Abstract
Genetic variability in cagL gene especially within the Helicobacter pylori CagL hypervariable motif (CagLHM) may affect the development of gastric cancer. Therefore, this study was conducted to investigate the association of CagL diversity with clinical outcomes and with H pylori virulence markers. A total of 126 patients with different gastric diseases including non‐ulcer dyspepsia (NUD), peptic ulcer disease (PUD), gastric erosion (GE), and gastric cancer (GC) were enrolled. H pylori was cultured from gastric biopsies, and the isolates were screened for the presence of cagL, cagA, vacA, babA2, sabA, and cagPAI integrity by PCR. The amino acid polymorphisms of cagL were analyzed using DNA sequencing. We isolated 61 (48.4%) H pylori strains from 36 NUD, eight PUD, 12 GE, and five GC patients. Almost all isolates were cagL positive (97%), and their RGD, RHS, and SKIIVK motifs were highly conserved. Among 10 CagLHM variants identified, NEIGQ and NKIGQ were detected as the most prevalent sequences. Interestingly, a significant association was found between the presence of NKMGK and PUD (P = 0.002). Notably, the NEIGQ isolates with multiple C‐type EPIYA repeat that carried intact cagPAI correlated with disease risk for PUD, GE, and GC (P = 0.021). In conclusion, we identified novel variants of H pylori CagLHM sequences in Iranian population such as NKMGK, which was associated with disease risk for PUD. Further studies using a large number of strains are required to better clarify the function of certain CagLHM motifs in gastric carcinogenesis and disease outcome.
Keywords: cagL, CagLHM, disease outcome, diversity, Helicobacter pylori, Iran, virulence factors
1. INTRODUCTION
The spiral‐shaped microaerophilic bacterium, Helicobacter pylori, is classified as a group I carcinogen currently regarded as the most common etiologic cause of infection‐related cancers.1, 2 H pylori‐infected individuals have an increased risk of developing gastroduodenal diseases, including chronic gastritis, peptic ulcers, gastric adenocarcinoma, and gastric lymphoma.3, 4 This highly adapted human gastric pathogen is one of the most genetically diverse bacterial species and displays remarkable genetic variability and microevolution even among closely related strains due to high rate of mutation and recombination events.5, 6, 7, 8 Dozens of bacterial factors have been identified to promote the pathogenesis of H pylori infections, including cytotoxin‐associated gene A protein (CagA), vacuolating cytotoxin A (VacA), outer inflammatory protein A (OipA), and several putative adherence factors such as the blood‐group antigen‐binding adhesin (BabA) and sialic acid‐binding adhesin (SabA).9, 10, 11, 12
A hallmark of the most virulent H pylori strains is the presence of an intact cag pathogenicity island (cagPAI), which is associated with severe gastric pathologies including gastric mucosal inflammation, atrophy, and cancer.13, 14, 15, 16 The cagPAI is approximately 40 kb long and contains 28‐31 genes encoding a multi‐component bacterial type IV secretion system (T4SS).17, 18 After bacterial attachment, the T4SS delivers the CagA oncogenic protein and also peptidoglycan into the host gastric epithelial cells.18, 19 Upon translocation into the host cell, CagA undergoes tyrosine phosphorylation at its carboxy‐terminal Glu‐Pro‐Ile‐Tyr‐Ala (EPIYA) motifs by a variety of cellular kinases. Consequently, translocated CagA interferes with various cell signaling cascades that regulate cell‐cell adhesion, cell proliferation, and elongation and induces host epithelial cell secretion of potent proinflammatory chemokines such as interleukin (IL)‐8.20, 21
T4SS‐mediated CagA translocation across the host cell membrane depends on a number of bacterial and host cofactors such as CagL and human integrin β1‐containing receptors, particularly integrin α5β1.18, 22 CagL protein is a pilin‐like component of T4SS encoded by the cagL gene (HP0539) and is proposed to be expressed on the surface of H pylori in a T4SS‐dependent manner.17, 23 The arginine‐glycine‐aspartate (RGD) tripeptide motif at residues 76‐78 of CagL and its neighboring surface‐exposed FEANE (Phe‐Glu‐Ala‐Asn‐Glu) motif, referred to as RGD helper sequence (RHS), maximize proposed to be essential for T4SS interaction with integrin receptors for translocation of bacterial effectors into the host cells.24 CagL itself can also trigger intracellular signaling pathways by RGD‐dependent binding to integrins and can induce cell proinflammatory responses independently of CagA translocation.25, 26, 27
Recent studies have shown that particular polymorphisms at amino acid residues 58‐62 upstream of the critical RGD motif, referred to as CagL hypervariable motif (CagLHM), may correlate with severe disease progression in a geographically dependent manner.28, 29 More specifically, CagL amino acid polymorphisms Y58/E59, D58/K59, and N58 may correlate with higher corpus inflammation and integrin α5β1 expression in the upper stomach, induction of hypochlorhydria vicious cycle, and subsequently with an increase in the risk of gastric carcinogenesis.30, 31, 32, 33 Given these findings, and those of our previous study showing a very high prevalence of cagL gene among Iranian H pylori strains,34 here we aimed to characterize the diversity of CagL sequence polymorphisms and investigate whether these polymorphisms associate with clinical outcomes in patients with different gastroduodenal diseases. Associations between CagL sequence polymorphisms and different H pylori virulence genotypes were also investigated.
2. MATERIALS AND METHODS
2.1. Patients and gastric biopsies
We enrolled 126 patients suffering from different gastroduodenal diseases who underwent standard upper gastrointestinal endoscopy at Research Institute for Gastroenterology and Liver Diseases, Tehran, Iran, from January 2011 to May 2012. Three gastric biopsies were taken from the antrum of the stomach of each patient for H pylori culture and histopathological examination. The biopsy specimen for isolation of H pylori strains were immediately kept in transport medium containing thioglycolate with 1.3 g/L agar (Merck, Germany) and 3% yeast extract (Oxoid Ltd., Basingstoke, UK). Written informed consent was obtained from all patients under a protocol approved by the Ethical Review Committee of the Gastroenterology and Liver Diseases Research Institute at Shahid Beheshti University of Medical Sciences.
2.2. H pylori growth conditions and identification
The fresh gastric biopsy samples were completely dissected, homogenized, and cultured on the surface of Brucella agar plates (Merck) supplemented with 7% (v/v) horse blood, 10% fetal calf serum (FCS), Campylobacter‐selective supplement (vancomycin 2.0 mg/L, polymyxin 0.05 mg/L, trimethoprim 1.0 mg/L), and amphotericin B (2.5 mg/L). The cultured plates were incubated at 37°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) in a CO2 incubator for 3‐7 days. Bacterial growth was identified as H pylori by colony morphology and Gram stain, as well as positive reactions for oxidase, catalase, and urease, and subsequently by species‐specific PCR assays as previously described.34, 35 Pure cultures from each strain were harvested and stored at −80°C in 0.5 mL of brain heart infusion (BHI) medium (Merck) containing 15% glycerol plus 20% FCS until further experiments.
2.3. DNA extraction and virulence genotyping
Genomic DNA was extracted from sweeps of primary H pylori colonies using the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) as per the manufacturer's instructions. Purified DNA samples were stored at −20°C until required for PCR analyses. The quality and specificity of DNA samples were confirmed using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). H pylori 16S rRNA‐ and glmM‐specific PCRs, which produced the expected amplicon sizes of 764 and 296 bp, were also performed for all strains. Virulence factor genotyping of each H pylori isolate was performed by cagL‐, cagA‐, babA2‐, sabA‐, and vacA‐specific PCRs using previously published primer pairs and PCR conditions.34 H pylori J99 (CCUG 47164) and a no‐template reaction served as positive and negative controls in each PCR experiment, respectively.
2.4. Determination of CagA EPIYA motifs and cagPAI integrity
For determining the CagA EPIYA motifs, the cagA gene 3′ variable region was amplified using specific primers 5′ TCCGTTAAAGATGTGATCATCAATC 3′ (cag3′F) and 5′ AGATTTTTGGA AACCACCTTTTG 3′ (cag3′R), as previously described.16 cagPAI integrity was investigated by multiple PCRs using eleven sets of specific oligonucleotide primers spanning the cagPAI locus as per our previously described scheme.16, 36 The cagPAI was defined as intact/complete when all the selected gene segments were present. Partially deleted cagPAI was defined as where some, but not all, of the cagPAI gene segments were present. Complete absence of the cagPAI was confirmed by a simple PCR amplification using Luni1 and R5280 primers yielding a 550 bp empty cagPAI site amplicon.37
2.5. Sequencing of cagL genes
For DNA sequencing of cagL genes, 25 µL PCRs using specific primers CagL‐B4 (5′ GCAGAATTCATAACAAGCGGCTTAAAG 3′) and CagL‐B5 (5′ ATTAGAATTCATAGCCTATCGTCTCAG 3′) generated 695 bp PCR amplicons. The PCR products were purified using the Silica Bead DNA Gel Extraction Kit (Thermo Scientific, Fermentas, USA). The partial nucleotide sequences of cagL genes from 46 strains characterized in this study were deposited in the GenBank database; accession numbers are shown in Table 1.
Table 1.
Distribution of 46 H pylori cagL‐positive isolates in relation to clinical status and demographic data of the respective patients
| No. | Strain | GenBank Accession No.a | Clinical status | Gender (F/M) | Age (years) | Ethnicity |
|---|---|---|---|---|---|---|
| 1 | HpOC179 | KC609279.1 | GC | F | 63 | Fars |
| 2 | HpNOC293 | KC609280.1 | GC | F | 52 | Turk |
| 3 | HpNOC560 | KC609281.1 | GC | F | 54 | Fars |
| 4 | HpOC485 | KC609282.1 | NUD | M | 54 | Fars |
| 5 | HpOC494 | KC609283.1 | NUD | F | 42 | Fars |
| 6 | HpOC557 | KC609284.1 | PUD | F | 50 | Fars |
| 7 | HpOC571 | KC609285.1 | NUD | F | 49 | Fars |
| 8 | HpOC573 | KC609286.1 | NUD | F | 36 | Turk |
| 9 | HpOC576 | KC621877.1 | NUD | F | 42 | Fars |
| 10 | HpOC606 | KC621878.1 | GE | M | 60 | Fars |
| 11 | HpOC639 | KC621879.1 | GE | M | 25 | Lur |
| 12 | HpOC656 | KC621880.1 | GE | M | 41 | Turk |
| 13 | HpOC658 | KC621881.1 | NUD | F | 33 | Fars |
| 14 | HpOC723 | KC621882.1 | NUD | M | 47 | Turk |
| 15 | HpOC728 | KC621883.1 | NUD | F | 23 | Turk |
| 16 | HpOC734 | KC621884.1 | NUD | M | 50 | Fars |
| 17 | HpOC743 | KC621885.1 | NUD | M | 60 | Turk |
| 18 | HpOC751 | KC621886.1 | NUD | F | 44 | Fars |
| 19 | HpOC770 | KC621887.1 | NUD | F | 73 | Fars |
| 20 | HpOC775 | KC621888.1 | GE | F | 39 | Fars |
| 21 | HpOC785 | KC621889.1 | NUD | M | 60 | Turk |
| 22 | HpOC790 | KC621890.1 | NUD | M | 26 | Fars |
| 23 | HpOC793 | KC621891.1 | NUD | F | 41 | Fars |
| 24 | HpOC796 | KC621892.1 | GE | F | 51 | Fars |
| 25 | HpOC797 | KC621893.1 | NUD | F | 28 | Fars |
| 26 | HpOC803 | KC621894.1 | NUD | M | 52 | Fars |
| 27 | HpOC805 | KC621895.1 | NUD | F | 48 | Fars |
| 28 | HpOC808 | KC621896.1 | NUD | F | 65 | Lur |
| 29 | HpOC810 | KC621897.1 | NUD | F | 53 | Lur |
| 30 | HpOC814 | KC621898.1 | PUD | F | 25 | Lur |
| 31 | HpOC815 | KC621899.1 | NUD | F | 34 | Fars |
| 32 | HpOC816 | KC621900.1 | NUD | M | 14 | Fars |
| 33 | HpOC819 | KC621901.1 | GE | F | 32 | Turk |
| 34 | HpOC824 | KC621902.1 | PUD | F | 43 | Turk |
| 35 | HpOC852 | KC621903.1 | NUD | M | 45 | Fars |
| 36 | HpOC854 | KC621904.1 | NUD | F | 71 | Fars |
| 37 | HpOC897 | KC621905.1 | PUD | F | 60 | Fars |
| 38 | HpOC912 | KC621906.1 | PUD | F | 64 | Fars |
| 39 | HpOC913 | KC621907.1 | PUD | M | 40 | Fars |
| 40 | HpOC937 | KC621908.1 | NUD | F | 48 | Fars |
| 41 | HpOC939 | KC621909.1 | PUD | M | 54 | Turk |
| 42 | HpOC975 | KC621910.1 | GE | F | 31 | Fars |
| 43 | HpOC996 | KC621911.1 | GE | F | 52 | Fars |
| 44 | HpOC1021 | KC621912.1 | GE | F | 50 | Fars |
| 45 | HpOC1028 | KC621913.1 | NUD | F | 27 | Turk |
| 46 | HpOC1031 | KC621914.1 | NUD | F | 52 | Fars |
GC, gastric cancer; GE, gastric erosion; NUD, non‐ulcer dyspepsia; PUD, peptic ulcer disease.
The accession numbers are deposited in GenBank database for cagL gene sequences of the H pylori strains in this study.
2.6. Sequence and phylogenetic analysis
DNA sequences were edited by Chromas Lite version 2.5.1 (Technelysium Pty Ltd, Australia) and BioEdit version 7.2.5 softwares.38 CagL peptide sequences were aligned to the sequence of H pylori strain P12 (GenBank: ACJ07700.1) as a reference sequence. The single nucleotide polymorphisms and codon usage of the cagL sequences were examined using BioEdit version 7.2.5 after in‐frame translation. Neighbor‐joining phylogenetic trees were constructed from both CagL nucleotide and peptide sequences of 46 H pylori isolates using Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7).39 Additionally, we compared the diversity and frequency of CagLHM sequences in this study with the available global motifs at this location by utilizing the 554 CagL amino acid sequences cited in Supplementary data of a study by Gorrell et al29 For clarification, this previously published dataset includes CagL sequences of the 46 H pylori isolates characterized in this current study.
2.7. Statistical analysis
Data were analyzed using IBM SPSS Statistics for Windows version 21.0 (IBM Corp., Armonk, NY). Significant associations between cagL amino acid polymorphisms and H pylori virulence genotypes in relationship to gastroduodenal diseases were assessed by Fisher's exact test. All statistical tests were two‐sided, and differences were considered statistically significant when P values <0.05.
3. RESULTS
3.1. Patient characteristics
Among the 126 patients that underwent upper gastrointestinal endoscopy, 61 (48.4%) patients had defined H pylori infection based on both positive histology and culture results. Detailed endoscopic diagnoses of these patients have been reported previously.34 Briefly, 36 had non‐ulcer dyspepsia (NUD), eight had peptic ulcer disease (PUD), 12 had gastric erosion (GE), and five had gastric cancer (GC). There was no significant difference in the age, gender, and ethnicity between these H pylori‐positive patients with different clinical outcomes (P > 0.05).
3.2. H pylori genotyping and vacA allelic diversity
Virulence genotypes previously determined for these isolates were cagL, cagA, babA2, and sabA carriage by 97% (59/61), 85% (52/61), 97% (59/61), and 84% (51/61) of the strains, respectively, and vacA genotypes s1m1 in 29% (18/61), s1m2 in 46% (28/61), and s2m2 in 25% (15/61) of the strains.34 No association was identified between virulence genotypes and clinical outcomes.
3.3. Diversity of CagA EPIYA motifs and cagPAI integrity
All 52 cagA‐positive H pylori isolates were found to contain the cagA 3'‐end region expressing distinct EPIYA motifs. Among these isolates, various cagA EPIYA types were detected as follows: ABC in 39, ABCC in 7, ABCCC in 1, and multiple EPIYA motifs of different sizes, indicating mixed infections, were detected in five strains. The East Asian CagA containing EPIYA‐D motif was not detected in the examined strains. Further PCR analysis was performed to evaluate the intactness of cagPAI locus from the 5′ to the 3′ end in all H pylori strains. Accordingly, 70% (43/61) of the isolates carried an intact cagPAI, 26% (16/61) carried a partial cagPAI and 3% (2/61) completely lacked the cagPAI genes. A significant association was found between patients infected with the isolates carrying intact cagPAI plus multiple C‐type EPIYA repeats and more severe clinical outcomes including PUD, GE, and GC (P = 0.013). In addition, patients ≥50 years infected with the isolates carrying intact cagPAI had a significant disease risk for PUD, GE, and GC (P = 0.038) than NUD.
3.4. CagL sequence diversity in disease outcomes
Although the expected 695 bp cagL amplicon was obtained from all 59 cagL‐genopositive H pylori clinical isolates, direct sequencing produced only 46 sequences of sufficient quality for cagL polymorphism analysis, including 27 NUD, 7 PUD, nine GE, and three GC isolates. Details of these strains and corresponding patients’ details, including clinical status, gender, age, and ethnicity, are presented in Table 1. The cagL sequences were manually edited and trimmed, and aligned with sequences available in the NCBI GenBank database. The cagL nucleotide sequences of our strains, which showed >97% homology with the cagL gene of the reference H pylori strain P12, were translated to amino acid sequences using BioEdit software.
The frequency of synonymous and non‐synonymous cagL nucleotide polymorphism for 46 H pylori strains are presented in Table 2 and Figure 1. The most variable codon usage was observed at residues 41, 62, 122, and 171 with amino acid polymorphisms including V/T/A, E/Q/K, K/N, and A/S, respectively. The frequency of V41 amino acid sequence polymorphism in patients with NUD (77.8%) and PUD (71.4%) was higher compared to GC patients in which V/T/A substitutions were equally distributed. However, the presence of V41 was found in all patients with GE (100%). The rate of D58 (7, 15.2%) amino acid substitution was significantly lower in comparison with N58 (39, 84.8%) among all patients with different gastric diseases. However, the occurrence rate of K59 vs E59 substitution was found to be approximately equal in different gastric diseases apart from GC patients with higher rate of E59. Moreover, all isolates (100%) with K62 substitution were PUD cases. The presence of N58 mostly accompanied E59 than K59 (22 vs 17 residue combinations), while the presence of D58 mostly accompanied K59 than E59 (6 vs 1 residue combinations). The combined residues N58E59 had higher rates in NUD (13/27, 48.1%), GE (5/9, 55.5%), and GC (2/3, 66.7%) patients compared to patients with PUD (2/7, 28.6%). In contrast, the N58K59 amino acid combination was more frequently found among PUD (3/7, 42.8%) patients than other disease outcomes. However, no significant associations were observed between these combined residue polymorphisms and clinical outcomes (P > 0.05).
Table 2.
Distribution of amino acid and nucleotide substitutions in CagL of 46 H pylori isolates from patients with different gastric diseases
| CagL polymorphic residuea | Amino acid diversity | Number of disease state‐associated H pylori isolates carrying each amino acid polymorphism | |||
|---|---|---|---|---|---|
|
NUD (n = 27) |
PUD (n = 7) |
GE (n = 9) |
GC (n = 3) |
||
| 41 | V/T/A | 21:3:3 | 5:2:0 | 9:0:0 | 1:1:1 |
| GT(G/A):ACG:GCGb | GTG:ACG | GT(G/A) | GTG:ACG:GCG | ||
| 56 | A/T | 24:3 | 7:0 | 9:0 | 3:0 |
| GCT:ACT | GCT | GCT | GCT | ||
| 58 | D/N | 3:24 | 2:5 | 2:7 | 0:3 |
| GAT:AAT | GAT:AAT | GAT:AAT | AAT | ||
| 59 | K/E | 14:13 | 4:3 | 4:5 | 1:2 |
| AAA:GAA | AAA:GAA | AAA:GAA | AAA:GAA | ||
| 60 | M/I | 3:24 | 3:4 | 2:7 | 0:3 |
| ATG:ATA | ATG:ATA | ATG:ATA | ATA | ||
| 61 | G/S | 25:2 | 7:0 | 9:0 | 3:0 |
| GGT:AGT | GGT | GGT | GGT | ||
| 62 | E/Q/K | 1:26:0 | 0:3:4 | 0:9:0 | 0:3:0 |
| GAA:CAA | CAA:AAA | CAA | CAA | ||
| 65 | A/T | 26:1 | 6:1 | 9:0 | 3:0 |
| GCT:ACT | GCT:ACT | GCT | GCT | ||
| 69 | K/E | 27:0 | 6:1 | 9:0 | 3:0 |
| AAA | AAA:GAA | AAA | AAA | ||
| 72 | A/T | 27:0 | 7:0 | 9:0 | 2:1 |
| GCC | GCC | GCC | GCC:ACC | ||
| 114 | I/Mc | 0:27 | 0:7 | 0:9 | 0:3 |
| ATG | ATG | ATG | ATG | ||
| 118 | P/L | 26:1 | 7:0 | 9:0 | 3:0 |
| CCC:CTC | CCC | CCC | CCC | ||
| 122 | K/N | 10:17 | 1:6 | 2:7 | 1:2 |
| AAG:AA(C/T) | AAG:AA(C/T) | AA(G/A):AA(C/T) | AAG:AA(C/T) | ||
| 134 | I/V | 9:18 | 1:6 | 3:6 | 1:2 |
| ATT:GTT | ATT:GTT | ATT:GTT | ATT:GTT | ||
| 171 | A/S | 26:1 | 7:0 | 8:1 | 3:0 |
| GCT:TCT | GCT | GCT:ACT | GCT | ||
| 175 | T/I | 27:0 | 7:0 | 8:1 | 3:0 |
| ACT | ACT | ACT:ATT | ACT | ||
| 194 | R/K | 23:4 | 7:0 | 8:1 | 3:0 |
| AGA:AAA | AGA | AGA:AAA | AGA | ||
| 200 | Q/H | 1:26 | 0:7 | 1:8 | 0:3 |
| CAA:CAC | CAC | CAA:CAC | CAC | ||
| 210 | E/K | 26:1 | 7:0 | 9:0 | 3:0 |
| GAG:AAG | GAG | GAG | GAG | ||
| 216 | R/I | 26:1 | 6:1 | 9:0 | 3:0 |
| AGA:ATA | AGA:ATA | AGA | AGA | ||
GC, gastric cancer; GE, gastric erosion; NUD, non‐ulcer dyspepsia; PUD, peptic ulcer disease.
Positions of amino acid residues correspond to the H pylori P12 reference strain.
Codon usage in italics; bolded nucleotides represent nucleotide polymorphisms.
Denotes that all of our strains had M (ATG) residue in this position in comparison to H pylori P12 that had I (ATA).
Figure 1.
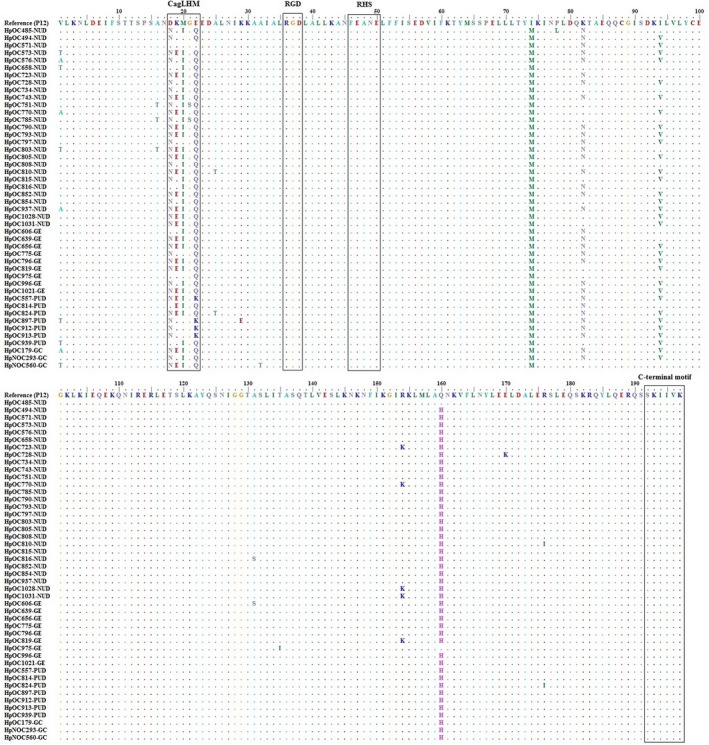
Partial amino acid sequence alignment of CagL from 46 H pylori clinical strains from patients with different gastric diseases. The CagL amino acid sequence of the H pylori reference strain P12 is shown on the top line. The clinical strains included 27 from non‐ulcer dyspepsia (NUD), seven from peptic ulcer disease (PUD), nine from gastric erosion (GE), and three from gastric cancer (GC) patients. Sequences of CagL hypervariable motif (CagLHM), conserved arginine‐glycine‐aspartate motifs (RGD), RGD helper sequence (RHS) motifs comprising the FEANE (Phe‐Glu‐Ala‐Asn‐Glu) sequence, and highly conserved C‐terminal hexapeptide motifs consisting of the SKIIVK (Ser‐Lys‐Ile‐Ile‐Val‐Lys) sequence are surrounded by boxes
3.5. High sequence conservation of RGD, RHS, and C‐terminal motifs of CagL
According to the nucleotide and amino acid sequence analysis, all of our H pylori strains expressed the RGD motif with no amino acid changes at residues 76‐78 (Figures 1 and 2). However, only a synonymous mutation (AGA to AGG transition) was detected in arginine residue of this tripeptide motif among 18/46 (39.1%) strains studied. Regarding the RHS motif, all of our strains also conserved the expression of FEANE pentapeptide motif with even no nucleotide polymorphisms at residues 86‐90. The C‐terminal SKIIVK (Ser‐Lys‐Ile‐Ile‐Val‐Lys) hexapeptide motif of CagL at residues 232‐237 was also conserved among all the strains in this study. However, three synonymous mutations were detected in serine (TCG to TCA), isoleucine (ATC to ATT), and valine (GTC to GTT) of this distal hexapeptide CagL motif among 46/46 (100%), 1/46 (2.2%), and 20/46 (43.5%) of the strains, respectively.
Figure 2.
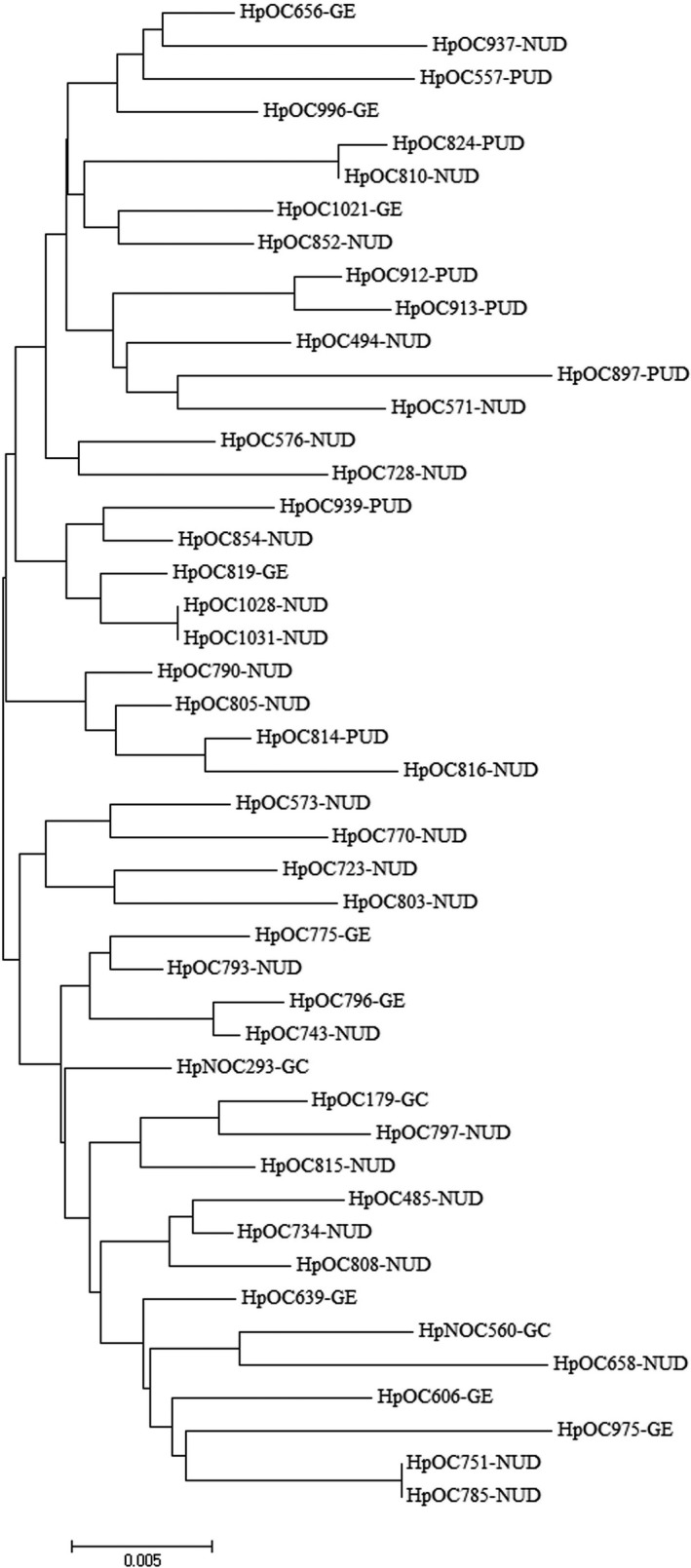
Phylogenetic tree of 46 H pylori clinical strains based on cagL nucleotide sequences. Neighbor‐joining tree of concatenated sequences was constructed using MEGA7 software with bootstrap method at 1000 replications. The evolutionary distances were computed using the Tamura 3‐parameter model
3.6. Association of CagLHM variants with disease outcomes and virulence genotypes
Based on the amino acid sequence comparison of CagLHM sequences, 10 motif variants were identified in the CagLHM located at residues 58‐62 in our studied strains (Table 3). Among these motifs, two common CagLHM sequences including NEIGQ and NKIGQ accounted for 21/46 (45.7%) and 9/46 (19.6%) of the sequences from the examined strains in this study. Moreover, most of the NUD (20/27, 74.1%), GE (6/9, 66.7%), and all of the GC (3/3, 100%) strains carried these two dominant motifs, whereas about half of the PUD strains (3/7, 42.8%) contained the NKMGK motif. Interestingly, a significant association was found between the isolates carrying NKMGK motif and PUD (P = 0.002). Three unique motifs for the CagLHM sequences including NKISQ (2/46, 4.3%), NKMGK (3/46, 6.5%), and DKMGQ (1/46, 2.2%) were identified among the translated CagL sequences in comparison with 508 sequences that were obtained from the previously deposited sequences in over‐mentioned databases.
Table 3.
CagLHM sequence types and gastric disease associations of 46 H pylori isolates included in this study
| CagLHM sequence |
Total (n = 46) |
Patient health status | |||
|---|---|---|---|---|---|
|
NUD (n = 27) |
PUD (n = 7) |
GE (n = 9) |
GC (n = 3) |
||
| NEIGQ | 21 | 13 | 1 | 5 | 2 |
| NKIGQ | 9 | 7 | — | 1 | 1 |
| DKIGQ | 4 | 2 | 1 | 1 | — |
| NKMGQ | 3 | 2 | — | 1 | — |
| NKMGK† | 3 | — | 3 | — | — |
| NKISQ† | 2 | 2 | — | — | — |
| NEIGK | 1 | — | 1 | — | — |
| DKMGE | 1 | 1 | — | — | — |
| DEIGQ | 1 | — | 1 | — | — |
| DKMGQ† | 1 | — | — | 1 | — |
| P Value* | 0.153 | 0.002 | 0.735 | 1.0 | |
CagLHM CagL hypervariable motif; GC, gastric cancer; GE, gastric erosion; NUD non‐ulcer dyspepsia; PUD peptic ulcer disease.
The “—” denotes none detected.
Denotes that these CagLHM sequence types are novel and uniquely identified in our H pylori strains in comparison to global strains.
P value indicates the significant difference between CagLHM diversity and different gastric diseases. The statistically significant relationships are calculated by two‐tailed Fisher's exact test and presented in bold.
Approximately half of the strains with vacA s1m2 allelic type (12/22, 54.5%) carried the NEIGQ, while about one‐third of the strains with vacA s1m1 genotype (5/14, 35.7%) contained this motif (Table 4). Additionally, most of the babA2‐positive strains (29/44, 65.9%) carried either NEIGQ or NKIGQ sequences. In contrast, nearly all of the sabA‐negative strains (7/8, 87.5%) had either NEIGQ or NKIGQ motifs in their CagLHM sequences. All cagA‐positive H pylori strains carrying ABC motif contained either one of the CagLHM variants identified, mostly NEIGQ (14/35, 40%) and NKIGQ (7/35, 20%) sequences. Among the strains with intact cagPAI structure, the most common CagLHM sequences were NEIGQ (14/33, 42.4%), NKIGQ (6/33, 18.2%), DKIGQ (4/33, 12.1%), NKMGK (3/33, 9.1%), and NKMGQ (2/33, 6.1%), respectively. All of the DKIGQ isolates carried an intact cagPAI. NEIGQ isolates having multiple C‐type EPIYA repeats and carrying intact cagPAI correlated with disease risk for PUD, GE, and GC (P = 0.021) than NUD. However, no significant association was found between other CagLHM sequences and virulence genotypes (P > 0.05).
Table 4.
Differences in polymorphisms of various CagLHM sequence types of 46 H pylori isolates in relation to virulence genotypes and cagPAI integrity
| Virulence genotypes | CagLHM sequence type polymorphism | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
NEIGQ (n = 21) |
NKIGQ (n = 9) |
DKIGQ (n = 4) |
NKMGQ (n = 3) |
NKMGK (n = 3) |
NKISQ (n = 2) |
NEIGK (n = 1) |
DKMGE (n = 1) |
DEIGQ (n = 1) |
DKMGQ (n = 1) |
Total (n = 46) |
|
| vacA s1m1 | 5 | 4 | 1 | 1 | 1 | — | — | — | 1 | 1 | 14 |
| vacA s1m2 | 12 | 2 | 3 | 1 | 2 | 1 | 1 | — | — | — | 22 |
| vacA s2m2 | 4 | 3 | — | 1 | — | 1 | — | 1 | — | — | 10 |
| babA2+ | 20 | 9 | 4 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 44 |
| babA2− | 1 | — | — | — | — | 1 | — | — | — | — | 2 |
| sabA+ | 16 | 7 | 3 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 38 |
| sabA− | 5 | 2 | 1 | — | — | — | — | — | — | — | 8 |
| EPIYA motifs | |||||||||||
| ABC | 14 | 7 | 3 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 35 |
| ABCC | 4 | 1 | 1 | — | — | — | — | — | — | — | 6 |
| ABCCC | 1 | — | — | — | — | — | — | — | — | — | 1 |
| Mixed typea | 1 | — | — | 1 | — | — | — | — | — | — | 2 |
| cagA− | 1 | 1 | — | — | — | — | — | — | — | — | 2 |
| cagPAI integrity | |||||||||||
| Intact cagPAI | 14 | 6 | 4 | 2 | 3 | 1 | — | 1 | 1 | 1 | 33 |
| Partial cagPAI | 7 | 3 | — | 1 | — | 1 | 1 | — | — | — | 13 |
CagLHM, CagL hypervariable motif; GC, gastric cancer; GE, gastric erosion; NUD, non‐ulcer dyspepsia; PUD peptic ulcer disease.
The “—” denotes none detected.
Denotes multiple cagA EPIYA motifs of different sizes, indicating mixed infections.
3.7. Phylogenetic analysis of cagL gene
The edited DNA and amino acid sequences were aligned against reference sequence using ClustalW multiple alignment. The constructed Neighbor‐joining trees from cagL nucleotide and amino acid sequences of 46 H pylori isolates are presented in Figures 2 and 3, respectively. In general, no characteristic clusters were observed between DNA and amino acid sequences of CagL and different disease outcomes.
Figure 3.

Phylogenetic tree of 46 H pylori clinical strains based on translated CagL amino acid sequences. Neighbor‐joining tree of concatenated sequences was constructed using MEGA7 software with bootstrap method at 1000 replications. The evolutionary distances were computed using the Poisson correction method
4. DISCUSSION
Functional cagPAI chromosomal DNA region, which is responsible for most of the H pylori‐related gastric pathologies and malignant phonotypes such as gastric atrophy and cancer, has been discovered in 1996.14, 40, 41 This most extensively studied part of the H pylori genome is present in approximately 95% of Asian isolates, whereas about 60% of Western isolates from low‐risk countries are cagPAI‐positive.16, 36, 42, 43 The cagPAI‐encoded T4SS is a multiprotein complex composed of homologs of Agrobacterium tumefaciens VirB/D proteins, which forms an extracellular pilus for injection of effector molecules into host target cells. CagL (HP0539), which was introduced as a putative VirB5 ortholog, is recruited to the surface of injection needle and binds to host cell β1 integrins via its surface‐exposed RGD motif essential for CagA translocation and the induction of IL‐8.23, 44, 45 Previous and some recent studies have shown that CagL is subject to genetic variations and positive or diversifying selection in some of its protein motifs that may affect its binding affinity to integrins.14, 29, 30, 31, 32, 46, 47 Some of these variations in amino acid sequences of CagL have been proposed to be involved in the cancer risk of infected patients.29, 30, 31, 32, 33, 47 Thus, in this study we determined the genetic variability in CagL on both nucleotide and amino acid sequence levels from H pylori strains isolated from patients with different clinical outcomes. We also examined the variations in H pylori CagLHM amino acid sequences in relation to other important virulence genotypes and different gastric diseases.
The results of the present study confirmed our previously published data indicating a very high frequency ofcagL gene (97%) in Iranian H pylori strains, although in a non‐statistically significant relationship with clinical outcomes.16, 34 The high prevalence of cagL genotype in our study is in agreement with the results obtained from Malaysia and Singapore (>85%), Taiwan (98.6%), and India (86.6%).31, 32, 48 Moreover, in order to assess whether the CagL amino acid sequence polymorphisms and codon usages correlate with clinical outcomes, the cagL genes of 46 H pylori strains were sequenced. Our findings showed that amino acid residues at positions 41, 62, 122, and 171 had the greatest variability in their codon usages and were mostly non‐synonymous. The majority of the variations arose from nucleotide substitutions at either the first or second position of the putative progenitor codons. At position 41, we had V/T/A substitutions in different disease groups, in which V41 variant was predominantly observed in most of the strains from NUD (77.8%) and PUD (71.4%) and in all GE (100%) patients. This is in agreement with previous reports from Taiwan and Japan, where V41 variant found to be predominant in GC and non‐GC isolates.32, 47 By contrast, in our study V/T/A variants occurred equally in H pylori strains from GC patients. In addition, A41 variant was merely observed among patients with NUD (3/27) and GC (1/3). This finding is contrary to the study performed by Ogawa et al47 from Japan where A41 was only detected among CagL variants in GC isolates (2/10, 20%), although in a not significant relationship vs non‐GC isolates. In addition, the majority of our strains had Asparagine (N) at position 122, which is in line with the data obtained by Shukla et al31 from India where all of the strains were found to have N122 variant. In contrast, K122 was predominantly observed among Japanese and Taiwanese isolates.32, 47 However, no significant difference was seen between these amino acid variations and clinical outcomes (P > 0.05).
Our results revealed that N58 variant occurred at higher rate than D58 among studied H pylori strains, and more importantly, all of the strains from GC patients carried this amino acid variant in their CagL protein. By contrast, in previous studies from Taiwan, India, and Japan D58 substitution was more frequent than N58 in all disease groups.31, 32, 47 Additionally, none of the amino acid variants including Y58, G58, and M58 occurred in our CagL sequences, which had been previously reported from aforementioned studies. Similar to studies from Taiwan and Japan, the rate of E59 variant was higher in GC strains than non‐GC strains.32, 47 Conversely, Shukla et al31 found higher rate of K59 variant in CagL sequence of strains isolated from GC patients. Our results showed that N58E59 and N58K59 combined variants were more common in H pylori strains from GC patients. However, three different studies from Taiwan, India, and Japan reported a higher rate of the Y58E59, D58K59, and D58E59 amino acid combinations in GC patients, respectively.31, 32, 47 In another study in a Mexican patient cohort, 74 gene polymorphisms were observed in the cagL, which out of 24 analyzed variations, four showed a differential distribution between cases of cancer and gastritis (P < 0.05).30 Among these polymorphisms G166A (amino acidic change of A56 to T56) and A172G (amino acidic change of N58 to D58) were non‐synonymous, and two mutations including (A228G and C516T) were synonymous. Moreover, Yeh et al concluded that H pylori isolates carrying Y58E59 variant possibly exert stronger acid suppression during chronic infection and have strong binding affinity for integrin α5β1, which significantly promotes CagA translocation and phosphorylation as compared to wild‐type CagL.32, 33 However, their findings were found to be contradictory to those obtained by Tegtmeyer et al49 suggesting that Y58E59 mutation in CagL turned off the function of the T4SS for delivery of CagA into host cells.
Recently, analysis of CagL crystal structures revealed an elongated four‐helix bundle that seems to be evolutionarily unrelated to the proposed VirB5 orthologs.45, 50 Previous studies have proposed that the RGD tripeptide is located within a long α2 helix and is a critical motif in the structure of H pylori CagL pilus protein, able to bind and activate integrin α5β1 receptor on gastric epithelial cells.22, 27, 45, 51, 52 It has been also observed that RGD motif is at least partly involved in the cell signaling pathways leading to secretion of IL‐8, despite some controversy in the literature indicating that during infection mutation of the RGD motif in CagL does not affect the CagA translocation and IL‐8 induction.22, 53 Consistent with previous reports, all H pylori strains in the current study expressed the RGD motif in CagL sequences.31, 32, 47 These data highlight the importance of the RGD‐integrin interaction mediated translocation of CagA and also CagL‐dependent signaling pathway for induction of proinflammatory cytokines such as IL‐8. In addition to RGD motif, CagL protein contains another motif named RHS pentapeptide or FEANE in spatial proximity to the RGD sequence, which is proposed to enhance the CagL interactions with β1 integrins.24 Notably, the RHS and the C‐terminal SKIIVK hexapeptide motifs were universally expressed in all of our strains, which is partly in line with previous reports from Taiwan and Japan.32, 47 As previously suggested, these findings underscore the significance and essential roles of aforementioned motifs especially the highly conserved SKIIVK sequence that is present nearly identical in other cagPAI components such as CagI and CagH, in the stability, subcellular transport of CagL, pilus formation, and biogenesis, and subsequently in the CagA translocation and IL‐8 induction from epithelial cells.54
Based on previous studies, certain variants of CagLHM sequence containing five hypervariable amino acid residues (58, 59, 60, 61, and 62), which is located upstream of the RGD motif have been associated with gastric carcinogenesis.28, 31, 32, 33, 47 Recently, a global analysis of geographical diversity and polymorphism was carried out within the CagLHM motif of more than 500 amino acid sequences of CagL in gastric cancer‐associated H pylori isolates worldwide.29 Accordingly, 33 CagLHM sequence combinations with diverse geographical prevalence have been identified in different regions of the world particularly among Asian countries showing extensive diversity with 20 out of 33 (60.6%) unique CagLHM motifs in this region. Additionally, four motifs including DKMGE, NEIGQ, NKIGQ, and DKIGK were identified as the most common CagLHM sequences and accounted for >75% of available sequences from H pylori strains worldwide. Interestingly, we detected 10 variants of CagLHM motif within the 46 CagL sequences revealing substantial diversity in Iranian strains. We also identified two common motifs including NEIGQ and NKIGQ accounted for 45.7% and 19.6% of the sequences, respectively, which is in agreement with data from European and West/Central/South Asian countries where these two motifs were predominant.29 These data once again highlight that Iranian H pylori strains shared ancestral origins with the European counterparts and were intermingled with strains assigned to the hpEurope population.55, 56 Our results showed that all of the strains isolated from GC patients carried one of the two over‐mentioned predominant motifs. We also found a strong association between the NKMGK CagLHM and PUD (P = 0.002) clinical outcome, where NKMGK was detected as dominant motif and none the isolates from other clinical outcomes contained this type of motif. More importantly, three unique motifs including NKMGK, NKISQ, and DKMGQ accounted for 6.5%, 4.3%, and 2.2% of the sequences, respectively, were identified among our strains that were not reported previously. In addition, in this study and for the first time we examined the co‐occurrence of specific CagLHM motifs with the main virulence‐associated genes of H pylori. Notably, our results revealed that the NEIGQ isolates with multiple C‐type EPIYA repeats that carried intact cagPAI correlated with increased disease risk for PUD, GE, and GC (P = 0.021) than NUD. This may indicate the critical importance of this CagLHM motif in the pathogenesis of H pylori strains in a synergistic relation to other virulence genotypes. Moreover, further studies are required to investigate the possible influence of specific CagLHM motifs on the CagL interaction with host cell integrins and also their impacts on expression and function of other components of H pylori T4SS.
5. CONCLUSIONS
In conclusion, we identified putative novel variants of CagL sequences especially in the CagLHM motif, which may have crucial effect on the activity and function of T4SS and its pilus formation. Among the ten different CagLHM variants identified, all of the DKIGQ isolates carried an intact cagPAI that may be concluded that this motif was mostly overrepresented by the hypervirulent strains, albeit not statistically significant. Moreover, our findings demonstrated that the majority of Iranian H pylori strains were cagL positive, and all such strains expressed the RGD motif, the so‐called RHS or FEANE pentapeptide and the C‐terminal SKIIVK motif within their CagL protein. The very high‐level genetic conservation seen in these sequences, possibly due to different evolutionary selection pressures, underscores the importance of aforementioned motifs in the RGD‐dependent and RGD‐accessory integrin binding of CagL protein, as well as in protein‐protein interaction with other T4SS components to facilitate the CagA injection into host cells. Taken together, more studies using a large number of H pylori strains from patients with different disease outcome are necessary to further define the function of specific CagL amino acid polymorphisms especially the novel CagLHM variants in certain intracellular signaling pathways and subsequently with respect to disease progression and clinical outcome.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGMENTS
The authors wish to thank the sample collection team and nursing staff of Behbood Research Center for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We are also grateful to Dr. Mahsa Molaei from Department of Pathology, Taleghani Hospital, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Yadegar A, Mohabati Mobarez A, Zali MR. Genetic diversity and amino acid sequence polymorphism in Helicobacter pylori CagL hypervariable motif and its association with virulence markers and gastroduodenal diseases. Cancer Med. 2019;8:1619–1632. 10.1002/cam4.1941
Contributor Information
Abbas Yadegar, Email: a.yadegar@sbmu.ac.ir.
Ashraf Mohabati Mobarez, Email: mmmobarez@modares.ac.ir.
REFERENCES
- 1. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1619‐241. [PMC free article] [PubMed] [Google Scholar]
- 2. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74‐108. [DOI] [PubMed] [Google Scholar]
- 3. Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175‐1186. [DOI] [PubMed] [Google Scholar]
- 5. Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119:2475‐2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bridge DR, Merrell DS. Polymorphism in the Helicobacter pylori CagA and VacA toxins and disease. Gut. Microbes. 2013;4:101‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuta Y, Konno M, Osaki T, et al. Microevolution of virulence‐related genes in Helicobacter pylori familial infection. PLoS ONE. 2015;10:e0127197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Linz B, Windsor HM, Gajewski JP, et al. Helicobacter pylori genomic microevolution during naturally occurring transmission between adults. PLoS ONE. 2013;8:e82187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerhard M, Lehn N, Neumayer N, et al. Clinical relevance of the Helicobacter pylori gene for blood‐group antigen‐binding adhesin. Proc Natl Acad Sci U S A. 1999;96:12778‐12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahdavi J, Sonden B, Hurtig M, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573‐578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miernyk K, Morris J, Bruden D, et al. Characterization of Helicobacter pylori cagA and vacA genotypes among Alaskans and their correlation with clinical disease. J Clin Microbiol. 2011;49:3114‐3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J, Qian J, Zhang X, Zou Q. Outer membrane inflammatory protein A, a new virulence factor involved in the pathogenesis of Helicobacter pylori . Mol Biol Rep. 2014;41:7807‐7814. [DOI] [PubMed] [Google Scholar]
- 13. Figueiredo C, Machado JC, Pharoah P, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high‐risk individuals for gastric carcinoma. J Natl Cancer Inst. 2002;94:1680‐1687. [DOI] [PubMed] [Google Scholar]
- 14. Olbermann P, Josenhans C, Moodley Y, et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiedemann T, Loell E, Mueller S, et al. Helicobacter pylori cag‐pathogenicity island‐dependent early immunological response triggers later precancerous gastric changes in Mongolian gerbils. PLoS ONE. 2009;4:e4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yadegar A, Alebouyeh M, Zali MR. Analysis of the intactness of Helicobacter pylori cag pathogenicity island in Iranian strains by a new PCR‐based strategy and its relationship with virulence genotypes and EPIYA motifs. Infect Genet Evol. 2015;35:19‐26. [DOI] [PubMed] [Google Scholar]
- 17. Schuelein R, Everingham P, Kwok T. Integrin‐mediated type IV secretion by Helicobacter: what makes it tick? Trends Microbiol. 2011;19:211‐216. [DOI] [PubMed] [Google Scholar]
- 18. Tegtmeyer N, Wessler S, Backert S. Role of the cag‐pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190‐1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hatakeyama M. Helicobacter pylori and gastric carcinogenesis. J Gastroenterol. 2009;44:239‐248. [DOI] [PubMed] [Google Scholar]
- 20. Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: the master key hypothesis. Helicobacter. 2010;15:163‐176. [DOI] [PubMed] [Google Scholar]
- 21. Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit‐and‐run carcinogenesis. Cell Host Microbe. 2014;15:306‐316. [DOI] [PubMed] [Google Scholar]
- 22. Kwok T, Zabler D, Urman S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862‐866. [DOI] [PubMed] [Google Scholar]
- 23. Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10:955‐965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conradi J, Tegtmeyer N, Wozna M, et al. An RGD helper sequence in CagL of Helicobacter pylori assists in interactions with integrins and injection of CagA. Front Cell Infect Microbiol. 2012;2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gorrell RJ, Guan J, Xin Y, et al. A novel NOD1‐ and CagA‐independent pathway of interleukin‐8 induction mediated by the Helicobacter pylori type IV secretion system. Cell Microbiol. 2013;15:554‐570. [DOI] [PubMed] [Google Scholar]
- 26. Saha A, Backert S, Hammond CE, Gooz M, Smolka AJ. Helicobacter pylori CagL activates ADAM17 to induce repression of the gastric H, K‐ATPase alpha subunit. Gastroenterology. 2010;139:239‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tegtmeyer N, Hartig R, Delahay RM, et al. A small fibronectin‐mimicking protein from bacteria induces cell spreading and focal adhesion formation. J Biol Chem. 2010;285:23515‐23526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tafreshi M, Zwickel N, Gorrell RJ, Kwok T. Preservation of Helicobacter pylori CagA translocation and host cell proinflammatory responses in the face of CagL hypervariability at amino acid residues 58/59. PLoS ONE. 2015;10:e0133531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gorrell RJ, Zwickel N, Reynolds J, Bulach D, Kwok T. Helicobacter pylori CagL hypervariable motif: a global analysis of geographical diversity and association with gastric cancer. J Infect Dis. 2016;213:1927‐1931. [DOI] [PubMed] [Google Scholar]
- 30. Rizzato C, Torres J, Plummer M, et al. Variations in Helicobacter pylori cytotoxin‐associated genes and their influence in progression to gastric cancer: implications for prevention. PLoS ONE. 2012;7:e29605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shukla SK, Prasad KN, Tripathi A, et al. Helicobacter pylori cagL amino acid polymorphisms and its association with gastroduodenal diseases. Gastric Cancer. 2013;16:435‐439. [DOI] [PubMed] [Google Scholar]
- 32. Yeh YC, Chang WL, Yang HB, Cheng HC, Wu JJ, Sheu B. pylori cagL amino acid sequence polymorphism Y58E59 induces a corpus shift of gastric integrin alpha5beta1 related with gastric carcinogenesis. Mol Carcinog. 2011;50:751‐759. [DOI] [PubMed] [Google Scholar]
- 33. Yeh YC, Cheng HC, Yang HB, Chang WL, Sheu B. pylori CagL‐Y58/E59 prime higher integrin alpha5beta1 in adverse pH condition to enhance hypochlorhydria vicious cycle for gastric carcinogenesis. PLoS ONE. 2013;8:e72735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yadegar A, Mobarez AM, Alebouyeh M, Mirzaei T, Kwok T, Zali MR. Clinical relevance of cagL gene and virulence genotypes with disease outcomes in a Helicobacter pylori infected population from Iran. World J Microbiol Biotechnol. 2014;30:2481‐2490. [DOI] [PubMed] [Google Scholar]
- 35. Farzi N, Yadegar A, Aghdaei HA, Yamaoka Y, Zali MR. Genetic diversity and functional analysis of oipA gene in association with other virulence factors among Helicobacter pylori isolates from Iranian patients with different gastric diseases. Infect Genet Evol. 2018;60:26‐34. [DOI] [PubMed] [Google Scholar]
- 36. Ahmadzadeh A, Ghalehnoei H, Farzi N, et al. Association of CagPAI integrity with severeness of Helicobacter pylori infection in patients with gastritis. Pathol Biol. 2015;63:252‐257. [DOI] [PubMed] [Google Scholar]
- 37. Mukhopadhyay AK, Kersulyte D, Jeong JY, et al. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182:3219‐3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hall TA.BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series c1979‐c2000 London: Information Retrieval Ltd., 95‐98. [Google Scholar]
- 39. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870‐1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Romano M, Ricci V, Zarrilli R. Mechanisms of disease: Helicobacter pylori‐related gastric carcinogenesis‐implications for chemoprevention. Nat Clin Pract Gastroenterol Hepatol. 2006;3:622‐632. [DOI] [PubMed] [Google Scholar]
- 41. Censini S, Lange C, Xiang Z, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I‐specific and disease‐associated virulence factors. Proc Natl Acad Sci U S A. 1996;93:14648‐14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nguyen LT, Uchida T, Tsukamoto Y, et al. Clinical relevance of cagPAI intactness in Helicobacter pylori isolates from Vietnam. Eur J Clin Microbiol Infect Dis. 2010;29:651‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peek RM Jr, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208:233‐248. [DOI] [PubMed] [Google Scholar]
- 44. Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451‐485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barden S, Lange S, Tegtmeyer N, et al. A helical RGD motif promoting cell adhesion: crystal structures of the Helicobacter pylori type IV secretion system pilus protein CagL. Structure. 2013;21:1931‐1941. [DOI] [PubMed] [Google Scholar]
- 46. Bönig T, Olbermann P, Bats SH, Fischer W, Josenhans C. Systematic site‐directed mutagenesis of the Helicobacter pylori CagL protein of the Cag type IV secretion system identifies novel functional domains. Sci Rep. 2016;6:38101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogawa H, Iwamoto A, Tanahashi T, et al. Genetic variants of Helicobacter pylori type IV secretion system components CagL and CagI and their association with clinical outcomes. Gut Pathogens. 2017;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmidt HM, Andres S, Nilsson C, et al. The cag PAI is intact and functional but HP0521 varies significantly in Helicobacter pylori isolates from Malaysia and Singapore. Eur J Clin Microbiol Infect Dis. 2010;29:439‐451. [DOI] [PubMed] [Google Scholar]
- 49. Tegtmeyer N, Lind J, Schmid B, Backert S. Helicobacter pylori CagL Y58/E59 mutation turns‐off type IV secretion‐dependent delivery of CagA into host cells. PLoS ONE. 2014;9:e97782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barden S, Schomburg B, Conradi J, Backert S, Sewald N, Niemann HH. Structure of a three‐dimensional domain‐swapped dimer of the Helicobacter pylori type IV secretion system pilus protein CagL. Acta Crystallogr D Biol Crystallogr. 2014;70:1391‐1400. [DOI] [PubMed] [Google Scholar]
- 51. Wiedemann T, Hofbaur S, Tegtmeyer N, et al. Helicobacter pylori CagL dependent induction of gastrin expression via a novel alphavbeta5‐integrin‐integrin linked kinase signalling complex. Gut. 2012;61:986‐996. [DOI] [PubMed] [Google Scholar]
- 52. Conradi J, Huber S, Gaus K, et al. Cyclic RGD peptides interfere with binding of the Helicobacter pylori protein CagL to integrins alphaVbeta3 and alpha5beta1. Amino Acids. 2012;43:219‐232. [DOI] [PubMed] [Google Scholar]
- 53. Jimenez‐Soto LF, Kutter S, Sewald X, et al. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD‐independent manner. PLoS Pathog.. 2009;5:e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shaffer CL, Gaddy JA, Loh JT, et al. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria‐host cell interface. PLoS Pathog.. 2011;7:e1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Latifi‐Navid S, Ghorashi SA, Siavoshi F, et al. Ethnic and geographic differentiation of Helicobacter pylori within Iran. PLoS ONE. 2010;5:e9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori . Infect Genet Evol. 2012;12:203‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]


