESGSLNFSKLEIQSQVDSQHVGHSVL2974 | 42 | 4835.3 |
| B3119 | 3119LKDFSLWEKTGLKEFLKTTKQSFDLSVKAQYKKNKHRHSITN3160 | 41 | 5024.8 |
| B3181 | 3181EKNRNNALDFVTKSYNETKIKFDKYKAEKSQDE3213 | 33 | 3982.3 |
| B3347 | 3347DALQYKLEGTTRLTRKRGLKLATALSLSNKFVEGSHNSTVSLT3389 | 42 | 4706.3 |
| E2 | 0129STEELRVRLASHLRKLRKRLLRDADDLQKRLAVYTQHG0166 | 38 | 4543.2 |
Analogue motifs in apo B100 of the LDL receptor ligand motifs in RAP, Receptor Associated Protein, are highlighted in bold and underlined. Peptides B3119 and B3347 contain the putative LDL receptor binding ligand sequences, Sites A and B, respectively. Apolipoprotein peptide E2 is unlabeled, note it contains two copies in tandem of RAP ligand 0306VKK analogue LRK and a less similar motif, QKR. Apo E sequence LRKRLLR is a Type I Cardin-Weintraub motif.
Chemicals.
All cell culture media, PBS (Media Tech, Inc.), Fetal Bovine Serum, Penicillin G-Sodium, streptomycin sulfate, Trypsin, and Corning Costar 12-well culture plates were purchased from Thermo-Fisher Scientific, Inc. Mevastatin and all other reagents used in this study were obtained from Sigma-Aldrich, Inc.
Water of 18 mega-Ohm resistivity (Barnstead, E-pure water purification system) was used throughout this project.
Fluorescence-labeled Peptides.
Fluorescein Isothiocyanate (FITC) -labeled synthetic peptides of selected regions of the apo E and apo B100 molecules (Table 1) were obtained from NEO Group, Inc. Cambridge, MA. All synthetic peptides were of 95% or greater purity assessed by HPLC analysis. Lyophilized peptides were solubilized prior to use in PBS-5 mM MgCl2 to yield 1 mg/mL. Peptide solutions were then stored in liquid N2.
Cells.
As reported previously (26), HeLa cells (human cervix epithelial adenocarcinoma cell line, CCL-2™) were obtained from American Type Culture Collection (ATCC). Cells were stored in a liquid N2 until needed. Cells were then thawed, seeded and propagated according to ATCC protocols in DMEM supplemented with 10% FBS, 100 units Penicillin G-Sodium and 100 units/mL Streptomycin sulfate (DMEM – P/S - FBS) at 37°C in an atmosphere of 5% CO2.
LDL Purification.
Highly purified preparations of low-density lipoproteins were obtained from single donor human plasma with sodium-EDTA added at time of collection (Innovative Research Corporation, Novi, MI). LDL was purified from human plasma by sequential ultracentrifugation in NaBr solutions within 1.019–1.05 g/mL density range using methods described previously (26). LDL preparations were then dialyzed in PBS-5 mM MgCl2, frozen drop-wise, and stored in liquid N2, in about 200 μL aliquots until needed. Purity was assessed using Coomassie Brilliant Blue-stained, native 0.5% agarose gel electrophoresis.
CM-DiI Labeling of LDL.
Purified human LDL, isolated as described above, was covalently labeled using CM-DiI. Briefly, 250 μL of ethanol was added to the vial containing 50 μg CM-DiI (C-7000), a thiol-reactive fluorescence compound from Invitrogen, Inc., to make a stock solution. Next, 1μL of CM-DiI stock solution was added to 1 mg (by protein) of purified LDL in 1 mL of PBS-5 mM MgCl2. The reaction mixture was allowed to stand for 30 minutes at ambient temperature then dialyzed overnight in 1 liter PBS-MgCl2 with three changes of the buffer.
FITC-Synthetic Peptide Cell Binding.
In cell binding/endocytosis experiments HeLa cells at ~70% confluence were preconditioned by incubating overnight in DMEM – P/S - 3% FBS medium containing 1.0 μM Mevastatin (47, 48). Cells were rinsed thrice with 3 mL PBS-5 mM MgCl2. Peptide-binding assays were performed at 37° C for 30 minutes in 0.5 mL solution containing 440 μL PBS-5 mM MgCl2 plus 60 μL the FITC-synthetic peptide. The assay solution was then removed, cells were rinsed as before, then incubated at ambient for 10 minutes in 1 mL 10% formaldehyde in PBS-MgCl2. Cells were next rinsed as previously and images were then obtained.
Dual Label Experiments.
In these assays FITC-labeled apo B synthetic peptides were used in combination with CM-DiI labeled purified LDL. HeLa cells were preconditioned as before then incubated at 37° C for 30 minutes in medium containing 340 μL PBS with 5 mM MgCl2, plus approximately 300 μg labeled-LDL and 60 μg labeled B peptide, then rinsed thrice with buffer, fixed in 1% formaldehyde; 1 sec exposure images were next obtained at 50 × and 630 × magnification.
Fluorescence Microscopy.
Microscopy was performed using a Zeiss Axiovert 25 equipped with Zeiss objectives, FITC/GFP (Zeiss HQ470/40× and HQ525/50m) and red (Chroma AT540/25× and AT605/55m) filter cubes, and a motorized x, y, z stage from Prior Scientific, Inc., Rockland, MA. Images were captured thru a 0.5× C-mount adapter using an Optronics Microfire CCD camera and Picture Frame software as described previously (26). FITC signal intensity obtained for 50× view field was used to assess cell binding and images obtained at 630× magnification were used to assess cell entry and location. For further processing, all green fluorescence images were obtained at identical exposure periods, 100, 173 and 1000 milliseconds. Adobe Photoshop CS Version 4 Extended was used to enhance images for presentation. Low signal was enhanced with the Brightness and Contrast algorithm under the Level tool command to detect all points emitting signal. Images were obtained and saved as *.jpg and *.bmp formats described previously (26).
Analysis and assessment of raw integrated densities, and cell number per field of view values were obtained using ImageJ (49). The following standard steps were used in image processing: removed background, auto-adjust Brightness/Contrast, reduce noise with Despeckle and Remove Outliers algorithms, then apply the Color Threshold command, select Original, and obtained data using Particle Analysis. This procedure was used to obtain cell count for view fields at 50 × magnification, total area covered by cell mass, percent of image area covered by cell mass, number of green or red fluorescence loci visible on corresponding, and total integrated densities for each color image. These results, shown in Figure 1, were then used to determine integrated density of FITC signal per cell, per locus, percent of cell area and percent of cells emitting signal. To determine the average values, standard deviations, and P-values for pairwise comparison of the binding characteristics of the peptides, statistical functions AVERAGE, STDEV.S, and T.TEST in Microsoft Excel 2013 were used.
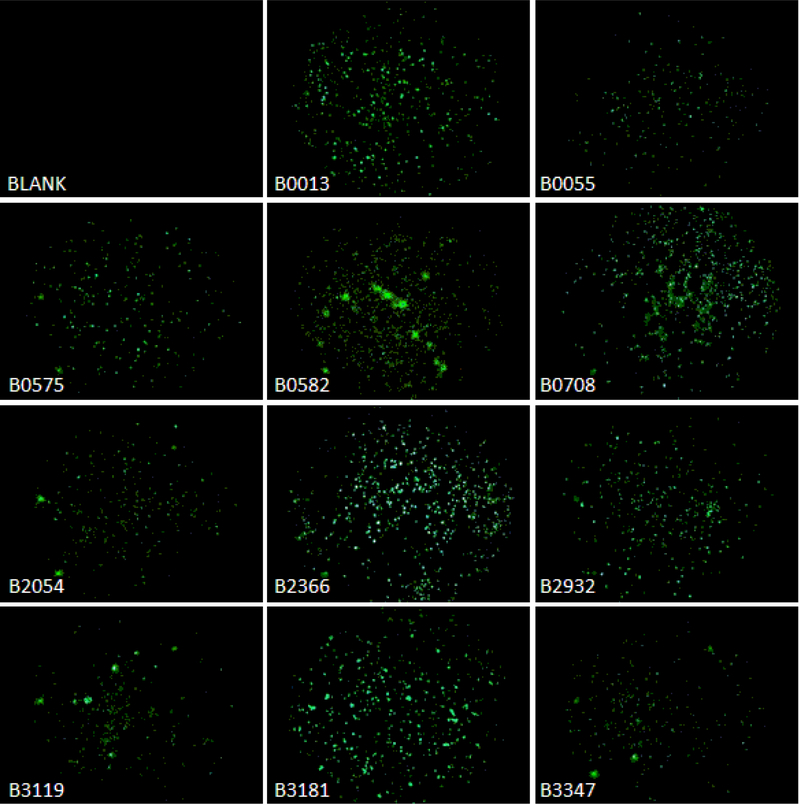
Figure 1.
Fluorescence micrographs of apo B100 FITC-labeled peptides binding to HeLa cells. Fluorescence images obtained for apo B100 FITC-peptides listed in Table 1 were captured at 50× magnification at 100mSec exposures using an Optronics Microfire CCD camera and Picture Frame software as described in Methods. Brightness and Contrast algorithms in ImageJ were used to enhance images.
Full-frame images of field of view, taken at the same magnification and equal in dimensions were used to create overlay images (26) in Photoshop. Bright Field images were also enhanced, first by converting from RGB to monochrome format. The image was next enhanced by applying Brightness and Contrast in Level menu. File was then converted backed to RGB format for overlay on green or red fluorescence image (as in Figures 2 and 3). Images obtained in dual label experiments were overlaid to create an image showing basic colors, green for FITC-peptide sites (white is seen for intense clusters) not in proximity to CM-DiI labeled LDL, red for labeled LDL (yellow is seen for concentrated LDL clusters) and co-located signals as a greenish/yellowish combination based on ratio. Next, an overlay with the corresponding Bright Field image and the dual signal overlay image was created (Figures 4–6).
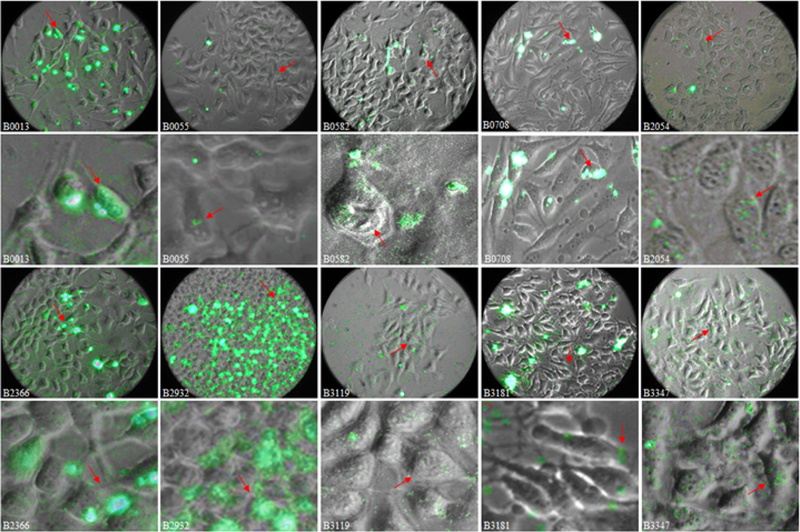
Figure 2.
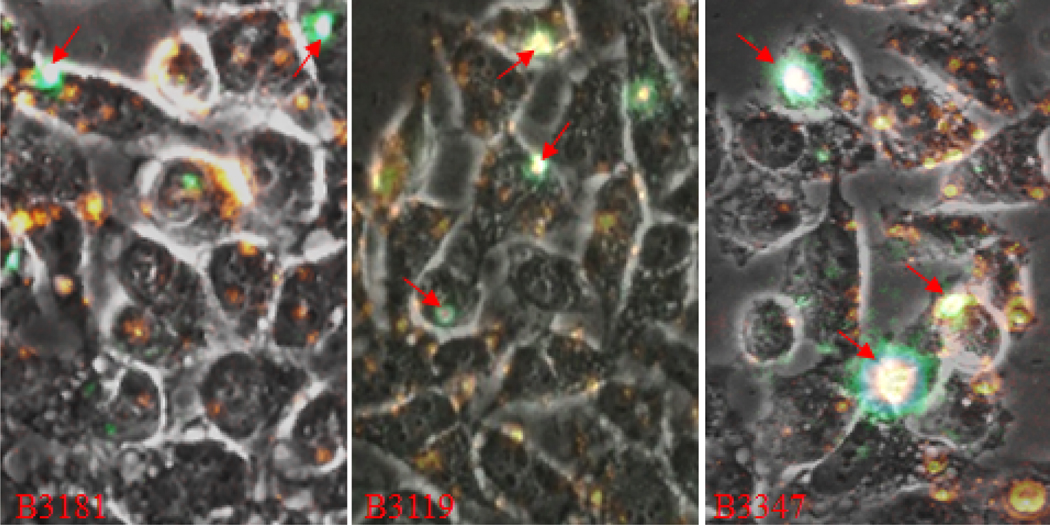
Overlay fluorescence micrographs of FITC-labeled apo B100 peptides interaction with HeLa cells. Overlays of Bright Field and fluorescence images for each peptide obtained at 630× magnification were used to assess cell entry and endoplasmic localization. Fluorescence signal for B0055, B0708, B2054, B3119 and B3347 images were amplified in Power Point for presentation only. Red arrows indicate regions enlarged and shown below corresponding whole view field images. Fluorescence signal seen in overlay images for B0013, B0582, B2054, B2366, B2932, B3119, and B3181 suggests presence of peptide in the endoplasmic spaces. Signal for B0708 peptide is emitted almost entirely from the cell surface and in some instance intense signal suggests cell apoptosis (red arrow).
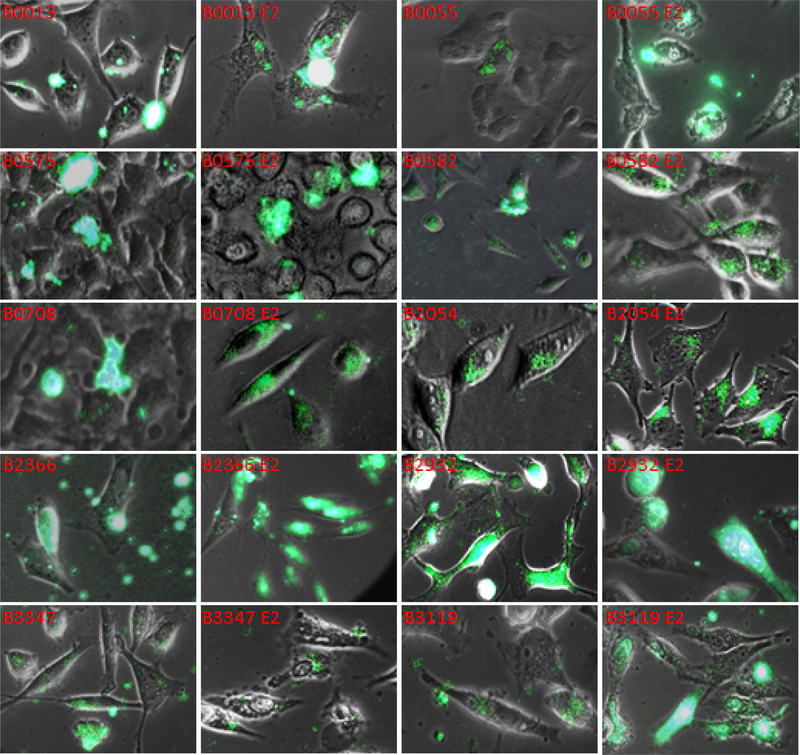
Figure 3.
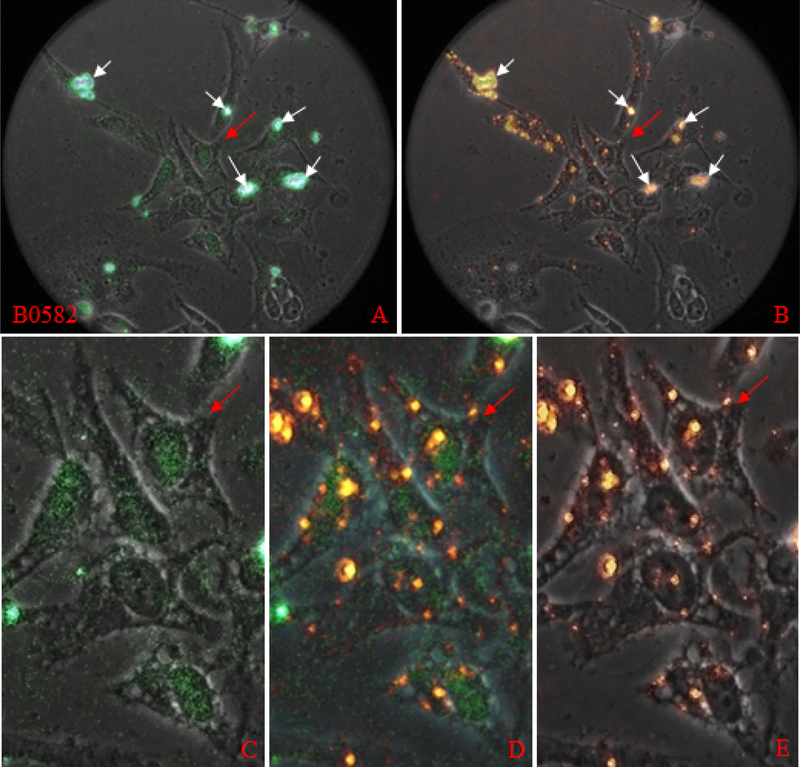
Qualitative Assessment of Influence of E2 Peptide on Cell Binding and Entry of FITC-B Peptides. Unlabeled Apo E2 peptide was added to HeLa cell assays to determine its effect on FITC-labeled apo B100 peptides shown in each frame above.
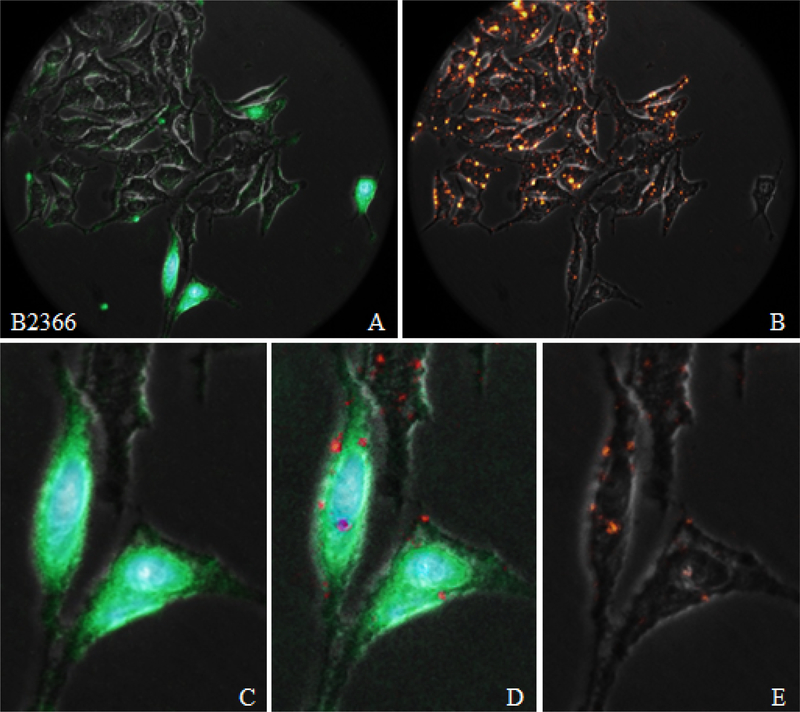
Figure 4.
Binding of peptide B2366 to HeLa cells in presence of DM-DiI LDL. Fluorescence and Bright Field overlay micrographs of FITC-B2366 (frames A and C) and labeled LDL (frames B and E) uptake by HeLa cells show non-colocalized signals. Frame C is an enlarged region of Frame A, and shows B2366 signal emanating from the ectodomain, endoplasm, perinuclear and nuclear space. Same region of the labeled LDL overlay (Frame B) was enlarged and is shown in Frame E. LDL signal appears on the cell surface with vague appears in the cell proper, perhaps due to the rather short incubation period (30 min). Overlay of Frames C and E is shown in Frame D, which reveals that CM-DiI LDL signal independent of FITC-B2366 signal.
Figure 6.
Binding of the peptides B3181, B3119 (Site A), and B3347 (Site B) to HeLa cells in presence of CM-DiI LDL. The triple overlay images (red and green signals plus Bright Field) recorded at 630× magnification are presented for B3181-LDL (Frames A, B), B3119-LDL (Frame C, D), and B3347-LDL (Frames E, F). The bottom row shows enlarged regions from the frames in the top row; red arrows serve as reference points. Loci that appear white are from the most intense signal (green, red, or both). Yellow-greenish color indicates colocalized signals.
RESULTS
Apo B100 contains one Type I, nine Type II, and twenty-three Type III receptor ligand analogues (Table 1 Supplement). FITC-labeled synthetic peptides of regions in the apo B100 sequence (Table 1) that contain at least one of these motifs were selected to determine cell surface binding, cell entry and intracellular location, and binding to extracellular matrix/debris based on 2D overlay Bright Field and fluorescence images. All synthetic peptides used in the present study were shown to bind HeLa cells, but with distinct avidities as well as different cell interaction pattern (Figure 1, Table 2).
Table 2.
Analysis of Green Fluorescence Signal for Viewed Fields at 50 × Magnification.
| Peptide | FITC Signal per Total Cell Area, % | Signal Loci Per Cell | Integrated Signal Density per Locus | Integrated Signal Density per Cell |
|---|---|---|---|---|
| B0013 | 13.0 ± 2.6 | 1.42 ± 0.70 | 2.7 ± 1.2 | 3.2 ± 1.2 |
| B0055 | 1.5 ± 0.6 | 0.12 ± 0.04 | 19.9 ± 24.2* | 1.4 ± 0.7 |
| B0575 | 2.2 ± 1.2 | 0.19 ± 0.08 | 14.7 ± 12.6 | 2.2 ± 1.1 |
| B0582 | 17.6 ± 2.4 | 0.15 ± 0.14 | 437.3 ± 165.0 | 51.9 ± 22.3 |
| B0708 | 11.3 ± 7.0 | 0.47 ± 0.08 | 6.6 ± 1.50 | 3.1 ± 0.6 |
| B2054 | 0.88 ± 0.25 | 0.11 ± 0.03 | 37.0 ± 20.9 | 3.6 ± 1.3 |
| B2366 | 11.5 ± 5.4 | 0.57 ± 0.34 | 12.1 ± 2.3 | 4.6 ± 2.3 |
| B2932 | 3.8 ± 0.7 | 0.35 ± 0.19 | 13.9 ± 10.5 | 4.1 ± 2.9 |
| B3119 | 1.5 ± 0.1 | 0.08 ± 0.02 | 25.3 ± 4.8 | 1.9 ± 0.4 |
| B3181 | 44.1 ± 13.0 | 0.13 ± 0.05 | 367.5 ± 59.1 | 46.3 ± 15.1 |
| B3347 | 1.8 ± 0.5 | 0.43 ± 0.16 | 2.9 ± 0.5 | 1.2 ± 0.2 |
Initial values for each image, FITC and Bright Field, were obtained using ImageJ software algorithms. Average and sample standard deviation values (STDEV.S) were generated using Excel 2013. FITC Signal per Total Cell Area and Integrated Signal Density per Locus values were x10, as initial images for peptides B0582 and B3181 were obtained at 0.1 sec exposure while images for all other peptides were obtained at 1 sec exposure.
Large variation was observed in number of signal loci.
Cell Binding Capacity.
Fluorescence images, in Figure 1, captured at 50 × magnification and 1 sec exposure were used to assess cell binding efficiency with ImageJ algorithms. Cell binding efficiency, i.e. number of cells emitting signal, number of signal loci per cell, signal density per cell, and signal density per locus were determined (see Methods). Forty-four Bright Field images were captured displaying 127,033 cells in toto, with 2,757 cells per view field on average. Multiple corresponding fluorescence images for each peptide and different magnifications were also obtained, except for peptides B0582 and B3181. For these peptides, green fluorescence images at 50 × were obtained at 100 and 173 milliseconds exposures, as 1 sec exposure times completely overwhelmed the view field. Images were then normalized to 1 sec exposures. Digital data was generated by analysis of multiple view field images, at least four for each peptide.
Analysis of the emitted signal for cells viewed at 50 × magnification is shown in Table 2. Results in column “Percent of Total Signal Area per Cell Area,” show peptides B0013, B0708, and B2366 bind to approximately 11–13% of the total cell area as determined using corresponding Bright Field images. Signal for B2932 and B3181 is seen for about 4.5% of the cell area; B0575 and B0582 signal represent about 2.3%; B0055, B3119 (Site A) and B3347 (Site B) signal corresponds to an average of 1.7%; and, B2054 signal emits from less than 1% of cell area. A different binding pattern is observed: an average of 1.5 loci emitting signal per cell was seen with B0013; while B0708 and B2366 signal was present on about half of the cells (0.47 loci/cell). Approximately 1 of 3 cells showed signal for B2932 and B3347; one of 8 cells bound peptides B0055, B0582, B2054, and B3181, and only one in 14 cells bound B3119. The highest signal RID per locus and per cell is seen for peptides B0582 and B3181 (Table 2); both contain two potential RAP receptor ligand motifs, 0583LKKLVKEVLKE and 0602FRKFSRN in B0582 (Table 1), and, 3181EKN, 3200IKFDKY and 3205YKAEKS in B3181, respectively, (Table 3). Underlined are the motifs similar to established receptor ligands in RAP, see the next section in Results.
Table 3.
Proposed LDLR and LRP Ligands in Apo B100 Based on Similar Sequences in Chaperone Proteins, RAP and Heat Shock Proteins.
| CAD62296 | 0020EERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEKEDKEEEK0066 |
| CAS62296 | 0001MGRGTKVILHLKEDQT*EYLEERRIKEIVKKHSQFIGYPITL*FVEKERDKEV0051 |
| NP003290 | 0252KEEASDYLELDTIKNLVKKYSQFINFPIYVWSSKTETVEEPM0293 |
| AMRP P30533 | 0238DKELEAFREELKHFEAKIEKHNHYQKQLEIAHEKLRHAESVGDGERVSRSREKH0290 |
| NP003290 | 0602EKTKESREAVEKEFEPLLNWMKDKALKDKIEKAVVSQRLTESP0644 |
| CAD62296 | 0230IRKNLVKKCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKK0279 |
| HS90B | 0040LRELISNASDALDKIRYESLT*DPSKLDSGKELKIDIIPNPQE0081 |
| NP003290 | 0596VKFDESEKTKESREAVEKEFEPLLNWMKDKALKDKIEKAVVS0637 |
| CAD62296 | 0548GLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVS0589 |
| BAB15121 | 0126GESKEQVANSAFVERVRKRGFEVVYMTEPIDEYCVQQL0163 |
Lysine and Arginine residues are highlighted in bold, potential receptor ligand motifs are in bold and underlined. Similar regions are underlined. apo B100 (CAA28420); Apo E (LPHUE); alpha-2-Macrogolublin Receptor-Associated Protein (AMRP P30533, RAP); Heat Shock Protein, HS90B (P08238); Endoplasmin (NP_003290); Heat shock proteins (AAC02679 – HSP75), (AAH09195 – HSP90B1), (BAB15121 – HSP90), (CAD62296 – HSP90), (NP_001017963 – HSP90 alpha isoform 1), (NP_005339 – HSP90kDa alpha), (NP_055969 – MESD); and, TNF receptor-associated protein 1 (AAH18950).
To assess cell entry and intracellular localization, images for each peptide were also obtained at 630 × magnification. Overlay images of green fluorescence and corresponding Bright Field images were developed and are shown in Figure 2. Enlarged regions of overlay images for each peptide displaying individual cells with associated peptide signal are shown under the corresponding frame in Figure 2.
Sequence comparisons of apo B100 peptides to proteins with similar short sequence motifs that may provide insights into other potential functions of the selected regions of apo B100 are presented in Tables 3 – 9. Sequence similarity is based on the location of potential receptor ligand motifs as well as coincidence of basic residues proximal to the ligand and/or presence of other motifs of known function, e.g. di-Lysine and a Lysine/Arginine NLS, a bipartite NLS.
Table 9.
Results of Control Assays for HeLa Cell Binding Experiments.
| Medium | Total Loci Emitting Signal | Total Area of Signal | Average Total Integrated Density | Average Total Cell Count* | % Cell Area Emitting Signal | Average Loci Per Cell (x100) | Integrated Density Per Cell |
|---|---|---|---|---|---|---|---|
| 10% FBS | 16 | 170 | 2291 | 1763 | 0.35 | 0.9 | 1.30 |
| 3 % FBS-M | 39 | 588 | 3847 | 1168 | 0.88 | 3.3 | 3.29 |
| PBS | 399 | 6,077 | 3835 | 2449 | 5.76 | 16.3 | 3.28 |
| B0582 0582DLKKLVKEVLKESQLPTVMDFRKFSRNYQ0610 | |||||||
| 10% FBS | 144 | 2,454 | 4373 | 1766 | 5.01 | 8.2 | 2.48 |
| 3 % FBS-M | 474 | 13,122 | 6861 | 1353 | 20.61 | 35.0 | 5.07 |
| PBS | 268 | 5,853 | 5328 | 1554 | 9.02 | 17.2 | 3.43 |
| B3181 3181EKNRNNALDFVTKSYNETKIKFDKYKAEKSQDE3213 | |||||||
| 3 % FBS-M | 238 | 9,722 | 9911 | 1518 | 16.54 | 15.8 | 6.53 |
| PBS | 142 | 3,419 | 6397 | 1933 | 4.16 | 7.3 | 4.86 |
| B2366 2366VGFIDDAVKKLNELSFKTFIEDVNKFLDMLIKKLKSFD YHQE2407 | |||||||
| 3 % FBS-M | 73 | 966 | 3378 | 1402 | 1.68 | 5.2 | 2.41 |
| PBS | 78 | 1,007 | 3179 | 1803 | 1.19 | 4.3 | 1.76 |
| B3119 3119LKDFSLWEKTGLKEFLKTTKQSFDLSVKAQYKKNKHRHSITN3160 | |||||||
| 3 % FBS-M | 171 | 1843 | 2727 | 1828 | 2.16 | 9.4 | 1.50 |
| PBS | 93 | 994 | 2739 | 1980 | 1.00 | 4.7 | 1.38 |
| B3347 3347DALQYKLEGTTRLTRKRGLKLATALSLSNKFVEGSHNSTVSLT3389 | |||||||
per view of Bright Field. Peptide B0582 contains RAP-ligand motifs and a ligand sequence in the GAG-binding region of FGF. Peptide B3181 contains three RAP-ligand motifs and one Type III Cardin-Weintraub motif. B2366 contain two RAP-ligand motifs, one Type II CW motifs, three potential endocytic motifs shown in italics. B3119, Site A, contains several Lysine-based motifs similar to RAP-ligand motifs. B3347, Site B, contains one Type I CW motif.
Apo B100 Regions Similar to Domains in Receptor Associated Protein, MESD, and Other Heat Shock Proteins.
Alignment of apo B100 sequences 0013Lys – Asn0090 (B0013, B0055) 0708Asp – Gly0786 (B0708), 2391Phe – Leu2478 (B2366), 2932Ser – Leu3009 (B2932), and 3166Cys – Tyr3244 (B3181) and regions of domains 1–3 of RAP is shown in Table 3. RAP is a resident endoplasmic reticulum protein and an intracellular chaperone for the LDL and VLDL receptors (LDLR and VLDLR, respectively) and 15 related receptors, such as LDLR related Proteins (LRPs) and alpha-2-macroglobulin receptor (37). All three domains in RAP (RAP1d, RAP2d, and RAP3d) were shown to have high binding affinity for LRP (50). Lysine residues 0060 in motifs WKKLK and 0191 in LKEKL of RAP Domains 1 and 2, respectively, were shown experimentally to bind LDLR related protein 1 (LRP-1) with high affinity (51). In RAP3d, residues Lys0253, Lys0257, Tyr0260, Lys0270, Arg0285, and Arg0296 were shown to interact with ligand adhesion modules LA3–4 of the LDLR and complement-like repeat CR5–6 of the LRP-1 (51). The following ligand motifs are established in RAP: EKL, at positions 0024 in RAP1d, positions 0119 and 0193 in RAP2d, and at 0270 in RAP3d; EKV at 0148, LKE at 0191 and DKL at 0137 in RAP2d; and, EKH at 0256 in RAP3d; these motifs are involved in binding to Ligand Adhesion Modules, LA3–4 of the LDLR and CR5–6 of LRP-1 (50–52). Motif VKK at positions 0305–0306 was also confirmed as a ligand for binding LRP (53).
Included in Table 3 are sequences of regions in MESD, mesoderm development protein and an LDLR chaperone (54), and in other heat shock proteins that are known to bind LDLR family (30), although mostly via unknown mechanisms. Potential Lysine-based receptor ligand motifs in these proteins are similar to motifs in the apo B100 sequence. Motifs in the B0013 region occur in HSPs BAB15121 and NP_001017963. We identify B0013 motif TRFKHLRKYTYNY as a strong candidate for binding to LDLR and perhaps other LRPs. Although B0055 sequence shares Lysine motifs with MESD and other HSP sequences, the motif KALLKKTK has not been shown to have the same HeLa cell binding capacity as B0013. Renditions of sequence LKKLVKEVLKESQ that is in B0575 and B0582 are in the HSP sequences shown in Table 3. B0708 contains motif TKDDKHEQD which appears as AKIEKHNHYQ in RAP. Motif SVEKLIKDLKSKE of B0708 shares similarities with SKEKNKTKQDKG of MESD and KDKALKDKIEK in HSP NP_003290. B2054 that shows low level HeLa binding capacity shares motifs ERNRQT and RKYRAALG with HSP NP_003290, which contains renditions EKTKES and KIEKAVVS, respectively. B2932 has three motifs that are similar those in HSPs shown, INS, YESGS, and NFSKLE. B2932 was selected for this study based on it similarity to B0013, potential cell binding was suspended on motif SKHLR. This peptide shows high binding capacity that may involve motifs, NKI, SKH, and SKL. The latter is in the HSP aligned with it in the table. B3119 is a high Lysine peptide and shares some similarity with HSP NP_003290. Renditions of sequence TKIKFDKYKAEKSQ, carboxyl end of B3181, are in RAP, HSPs, and MESD. Residues highlighted in yellow in the MESD sequence are shown to be located in a loop on the surface of the molecule (2KGL – CN3D) and may be the ligand region for the LDLR (54). High sequence similarity between B3181 and these chaperone proteins support a role for this region of apo B100 in binding to the LDLR. Although the motif RLTRKRGLK, Type I C-W, in B3347 is very similar to RKLRKRLLR in apo E and KVIRKKLVRK in HSP NP_003290, it different from these sequences in that it lacks the LRK of apo E which is present in the HSP AS IRK, VRK, and IKK. This deficiency may attribute to its poor binding capacity to HeLa cells.
Similarity of our synthetic peptide sequences to RAP, MESD, and heat shock proteins suggests that Apo B100 contains several potential receptor ligands.
Assessment of B3181 HeLa Cell Interaction.
Total Integrated Signal Density and ISD per cell for B3181 are significantly different (P < 0.05) compared to all other peptides except B0582. We identify B3181 as the principal candidate for potential receptor ligand based on its similarity to RAP, MESD, and other heat shock proteins, as discussed above. Interestingly, this region of Apo B100 is located between Sites A and B, the putative ligand regions for the LDLR. Large, high intensity clusters of FITC signal appear throughout the view field of B3181 images in Figure 2. Extracellular space appears to be devoid of signal.
Assessment of B0013/B0055 HeLa Cell Interaction.
Apo B100 N-terminal region spanning residues 0013Lys - Phe0094, which includes peptides B0013 and B0055, has the capacity to bind plasmid DNA and deliver it to the cell nucleus (25, 26). In the present study, experiments were designed to assess the role that each sequence may have in to binding HeLa cells and negotiating entry. Results in Figure 2 show B0013 binds to most cells in view field; signal clusters and multiple fluorescence points are seen in the top frame. Red arrow in lower frame with enlarged region denotes signal that appears to be in the cytoplasm. In Bright Field images some clusters appear as dull golden speckles on the cell surface. A scatter of weak signal is apparent in the extracellular space with some faint points on the individual cell filopodia. Integrated density in some loci is relatively strong as indicated by red arrow in the enlarged frame, although average signal RID per locus is the lowest for this peptide (Table 2) due to many faint loci in the view field.
In Figure 2, top row, column 2, B0055 signal is seen only at discrete loci on about 12% of the cells in the view field, as multiple fluorescence points and brighter clusters; the image of an enlarged region shows mostly the faint signal.
LDL Receptor Binding: Similarity of B0013 and B0055 to RAP Domain 2.
Similarities between the apo B100 sequence and RAP2d are highlighted in Table 3. Presented sequence comparison suggests that both peptides contain potential receptor ligands, 0018FKH and 0021LRK in B0013, 0071LKE and 0085LKKTKN in B0055.
Potential Function: Similarity of B0013 and B0055 to DNA-binding Domain of IRF Proteins.
The similarity between the first 110 residues of the N-terminus of apo B100 and the DNA-binding domain of IRF 9 was reported previously (21, 25). The apo B100 region, 0013Glu – Arg0100 contains motifs common to the DNA-binding domains in IRF proteins (Table 4). Sequence alignment is based on a putative NLS cluster in the N-termini of IRF proteins, a polar amino acid stretch, and a Proline-Glycine dipeptide, followed by motifs that are analogous to RSATRINCK, SFILK, KALLK, and KKTKNSEEF in apo B100. This sequence comparison shows that the N-terminus of apo B100 is most like the DNA-binding domains of IRFs 9, 8 and 4.
Table 4.
Potential Nucleic Acid Binding Regions in Apo B100 Compared to DNA-Binding Domains of Interferon Regulatory Factors.
| Polar GXXG | >~< |
| 0013 DATRFKHLRKYTSYNYEAESS*GVPGTA**DS*RSATRINCKV**ELEVPQLC*SFILKTSQCT***LKEVY**GFNPEGKALLKK**TKNSEEFAAAMSR | |
| IRF9 RARCTRKLRNWVV*EQVESGQ**FPGVCWDDTAKTMFRIPWKHAGKQDFREDQDAAFFKAWAIFKGKYKEGDTGG*PAVWKTRLRCALNKSSE*FKEVPER | |
| IRF8 DRNGGRRLRQWLI*EQIDSS**MYPGLIWENEEKSMFRIPWKHAGKQDYNQEVDASIFKAWAVFKGKFKEGDKA*EPATWKTRLRCALNKSPD*FEEVTDR | |
| IRF4 VSCGNGKLRQWLI*DQIDSG**KYPGLVWENEEKSIFRIPWKHAGKQDYNREEDAALFKAWALFKGKFREGIDKPDPPTWKTRLRCALNKSND*FEELVER | |
| 0705 LVDHFGYTKDDKHEQDMVNGIMLSVEKLIK*DLKSKEVPEARAYLRILGEELGFASLHDLQLLGKLLLMGARTLQGIPQMIGEVIRKGSKNDFFLHYIFME | |
| VTG2 HIEPVFSESKISVYNYEAVILNGFPE***SGLSRAGIKINCKVEISAYAQR***SYFLKIQSPE***IKEYN***GVWP**KDPFTRSSKLTQALAEQLTK | |
| 2932 SNKINSKHLRVNQNLVYESGSLNFSKLEIQSQVDSQHVGHSVLTAKGMALFGEGKAEFTGRHDAHLNGKVIGTLKNSLFFSAQPFEITASTNNEGNLKVR | |
| IRF1 *MPITRMRMRPWLEM*QINSN*QIPGLIWINKEEMIFQIPWKHAAKHGWDIN**KDACLFRSWAIHTGRYKAGEKEPDPKTWKANFRCAMNSLPDIEEVK | |
| IRF4 AVSCGNGKLRQWLI*DQIDSG*KYPGLVWENEEKSIFRIPWKHAGKQDYNREEDAALFKAWALFKGKFREGIDKPDPPTWKTRLRCALNKSNDFEELVER | |
0013, B100 (0013–0100); IRF9 (0004–0101); IRF8 (0002–0099); IRF4 (0016–0113); 0705, B100 (0705–0805); IRF2 (0001–0109); IRF3 (0003–0109); 2932, B100 (2932–3031); IRF1 (0001–0097); VTG2 (0019–0105); Polar - amino acid stretch; GXXG - K homology RNA-binding Domain; Motif RSATRINCK was used in sequence alignment; >~< indicates a non-RGD integrin-binding tripeptide motif.
The first basic residue cluster in apo B100, 0013KDATRFKHLRK, is similar to RARCTRKLR, an NLS motif in IRF9; and, in apo B100 it appears to be a new potential receptor ligand motif. An endocytic motif, FKHL, occurs in this sequence. An extensive sequence of polar residues, YTSYNYEAESS, in apo B100 is perhaps analogous to sequence NWVVEQVESGQ in IRF9. In Apo B100, this stretch is followed by 0035GVPG (included in B0013), a motif characteristic of the heterogeneous nuclear ribonucleoprotein K, the K homology (KH) motif (27, 28). The Proline-Glycine dipeptide is also common in a similar location in most DNA-binding domains of IRF proteins. Renditions of the KH motif appear in IRF4 in the sequence “0005GGGRGGEFG” and in IRF9 as 0072GDTG. The B100 sequence 0043RSATRINCK that follows is analogous to KSIFRIPWK, a motif conserved in IRFs 4, 7, 8 and 9. The carboxyl region of apo B100 DNA-binding domain sequence, 0059Leu – Leu0105 (26), contains motifs present in IRF proteins 4, 8 and 9; for example, 0060CSFILKTS of B0055 occurs in tandem in IRFs, as DAALFK in 4, ASIFK in 8, and AALFK in 9. The KTS motif in B0055 may function as an analogue of a potential anti-integrin, non-RGD adhesion motif (55). Another motif in B0055 is KALLK, which appears as KTRLR in IRFs 4, 8 and 9. Next, apo B100 sequence 0088TKNSEEFAAAMSR is an analogue of NKSNDFEELVER in IRF 4 and 0089NKSSEFKEVPER in IRF 9. The B sequence FAAAM may be an endopeptidase, i.e. neutrophil elastase, cleavage site (56). Also, B0055 contains an endocytic motif, 0080EGKALL, and a true Type II receptor ligand motif, 0085LKKTKN that does not impart cell binding or entry for B0055. Contact points with Phosphate moieties of the DNA backbone identified for IRFs (57), are also in proposed IRF-like DNA-binding regions of apo B100.
Assessment of B0708 HeLa Cell Binding Capacity.
The contrast-enhanced, high magnification image of B0708 fluorescence (Figure 2 top row, column 4), shows a diffuse signal emitted across the field of view and suggests that signal is located only on the cell surface. There are also several clusters of intense fluorescence; the arrow denotes one of such clusters. An enlargement of the cluster shows that the peptide is on the surface, and not internalized. Clusters appear to be associated with an apoptotic cell.
LDL Receptor Binding: Similarity of B0708 Sequence to RAP Domain 3.
Sequence comparison of B0708 region spanning 0705Val – Glu0802 to RAP and the DNA-binding domains of IRF proteins 2 and 3 are shown in Table 3 and 4, respectively. This region of apo B100 is similar to RAP3d sequence 0247Leu – Leu0321. Motifs AKIEKH and EKL in RAP3d occur as TKDDKH and EKL in B0708. Also, B0708 has motifs IKD, LKS, and SKE, which may also be involved in receptor binding. In RAP3d, AKIE0257KH and E0270KL are involved in binding Ligand Adhesion Modules, LA3 – 4 of the LDLR and CR5 – 6 of LRP-1 (50–53).
Potential Function: Similarity of B0708 Sequence to IRF DNA-Binding Domain.
Apo B100 region, 0705Val – Glu0805 (Table 4), which includes B0708, does not contain a potential NLS motif, but it does contain stretches/motifs that align well with IRFs 4, 8 and 9. The polar region of IRF 9, EQVESGQ, is conserved to a degree as 0718HEQDMVNG0725 in B0708. The Proline-Glycine dipeptide is absent. Short motifs such as DLKS, EELGFASL, LQLLGK, and SKNDFF in B0708 are analogues of motifs EEKS, EEDAALF, AWALFK, and NKSNDF in IRF 4. These similarities are shown and highlighted in Table 4. DNA binding capacity has not been confirmed experimentally.
Assessment of B2366 HeLa Cell Binding and Entry Capacity.
In Figure 2 (row 3, column 1), the 630× overlay images for B2366 shows fluorescence signal dusted over most of the view field. The enlarged region of the image (row 4, column 1) shows signal loci on the membrane perhaps in the cytoplasm. Another enlarged area of the overlay image of B2366 (Figure 3 top row, left column) shows that signal appears at cell surface (white arrow), with yellow arrow indicating signal at organelle structures in the cytoplasm. Orange arrow shows fluorescence signal from extracellular vesicles and very weak signal emanates from other extracellular debris. There appears to be an increase in extracellular debris, including bleb formation and small vesicles. Signal does not appear to emanate from the nuclear space nor at the nuclear envelope.
LDL Receptor Binding: Similarity of B2366 Sequence to RAP.
The B2366 peptide that spans 2366Val – Phe2407 has five potential endocytic/sorting motifs, LNEL, FKTF, FLDM, LDMLI, and YHQF (38, 39). A potential bipartite NLS spanning 2374KK – 2397KKLK appears to be non-functional. Apo B100 sequence spanning 2391Phe – Leu2478 contains K/R motifs similar to those scattered throughout the RAP sequence (Table 3). For example, motif “ΨKKΨK” appears as 0059WKKLKL in RAP1d occurs as 2396IKKLKS in B2391 (and, hence, in B2366). Apo B100 sequence 2394Met – Arg2416 is similar to RAP3d 0250Phe – Arg0272. Comparison this apo B100 sequence to RAP domains is shown in Table 3. In addition, B2366 contains motif 2373VKK, also appearing in sequence 0304–0306 in RAP3d. Overall, apo B100 sequence spanning residues 2110 to 2410 is rich in the receptor ligand motifs of RAP.
Potential Function:
Molecular models generated using Phyre 2 suggest that this region has DNA-binding capacity; however, confidence value is low. Ramachandran plot analysis by Rampage software, http://mordred.bioc.cam.ac.uk/~rapper/rampage.php, indicates 7.6% of residues are in outlier region, 77.1% in favored region, and 15.3% in allowed region.
Assessment of B2932 Cell Binding and Entry Capacity.
Overlay image obtained at 630 × magnification shows high intensity signal across the view field (Figure 2, rows 3–4. Column 2). B2932 signal emanates from cell surface as clusters, and endosome formation is suggested (Figure 3, 2nd row, left column). There is scant extracellular signal.
LDL Receptor Binding: Similarity of B2932 Sequence to RAP Domain 3.
As stated above, residues shown to interact with Ligand Adhesion Modules LA3–4 of the LDLR and complement-like repeats in LRP-1, AKIE0257KHNHY and E0270KL in RAP, are conserved as SKHLRVNQN and SKL, respectively, in B2932.
Potential Function: Similarity of B2932 Sequence to B0013 and IRF DNA-binding Domain.
Testing of B2932, included in sequence 2932Ser – Arg3031 (Line 12, Table 3), was based on the presence of the 2938SKHLR motif, homologue of 0018FKHLR in B0013 and TRKLR in IRF9, and other short, like sequences such as 2948YESGS in B2932 occur as YEAES in B0013. B2932 also shares some similarity with IRFs 1 and 2 DNA binding domains. Examples include: B2932 motif VNQN occurs as INSN in IRFs 1 and 2, and as VNSC in IRF5; and, B2932 motifs HVG and MALF are analogues of HAA and APLF, respectively, in IRF2.
B0582 a Potential Proteoglycan/GAG Ligand.
Overlay image of B0582 shown in Figure 2 (top row, 3rd column) was generated with a 173 millisecond green filter exposure image. B0582 signal appears indiscriminately throughout the view field, i.e. it appears bound to components of the extracellular matrix, cell membrane, cytoplasmic spaces, organelles, including nucleus.
Corresponding enlarged image of the area indicated by red arrow (Figure 3, row 2, column 3), shows signal on the cell surface, in the cytoplasm, and at the nuclear envelope. Discrete points of signal appear attached to extracellular debris and associated to apparent cellular material outside the focal plane. Few aggregates are present. Fluorescence images for the shorter peptide B0575 were obtained at 1 sec exposure. B0575 signal appears associated predominantly with cells and integrated density of signal per locus and per cell is about 3% and 4%, respectively, of that determined for B0582. Signal area, integrated signal density per locus and per cell (Table 2) are significantly different for these peptides (P < 0.05). Although B0575 contains potential ligand motifs, 0583LKK, 0587VKE, and 0591LKE, it contain four glutamates and two aspartates in a cluster similar to those contained in the receptors, which may influence binding, e.g. via formation of homodimers.
Comparison of B0582 Sequence and Human Fibroblast Growth Factor 1 GAG Ligand Motif.
Apo B100 sequence spanning residues 0573 to 0625, which contains peptides B0575 and B0582, is similar to the heparin-binding site of the human fibroblast growth factor 1 (Table 5), but it lacks the “Glycine box” (58). Motifs 0583LKKLVKE, LKE and 0604FRKFSR are potential ligands for the ligand adhesion modules LDLR family of proteins and may function also in binding proteoglycans/GAGs (32–34). Sequence spanning 0584KK to 0605RKFSR presents a legitimate bipartite NLS motif as well. B0575 contains one mono-NLS cluster, 0584KKLVK.
Table 5.
Comparison of Apo B100 Peptide B0582 and Heparin Binding Region of Fibroblast Growth Factor 1
| Accession | Protein | Potential Proteoglycan/GAG Ligand |
|---|---|---|
| AC010872 | B100 | 0573LNSEELDIQDLKKLVKEALKESQLPTVMDFRKFSRNYQLYKSVSLPSLDPASA0625 |
| NP000791 | FGF1 | 0104LEENHYNTYISKKHAEKNWFVG*LKKNGSCKRGPRTHYGQKAILFLPLPVSSD0155 |
Lysine and Arginine residues are highlighted in bold. Similar residues located in like regions of the sequences are underlined.
B2054 and Potential RNA Recognition Motifs (RRM) in Apo B100.
B2054 image in Figure 1 show low density signal that is well dispersed in the view field. Images in Figure 2 at 630× magnification and enlarged region show sparse, discrete signal in loci mostly in the extracellular space; no large fluorescing aggregates are seen. Sparse signal emanates from the cytoplasm as indicated by arrow and only a few clusters are apparent. Peptide B2054 contains only one potential ligand motif, LKH; which may be reflected by the low level of fluorescence signal. B2054 contains a potential bi-partite NLS formed by clusters 2055RNR and 2081VRKYR, the latter sequence is a legitimate, low affinity Type II LDLR/GAG ligand and a mono-NLS.
Apo B100 Regions with Similarity to RRM Motifs in ApoBEC-1 Complementation Factor and MSX2-Interacting Nuclear Protein.
Apo B100 sequences spanning 2015Arg – Lys2089 (potential RNA Recognition Motif, BRRM1), which contains peptide B2054, and 2237Lys – Glu2315 (BRRM2) share short motifs with known RRM domains in RNA-binding proteins, msx2-IP (NP_055816; also known as MINT: Muscle Segment Homeobox Interacting Nuclear Target) and APOBEC-1 complementation factor (ACF) (AAF76221). A comparison of these sequences is shown in Table 6. Apo B100 region 2015Arg – Lys2089 is represented as BRRM1; and, Apo B100 sequence 2237Lys – Glu2315 is shown as BRRM2.
Table 6.
Comparison of Potential RNA Recognition Motifs in Apo B100, Putative RRM Sequences in ApoBEC-1 Complementation Protein and MSX2-Interacting Nuclear Protein (MINT) and Flavivirus NS5 Proteins.
| BRRM1 | 2015RDAVEKPQEFTI*****VAFVKYDKNQDVHSI*NLPFFETLQEYFERNRQTIIVVLE********NVQRNLKHINIDQ*FVRKYRAALGK2089 |
| MRRM3 | 0423RPLDERIDEFHPKATRTLFIGNLEKTTTYHDLRNIFQRFGEIVDIDIKKVNGVPQYAFLQYCDIASVCKAIKKMDGEYLGNNRLKLGFGK0512 |
| ACF | 0230KILYVRNLMLSTSEEMIEKEFNNIKPGAVERVKKIRDYAFVHFSN*RKDAVEAMKALNGKVLDGSPIEVTLAKPVDKDSYVRYTRGTGGR0320 |
| BRRM2 | 2237KLQQLKRHIQNIDIQH**LAGK*LKQHIEAID***********VRVLLDQLGTTISFERINDVLEHVKHFVINLIGDFEVAEKINAFRAK2315 |
| MRRM4 | 0507KLGFGKSMPTNCVWL**DGLSSNV*SDQYLT*RHFCRYGPV**VKVVFDRLKGMALVLY*SEIED******AQAAVKETKGRKIGGNKIK0583 |
Overall, BRRM1 and BRRM2 sequences share rather low-level similarity with these RRM proteins; however, several highly similar motifs are present. The motifs 2015RDAVEK2020 and 2084RAALGK2089 of apo B100 and 0230KILYVR0236 and 0314RGTGGR0319, present in ACF, were used to align both regions of apo B100 with the known RRM domains. Relatively high sequence similarity is seen between residues 2015Arg – Ile2026, 2027Val – Pro2045, and 2063Val – Gln2093, in BRRM1 region and the msx2-IP RRM spanning 0423Arg – Thr0516.
APOBEC-1 Stimulating Protein (ASP) and ACF also show similar short sequences. ACF and ASP are heterogeneous nuclear ribonucleoproteins that are known to bind APOBEC-1 and stimulate the APOBEC-1 mediated mRNA editing (59, 60). Msx2-IP, a potent transcriptional repressor (61, 62), contains RNA recognition motif domains. Msx2-IP contains three N-terminal RNA recognition motifs, 0323Lys – Val0408, 0423Arg – Lys0512, and 0507Lys – Lys0583, and the Msx2 binding region, 2130Glu – Asp2464. The RNA recognition domain of Msx2-IP binds Runx2 (Runt-related transcription factor 2) to enhance expression of the osteocalcin gene by binding to its control element, the fibroblast growth factor responsive element (OCFRE). Osteocalcin is a non-collagen protein of bone extracellular matrix. Sequence similarity between these proteins and regions in Apo B100 suggests a similar function.
B3119 LDL Receptor Ligand Site A is a Potential Procoagulant Inhibitor.
Sequence B3119 contains a true Type II ligand, 3149SKKNKHRH, and is historically known as Site A. This short motif is also a true NLS. B3119 contains two motifs, LKD and LKE, analogues of a receptor ligand motif in RAP, 0190LKE. Overlay images of B3119 in Figure 2, rows 3 and 4, show low-level binding on the cell surface and not in the cytoplasm proper. Cell surface aggregates are not apparent. Approximately 7% of cell area in view field binds B3119, forming clusters with relatively high integrated density per locus (Table 2). Signal at discrete loci in the extracellular space is significant as seen in original size overlay image (Figure 2). These results indicate that Site A in apo B100 may not be a major ligand for an LDL receptor, and perhaps has a different function.
Apo B100 Site A Region as a Procoagulant Inhibitor.
Previous reports suggest that this region of apo B100 plays a part in hemostasis as an anticoagulant (63, 64). Synthetic peptide KRAD-14 spanning residues 3146–3159 of apo B100 (included at the carboxyl end of our peptide B3119), and its mutated form, Mutant-4, were shown to inhibit procoagulant activity of tissue factor, 78.5% and 95.5% of Control assay, respectively (63). The functional motif in the native form is then KAQYKKNKHRHSTT (KRAD-14), which is included in B3119. In Mutant-4, functionally important is substitution of Lysine in position 5 of the underlined motif with Alanine; and sequence KAQSKKNA5HRHSTT (Mutant-4) appears to be a more effective procoagulation inhibitor. These studies showed that LDL, recombinant peptides of apo B100 spanning 3121Asp – Thr3217 (KRAD-98), KRAD-14, and Mutant-4 influenced procoagulant activity of tissue factor by preventing Factor VII activation (64). Peptide KRAD-14 also prevents prothrombin activation by interacting with Factors Xa and V. Hence, the Lysine-rich Site A ligand region imparts LDL, and other LLPs, with the capacity to regulate hemostasis.
Sequence Similarity of Apo B100 Site A Region and Known Inhibitors of Coagulation.
Further evidence that suggests anticoagulant function for Site A is found in pit viper venom proteins. A comparison of apo B100 sequence spanning residues 3119 to 3170 to snake venom anticoagulant proteins of Deinagkistrodon acutus (1IOD_B), Protobothrops flavoviridis (BAA11888.1), Trimeresurus stejnegeri (Q71RQ9.1 and Q71RR1.1), and Crotalus oregano helleri (AEU60005.1) is shown in Table 7. The D. acutus anticoagulant B protein binds Factor X and shares several motifs with Site A. Blood factor IX/factor X Binding Protein B chain of P. flavoviridis, contains a C-type lectin (γ-carboxyglutamic acid recognition) GLA-binding sequence, 0121YFKSTNNKWRS. These residues are highlighted in bold in sequence identified as fIXBP in Table 7. Analogues occur as MVKKHKENLRTW in snake venom Zinc metalloproteinase-disintegrin-like endopeptidases (ADAMS) (65, 66), and as QYKKNKHRHSITN in apo B100 (a part of Site A). This region of apo B100 shares several motifs with its analogues in pit viper venom proteins, notably LSVK, QYKKN, HRH, SIT, and VLCEFIS. Variations of these motifs appear in other venom proteins as well and are indicated in Table 7.
Table 7.
Comparison of Apo B100 Potential Procoagulant Inhibitor Region to Sequences in Snake Venom ADAMS Proteins.
| Source | Protein | Procoagulant Inhibitor Region |
|---|---|---|
| Ettelaie | KRAD-14 | 3146KAQSKKNKHRHSTT |
| Ettelaie | Mut-4 | 0000KAQSKKNAHRHSTT |
| AC010872 | ApoB100 | 3119LKDFSLWEKTGLKEFLKTTKQSFDLSVKAQYKKNKHRHSITNPLAVLCEFIS3170 |
| 1IOD_B | fXBP | 0070WQCNWQWSNAAMLKYTDWAEESYCVYFKSTNNKWRSITCRMIAN*FVCEFQA0123 |
| BAA11888 | fIXBP | 0096NQCNWQWSNAAMLRYKAWAEESYCVYFKSTNNKWRSRACRMMAQ*FVCEFQA0146 |
| Q71RQ9 | SLBB1 | 0096NACKLQWSDGTELKYNAWSAESECITSKSIDNQWFTRSCSQTYP*FVCKFQA0146 |
| Q71RR1 | SL9B2 | 0096NQCNWQWSNAAMLRYKAWAEESYCVYFKSTNNKWRSRSCRMMA*NFVCEFQV0146 |
| AEU60005 | CROTALUS | 0096NKCSWQWSDGAMLKYEDWAEESYCVYFKSTNNKWRSRACRMDA*HFVCEFQA0146 |
| 0116ESYCVYFKSTNNKWRSRSCRMMA*N | ||
| AC010872 | ApoB100 | 3118PLKDFSLWEKTGLKEFLKTTKQSFDLSVKAQYKKNKHRHSITNPLAVLCEF3168 |
| Q3HTN2 | ADAM III | 0182KASQLVVTAEQQRFPRRYVKLAIVADHRMYTKHKKNLKPWVFQMVNSVHQI0232 |
| BAP39921 | ADAM II | 0004KASQLVVTAEQQRFPRRYVKLAIVADHRMVKKHKENLRTWVFQMVNSVNQM0054 |
| AAP23053 | ADAM II | 0181KASQLYLTPEQQRFPQRHIELAIVVDHGMYTKYSSNFKKIRKRVHQMVSNI0231 |
| 0116ESYCVYFKSTNNKWRSRSCRMMAN |
Peptides KRAD-14, Site A of apo B100, Mut-4, a mutate version, and test peptide B3119 (AC010872), used in the present study, are compared to similar sequences in snake venom anticoagulant proteins of Deinaqkistrodon acutus (1IOD_B), Factor IX/factor X binding protein B chain of Protobothrops flavoviridis (BAA11888), hemostasis impairing toxin of Trimeresurus stejnegeri (Q71RQ9.1, Stejneger’s pit viper), Stejneger’s Coagulation Factor IX/factor X-binding protein (Q71RR1.1), C-type lectin 3 of Crotalus oreganus helleri (AEU60005), cell adhesion impairing toxin of T. stejnegeri (Q3HTN2), and metalloproteases of P. elegans (BAP39921 and AAP23053).
B3347 - Apo B100 LDL Receptor Ligand Site B.
Peptide B3347 that spans residues 3347 to 3389 contains a Type I motif, TRLTRKRGLK that was initially designated as Site B ligand (7, 8, 19). B3347 peptide displays low binding avidity, and signal appears on cell surface and in the extracellular spaces. A labeled synthetic peptide of Site B was shown to bind heparin and dextran sulfate cellulose; hence, this region may bind GAGs and highly sulfated proteoglycans (43).
Comparison of B3347 sequence to the established LDLR ligand sequence in apo E and similar sequences in unrelated proteins is shown in Table 8. In apo E, Lysine residues in positions 0143 and 0146 (KXXK) are critical in binding LA modules 4 and 5 of the LDLR (32) and are in a Type I motif. Although Site B of B100 and the other proteins listed in Table 8 contain a Type I receptor ligand motif, the Lysine residues in equivalent positions are not conserved. The slight difference between Site B in apo B100 (hence, in B3347) and the apo E ligand sequence is reflected by their significant difference in cell binding.
Table 8.
Sequence Comparison of RKR Motif Region of Apo B100 Site B to Similar Sequences in Apo E, Msx2-IP, Dengue Virus Capsid and Helicase Proteins.
| Accession | Protein | Site B - Apo B Putative Receptor Ligand |
|---|---|---|
| AC010872 | apo B100 | 3347DALQYKLEGTTRLTRKRGLKLATALSLSNKFV3378 |
| LPHUE | apo E | 0131EELRVRLASHLRKLRKRLLRDADDLQKRLAVY0163 |
| NP055816 | Msx2IP | 1919RSPVKEPVEQPRVTRKRLERELQEAAAVPTTP1948 |
| NP055816 | MMsx2IP | 1352DVSFPNSIIKRDSLRKRSVRDLEPGEVPSDSD1383 |
| ADI80662 | dnv3CP | 0001MNNQRKKTGKPSINMLKRVRNR0022 |
| AC010872 | apo b100 | 3347DALQYKLEGTTRLTRKRGLKLATALSLSNKFVEGS3378 |
| BAD42414 | dnvl Helil | 1675KTRRYLPAIVREAIKRK*LRTLVLAPT**RVVASE1706 |
| ACY70846 | dnv2 Helil | 1674KTKRYLPAIVREAIKR*GLRTLILAPT**RVVAAE1705 |
| ADI80662 | dnv3 Helil | 1673KTRKYLPAIVREAIKRR*LRTLILAPT**RVVAAE1704 |
| AAW51421 | dnv4 Helil | 1669KTKRILPSIVREALKRR*LRTLILAPT**RVVAAE1700 |
Regions of high similarity with Site B are underlined. The LRK motif, present in tandem in apo E, and occurs as LKR in the dengue capsid proteins. The RKR motif of apo B100, apo E and Msx2-IP is not identical in the Dengue Helicases; however, sequence of the region that follows is highly similar to that in apo B100. This similarity is not shared with apo E or the Msx2-IP sequences.
Control Assays – Influence of PBS, 10% FBS, and 3% FBS-mevastatin.
Three sets of control cell binding assays were included to distinguish between receptor and non-receptor binding for peptides B0582, B3181, B2366, B3119 (Site A), and B3347 (Site B). Cells were propagated in six well trays as described in Methods. One set of trays was preconditioned in 3% FBS-DMEM plus mevastatin and two sets were maintained in 10% FBS-DMEM less mevastatin. Mevastatin is known to cause upregulation of LDL receptors (47, 48). Cultures were rinsed thrice with PBS then incubated in3%FBS-DMEM + B peptide (for mevastatin-treated cells only), and 10% FBS-DMEM + B peptide or PBS + B peptide. Results are shown in Table 9 are based on average values obtained from 3–4 different view fields processed using ImageJ as described in Methods.
B0582 Control Assays.
Values for Total Loci/ cell and % signal area seen in FBS-containing assays are approximately 3.7 and 2.5 times higher (respectively) in mevastatin-treated versus untreated cells. PBS alone assays show substantial increase in % signal area (approximately 6.5 and 16.6-fold increase compared to mevastatin-treated and untreated cells, respectively). B0582 is similar to the heparin binding region of FGF1; however, it also contains three potential LDL receptor ligand motifs, LKK, VKE, and LKE, analogues of receptor ligand motifs in RAP. Hence, both properties, GAG-binding and Receptor binding motifs, may contribute to binding in these assays. Pre-treatment with mevastatin may enhance binding via the proposed receptor ligand motifs in the sequence.
B3181 Control Assays.
In assays with FBS, the total loci number and the %signal area per cell are substantially (approximately 4 times) higher in mevastatin-treated cells. In absence of mevastatin treatment, these values are approximately 1.8–2.1 times higher in PBS alone assays compared to 10%FBS assays. B3181 is a Lysine-rich sequence that contains three RAP-like ligand motifs and one Type III motif. Cell binding characteristics observed for B3181 in the 3% FBS-mevastatin assays are the highest values obtained with other B peptides used in these control studies. Strong influence of mevastatin treatment on B3181 binding suggests involvement of the LDL or related receptors interaction of this peptide with the cells.
B2366 Control Assays.
Treatment of cells with mevastatin enhances binding of peptide B2366 in FBS-containing assays (approximately 4- and 2-fold, using the %area and the number of loci per cell, respectively). This peptide contains RAP-like receptor ligands motifs, VKK and LKK, and binds HeLa cells in 3%FBS-mevastatin and PBS media almost as well as B3181. The data suggest involvement of LDL receptor in interaction of this peptide with the cells.
B3119 Control Assays.
The effect of mevastatin on binding of B3119 (site A) was not significant. Values for FITC loci per cell, % signal area and IDS per cell are only slightly higher in the mevastatin-treated cells compared to PBS alone assays.
B3347 Control Assays.
Treatment with mevastatin enhances binding of B3347 (2.0- and 2.2-fold increase in loci number per cell and % signal area, respectively, compared to untreated cells), however, the integrated signal density per cell does not change significantly. This peptide contains a Type I receptor/GAG binding motif but lacks motifs present in the receptor ligand region of apo E or any rendition of a RAP receptor ligand.
Influence of E2 synthetic peptide on binding and internalization of B-peptides.
The LDL receptor-related protein gene family is vast. Each member protein contains multiple ligand adhesion modules (LA), also complement-type repeats. The ligand moiety in apo E binds ligand modules 3–5 of LDLR (67) while apo B100 putatively binds LRs 3, 4, 5, 6 and perhaps 7 of the LDLR (68). HeLa cell assays were used to assess qualitatively the influence of unlabeled synthetic E2 peptide on binding of apo B peptides. E2 peptide spans residues 0129Ser – Gly0165 of apolipoprotein E and includes two in tandem copies of LRK, an analogue of RAP motif 0304VKK. In these experiments, the 100 μL cell-binding assay was prepared using 15 μg of E2 peptide, and 10 μg of apo B peptide in PBS with 5 mM MgCl2. Control assays without E2 were included for each apo B peptide. Cells were incubated, fixed and images were obtained as described in Methods. Images were subjected to analysis using ImageJ as before. Bright Field and fluorescence overlay images in Figure 3 show results obtained in assays that included E2 with peptides all peptides listed in Table 1 except for B3181 (depleted supply).
E2 peptide and B0013-FITC peptide HeLa cell Interaction.
B0013 contains on analogue of the RAP receptor ligand motif VKK as LRK identical to that in E2 which contains two copies; hence, the unlabeled E2 and B0013 ratio is approximately 3 to 1. Addition of E2 peptide reduced the number of signal emitting sites by 70%, while total area of signal was only reduced by ~9%, average size of area emitting signal and integrated density were correspondingly increased by 3.2 fold. In both assays signal appears to be in clusters and on the cell surface.
E2 peptide and B0055-FITC peptide HeLa cell Interaction.
Addition of E2 to assays with B0055-FITC increased the number of sites emitting signal by approximately 2.7 and total area of signal by 4.4 fold while total integrated density was increased by only 5%. In both overlay images shown in Figure 3, FITC signal appears to be on the cell surface and the presence of E2 appears to increase the size of the clusters but not the intensity of signal.
E2 peptide and B0575-FITC peptide HeLa cell Interaction.
Peptide B0575 includes three analogues of RAP receptor ligand motifs. Although E2 peptide increase the number of sites emitting B0575 signal 6 fold the average locus size and total integrated density values were deceased by 86%. Enlarged regions of corresponding overlay images in Figure 3 show that in both assays the signal appears on the cell surface.
E2 peptide and B0582-FITC peptide HeLa cell Interaction.
Fluorescence images of B0582, which includes B0575 receptor ligand analogue motifs and a potential endocytic motif as FRKF, show a relatively high background on the extracellular matrix, signal is also emitted from the cell surface and in the cytoplasmic space. These are characteristic in images of B0582 and B0582 E2. E2 increased the number of fluorescence loci 3 fold on B0582 alone and the average size of the loci and integrated density values in B0582 E2 assay are about 33% of the B0582 assay. Hence, E2 appears to compete with both B0575 and B0582.
E2 peptide and B0708-FITC peptide HeLa cell Interaction.
B0708 has 0716DKH, 0731EKL, 0734IKD, and 0737LKS that are corresponding analogues in RAP, but this apo B sequence does not appear to contain a functional endocytic motif. Although E2 does not increase binding of B0708, its presence mediates endocytosis. In Figure 3, B0708 E2 frame, signal is seen in the extracellular space at discrete loci, on the cell surface, in the cytoplasm, and perinuclear.
E2 peptide and B2054-FITC peptide HeLa cell Interaction.
Potential receptor ligand motifs LKH and VRK are in B2054. This is low binding peptide covering only about 0.1% of the total area; E2 reduces this value by 70%. Average size and integrated density of loci are reduced by 77%. E2 does not appear to enhance cell entry for B2054.
E2 peptide and B2366-FITC peptide HeLa cell Interaction.
B2366 is replete with two RAP receptor ligand, VKK and IKK, and several types of endocytic motifs. Addition of E2 enhances binding, uptake and intracellular distribution of the signal changes substantially (Figure 3). Concomitantly signal loci and total area of signal are slightly reduced by 15% and 5%, respectively; while the average size of loci and integrate density are increased by 10%. In a representative overlay image B2366 E2 shows signal from apparent cell surface, in endosome-like structures in the cytoplasm and periphery of the nucleus, and in the nucleus. A trace of FITC signal appears associated with extracellular materials.
E2 peptide and B2932-FITC peptide HeLa cell Interaction.
B2932 contains motif 2937SKHLRV, an analogue of B0013 motif, 0018FKHLRK (see Table 1). Three renditions of the KHL motif occur in RAP. In the control assay, B2932 overlay, discrete microvesicle-like structures are seen in the extracellular space. B2932 E2 overlay also shows signal emanating from extracellular regions. Interestingly, approximately 30% of cells emitting signal in the related 630× magnification view field of the B2932 E2 overlay appear to be undergoing apoptosis, two are seen at the upper left corner of the corresponding frame in Figure 3. This effect was not evident in B2932 alone assays. B2932 E2 frame also shows intense signal associated with what appears to be the nucleolus and chromatin, and appears as spheres at the nuclear envelope. E2 reduced the number of signal loci by half. Total area emitting signal and integrated density values were increased by 81%.
E2 peptide and B3119-FITC peptide HeLa cell Interaction.
E2 increased integrated density of signal in the view field of B3119 as about 8.9 fold. B3119 overlay in Figure 3 shows signal in the cell cytoplasm in discrete endosome-like structures, with a few located at the nuclear periphery. Addition of E2 peptide enhanced intracellular uptake and altered discrete site distribution with vibrant signal in cytoplasm, organelles, and in the nucleus appears associated with chromatin and nucleolus. B3119 E2 frame also shows signal emanating from vesicles in the extracellular space.
E2 peptide and B3347-FITC peptide HeLa cell Interaction.
In Figure 3 frames for B3347 and B3347 E2 show that E2 appears to influence binding of B3347 to HeLa cells but cell entry is not discernable in either frame. E2 increases number of signal loci by 8.6 fold and total signal area by almost 2 fold. Average size of loci and integrated density values are decreased 78%.
B3347 peptide lacks a legitimate RAP receptor ligand; however, it contains a Cardin-Weintraub Type I GAG binding motifs (32, 33). It is possible that E2 opens an increased number of receptors providing more sites for B3347 binding. Another consideration is that motifs, YKL, TRK, and NKF, are actually weak binding ligands.
Co-localization of CM-DiI LDL and Peptides B0582, B3181, B2366, B3119 and B3347.
Experiments were performed to characterize binding of FITC-labelled peptides in presence of intact cm-DiI-labelled LDL particles, excitation/emission 553/575nm, expected possible scenarios included competition for receptors, sharing of receptors, or binding to receptors independently. Each image was then processed as described previously. Results of these experiments are shown in Figures 4, 5, and 6.
Figure 5.
Binding of peptide B0582 to HeLa cells in the presence of CM-DiI LDL. Top row: overlay images of FITC-B0582/Bright Field and CM-DiI LDL/Bright Field are shown in Frames A and B, respectively. White arrows in these frames indicate colocalized signals in clusters that are apparently located on the cell surface. These appear white in Frame A and yellow-greenish in Frame B due to intensity of fluorescence enhance by ImageJ. Bottom row: Enlarged region of Frames A and B was selected to show non-colocalized signals indicated by red arrow in Frames C and E. FITC-B0582 signal in Frame C appears on cell surface, in apparent endosome-like vesicles, and in the extra-cellular spaces. In contrast, CM-DiI LDL signal in Frame E appears in clusters on the cell surface and in the cytoplasm. Frame D, a tri-overlay image, clearly shows green fluorescence signal of FITC-B0582 peptide is apart from intense, yellowish signal emitted by CM-DiI LDL.
CM-DiI LDL and FITC-B2366 HeLa cell Interaction.
Figure 4 shows B2366 peptide signal (green) and the LDL signal (red and yellowish-orange for intense fluorescence), in Frames A and Frame B, respectively. Green FITC signal is emanating from the cell surface, cytoplasm and nucleus. Enhanced contrast methods, e.g. auto contrast command in ImageJ, show signal on the surface boundary of practically all cells in the view field. Clusters of the peptide appear on the surface of a few cells. Enlarged regions of B2366 image, in Frame C, show green signal emanating from locations in the cytoplasm, peripheral to nuclear envelope, and from the nucleus. Frames E, the corresponding overlay of the Bright Field and the CM-DiI LDL images, shows red signal at discrete loci on the cell surface in the cytoplasm. The green/red/Bright Field overlay image in Frame D shows B2366 and LDL signals occur separately, with LDL signal appearing in clusters on the membrane as well as in a few instances inside the cell.
CM-DiI LDL and FITC-B0582 HeLa cell Interaction.
Figure 5 shows results obtained for B0582 with LDL; Frame A and Frame B show green and red signals, respectively. Frame C shows an enlarged region from Frame A with white arrow for reference. Here FITC signal is shown in distinct endosome-like vesicles in the cytoplasm and in the nucleus. The corresponding CM-DiI signal for LDL is shown in Frame E. This signal occurs in large clusters on the cell surface with high intensity as well as in the cytoplasm and nucleus. The overlay of the Bright Field, red and green fluorescence images (Frame D) shows that most of the B0582 signal appears separated from LDL clusters on the cell surface and from the endosomes in the cell proper. Co-localized signals in frames A and B are indicated by red arrows. White arrow indicates enlarged region shown in frames C, D, and E.
CM-DiI LDL and FITC B Peptides 3181, B3119 and B3347 HeLa cell Interaction.
Results for B3181, B3119 and B3347 are shown as triple image overlays, Bright Field on FITC on CM-DiI, in Figure 6. These peptides showed low binding capacity with few sites emanating signal. In Frame B3181 signal appears as large, discrete aggregates (red arrows) on the cell surface and as endosome-like structures in the cytoplasm as seen also in Figure 2. Co-location of B3181 and LDL signals is seen at few sites and appear to be only outside the cell. B3119 peptide also appears in large, high intensity signal aggregates alone as well as co-located with LDL signal. The LDL signal emanates from the majority of the cells in the view field while B3119 signal is not. Signal for B3347 appears in a few high density aggregates and small discrete points on the cell surface and in a few instances also appears to be in the cytoplasm. There is a trace of signal for this peptide in the extracellular spaces. LDL signal is throughout the view field on the cell surface, in the cytoplasm and outside the cell proper. Red arrows indicate co-localized signals in the three frames of Figure 6.
DISCUSSION
Hydrophilic synthetic peptides of regions in apoB100 sequence are useful in establishing its potential functional domains. Our initial aim was to identify the regions in apo B100 that impart receptor binding capacity to the LDL particles. The apo B100 sequence contains 148 Arginine residues and 356 Lysine residues, roughly 11% of the molecule. All peptides used in the present study are rich in basic amino acids and therefore all bind non-specifically to dextran sulfate cellulose beads. Arginine residues bind 2.5× more tightly to GAGs than Lysine (34). In apo E Lysines in positions 143 and 146 are critical residues for high affinity binding to LDL B/E receptor (35, 36).
Previously, Cardin-Weintraub ligand motifs were used to identify potential GAG/receptor ligands (32 – 34) in apo B100, Site A, 3145VKAQYKKNKHRHS3157 and Site B, 3351YKLEGTTRLTRKRGLKL3367 (7, 8, 18, 19, 42). Peptides obtained using proteolytic digestion showed two sites, 3134Leu-Lys3209 and 3356Thr-Ala3489, with high binding affinity for heparin; and, hence, were considered to possess LDL receptor binding capacity also (33). A separate study showed four murine monoclonal antibodies, Mabs 2A, 5A, 7A, and 9A, to epitopes at three different regions in apo B100, reduced LDL interaction with the receptor by “up to 95%” (44). These apo B100 regions are defined as follows: 5A, 1480Asp-Val1693; 2A and 7A, 2152Gln-Asn2377; and 9A, 2657Gln-Lys3248. There is only one motif, DKI, in the sequence spanning 1480Asp-Val1693 and covered by Mab 5A, which is also in RAP. However, the sequence 2152Gln-Asn2377, Mab 7A epitope, contains numerous analogues of this RAP motif. This region of apo B100 includes motifs 2331DKL and 2342LKE, present in RAP, positions 0136–0138 and 0190–0192. Sequence 2657Gln-Lys3248, Mab 9A epitope, includes our peptides B2932, B3119 and B3181. Therefore, our findings indirectly support the results of the Mabs study. A major drawback is that the monoclonal antibody “footprint”, i.e. molecular size, may obscure the actual ligand site residues that are remote to the epitope.
Proposed Receptor Binding Regions in Apo B100.
Six short stretches of the apo B100 sequence, B0013, B0582, B0708, B2366, B2932 and B3181, bind HeLa cells with high affinity. The sequence comparison in Table 3 shows many similar motifs and short sequences shared between these stretches of apo B100 and RAP. RAP is an ER resident chaperone protein that is involved in the proper intracellular folding of the LDL receptor and related proteins (69). RAP putatively binds to all members of the LDL receptor family (55). RAP is comprised of 357 amino acids in three distinct, 100-residue domains. Each domain contains three helical structures. Three residue Lysine-based motifs, e.g. DKL, EKH and VKK, in RAP that are involved in binding ligand adhesion modules, are common in apo B100. Different renditions of the polar sequence, 0024YTYNYEAESSS in apo B100, are also in apo E2 peptide, RAP and the DNA-binding domains of the IRF proteins. Effects of E2 peptide, containing known receptor ligands, on B peptides interaction with HeLa cells, both competition and enhancement of binding, support presence of functional receptor ligand motifs in the B peptides.
Four regions in apo B100, residues 0013–0094, 0705–0805, 2932–3031 and 3120–3216, may also function as nucleic acid binding domains. Each region contains motifs that are present in the DNA binding domain of the IRF proteins. Previously, highly purified LDL was shown to bind plasmid DNA and RNA (25), and DNA binding capacity for the N-terminal region of apo B100 was confirmed experimentally using synthetic peptides for this region (26).
Apo B100 sequence also contains eleven K homology (KH) modules typical in RNA-binding proteins including vigilin (Q00341), which are identified by motif GXXG. Herein, we identify two other regions that appear to be RNA Recognition Motifs based on the similarity of the amino- and carboxyl- termini of known RRM domains in MINT and apoBEC-1 complementation factor.
Potential Anticoagulant Function of Apo B100 Site A, KAQYKKNKHRHSITN.
Anticoagulant function for this region of apo B100 was proposed previously by Ettelaie, et al (63, 64). Those studies showed that a synthetic peptide spanning apo B100 region 3146Lys- and Thr3160 inhibits Tissue Factor - Factor VII/VIIa procoagulant activity. Peptide KRAD-14 and Mutant 4 (63) were shown to inhibit procoagulant activity by 98–99% and suggest “KKN” and “HRHS” as essential motifs in the process. Analogues of both motifs are present in snake venom Blood Factors IX and X binding proteins and in ADAMS proteins that are associated with hemorrhagic phenomena. We surmise that upon injury to the arterial wall and/or endothelium, neutrophils and keratinocytes release elastase, which cleaves the molecule at Val3145 or Ala3147, and Ala3163 or Val3164 to liberate a functional anticoagulant Site A peptide.
What is the actual function of Apo B100 Site B, TRKRGLKL?
Apo B100 sequence spanning 3347Asp – Lys3366 is highly similar to apo E receptor ligand sequence 0131Glu - Arg0150, both sequences contain a Type I ligand motif. Reproducible experimental evidence indicates that Site B100 is not the preferred ligand for the receptors in HeLa cells. The comparison of these sequences in Table 8 reveals two differences that suggest different functions. Apo E contains two copies of the motif LRK in tandem. Site B lacks this motif and does not contain any of the other Lysine-based ligand motifs present in RAP. The apo B100 sequence that follows, 3367Leu - Val3378, is significantly different from 0151Asp – Tyr0163 that trails the receptor ligand sequence in apo E, and is more similar to the sequence, 1692Leu – Glu1706 of the NS3 helicase region of the dengue polyprotein (BAD42414). This difference in primary structure suggests a difference in the secondary structure of Site B region, i.e. this segment is part of an amphipathic helix in apo E but is a β-strand in the dengue (70). Synthetic peptides that include Site B were previously shown to bind plasmid DNA (26). These observations suggest this apo B100 region may function as a helicase.
The RKR motif of apo B100 Site B is also contained twice in MINT, a DNA-binding protein, and twice in dengue 1 polyprotein, at the Carboxyl-termini of the capsid protein and NS3 peptidase region. This motif may also indicate a separate function in both dengue and apo B100. It also occurs at the N-terminal region of human intermediate conductance calcium-activated potassium channel, hIK1 (AAC23541), and plays a role in its function (71). The RKR motif, located at the C-terminus of the ATP-sensitive inward rectifier potassium channel 11 (KCJ11_HUMAN), is an ER localization signal that prevents ER exit of unassembled membrane protein complexes (72). Also, intriguing is the potential that RKR and related analogues, KKK, KRK, and RKR in apo B100, and carboxyl-terminal KKXX motifs, two occur in apo B100 at positions 4457 and 4512, are intracellular signaling motifs, i.e., endoplasmic reticulum localization signals (73). These motifs function in partitioning of proteins to designated subcellular compartments (74).
Are Apo B100 lipoparticles involved in the immune response?
Apo B100 is a member of the Large Lipid Transfer Protein (LLTP) superfamily, which includes vitellogenin-like LLTPs, vigilin, microsomal triglyceride transfer protein - MTPs (9, 10), and insect apolipophorins (11, 12). LLTP molecules are involved in extracellular transport of hydrophobic lipids and triglycerides (9, 10), binding nucleic acids (15–17), and appear to be involved in the immune response (12–14). LLTPs, including apo B100, share a common ancestor (9) and therefore contain similar primary structures that impart similar functions. For example, Vigilin contains 13 K-homology nucleic acid binding domains as compared to 11 similar sequences in apo B100, both molecules have been shown to bind RNA (25, 15–17). VTG A2 also contains 10 K-homology RNA binding motifs. K-homology nucleic acid binding domains are characterized by the “GXXG” motif. A motif contained in Vitellogenin, Vigilin and apo B100 is “GXXGXG”, 0680IEIGIRGEGIE, 0986YVIGQKGSGIR, and 0660IEIGLEGKGFE, respectively.
There is also significant sequence similarity of N-termini of apo B100, vitellogenin and the DNA-binding domains of the IRF proteins. The N-terminal sequence of Vitellogenin-A2, VTG A2, is compared to the DNA-binding domains of the IRF proteins and the N-terminal region of apo B100 in Table 4. These observations support the concept that LLTPs play a role in the immune response.
Apo B100 is likely a component of a circulating acute-phase response mechanism, an element of an emergency control and repair unit/package, always ready on site to address any injury/assault to the tissue. A multi-functional apo B100 may work in concert with apo E, and other “riders” lipids, phospholipids, cholesterol, steroids, vitamins, and other metabolites necessary to maintain the integrity of the organism by repairing tissue, removing foreign matter, and alerting the immune system. We speculate that lipids, phospholipids and cholesterol may facilitate tertiary structural transformation of apo B100 into a conformation necessary to address a specific need. We surmise that upon injury endopeptidases present on site and/or secreted by keratinocytes and neutrophils may release functional regions of apo B100.
Supplementary Material
ACKNOWLEDGEMENTS AND GRANT SUPPORT
All phases of this study were performed by the authors at the Biophysics Research Laboratory at UTB and subsequent UTRGV. Studies were supported in part by grants F49620–99–1–0327 (AFOSR, PI – J. Guevara), and FA 9550–05–1–0472 (AFOSR, PI - A. Hanke), and SC3GM099637 (NIH, PI – N. Guevara). Mr. Ernesto Hernandez donated his time and effort on this project. Mr. Jaime Romo, Jr., was supported by R25GM083755 (NIH, PIs – J. Facelli, S. Nair).
Funding: All phases of this study were performed by the authors at the Biophysics Research Laboratory at UTB and subsequent UTRGV. Studies were supported in part by the following grants:
F49620–99–1–0327 (AFOSR, PI – J. Guevara), and FA 9550–05–1–0472 (AFOSR, PI - A. Hanke) – contributed to purchase of equipment used in the studies; SC3GM099637 (NIH, PI – N. Guevara) – contributed to salaries (N.Guevara, J.Guevara), and purchase of supplies.
Mr. Ernesto Hernandez donated his time and effort on this project.
Mr. Jaime Romo, Jr., was supported by R25GM083755 (NIH, PIs – J. Facelli, S. Nair).
Abbreviations:
- ACF
apo BEC complementation factor
- ADAMS
A Disintegrin-like And Metalloprotease motifs
- Apo BEC
apolipoprotein B editing complex
- ASP
APOBEC-1 Stimulating Protein
- CCD
charge-coupled device, e.g. digital camera
- CM-DiI
C68H105Cl2N3O – red fluorescent dye
- GAG
glycosaminoglycan
- GLA
gamma-carboxyglutamic domain
- HSP
heat shock protein
- IRF
interferon regulatory factor
- JAK1
Janus Kinase 1
- LRP
Low Density Lipoprotein Receptor-related Protein
- LLP
Low Density Lipoprotein Particle
- MESD
mesoderm development protein
- MINT
Muscle Segment Homeobox Interacting Nuclear Target
- MSX2
Muscle Segment Homeobox DNA-binding transcriptional repressor
- OCFRE
Osteocalcin Fibroblast Growth Factor Responsive Element
- RAP
LDL Receptor-Associated Protein
- RRM
RNA-Recognition Motif
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
- 1.Chapman MJ 1980. Review. Animal lipoproteins: chemistry, structure, and comparative aspects. J. Lipid Res 21:789–853. [PubMed] [Google Scholar]
- 2.Prassl R, and Laggner P. 2012. Lipoprotein Structure and Dynamics: Low Density Lipoprotein Viewed as a Highly Dynamic and Flexible Nanoparticle. Chapter 1. In Lipoproteins – Role in Health and Diseases Frank S and Kostner G, editors. INTECH Biochemistry, Genetics and Molecular Biology, OPEN ACCESS 3–20. 10.5772/48145 [DOI] [Google Scholar]
- 3.Chapman MJ, Goldstein S, Lagrange D, and Laplaud PM. 1981. A density gradient unltracentrifugal procedure for the isolation of the major lipoprotein classes from human serum. J. Lipid Res 22:339–358. [PubMed] [Google Scholar]
- 4.Karlsson H, Leanderson P, Tagesson C, and Lindahl M. 2005. Lipoproteomics I: Mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 5:551–565. [DOI] [PubMed] [Google Scholar]
- 5.Sun H-Y, Chen S-F, Lai M-D, Chang T-T, Li P-Y, Shieh D-B, and Young K-C. 2010. Comparative proteomic profiling of plasma very-low-density and low-density lipoproteins. Clinica Chimica Acta 411:336–344. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein JL, Anderson RGW, and Brown MS. 1979. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 279:679–685. [DOI] [PubMed] [Google Scholar]
- 7.Knott TJ, Pease RJ, Powell LM, Wallis SC, Rall SC Jr., Innerarity TL, Blackhart B, Taylor WH, Marcel Y, Milne R, Johnson D, Fuller M, Lusis AJ, McCarthy BJ, Mahley RW, Levy-Wilson B, and Scott J. 1986. Complete protein sequence and identification of structural domains of human apolipoprotein B. Nature 323:734–738. [DOI] [PubMed] [Google Scholar]
- 8.Yang C-Y, Chen S-H, Gianturco SH, Bradley WA, Sparrow JT, Tanimura M, Li W-H, Sparrow DA, DeLoof H, Rosseneu M, Lee F-S, Gu Z-W, Gotto AM Jr., and Chan L. 1986. Sequence, structure, receptor-binding domains and internal repeats of human apolipoprotein B-100. Nature 323:738–742. [DOI] [PubMed] [Google Scholar]
- 9.Babin PJ, Bogerd J, Kooiman FP, Van Marrewijk WJA, and Van der Horst DJ. 1999. Apolipophorin II/I, Apolipoprotein B, Vitellogenin, and Microsomal Triglyceride Transfer Protein Genes Are Derived from a Common Ancestor. J. Mol. Evol 49:150–160. [DOI] [PubMed] [Google Scholar]
- 10.Smolenaars MMW, Madsen O, Rodenburg KW, and Van der Horst DJ. 2007. Molecular diversity and evolution of the large lipid transfer protein superfamily. Review. J. Lipid Research 48:489–502. [DOI] [PubMed] [Google Scholar]
- 11.Avarre J-C, Lubzens E, and Babin PJ. 2007. Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. BMC Evol. Biol 7:3 Doi: 10.1186/1471-2148-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zdybicka-Barabas A and Cytrynska M. 2013. Apolipophorins and insects immune response. Invertebrate Survival J 10:58–68. [Google Scholar]
- 13.Li Z, Zhang S, Zhang J, Liu M, and Liu Z. 2009. Vitellogenin is a cidal factor capable of killing bacteria via interaction with lipopolysaccharide and lipoteichoic acid. Mol. Immun 46:3232–3239. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Wang S, Li H, and Li L. 2011. Vitellogenin, a multivalent sensor and an antimicrobial effector. Internat. J. Bioch. & Cell Biol 43:303–305. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Zhang Z, Blackwell K, and Carmichael GG. 2005. Vigilins Bind to Promiscuously A-to-I-Edited RNAs and Are Involved in the Formation of Heterochromatin. Current Biol 15:384–391. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Wang Q, Chen L-L, and Carmichael GG. 2008. On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA 14:1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mobin MB, Gerstberger S, Teupser D, Campana B, Charisse K, Heim MH, Manoharan M, Tuschi T, and Stoffel M. 2016. The RNA-binding protein vigilin regulates VLDL secretion through modulation of Apob mRNA translation. Nature Comm DOI: 10.1038/ncomms12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knott TJ, Rall SC Jr., Innerarity TL, Jacobson SF, Urdea MS, Levy-Wilson B, Powell LM, Pease RJ, Eddy R, Nakai H, Byers M, Priestley LM, Robertson E, Rall LB, Betsholtz C, Shows TB, Mahley RW, and Scott J. 1985. Human Apolipoprotein B: Structure of Carboxyl-Terminal Domains, Sites of Gene Expression, and Chromosomal Localization. Science 230 (4721):37–43. [DOI] [PubMed] [Google Scholar]
- 19.Law SW, Grant SM, Higuchi K, Hospattankar A, Lackner K, Lee N, and Brewer HB Jr. 1986. Human liver apolipoprotein B-100: complete nucleic acid and derived amino acid sequence. Proc. Natl. Acad. Sci. USA 83:8142–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guevara J Jr., Walch ET, Epstein HF, Sparrow JT, and Valentinova NV Evidence That Apo B-100 of Low Density Lipoprotein is A Novel Src-related Protein Kinase. J. Protein Chemistry 14(7): 627–631, 1995. [DOI] [PubMed] [Google Scholar]
- 21.Guevara JG Jr., Valentinova NV, Yang C. y., Davison D, and Morrisett JD. 1995. Human lipoprotein[a]: Regions in sequences of apolipoproteins similar to domains in signal transduction proteins. J. Endocrine Practice 1(6):440–448. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Lin JH-C, Liao H-L, Friedli O Jr., Verna L, et al. , 1998. LDL Induces Transcription Factor Activator Protein-1 in Human Endothelial Cells. Arterioscler. Thromb. Vasc. Biol 18:473–480. [DOI] [PubMed] [Google Scholar]
- 23.Voyno-Yasenetskaya TA, Dobbs LG, Erickson SK, and Hamiltorn RL. 1993. Proc. Natl. Acad. Sci. USA 90:4256–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Relou IAM, Bax LAB, van Rijn HJM, and Akkerman J-WN. 2003. Site-specific phosphorylation of platelet focal adhesion kinase by low-density lipoprotein. Biochem. J 369:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guevara JG Jr., Kang D-c., and Moore JP. 1999. Nucleic Acid-Binding Properties of Low-Density Lipoproteins: LDL as a Natural Gene Vector. J. Prot. Chem 8 (18):845–857. [DOI] [PubMed] [Google Scholar]
- 26.Guevara JG Jr., Prashad N, Ermolinsky B, Gaubatz JW, Kang D-c., Schwarzbach AE, Loose DS, and Guevara NV. 2010. Apo B100 similarities to viral proteins suggest basis for LDL-DNA binding and transfection capacity. J. Lipid Res 51:1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, and Burley SK. 1999. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure 7:191–203. [DOI] [PubMed] [Google Scholar]
- 28.Grishin NV 2001. KH domain: one motif, two folds. Nucleic Acids Res 29 (3):638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Cam J, Bu G. 2001. Low-density lipoprotein receptor family; endocytosis and signal transduction. Mol. Neurobiol 23(1):53–67. [DOI] [PubMed] [Google Scholar]
- 30.Gent J, Braakman I 2004. Low-density lipoprotein receptor structure and folding. Cell. Mol. Life Sci 61:2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieckmann M, Dietrich MF, Herz J. 2010. Lipoprotein receptors - an evolutionarily ancient multi-functional receptor family. Biol Chem 11:1341–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardin AD, Hirose N, Blankenship DT, Jackson RL, and Harmony JAK. 1986. Binding of High Reactive Heparin to Human Apolipoprotein E: Identification of Two Heparin-Binding Domains. Biochem. Biophys. Res. Comm 134 (2):783–789. [DOI] [PubMed] [Google Scholar]
- 33.Cardin AD, and Weintraub HJR. 1989. Molecular Modeling of Protein-Glycosaminoglycan Interactions. Arterioscl 9:21–32. [DOI] [PubMed] [Google Scholar]
- 34.Hileman RE, Fromm JR, Weiler JM, and Linhardt RJ. 1998. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Review. BioEssays 20:156–167. [DOI] [PubMed] [Google Scholar]
- 35.Zaiou M, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH, Segall ML, Phillips MC, and Lund-Katz S. 2000. Apolipoprotein E – low density lipoprotein receptor interaction: influences of basic residue and amphipathic α-helix organization in the ligand. J. Lipid Res 41:1087–1095. [PubMed] [Google Scholar]
- 36.Lalazar A, Weisgraber KH, Rall SC Jr., Giladi H, Innerarity TL, Levanon AZ, Boyles JK, Amit B, Gorecki M, Mahley RW, and Vogel T. 1988. Site-specific Mutagenesis of Human Apolipoprotein E. J. Biol. Chem 263(8): 3542 – 3545. [PubMed] [Google Scholar]
- 37.Lee D, Walsh JD, Migliorini M, Yu P, Cai T, Schwieters CD, Krueger S, Strickland DD, and Wang Y-X. 2007. The structure of receptor-associated protein (RAP). Protein Sci 16: 1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonifacino JS, and Traub LM. 2003. Signal for Sorting of Transmembrane Proteins to Endosomes and Lysosomes. Annu. Rev. Biochem 72:395–447. [DOI] [PubMed] [Google Scholar]
- 39.Kozik P, Francis RW, Seaman MNJ, and Robinson MS. 2010. A Screen for Endocytic Motifs. Traffic 11:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strickland DK, Gonias SL, and Argraves WS. 2002. Diverse roles for the LDL receptor family. Review. Trends Endocrin. Metab. 13 (2):66–74. [DOI] [PubMed] [Google Scholar]
- 41.Thompson GR, et al. 2010. Review. Efficacy criteria and cholesterol targets for LDL apheresis. Atheroscl 208:317–321. [DOI] [PubMed] [Google Scholar]
- 42.Hospattankar AV, Law SW, Lackner K, and Brewer HB Jr., 1986. Identification of Low Density Lipoprotein Receptor Binding Domains of Human Apolipoprotein B-100: A Proposed Consensus LDL Receptor Binding Sequence of Apo B-100. Biochem. Biophys. Res. Comm 139 (3):1078–1085. [DOI] [PubMed] [Google Scholar]
- 43.Weisgraber KH, and Rall SC Jr. 1987. Human Apolipoprotein B-100 Heparin-binding Sites. J. Biol. Chem 262 (23):11097–11103. [PubMed] [Google Scholar]
- 44.Fantappiè S, Corsini A, Sidoli A, Uboldi P, Granata A, Zanelli T, Rossi P, Marcovina S, Fumagalli R, and Catapano AL. 1992. Monoclonal antibodies to human low density lipoprotein identify distinct areas on apolipoprotein B-100 relevant to the low density lipoprotein-receptor interaction. J. Lipid Res 33:1111–1121. [PubMed] [Google Scholar]
- 45.Ali BR, Nouvel I, Leung KF, Hume AN, Seabra MC. 2010. A novel statin-mediated “prenylation block-and-release” assay provides insight into the membrane targeting mechanisms of small GTPases. BBRC 397:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen B, Piel WH, Gui L, Bruford E, Monteiro A. 2005. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 86:627–637. [DOI] [PubMed] [Google Scholar]
- 47.Bashmakov YK, Zigangirova NA, Pashko YP, Kapotina LN, and Petyaev IM. 2010. Chlamydia trachomatis inhibition and restoration of LDL-receptor level in HepG2 cells treated with mevastatin. Comp. Hepatol 9:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinson-Daza ML, Garzon R, Couraud PO, Romero IA, Weksler B, Ghigo D, Bosia A, and Riganti C. 2012. The association of statins plus LDL receptor-targeted liposome-encapsulated doxorubicin increases in vitro drug delivery across blood-brain barrier cells. British J. Pharma 167:1431–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindelin J, Rueden CT, Hiner MC, and Eliceiri KW. 2015. The ImageJ Ecosystem: An Open Platform for Biomedical Image Analysis. Mol. Reprod. Dev 82:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen JK, Dolmer K, Schar C, and Gettins PGW. 2009. Receptor-associated protein (RAP) has two high-affinity binding sites for the low-density lipoprotein receptor-related protein (LRP): consequences for the chaperone functions of RAP. Biochem. J 421: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasad JM, Young PA, and Strickland DK. 2016. High Affinity Binding of the Receptor-associated Protein D1D2 Domains with the Low Density Lipoprotein Receptor-related Protein (LRP1) Involves Bivalent Complex Formation. CRITICAL ROLES OF LYSINE 60 AND 191. J Biol. Chem 291(35): 18430–18439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher C, Beglova N, and Blacklow SC. 2006. Structure of an LDL-R-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors. Mol. Cell 22: 277–283. [DOI] [PubMed] [Google Scholar]
- 53.Bloem E, Ebberink EHTM, van den Biggelaar M, van der Zwaan C, Mertens K, and Meijer AB. 2015. A novel chemical footprinting approach identifies critical lysine residues involved in the binding of receptor-associated protein to cluster II of LDL receptor-related protein. Biochem. J 468: 65–72. [DOI] [PubMed] [Google Scholar]
- 54.Kohler C, Lighthouse JK, Werther T, Andersen O, Diehl A, Schmieder P, et al. 2011, The Structure of MESD45–184 Brings Light into the Mechanism of LDLR Family Folding. Structure 19:337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Juarez P, Comas I, Gonzalez-Candelas F, and Calvete JJ. 2008. Evolution of Snake Venom Disintegrins by Positive Darwinian Selection. Mol. Biol. Evol 25(11): 2391–2407. [DOI] [PubMed] [Google Scholar]
- 56.Korkmaz B, Horwitz MS, Jenne DE, and Gauthier F. 2010. Neutrophil Elastase, Proteinase 3, and Cathepsin G as Therapeutic Targets in Human Diseases. Pharma. Reviews 62(4): 726–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, and Hakoshima T. 1999. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J 18(18):5028–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y, Lu W, Mohamedali KA, Jang J-H, Jones RB, Gabriel JL, Kan M, McKeehan WL. 1998. The Glycine Box: A Determinant of Specificity for Fibroblast Growth Factor. Biochem 37:16506–16515. [DOI] [PubMed] [Google Scholar]
- 59.Blanc V, Henderson JO, Kennedy S, and Davidson NO. 2001. Mutagenesis of Apobec-1 Complementation Factor Reveals Distinct Domains That Modulate RNA Binding Protein-Protein Interaction with Apobec-1 and Complementation of C to U RNA-editing Activity. J. Biol. Chem 276(49):46386–46393. [DOI] [PubMed] [Google Scholar]
- 60.Mehta A, and Driscoll DM. 2002. Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing. RNA 8:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newberry EP, Latifi T, and Towler DA. 1999. The RRM Domain of MINT, a Novel Msx2 Binding Protein, Recognizes and Regulates the Rat Osteocalcin Promoter. Biochem 38:10678–10690. [DOI] [PubMed] [Google Scholar]
- 62.Sierra OL, Cheng S-L, Loewy AP, Charlton-Kachigian N, and Towler DA. 2004. MINT, the Msx2 Interacting Nuclear Matrix Target, Enhances Runx2-dependent Activation of the Osteocalcin Fibroblast Growth Factor Response Element. J. Biol. Chem 279(31):32913–32923. [DOI] [PubMed] [Google Scholar]
- 63.Ettelaie C, James NJ, Adam JM, Nicola KP, Wilbourn BR, and Bruckdorfer KR. 1998. Identification of a domain in apolipoprotein B-100 that inhibits the procoagulant activity of tissue factor. Biochem. J 333:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ettelaie C, Wibourn BR, Adam JM, James NJ, and Bruckdorfer KR. 1999. Comparison of the Inhibitory Effects of ApoB100 and Tissue Factor Pathway Inhibitor on Tissue Factor and the Influence of Lipoprotein Oxidation. Arterioscler. Thromb. Vasc. Biol 19:1784–1790. [DOI] [PubMed] [Google Scholar]
- 65.Takeda S, Takeya H, and Iwanaga S. 2012. Snake venom metalloproteinases: Structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta 1824:164–176. [DOI] [PubMed] [Google Scholar]
- 66.Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez P, and Sanz L. 2005. Snake venom disintegrins: evolution of structure and function. Toxicon 45:1063–1074. [DOI] [PubMed] [Google Scholar]
- 67.Guttman M, Prieto JH, Croy JE, and Komives EA. 2010. Decoding of Lipoprotein-Receptor Interactions: Properties of Ligand Binding Modules Governing Interactions with Apolipoprotein E. Biochem 49:1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Russell DW, Brown MS, and Goldstein JL. 1989. Different Combinations of Cysteine-rich Repeats Mediate Binding of Low Density Lipoprotein Receptor to Two Different Proteins. J. Biol. Chem 264(36):21682–21688. [PubMed] [Google Scholar]
- 69.Bu G, Geuze HJ, Strous GJ, and Schwartz AL. 1995. 39kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J 14(10): 2269–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, and Lescar J. 2008. Crystal Structure of the NS3 Protease-Helicase from Dengue Virus. J. Virol 82(1): 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones HM, Bailey MA, Baty CJ, MacGrego GG, Syme CA, Hamilton KL, and Devor DC. 2007. An NH2-Terminal Multi-Basic RKR Motif Is Required for the ATP-Dependent Regulation of hIK1. Channels 1(2): 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan H, Michelsen K, and Schwappach B. 2003. 14–3–3 Dimers Probe the Assembly Status of Multimeric Membrane Proteins. Curr. Biol 13: 638–646. [DOI] [PubMed] [Google Scholar]
- 73.Shikano S, and Li M. 2003. Membrane receptor trafficking: Evidence of proximal and distal zones conferred by two independent endoplasmic reticulum localization signals. Proc. Nat’l Acad. Sci. USA 100(10): 5783–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shikano S, Boblitz B, Sun H, and Li M. 2005. Genetic isolation of transport signals directing cell surface expression. Nature Cell Biol 7(10): 985–992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.