Abstract
Insight into how plants simultaneously cope with multiple stresses, for example, when challenged with biotic stress from pathogen infection and abiotic stress from drought, is important both for understanding evolutionary trade-offs and optimizing crop responses to these stresses. Mechanisms by which initial plant immune signaling antagonizes abscisic acid (ABA) signal transduction requires further investigation. Using a chemical genetics approach, the small molecule [5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione (DFPM) has previously been identified due to its ability to suppress ABA signaling via plant immune signaling components. Here, we have used forward chemical genetics screening to identify DFPM-insensitive loci by monitoring the activity of ABA-inducible pRAB18::GFP in the presence of DFPM and ABA. The ability of DFPM to attenuate ABA signaling was reduced in rda mutants (resistant to DFPM inhibition of ABA signaling). One of the mutants, rda2, was mapped and is defective in a gene encoding a lectin receptor kinase. RDA2 functions in DFPM-mediated inhibition of ABA-mediated reporter expression. RDA2 is required for DFPM-mediated activation of immune signaling including phosphorylation of MAP kinase 3 (MPK3) and MPK6 and induction of immunity marker genes. Our study identifies a previously uncharacterized receptor kinase gene that is important for DFPM-mediated immune signaling and inhibition of abscisic acid signaling. We demonstrate that the lectin receptor kinase RDA2 is essential for perceiving the DFPM signal and activating MAP kinases, and that MKK4 and MKK5 are required for DFPM interference with ABA signal transduction.
Keywords: Abscisic acid, cross-interference, lectin receptor kinase, DFPM, chemical genetics, plant defense, immune signaling, MAPK, Arabidopsis thaliana
Significance Statement
By activating plant immune signaling, the synthetic small molecule [5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione (DFPM) inhibits signaling by the drought-response hormone abscisic acid (ABA). DFPM thus provides a powerful tool for dissecting mechanisms that enable initial biotic signals to interfere with ABA signal transduction. We have used DFPM to identify, by a forward chemical genetics screen, a lectin receptor kinase that is essential to this process.
Introduction
Plants are continuously challenged with environmental stresses that result in significant impact on plant growth and crop yields. Plants have evolved sophisticated stress tolerance mechanisms to minimize damage and to conserve resources for growth and reproduction. To investigate mechanisms of stress tolerance in plants, traditional research has focused on single stress conditions, for example by imposing individual stresses (Mittler et al., 2010). However, when plants face more than one stress simultaneously, their responses are not easily predictable based on responses to single stresses (Fujita et al., 2006, Atkinson et al., 2012, Ramegowda et al., 2015). Different pairs of environmental stresses result in different physiological and molecular responses, suggesting that plants utilize specific tolerance mechanisms depending on different stress combinations, which cannot be directly inferred from individual stress studies (Mittler et al., 2010, Suzuki et al., 2014). Therefore, detailed investigation of stress tolerance signaling pathways in defined combinations of stresses will be required.
Drought and pathogen infections are two major types of stresses that result in significant reduction in plant growth and yield (Wang et al., 2003, Bruce, 2010, Maxmen, 2013, Hatfield et al., 2015). Agricultural production will increasingly suffer from abiotic stresses including drought (Melillo et al., 2014) and biotic stresses (Oerke, 2005, Fischer et al., 2009). When plants are exposed to drought and pathogens at the same time, overall plant resistance is more reduced than by single stress conditions (Suzuki et al., 2014, Deutsch et al., 2018) and tolerance mechanisms against the two stresses often negatively regulate each other (Mauch-Mani et al., 2005, Prasch et al., 2013).
The phytohormone abscisic acid (ABA) is a major stress hormone that regulates plant responses to abiotic stresses including drought (Cutler et al., 2010, Finkelstein, 2013, Hauser et al., 2017). Roles of ABA and components of ABA signal transduction on regulating plant defense signaling have been discovered (Mauch-Mani et al., 2005, Yasuda et al., 2008, Ton et al., 2009, Bostock et al., 2014). ABA treatment increases pathogen susceptibility (Audenaert et al., 2002, Mohr et al., 2003, Asselbergh et al., 2008a, Fan et al., 2009, Ulferts et al., 2015, Buhrow et al., 2016). Negative roles of ABA in pathogen defense were also suggested by Arabidopsis and tomato mutants deficient in ABA synthesis (Asselbergh et al., 2008a, Fan et al., 2009). Abiotic stress signaling pathways often act antagonistically on pathogen defense signaling under combinations of stresses (Asselbergh et al., 2008b).
Conversely, pathogen infections of plants often result in increased susceptibility to drought (Olson et al., 1990, Atkinson et al., 2015) and reduced responsiveness to ABA (Kim et al., 2011). Plants with increased levels of salicylic acid (SA), an important hormone for plant innate immunity and systemic acquired resistance (SAR), exhibited decreased drought resistance and ABA signal transduction (Németh et al., 2002, Yasuda et al., 2008, Mosher et al., 2010). However, other reports suggest that SA signaling enhances ABA signaling (Hayat et al., 2010, Khan et al., 2015). Therefore, this antagonistic interaction remains very complex and the underlying molecular mechanisms by which immune signaling pathways can interfere with ABA signal transduction remain largely elusive (Atkinson et al., 2015). Since pathogen infections induce complex, layered signaling pathways in plants depending on the pathogen species and time after infection (Fujita et al., 2006), imitating natural conditions to investigate interactions of tolerance mechanisms under drought and pathogen infection events has been difficult.
Chemical genetics is a powerful approach for dissecting signal transduction pathways through bypassing genetic redundancy, mutant lethality and network robustness (Park et al., 2009, Toth et al., 2010). The small molecule DFPM ([5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione) was isolated in a chemical genetics screen and interferes with ABA signal transduction by activating immune signaling (Kim et al., 2011). DFPM rapidly represses ABA-mediated gene expression, activation of slow-type anion channels, and ABA-induced stomatal closing (Kim et al., 2011). Early ABA signaling, including the interaction between the ABA receptor PYRABACTIN RESISTANCE1 (PYR1) and the protein phosphatase type 2C (PP2C) ABA INSENSITIVE1 (ABI1), and ABA-dependent activation of the protein kinase OPEN STOMATA1 (OST1), was not affected by DFPM (Kim et al., 2011). DFPM requires core components of effector-triggered immunity including EDS1 and PAD4 (Bhattacharjee et al., 2011, Heidrich et al., 2011) for DFPM-mediated inhibition of ABA signal transduction (Kim et al., 2011) and for an effector-triggered immune signaling components-requiring growth arrest response that is specific to roots (Kim et al., 2012, Kunz et al., 2016). The co-chaperons RAR1 and SGT1b are also important for DFPM-mediated ABA interference signaling (Kim et al., 2011, Kim et al., 2012). Moreover, a Toll-Interleukin1 Receptor–nucleotide binding–Leucine-rich repeat (TIR-NB-LRR) protein VICTR is essential for DFPM-mediated root growth arrest (Kim et al., 2012, Kunz et al., 2016). Studies using DFPM provide evidence for a ‘cross-interference’ signal, which occurs from biotic stress signaling to inhibit ABA signal transduction and a platform to further dissect the molecular mechanisms for biotic-to-abscisic acid interference signaling.
Here we performed a forward genetic screen to find mutants with reduced levels of ABA signal transduction interference in response to DFPM. resistant to DFPM-inhibition of ABA signaling (rda) mutant plants showing decreased sensitivity to DFPM in inhibiting ABA reporter expression were isolated. We mapped one of the rda mutants, rda2, to a previously uncharacterized lectin receptor-like kinase. RDA2 plays an important role in activating immune signaling and inhibits ABA signal transduction. RDA2 is required for perceiving a signal derived from DFPM and subsequently transducing immune signaling. Furthermore, components of MAPK cascades including MAP kinase kinase 4 (MKK4) and MKK5 are involved in DFPM-mediated interference signaling.
Results
Chemical genetic screening for DFPM hypo-sensitive mutants
The small molecule DFPM ([5-(3,4-dichlorophenyl)furan-2-yl]- piperidine-1-ylmethanethione) was identified through its ability to rapidly inhibit ABA signal transduction by activating an immune signaling pathway (Kim et al., 2011). To dissect DFPM-mediated ABA interference signaling and to find genetic elements involved in this interference signaling, a forward-genetic screen was performed using DFPM (Figure S1). Columbia-0 (Col-0; wild-type) Arabidopsis seeds expressing the ABA response reporter pRAB18::GFP (Kim et al., 2011) were mutagenized with 1.5 to 3 % ethyl methanesulfonate (EMS). Two to three week-old M2 EMS mutant plants were exogenously treated with ABA and DFPM in which ABA-mediated induction of pRAB18::GFP fluorescence signal was inhibited in wild-type plants (Figure S1). Over 26,000 M2 lines were screened to isolate mutant plants which showed an ABA-induced pRAB18::GFP fluorescence signal in leaves despite treatment with 10 μM DFPM. During the first round of screening, 62 putative mutants were isolated and they were further tested in a second round of screening. Several criteria were used to select mutants for further studies: i) plants exhibited a wild-type level of ABA-mediated pRAB18::GFP reporter expression; ii) plants showed less sensitivity to DFPM inhibition of ABA reporter expression over 3 generations and iii) plants did not show a severe growth defect or morphological alteration throughout developmental stages. Isolated mutant plants were named resistant to DFPM-inhibition of ABA signaling (rda) mutants. Among three rda mutants rda1, rda2 and rda3 that were isolated during the screen, rda2 showed the highest reduction in sensitivity to DFPM in leaf epidermal cells (Figure S2).
rda2 is tolerant to DFPM inhibition of ABA signal transduction
rda2 exhibits a reduced sensitivity to DFPM in inhibiting ABA-induced reporter expression (Figure 1). When treated with DFPM and ABA, rda2 plants showed increased levels of pRAB18::GFP fluorescence signal compared to wild-type controls. Confocal microscope images showed enhanced fluorescence in the epidermal layer of true leaves in rda2 (Figure 1A). The difference between rda2 and wild-type leaves was clearly visible in epidermal pavement cells where wild-type leaves showed inhibition of ABA-mediated pRAB18::GFP fluorescence expression in response to DFPM (Figure 1A, B). In the absence of DFPM, ABA-mediated reporter expression was indistinguishable from rda2 and wild type leaves. The strong pRAB18::GFP fluorescence in guard cells (Figure 1 A, B) has been suggested to result from elevated basal ABA concentration in this cell type (Lahr et al., 1988, Waadt et al., 2014, Waadt et al., 2015, Hsu et al., 2018). The intensity of fluorescence signals was measured from multiple images of leaf epidermal layers showing that rda2 has a reduced sensitivity to DFPM (Figure 1C).
Figure 1. rda2 exhibits reduced sensitivity to DFPM.
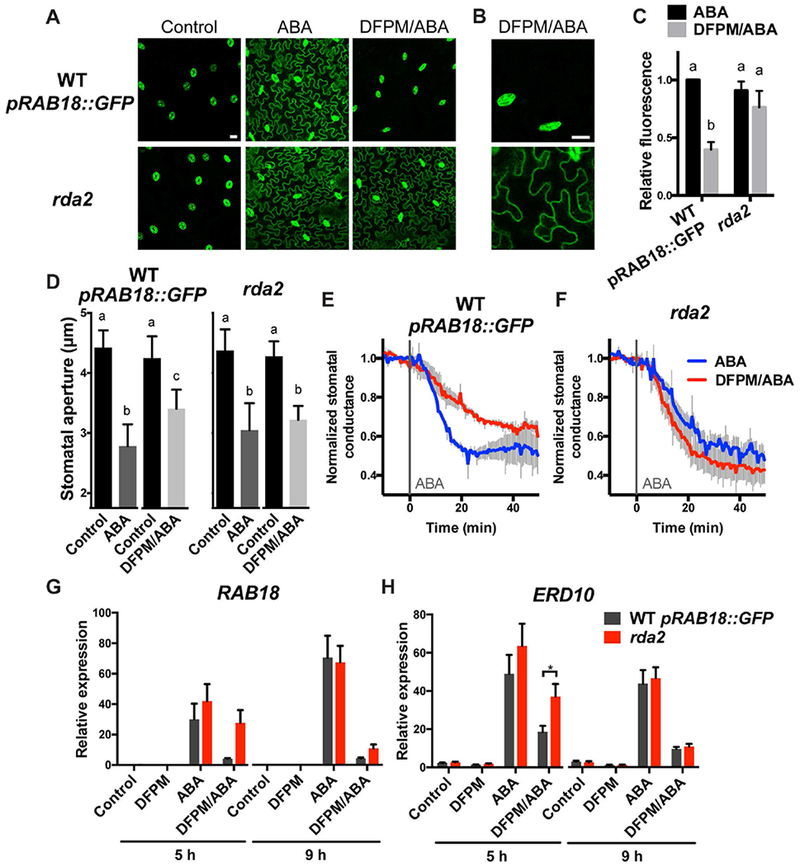
(A) ABA-mediated pRAB18::GFP fluorescence levels were less inhibited by DFPM in epidermal pavement cells of rda2 leaves, when compared to those of wild-type Col-0 expressing pRAB18::GFP (WT pRAB18::GFP). First and second true leaves of two week-old plants were pre-treated with 10 μM DFPM or solvent control for 1 hour, and then 20 μM ABA or solvent control was added. GFP fluorescence was observed from abaxial epidermal layers of leaves using a confocal microscope after 24-26 hours of ABA addition. Scale bar, 20 μm. Representative images from at least 5 biological repeats are shown. (B) Enlarged images of leaf epidermal layers treated with ABA and DFPM. Scale bar, 20 μm. (C) The intensity of pRAB18::GFP fluorescence signal in rda2 and wild type treated with ABA and/or DFPM was quantified. Averages of 5-8 independent experiments, and each are averages of 5-27 images of 3-9 plants per condition per genotype. Data were normalized to the fluorescence intensity of the wild type treated with ABA for each experiment. Error bars = SEM. Asterisk represents P < 0.05 based on two-way ANOVA and Sidak’s test. (D) Effect of DFPM in ABA-induced stomatal closing. Stomatal apertures were measured before (Control) and after 30 min treatment of solvent control or 30 μM DFPM followed with additional 1.5 h treatment of 10 μM ABA (ABA or DFPM/ABA, respectively). Each control bar and their corresponding right bar represent the average widths of the same apertures before and after the chemical treatment, respectively. n = 4 independent experiments, each experiments: 10-27 stomates. Error bars = SEM. Means with different letters are grouped based on two-way ANOVA and Sidak’s test, P < 0.05. (E-F) Time resolved stomatal conductance of wild type (E) and rda2 (F) in response to DFPM and ABA. Leaves were pre-treated with 30 μM DFPM or DMSO solvent control via the transpiration stream, and after 60 min 2 μM ABA was added at time=0 (grey line). Stomatal conductance was measured for an additional 50 min. Data are averages of three independent biological repeats, and error bars are SEM. Stomatal conductance was normalized by averaging 10 min steady-state stomatal conductance before ABA addition. (G-H) ABA-mediated gene expression was less inhibited by DFPM in rda2. Expression levels of ABA responsive RAB18 (G) and ERD10 (H) in wild-type and rda2 plants were monitored via real-time qPCR after pre-treated with 30 μM DFPM or solvent control for 1 hour and then incubated in 10 μM ABA or solvent control for the indicated time points. Expression levels were normalized with the housekeeping gene PDF2. Results are averages of n=4 biological replicates. Asterisk represents P < 0.05 based on two-way ANOVA and Sidak’s test.
In addition to the ABA response reporter, other ABA responses of rda2 were investigated. Treatment of ABA resulted in stomatal closure, whereas pre-exposure to DFPM for 30 min followed by ABA exposure slightly inhibited ABA-mediated stomatal closing in wild-type leaves at 90 min after ABA treatment (Figure 1D). Time-resolved stomatal movement analyses in intact leaves provide a more robust and kinetic method for investigating stomatal responses (Hsu et al., 2018). Time-resolved stomatal conductance was monitored in intact leaves in the presence of DFPM and ABA by whole leaf gas exchange experiments (Figure 1E). Addition of 2 μM ABA to the transpiration stream via petioles resulted in a rapid decrease in stomatal conductance in wild-type leaves. When wild-type leaves were pre-exposed to DFPM via the transpiration stream for 60 min followed by ABA treatment, stomatal closing in response to ABA was partially inhibited (Figure 1E). Average maximum rates in stomatal conductance reduction in figure 1E were −0.0385 min−1 for wild-type leaves treated with ABA, and −0.0151 min−1 for wild type with DFPM/ABA (P = 0.002, two-way ANOVA and Sidak’s multiple comparison test). The relatively weaker effect of DFPM in stomatal aperture assays (Figure 1D) is consistent with the effect of DFPM on the kinetics of stomatal closing. In rda2 mutant leaves, pre-incubation in DFPM did not result in a clear inhibition of ABA-mediated stomatal closing (Figure 1F). Average maximum rates in stomatal conductance reduction were −0.0232 min−1 for rda2 leaves treated with ABA, and −0.0306 min−1 for rda2 with DFPM/ABA (P = 0.279, two-way ANOVA and Sidak’s multiple comparison test), showing reduced sensitivity of rda2 to DFPM.
ABA-induced gene expression was examined in 2 week-old wild-type and rda2 plants in the presence or absence of DFPM using real-time quantitative PCR (Figure 1G, H). Pre-exposure of wild-type seedlings to 10 μM DFPM for 1 hour followed by 10 μM ABA exposure for 5 hours or 9 hours inhibited ABA-mediated increase in gene expression (Figure 1G, H). In contrast, rda2 seedlings exhibited a less severe DFPM inhibition of ABA-induced RESPONSIVE TO ABA 18 (RAB18) and EARLY RESPONSIVE TO DEHYDRATION10 (ERD10) expression at 5 hours after ABA treatment, while there was no clear change at 9 hours after ABA treatment (Figure 1G, H). The described results demonstrate that rda2 mutants are not impaired in ABA responses, but altered in their ABA responses after DFPM exposure.
rda2 carries a recessive mutation, as F1 generation plants after backcrossing rda2 to the parental (pRAB18::GFP) wild type plants exhibited wild type-like DFPM sensitivity. When self-pollinating F1 progeny, 88 F2 plants among 387 (22.7%, χ2=1.055) showed rda2-like reduced DFPM sensitivity suggesting that rda2 carries a single recessive mutation. The sequence of the promoter region in the ABA responsive reporter pRAB18::GFP was also determined in rda2 and showed no mutation. This suggests the integrity of the reporter expression construct in rda2. rda2 was not allelic with the other rda mutants, rda1 and rda3. Crossing rda2 with rda1 resulted in F1 progeny with a wild-type level DFPM response. As rda3 was isolated after rda2 was mapped, the genomic DNA region of the RDA2 gene was sequenced in rda3 and no mutation was found.
We investigated whether rda2 carries any mutation in genes that are necessary for DFPM signaling including core components of diseases resistance signaling, EDS1 and PAD4, co-chaperone genes SGT1b and RAR1, and the TIR-NB-LRR gene encoding VICTR (Kim et al., 2011, Kim et al., 2012). Genomic DNA regions of these 5 genes were PCR-amplified in the rda2 mutant and sequenced. rda2 does not carry mutations in EDS1, PAD4, SGT1b, RAR1 and VICTR1 suggesting that it harbors a mutation in a novel genetic component that is required for the interference signal that inhibits ABA signal transduction by DFPM.
Previous studies showed that eds1, pad4, sgt1b, and rar1 mutants exhibited reduced sensitivity to DFPM in inhibiting ABA-mediated gene expression in whole seedlings (Kim et al., 2011). Note that victr mutants have not yet been analyzed for DFPM sensitivity in inhibiting ABA-induced gene expression. VICTR expression is strongly induced in roots and VICTR is a member of four homologous tandem repeat TIR-NB-LRR genes (Kim et al., 2012). To examine DFPM sensitivity with ABA reporter expression in leaves, mutant plants of EDS1, PAD4, SGT1b, RAR1, VICTR1 and its closest homolog gene VICTR Like 1 (VICTL1) (Kim et al., 2011, Kim et al., 2012) were introduced with the pRAB18::GFP reporter by cross-pollination. The mutant plants showed inhibition of ABA-mediated pRAB18::GFP fluorescence levels in epidermal pavement cells in response to DFPM, similarly to the corresponding wild type (Figure S3), showing a difference in whole seedling RT-qPCR analyses of RAB18 expression (Kim et al., 2011) and leaf pavement cell pRAB18::GFP reporter protein activity. These results suggest that EDS1, PAD4, SGT1b, RAR1, VICTR and VICTL1 are not primarily necessary for DFPM inhibition of ABA-mediated pRAB18::GFP expression at least in leaf epidermal pavement cells. rda2 is a single locus exhibiting the strongest DFPM-insensitive mutant phenotype in leaves, whereas VICTR and early ETI signaling components are required for the DFPM response in roots.
Mapping of rda2 via whole genome sequencing
To map the causative mutation for reduced DFPM sensitivity in rda2, the rda2 mutant plants (named rda2-1 from here onwards) were backcrossed into the wild-type parent line expressing pRAB18::GFP. F2 plants from self-pollinating F1 progeny were screened for individuals with an rda2-like phenotype showing higher ABA-mediated pRAB18::GFP fluorescence levels in leaves pre-treated with DFPM. Eighty-six F2 plants with reduced sensitivity to DFPM were selected for a mapping population.
Genomic DNA pooled from the 86 F2 lines of the mapping population was used for whole genome sequencing. The parental Col-0 plants that express the ABA responsive reporter pRAB18::GFP and the non-allelic rda1 mutant were also used for whole genome sequencing for comparison. Single nucleotide polymorphisms (SNPs) of rda2 also found in the parental line and rda1 were excluded. Occurrence ratios of rda2-specific SNPs and reference type bases for each base position were plotted throughout the five chromosomes of Arabidopsis thaliana (Figure 2)(Schneeberger et al., 2009), and showed a clear single peak with high density of rda2 SNPs in chromosome 1, ranging from 2.2 to 4.8 Mb. This region contains 26 EMS-type C-to-T or G-to-A mutations, and among them 16 mutations resulted amino acid substitutions, premature stop codons or predicted splicing errors (Figure 2, Table S1). None of the 16 genes which carry rda2-specific SNPs was reported previously to have a function in ABA signal transduction or immune responses.
Figure 2. Genome-wide SNPs of rda2 identified by whole genome sequencing-based mapping.
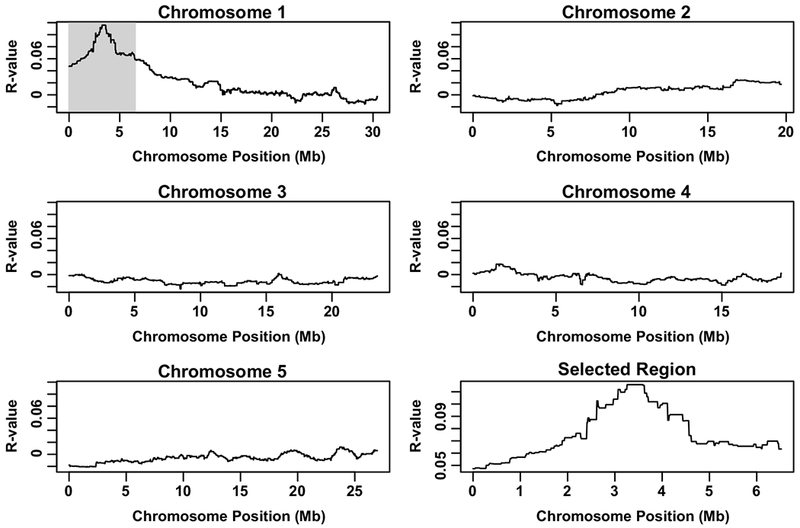
rda2-specific single nucleotide polymorphisms (SNPs) were identified from whole genome sequencing of rda2 mapping population and the corresponding wild type Col-0 expressing pRAB18::GFP. Occurrence ratios of rda2-specific SNPs and wild-type reference bases (TAIR 10) for each base position were plotted through the five Arabidopsis thaliana chromosomes. Note that the gray area of chromosome 1 shows a peak with higher ratios of rda2-specific SNPs. This area is shown in the right bottom graph in detail. Y axis: R-values of each base position. R-values are calculated by dividing the number of reads with a non-reference base by [the number of reads with a reference base multiplied by the total read number at each position] as previously reported (Schneeberger et al., 2009).
T-DNA insertion mutant lines for the 16 genes containing SNPs were used to identify the causative mutation for rda2. Homozygous T-DNA knockout lines were tested for DFPM sensitivity in inhibiting ABA-mediated gene expression. We were not able to isolate homozygous mutants in two of the T-DNA lines, At1g11330 (SALK_143489) and At1g09750 (GABI_565C02). Instead, their heterozygous lines were crossed with the rda2 mutant to determine allelism between the causative mutation of rda2 and the T-DNAs (Figure S4). If any T-DNA is allelic with the rda2 mutant, F1 progeny after these crosses would include 50% of plants exhibiting an rda2-like DFPM response. When heterozygous plants of the GABI_565C02 T-DNA insertion line, mutated in At1g09750 were crossed with rda2, all F1 generation plants showed a WT-like DFPM response (Figure S4B). However, half of the F1 population from the cross between heterozygous SALK_143489 and rda2 exhibited a phenotype similar to rda2, showing a reduced sensitivity to DFPM (Figure S4A). Genotyping of the F1 lines revealed that half of the F1 lines showing the rda2-like DFPM response carried a T-DNA insertion in At1g11330. As a control, the same heterozygous T-DNA insertion line plants (SALK_143489) were crossed with the wild-type parent pRAB18::GFP line, and all F1 plants tested showed wild type-like DFPM sensitivity (Figure S4A). These results suggest that the T-DNA insertion line SALK_143489 in the At1g11330 gene is allelic to rda2. The SALK_143489 line was later named as rda2-3.
RDA2 encodes a putative lectin receptor kinase
In rda2, a C-to-T mutation in At1g11330 resulted in change of Glutamine 574 to a premature stop codon (Q574*) (Figure 3A, Table S1). To independently investigate whether At1g11330 is responsible for the observed phenotypes of the rda2-1 mutant, an additional homozygous T-DNA insertion line rda2-2 (SALK_122993) was isolated (Figure 3A). This suggests that the inability to isolate homozygous rda2-3 (SALK_143489) may result from a linked second site mutation. rda2-2 was transformed with the ABA response marker pRAB18::GFP to examine the DFPM response in inhibiting ABA-mediated reporter expression. Similarly to the rda2-1 EMS mutant, leaves of the rda2-2 T-DNA insertion mutant exhibited a wild-type level of ABA-mediated reporter expression (Figure 3B). When treated with DFPM, rda2-2 showed a reduced sensitivity in inhibiting the ABA-mediated GFP fluorescence signal in epidermal pavement cells of leaves, when compared to wild-type Col-0 plants transformed with the pRAB18::GFP reporter (Figure 3B). The ABA-mediated decrease in stomatal conductance was examined in intact wild-type and rda2-2 leaves after pre-exposure to DFPM or a solvent control (Figure 3C, D). In DFPM-treated wild-type leaves, stomatal closing in response to ABA was partially inhibited compared to the solvent control (Figure 3C). In rda2-2 leaves, DFPM did not result in a strong inhibition in ABA-induced stomatal closing (Figure 3D). DFPM sensitivity of rda2-2 was also tested in ABA-mediated gene expression by real-time qPCR. In the wild type, ABA-induced expression of RAB18 and ERD10 was inhibited by DFPM (Figure 3E, F). In rda2-2, the decrease in ABA-mediated gene expression levels by DFPM was smaller than in the wild type after 1 hour of DFPM pre-treatment followed by 5 hours of ABA treatment (Figure 3E, F). A clear difference was not observed at 9 hours after ABA treatment. Together, these results showed that the rda2-2 T-DNA insertion line resembles reduced DFPM sensitivity of the rda2-1 EMS mutant, and suggest that the mutations in the At1g11330 gene are responsible for the rda2 phenotypes.
Figure 3. Identification of RDA2 as a putative lectin receptor kinase gene.
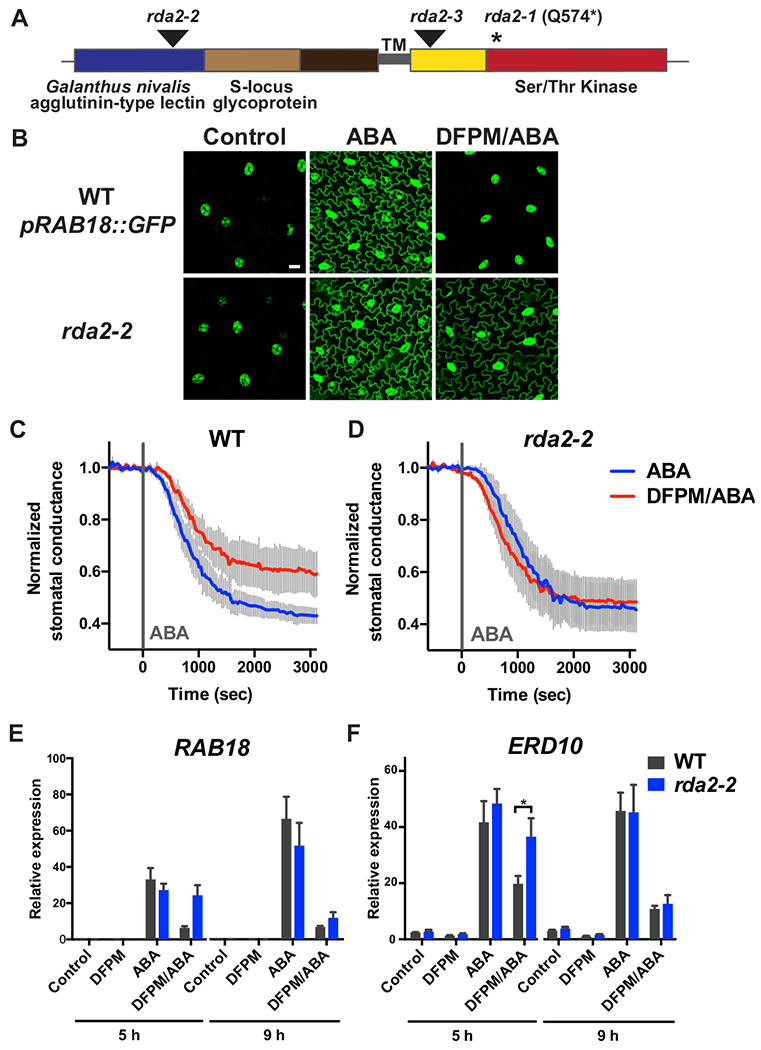
(A) Location of the causative mutation in rda2-1 (*) and T-DNA insertion (arrowheads) in rda2-2 and rda2-3 mutant alleles. Predicted protein domains including a lectin domain and a protein kinase domain in RDA2 are also presented. (B) DFPM-mediated inhibition of ABA reporter pRAB18::GFP expression was less pronounced in leaf epidermal pavement cells of rda2-2 compared to the wild type Col-0 expressing pRAB18::GFP (WT pRAB18::GFP). First and second true leaves of 2 week-old plants expressing pRAB18::GFP were pre-treated with DFPM or solvent control for 1 hour and then 20 μM ABA or solvent control was added. After 24-26 hours of ABA addition, GFP fluorescence signals were observed from abaxial epidermal layers of leaves using a confocal microscope. Scale bar, 20 μm. Representative images from at least 3 biological repeats are shown. (C-D) rda2-2 exhibited reduced sensitivity to DFPM-mediated inhibition of ABA-induced stomatal closing. Leaves of Col-0 wild type (C) and rda2-2 (D) were pre-treated with 30 μM DFPM or solvent control for 60 min, and then treated with 2 μM ABA treatment at time=0 (grey line) via the transpiration stream. Stomatal conductance was measured for an additional 50 min. Data are averages of 3-4 independent biological replicates, and error bars are SEM. Stomatal conductance is normalized by averages of 10 min steady-state stomatal conductance before ABA addition. (E-F) ABA-induced expression levels of RAB18 (E) and ERD10 (F) were less suppressed by DFPM in rda2-2 mutants than in wild type plants. For real-time qPCR, plants were pre-treated with 30 μM DFPM or solvent control for 1 h and then incubated in 10 μM ABA or solvent control for the indicated time points. Expression levels were normalized with the housekeeping gene PDF2. Results are averages of n=4 biological repeats. Error bars = SEM. Asterisk represents P < 0.05 based on two-way ANOVA and Sidak’s test.
DFPM-mediated immune MAPK activation requires RDA2
RDA2 encodes a putative lectin receptor kinase (LecRK) (Figure 3A). Lectin receptor kinases are composed of an extracellular lectin domain known to bind carbohydrates and an intracellular protein kinase domain, separated by a single hydrophobic transmembrane domain (Figure 3A) (Bellande et al., 2017). In Arabidopsis thaliana and other plant species, LecRKs play a role in pathogen-associated molecular pattern (PAMP)-triggered immune signaling (PTI) by perceiving PAMPs or molecules released to the apoplast during pathogen infection (Chrispeels et al., 1991, Chen et al., 2006, Singh et al., 2013, Vaid et al., 2013, Lannoo et al., 2014, Ranf et al., 2015, Bellande et al., 2017, Chen et al., 2017, Wang et al., 2017).
Plants exposed to DFPM activate immune signaling including expression of Pathogenesis-related 5 (PR5) gene in an EDS1 and PAD4-dependent manner (Kim et al., 2011, Kim et al., 2012). Note that PR5 expression is also induced by PAMPs including flg22 in the PTI response (Asai et al., 2002). Therefore, we analyzed whether DFPM treatment can activate mitogen-activated protein kinases (MAP kinases), which is one of the earliest cellular responses in PTI signaling (Meng et al., 2013, Frei dit Frey et al., 2014). Wild-type plants treated with flagellin 22 (flg22) for 15 min to 2 hours phosphorylated MPK3 and MPK6 at the highest levels at 15 and 30 min after flg22 treatment (Figure 4A). Levels of phosphorylated MPK3 and MPK6 were then reduced (Figure 4A), similarly to previously reported findings for PTI signaling responses (Frei dit Frey et al., 2014). When wild-type Col-0 plants were exposed to DFPM for the same time periods, phosphorylation levels of MAP kinases increased to the highest level at 15 and 30 min after DFPM treatment and then decreased (Figure 4A). The DFPM-mediated activation of MAP kinases was weaker than that mediated by flg22, but the temporal dynamics were comparable (Figure 4A). The lower band of the two phosphorylated MAP kinase bands by DFPM was significantly reduced in the mpk3-1 mutant allele (Figure 4B) (Wang et al., 2007), whereas the upper band was reduced in the mpk6-2 mutant (Figure 4B) (Liu et al., 2004) supporting that the two bands are MPK3 and MPK6. These results suggest that DFPM could activate the MAP kinases MPK3 and MPK6 with a time-dependence similar to flg22-triggered MAPK activation.
Figure 4. Involvement of RDA2 in DFPM-mediated PAMP-triggered immune signaling.
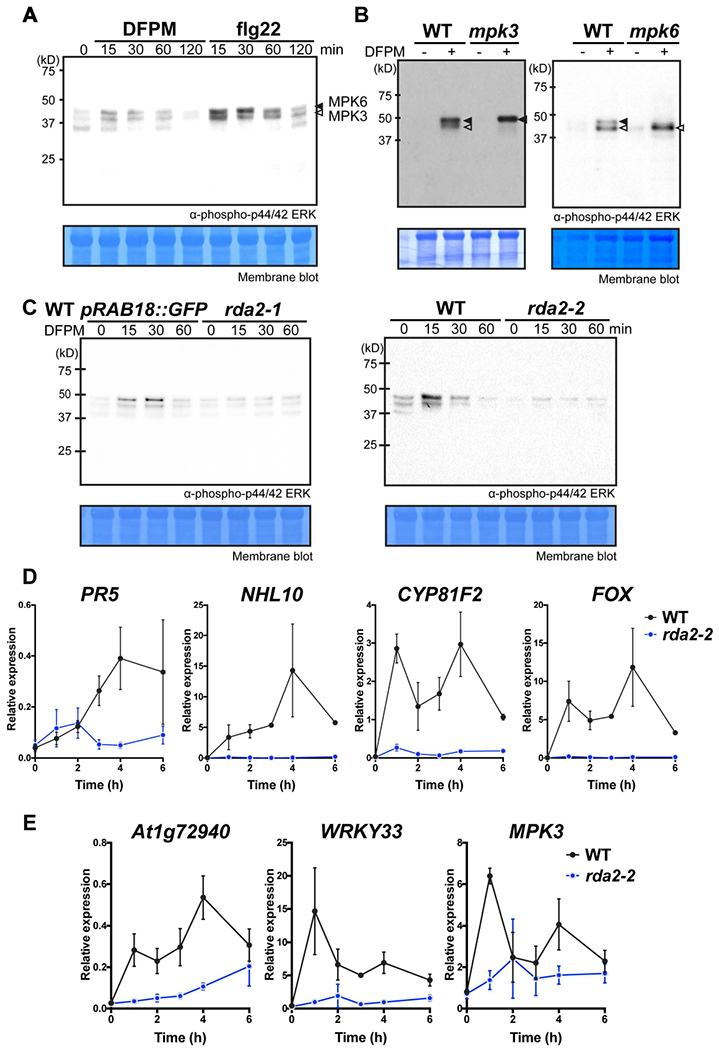
(A) DFPM activates MAP kinases (MPKs) within 15 min in wild-type plants. Arrowheads represent two MAP kinases, MPK3 and MPK6, which were activated by flg22. Wild-type Col-0 plants were treated with 30 μM DFPM or 1 μM flagellin 22 (flg22) for the indicated time periods. Whole proteins were used for Western blotting with anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (α-phospho-p44/42 ERK). Loading controls: coomassie blue staining of the membrane. A representative blot from three biological repeats is shown. (B) The lower band of the two bands (open arrowheads) shown in DFPM-treated wild type plants was reduced in the mpk3-1 mutant. The upper band (filled arrowheads) was weaker in the mpk6-2 mutant. Plants were treated with 30 μM DFPM or solvent control for 30 min. (C) DFPM-mediated activation of MAP kinases was diminished in rda2-1 and rda2-2 mutants compared to the corresponding wild-type plants. Col-0 expressing pRAB18::GFP (WT pRAB18::GFP) was used as a wild-type control for rda2-1 and Col-0 (WT) was used as a wild-type control for rda2-2. Plants were treated with 30 μM DFPM for the indicated time periods. (D) Transcription of PAMP-responsive genes PR5, NHL10, CYP81F2 and FOX was induced by DFPM in wild type plants. DFPM-induced gene expression was strongly impaired in the rda2-2 mutant. For real-time qPCR, plants were treated with 30 μM DFPM for 1, 2, 3, 4 and 6 hours. Expression levels were normalized with the housekeeping gene PDF2. n=3 biological replicates. Error bars = SEM. (E) Expression of ETI-specific genes (Mine et al., 2018) a TIR-NB-LRR gene At1g72940, WRKY33 and MPK3 was induced by DFPM in the wild type. DFPM-mediated induction of these genes is partly impaired in rda2-2. Real-time qPCR was performed as in (D). n=3 biological replicates. Error bars = SEM.
To determine whether RDA2 functions in DFPM-mediated activation of MAP kinases, rda2 mutant plants exposed to DFPM were examined along with wild-type plants. Levels of phosphorylated MPK3 and MPK6 were clearly reduced in the rda2-1 and rda2-2 mutants treated with DFPM, when compared to wild type (Figures 4C, S5). In some experiments, DFPM activation of MAP kinase phosphorylation remained high in wild type between 15 and 60 minutes (Figure S5). But in these experiments, rda2 exhibited strong reduction in DFPM-mediated MAP kinase phosphorylation (Figure S5). To examine whether rda2 mutants are capable of activating MAP kinases upon exposure to other stimuli, rda2 mutant plants were treated with the pathogen elicitor flg22 (Figure S6A). rda2 mutant plants showed flg22-mediated phosphorylation in MAP kinases, similar to wild type plants (Figure S6A). These results together show that rda2 mutants are defective in DFPM-mediated activation of MAP kinases.
To further dissect whether DFPM-mediated activation of MAPKs via RDA2 is affected by genes required for DFPM signaling (Kim et al., 2011, Kim et al., 2012), we investigated mutant plants in eds1, pad4, eds1 pad4, sgt1b, rar1 and victr1 (Kim et al., 2012) (Figure 5). The near-isogenic line NIL-Bu-5, which lacks VICTR, VICTL1 and two tandem homologous TIR-NB-LRR genes, was generated by backcrossing Arabidopsis thaliana accession Bu-5 with Col-0 five times (Ariga et al., 2017) and analyzed for DFPM-mediated MAPK activation with the corresponding control (NIL-Col-0) (Ariga et al., 2017). All tested mutants showed phosphorylation of the MAP kinases in response to DFPM, which were comparable to the wild type (Figure 5). Also, the NADPH oxidase genes RbohD and RbohF that are required for rapid production of ROS upon pathogen infection (Couto et al., 2016) were examined for their involvement in DFPM-mediated MAPK activation. A double mutant allele rbohD rbohF-F3 (Kwak et al., 2003) did not exhibit a strong reduction in levels of phosphorylated MAP kinases in response to DFPM, when compared to the wild type (Figure 5C). Therefore, EDS1, PAD4, SGT1B, RAR1, VICTR, RbohD and RbohF appear not to be necessary for DFPM-mediated activation of MAP kinases.
Figure 5. DFPM-mediated activation of MAP kinases in immune-deficient and DFPM signaling-deficient mutants.
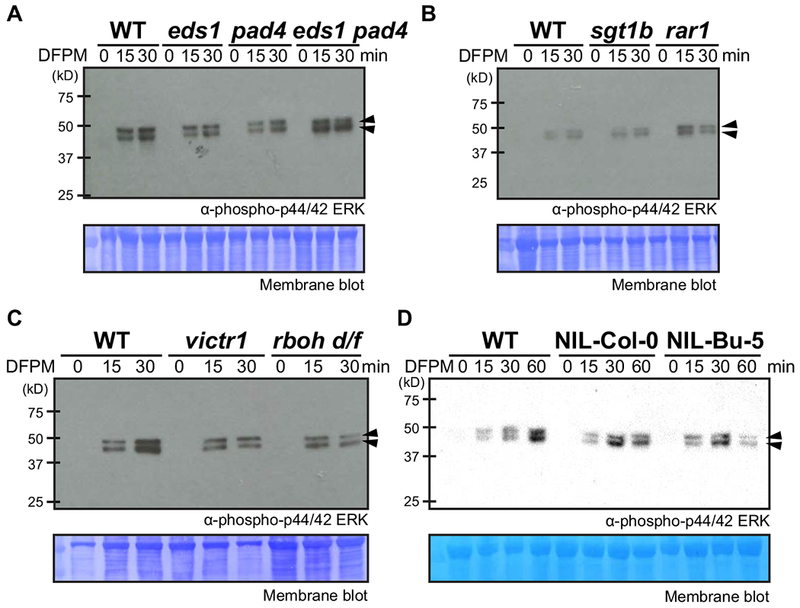
DFPM-mediated activation of MAP kinases was examined in eds1, pad4, eds1 pad4 (A), sgt1b, rar1 (B), victr1, and rbohd rbohf (C) mutants. A near-isogenic line of Col-0 and Bu-5 that lacks VICTR and 3 homolog TIR-NB-LRR genes (NIL-Bu-5) and its corresponding control (NIL-Col-0) (D) plants were also tested. Col-0 was used as wild type (WT). None of the tested mutants exhibited a clear alteration in DFPM-mediated activation of MAP kinases (arrowheads). Plants were treated with 30 μM DFPM for the indicated time points and used for whole protein extraction and subsequent Western blotting. Phosphorylated MAP kinases were detected using anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody. Loading controls: coomassie blue staining of membranes. Representative results from three independent repeats are shown.
To examine whether DFPM mediates other types of PTI responses, reactive oxygen species (ROS) production in response to DFPM was analyzed, as ROS production is one of the first physiological outputs after PAMP perception (Couto et al., 2016). When leaves of wild type plants were treated with DFPM, levels of ROS production were not clearly distinguishable from ones treated with the solvent control (Figure S6B). Likewise, there was no clear change in ROS production in rda2 mutant plants treated with DFPM (Figure S6B) suggesting that DFPM does not induce a strong oxidative burst in leaves. We further analyzed the PAMP-induced rapid burst of ROS in rda2 mutants in response to flg22 or the endogenous Plant Elicitor Peptide 1 (PEP1) (Yamaguchi et al., 2011, Bartels et al., 2015). In rda2-1 and rda2-2 mutant plants, ROS production in response to flg22 or PEP1 was not different from that of wild-type plants (Figure S6C). These data provide positive controls for PAMP-induced ROS burst and suggest that RDA2 is not involved in perception of flg22 and PEP1.
To further investigate whether RDA2 is required for DFPM-induced immune-responsive gene expression, transcript levels of PAMP-responsive genes including PR5, NDR1/HIN-LIKE10 (NHL10), cytochrome P450 monooxygenase CYP81F2 and FAD-LINKED OXIREDUCTASE (FOX) (Boudsocq et al., 2010, Couto et al., 2016) were examined in wild-type and rda2-2 mutant plants treated with DFPM (Figure 4D). After 1 to 6 hours of DFPM treatment, the transcript levels of all four genes were induced by DFPM in wild type seedlings. In rda2-2 seedlings, DFPM-mediated expression of NHL10, CYP81F2 and FOX was severely impaired (Figure 4D). The expression levels of the PR5 transcript in rda2-2 were compromised after 3 hours of DFPM treatment, compared to wild type controls (Figure 4D). These results suggest that DFPM induces expression of PAMP-responsive genes, and RDA2 is important for DFPM-induced expression of PAMP-responsive genes.
In the same time periods, we also investigated whether DFPM induces expression of genes reported to be specifically induced in response to effector-triggered immune (ETI) stimulation (Mine et al., 2018) (Figure 4E). Expression levels of a TIR-NB-LRR gene At1g72940, WRKY33 and MPK3 were increased in wild type plants treated with DFPM (Figure 4E). In the rda2-2 mutant, DFPM-mediated expression of At1g72920 and WRKY33 was lower than in the wild type at 1 to 4 hours of DFPM treatment. A difference in DFPM-mediated MPK3 expression between wild type and rda2-2 plants was observed at 1 and 4 hours after DFPM exposure (Figure 4E). These results suggest that DFPM-mediated induction of the ETI-linked transcript levels At1g72940, WRKY33 and MPK3 is partly dependent on RDA2.
To assess whether RDA2 is involved in bacterial resistance, leaves of rda2-1 and wild type plants were infiltrated with Pseudomonas syringae pv. tomato strain DC3000 (Pst). Pst growth at 2 days after infection was not different between rda2 and the wild type (Figure 6A) suggesting that loss-of-function in RDA2 alone does not affect Pst tolerance in plants. We also tested whether RDA2 is involved in Pst tolerance when leaves are pre-treated with flagellin 22 (flg22). Wild type plants pre-treated with flg22 exhibited increased tolerance to Pst. A mutant allele of FLAGELLIN-SENSITIVE 2 (FLS2) that encodes a receptor for flg22 did not show increased tolerance by flg22 pre-treatment (Figure 6B), as described previously (Zipfel et al., 2004). In response to pre-treatment with flg22, rda2 showed increased tolerance to Pst compared to the non-pretreatment control, similarly to the wild type control (Figure 6B), suggesting that RDA2 is not responsible for FLS2-mediated immune signaling. Furthermore, we examined whether DFPM treatment can improve pathogen tolerance in planta. Leaves of wild-type and rda2 plants were infiltrated with DFPM or solvent control at the time as Pst treatment (Figure 6C). Wild type plants treated with DFPM did not show a clear change in Pst tolerance at 2 and 5 days after infection compared to solvent control (Figure 6C). rda2 also showed no apparent change in Pst growth between DFPM and solvent treatment conditions. The results suggest that DFPM does not enhance plant tolerance to Pseudomonas syringae DC 3000 (Pst), when infiltrated into leaves. DFPM causes a substantial immune signaling effect in roots (Kim et al., 2012) and future research with diverse root pathogens would be of interest.
Figure 6. Growth of syringe-infiltrated Pseudomonas syringae pv. tomato DC3000 (Pst) in rosette leaves.
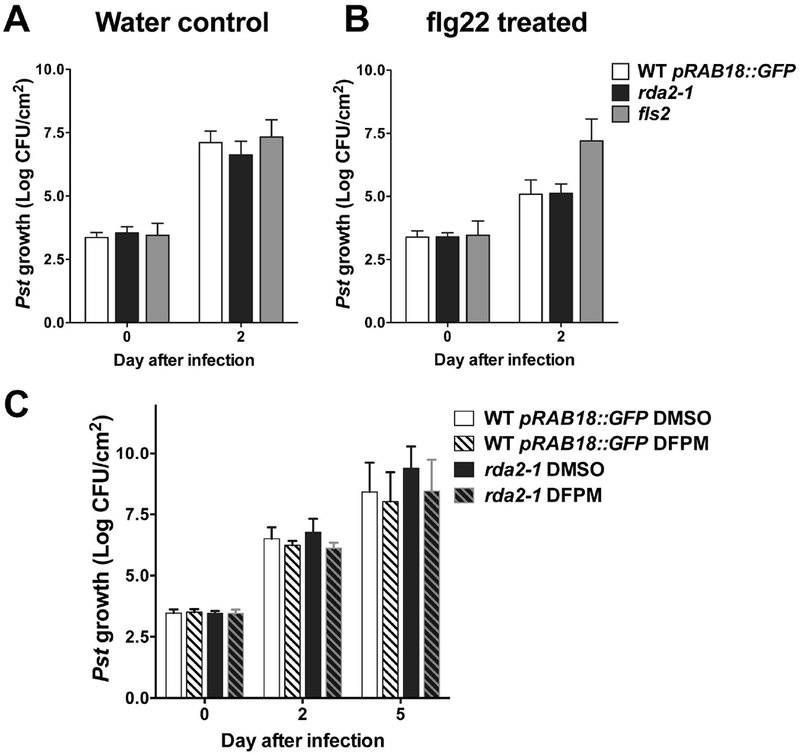
(A-B) Leaves of rda2-1, wild-type and fls2 plants were infiltrated with 1 μM flg22 (B) or water control (A), and after one day the same leaves were infiltrated with Pst. Growth of bacteria was determined at the indicated time points. n=4 independent experiments for WT and rda2-1 and n=3 for fls2. Each experiment contains 7-8 plants per genotype. Error bars = SEM. (C) Leaves of rda2-1 and wild-type plants were infiltrated with 30 μM DFPM or solvent control when treated with Pst. Growth of bacteria was determined at the indicated time points. Two-way ANOVA and Tukey’s test showed no difference between genotypes. n=3 independent experiments which contain 8 plants per genotype. Error bars = SEM. CFU: colony-forming units.
Protein kinase activity of RDA2
To investigate whether the putative lectin receptor kinase RDA2 has a protein kinase activity, the intracellular domain that contains the protein kinase domain of RDA2 was fused with an N-terminal StrepII tag and a C-terminal GST tag, and expressed and purified from Escherichia coli (Figure 7). In vitro kinase analyses revealed that the recombinant intracellular kinase domain of RDA2 exhibits ATP-dependent auto-phosphorylation and trans-phosphorylation activity of the synthetic substrate, myelin basic protein (MyBP) (Figure 7). The protein kinase activity of RDA2 was greatly reduced when the lysine residue K552 in the ATP-binding domain that is conserved within RDA2 and the receptor kinases FLS2 and BRI1-ASSOCIATED RECEPTOR KINASE (BAK1) (Li et al., 2014), was mutated to a glutamic acid (K552E) (Figure 7). The protein kinase activity of the recombinant RDA2 intracellular domain was also reduced by kinase inhibitors K252a (10 μM) and staurosporine (10 μM) (Figure 7). To investigate whether DFPM affects the protein kinase activity of the intracellular kinase domain of RDA2, 50 to 200 μM DFPM was added to protein phosphorylation reactions. DFPM did not increase or decrease the in vitro protein kinase activity of the recombinant intracellular kinase domain of RDA2 (Figure 7). This result suggests that the protein kinase activity of the intracellular domain of RDA2 does not directly respond to DFPM.
Figure 7. Protein kinase activity of RDA2.
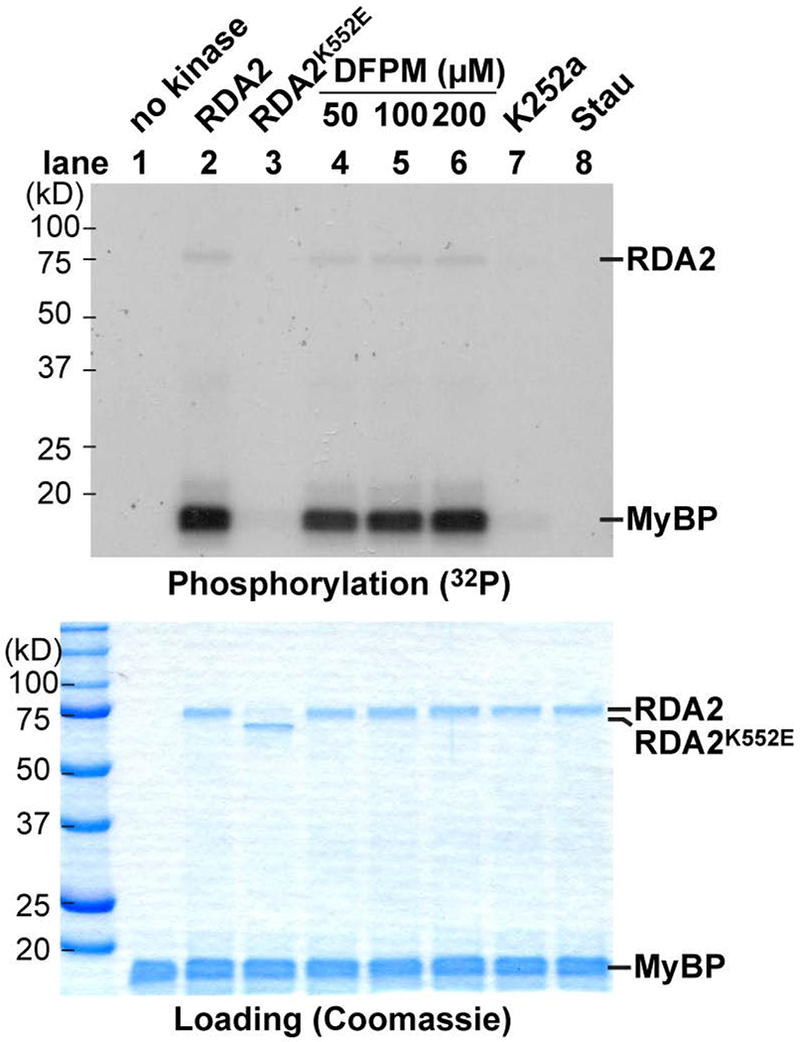
(Lane 1-2) The recombinant intracellular kinase domain of RDA2 exhibited an autophosphorylation activity and a trans-phosphorylation activity to a synthetic substrate myelin basic proteins (MyBP). The intracellular kinase domain of RDA2 was expressed in E. coli, purified and used for in vitro phosphorylation assay with [γ−32P]-ATP. Phosphorylated proteins were detected by autoradiography after SDS-PAGE. (Lane 3) The kinase activity of the recombinant RDA2 protein was strongly diminished, when a conserved lysine residue in the ATP binding domain was mutated to a glutamic acid (K552E). Note that K552E mutation resulted in a slight shift of the recombinant protein on SDS-PAGE gel. (Lanes 4-6) Addition of 50 to 200 μM DFPM to phosphorylation assay reactions did not alter the kinase activity of the recombinant intracellular kinase domain of RDA2. (Lanes 7-8) Pre-incubation with protein kinases K252a and staurosporine (Stau) for 30 min before phosphorylation reactions suppressed the kinase activity of RDA2. Loading control: coomassie blue staining of SDS-PAGE gel.
To analyze involvement of MAP kinase kinase kinase (MAPKKK) and MAP kinase kinase (MAPKK) in DFPM signaling, DFPM-mediated MAPK activation was examined in the presence of inhibitors for MAPKKKs and MAPKKs (Figure 8A). Wild-type plants were treated with DFPM in the presence or absence of GW5074, Sorafenib or PD098059. Sorafenib, an inhibitor for Raf-type MAPKKKs, inhibited DFPM-mediated activation of MPK3 and MPK6 in wild type plants, while another Raf-type MAPKKK inhibitor GW5074 did not show a clear effect (Figure 8A). The PD098059 MAPKK inhibitor did not abolish DFPM activation of the MAP kinases (Figure 8A). These results suggest that Sorafenib-sensitive Raf-type MAPKKKs may be involved in DFPM-mediated MAP kinase activation.
Figure 8. Involvement of MAP kinase cascades in DFPM signaling.
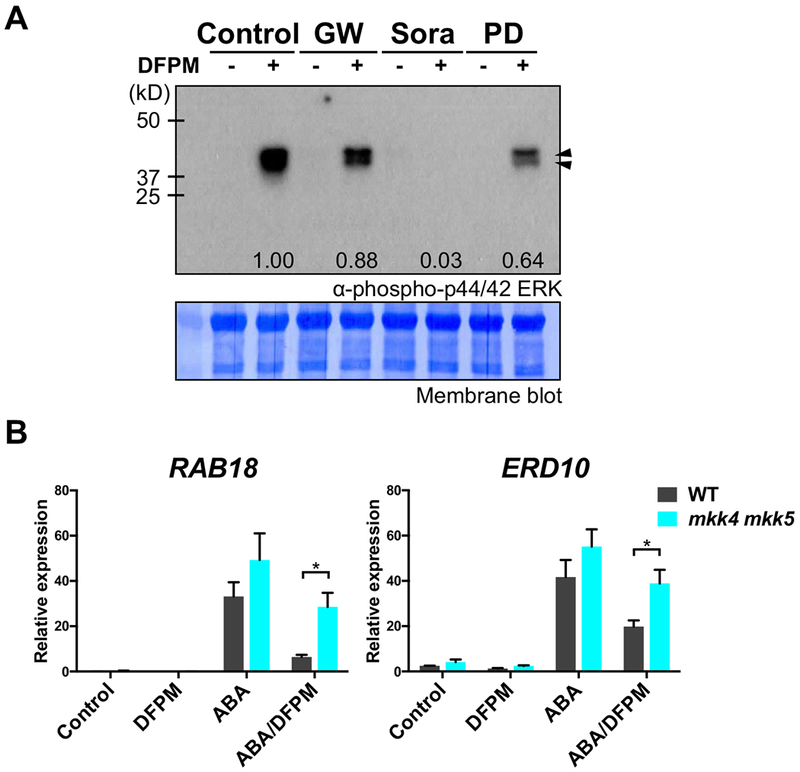
(A) Sorafenib, an inhibitor for Raf-type MAPKKKs suppressed DFPM-mediated activation of MAP kinases (arrowheads), while other MAPK cascade inhibitors GW5074 and PD098059 did not exhibit a clear inhibition. Wild-type Col-0 plants were treated with 30 μM DFPM or solvent control in the presence of 10 μM of inhibitors for MAPKKs and MAPKKKs, GW5074 (GW), Sorafenib (Sora) or PD098059 (PD). Phosphorylated MAPKs were detected by Western blotting using anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody. A representative result from two biological repeats is shown. The intensity of phosphorylated MPK3 and MPK6 bands was quantified and is presented as ratios to the DFPM control without inhibitor treatment.
(B) DFPM-mediated inhibition of ABA marker gene expression was less pronounced in mkk4 mkk5 mutant plants when compared to Col-0 wild type plants (WT). Plants were pre-treated with 30 μM DFPM or solvent control for 1 hour and then incubated in 10 μM ABA or solvent control for additional 5 hours. Expression levels were normalized with the housekeeping gene PDF2. Averages of 4 biological replicates are shown. Error bars = SEM. Asterisks represent P < 0.05 based on two-way ANOVA and Sidak’s test.
As DFPM enhanced phosphorylation of MPK3 and MPK6 in an RDA2-dependent manner (Figure 4) and RDA2 is required for DFPM inhibition of ABA signaling (Figures 1, 3), we explored whether MAPK cascades have a role in interfering with ABA signaling by DFPM. A double mutant allele in the MAP kinase kinases MKK4 and MKK5 which function upstream of MPK3 and MPK6 (Zhang et al., 2018) was used. When mkk4-18 mkk5-18 mutant plants (Zhao et al., 2014) were pre-treated with DFPM for 1 hour and treated with ABA for additional 5 hours, ABA-induced transcription of RAB18 and ERD10 was less inhibited by DFPM compared to the wild type (Figure 8B). This reduced sensitivity to DFPM suggests roles of MKK4 and MKK5 in DFPM-mediated inhibition in ABA signal transduction.
Discussion
The small molecule DFPM activates plant immune signaling and thereby interferes with the ABA signaling pathway (Kim et al., 2011). We aimed to gain further insight into the molecular basis for the DFPM-mediated immune signaling and inhibition in ABA signaling in this study. By screening EMS mutant plants for reduced sensitivity to DFPM in inhibiting ABA-mediated pRAB18::GFP reporter protein expression, we identified RDA2 that is required for inhibiting ABA signal transduction by DFPM. Two alleles of rda2 mutant plants exhibited higher levels of ABA-mediated reporter expression in leaves in response to DFPM than the wild type, while responses to ABA were not altered in rda2 (Figures 1, 3). The rda2 mutant plants also showed a reduced sensitivity to DFPM in inhibiting ABA responses including ABA-induced stomatal closure and gene expression (Figures 1, 3). Whole genome sequencing based mapping revealed that RDA2 encodes a previously uncharacterized Galanthus nivalis agglutinin-type (G-type) (Bellande et al., 2017) lectin receptor kinase.
DFPM causes signaling in an EDS1/PAD4/SGT1b/RAR1-dependent manner (Kim et al., 2011, Kim et al., 2012). Additionally, here we show that DFPM also induces activation of the MAP kinases MPK3 and MPK6 with temporal dynamics similar to flg22-mediated MAPK activation (Figure 4). Activation of MPK3 and MPK6 is one of the earliest signaling responses upon pathogen perception in PAMP-triggered immunity (PTI) (Meng et al., 2013). It is noteworthy that DFPM induces PTI-like responses including MAP kinase activation and gene expression (Figure 4), while DFPM did not induce a rapid ROS burst (Figure S6B). These data suggest that DFPM induces an atypical PTI response. DFPM-mediated MAPK activation was not altered in eds1, pad4, eds1 pad4, sgt1b, rar1 and victr mutants (Figure 5). EDS1, PAD4, SGT1b, RAR1 and VICTR are required for DFPM-mediated ETI signaling, as mutant in these five genes were insensitive to DFPM in inhibiting root growth (Kim et al., 2012, Kunz et al., 2016). Our results suggest that DFPM activation of MAP kinases does not require signaling components of effector-triggered immunity (ETI) mediated immune signaling, and DFPM also mediates components of immune signaling that are not mediated by EDS1/PAD4-dependent ETI signaling. DFPM activation of ETI mechanisms appears to lie downstream of or parallel to MAPK activation.
Furthermore, we observed that mutant plants of EDS1, PAD4, SGT1b, RAR1, VICTR and VICTL1 showed wild-type levels of DFPM sensitivity in inhibiting ABA-mediated pRAB18::GFP reporter protein levels in leaves (Figure S3), which differs from transcript level analyses in whole seedlings (Kim et al., 2011). It is conceivable that the effect of the ETI signaling components is weaker in terms of interference with ABA signaling in leaf pavement cells and ETI signaling may be less central to the DFPM-ABA response. However, a major ETI dependent response of DFPM occurs in roots (Kim et al., 2012, Kunz et al., 2016). Therefore, the present findings in leaves show difference in these responses. DFPM further strongly upregulates the expression of VICTR specifically in roots which itself regulates the native VICTR gene (Kim et al., 2012, Kunz et al., 2016).
In rda2 mutants, DFPM-mediated MAPK activation and induction of PAMP-responsive genes were severely abolished (Figure 4C and D). These results suggest that the lectin receptor kinase RDA2 functions in early DFPM signaling thus transducing the signal to downstream pathways including MAPK activation and transcriptional regulation. DFPM-mediated expression of ETI-specific genes was partly dependent on RDA2 (Figure 4E). Interestingly, rda2 mutant plants showed a wild-type response to DFPM in inhibiting root growth (Figure S7) suggesting that RDA2 may not be fully involved in DFPM-mediated ETI signaling (Kim et al., 2011, Kim et al., 2012, Kunz et al., 2016). Note however that RDA2 is a member of 3 tandemly repeated lectin receptor kinase (LecRK) genes and higher order LecRK mutants may need to be investigated in future analyses.
Lectin receptor kinases (LecRKs) activate plant immune signaling by perceiving and sensing molecules in the apoplast that are released from pathogens or released during pathogen infection (Van Holle et al., 2018). There is only a handful of LecRKs whose carbohydrate binding activity has been identified thus far (Willmann et al., 2011, Liu et al., 2012, Kouzai et al., 2014, Ranf et al., 2015). G-type LecRK LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION (LORE) was shown that it can sense lipopolysaccharides (LPS) (Ranf et al., 2015). Recent studies have also shown that LecRK can recognize extracellular ATP and nicotinamide adenine dinucleotide (NAD+) that could be a sign of pathogen infection (Choi et al., 2014, Wang et al., 2017). This binding activity of LecRKs to non-carbohydrate ligands has been suggested as a result of neofunctionalization of duplicated LecRK genes (Van Holle et al., 2018). Whether RDA2 can perceive the DFPM signal and whether DFPM or its metabolized products can directly bind to RDA2 will require further investigation.
Plasma membrane localization of RDA2 has been suggested by experimental evidence including plasma membrane proteomic studies (Elmore et al., 2012, de Michele et al., 2016, Hooper et al., 2017), and supports a role of RDA2 in perceiving exogenous cues. Our experiments to examine the subcellular localization of the RDA2-GFP fusion protein were not successful due to possible mis-localization at endosomal organelles, because of high expression of the ectopically expressed proteins. A study with a lectin receptor kinase LecRK-I.8 failed to observe LecRK-I.8 fused with GFP in stably transformed plant lines, suggesting tight regulation of LecRK protein expression (Wang et al., 2017). Furthermore, examining natural ligands of RDA2 will open deeper insights into roles of the LecRK in immune signaling and biotic-to-abscisic acid cross interference signaling.
MAPK cascades transduce and amplify immune signals from receptor kinases (Tena et al., 2011). Here we show that mkk4 mkk5 double mutant plants exhibited reduced sensitivity to DFPM-mediated inhibition of ABA marker gene expression (Figure 8B). The results suggest that MKK4 and MKK5 are involved in DFPM-mediated inhibition of ABA signal transduction. Further, DFPM-mediated activation of MPK3 and MPK6 was strongly impaired in rda2 mutant alleles (Figure 4C), which indicates that the lectin receptor kinase RDA2 activates MAPKs MPK3 and MPK6 in response to DFPM. Receptor-like kinases including FLS2 and EF-TU RECEPTOR (EFR) were shown to function upstream of MPK3 and MPK6 (Xu et al., 2015). Rice LecRK SALT INTOLERANCE1 (SIT1) binds and directly phosphorylates rice MAP kinases MPK3 and MPK6 (Li et al., 2014). Rice lysine motif (LysM)-containing receptor kinase OsCERK1 interacts with the receptor-like cytoplasmic kinase RLCK185 which activates rice MPK3 and MPK6 (Kishi-Kaboshi et al., 2010, Yamaguchi et al., 2013). The receptor-like cytoplasmic kinase VII (RLCK VII) together with MAPKKK3 and MAPKKK5 is important for PAMP-induced activation of MPK3 and MPK6 via receptor kinases (Bi et al., 2018). The RDA2 lectin receptor kinase could transduce immune signals to downstream MAPK cascades, and may work with receptor-like cytoplasmic kinases (RLCKs) (Jurca et al., 2008, Bi et al., 2018).
MAPK cascades including MPK3 and MPK6 are thought to be convergence points of PAMP signaling and other immune signaling, since both FLS2- and EFR-mediated PAMP responses and the damage-associated molecular pattern (DAMP) oligogalacturonic acids (OGs) activate MAP kinases (Meng et al., 2013). The exact mechanisms by which MKK4 and MKK5 are involved in biotic-to-abscisic acid interference signaling are yet to be determined. Further investigation using viable loss-of-function mutants of MPK3 and MPK6 would help examine their role in this response. Since rbohd rbohf double mutant plants did not show an altered response in DFPM-mediated MAPK activation, apoplastic ROS may not function upstream of DFPM-mediated activation of MAP kinases. However, we cannot rule out roles of ROS and NADPH oxidases in cross interference signaling between biotic and abiotic stress signaling transduction in tissues other than those tested in this study. Further characterization of the genes defective in other rda mutants will provide additional insights into the molecular mechanisms of DFPM-mediated inhibition of the ABA signal transduction.
In conclusion, the present study identifies the lectin receptor kinase RDA2, which is important for DFPM-mediated immune signaling and interference signaling that inhibits ABA signal transduction. RDA2 is required for transducing the DFPM-mediated signal to activate MAP kinases and subsequently suppress ABA-mediated responses.
Experimental procedures
Plant materials, growth conditions and chemicals
The Arabidopsis thaliana accession Columbia-0 (Col-0) was used throughout this study unless indicated otherwise. Arabidopsis seeds were surface-sterilized in 70% EtOH and 0.004% SDS for 15 min, then rinsed with 100% EtOH 5 times and sown on 1/2 Murashige and Skoog (MS) media (pH 5.8) supplemented with 1% sucrose and 0.8% Phyto Agar. Plates with sterilized seeds were stratified in the dark for 2 to 3 days at 4 °C and then transferred to a growth chamber under a 16/8 h light/dark cycle at 60-80 μmol/m2·s1 light intensity at 19-21 °C.
To generate an ethyl methanesulphonate (EMS)-mutagenized pool, seeds of Col-0 expressing pRAB18::GFP were incubated in 1.5 to 3 % EMS overnight and thereafter M2 plants were used for forward genetic screening. mpk3-1 (SALK_151594), mpk6-2 (SALK_073907) and mkk4-18 mpk5-18 were gifts of Prof. Shuqun Zhang (University of Missouri). NIL-Bu-5 and NIL-Col-0 seeds were gifts from Prof. Teruaki Taji (Tokyo University of Agriculture, Japan). eds1-2, pad4-1, eds1-2 pad4-1, sgt1b (eta3), rar1-21, victr-1, victl1-1 (Kim et al., 2012) and rbohd rbohf-f3 (Kwak et al., 2003) were previously reported. To obtain eds1-2, pad4-1, eds1-2 pad4-1, sgt1b (eta3), rar1-21, victr-1, victl1-1 mutant plants expressing pRAB18::GFP, the mutant plants were crossed with wild-type Col-0 expressing pRAB18::GFP. rda2-2 (SALK_122993), rda2-3 (SALK_143489), GABI_565C02 and other T-DNA insertion lines were ordered from the Arabidopsis Biological Resource Center. To obtain rda2-2 expressing pRAB18::GFP, rda2-2 plants were transformed with pRAB18::GFP via Agrobacterium-mediated floral dipping. For comparison, wild-type Col-0 plants were also transformed with pRAB18::GFP at the same time.
DFPM ([5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione) was purchased from ChemBridge (chemical ID 6015316) and Innovapharm (Ukraine, chemical ID STT-00334837). A 23 amino acid peptide corresponding to AtPep1 active sequence (PEP1) was purchased from Genscript.
ABA response reporter pRAB18::GFP microscopy
Arabidopsis plants expressing pRAB18::GFP were germinated in ½ MS plates with 1% sucrose and 0.8% Phyto agar, and grown in the growth chamber at 60-80 μmol/m2·s1 light intensity at 19-21 °C for 7 days. Plants were then transferred to soil and grown in a growth chamber under a 12/12 h light/dark cycle at 60-80 μmol/m2·s1 light intensity at 19-21 °C for additional 7 days. First and second true leaves, the first pair of true leaves developed after cotyledons, were detached and floated on ½ MS liquid medium with 1% sucrose. Each leaf was placed in one well of 96-well plates with its adaxial side up. DFPM was added to a final concentration of 10 μM, and after one hour ABA was added to a final concentration of 20 μM. DMSO was used as a solvent control for DFPM and ethanol (EtOH) was used as a solvent control for ABA. After 24-26 hours of ABA addition, GFP fluorescence signals were observed from abaxial epidermal layers of leaves using confocal microscopes, LSM 800, LSM 710 (Zeiss), or Eclipse TE2000-U (Nikon). Constant gain and pinhole values, or exposure time were used for each experiment and comparisons of genotypes. Average fluorescence intensity of each image was calculated using Image J software in a genotype and chemical treatment blind manner. Statistical analyses were performed with Prism (Graphpad).
Whole genome sequencing and mapping
Genomic DNA from 86 independent F2 backcrossed lines displaying the rda2 phenotype was isolated by DNeasy Plant kit (Qiagen). The concentration of gDNA of each F2 line was quantified using Qubit™ dsDNA HS Assay kit and Qubit® Fluorometer (Invitrogen) by the manufacturer’s protocol, and equal amounts of gDNA from individual lines were pooled for whole genome sequencing. One μg genomic DNA of the Col-0 pRAB18::GFP parental line and of the two rda mutants rda1 and rda2 was used to construct ~450 bp insert size sequencing libraries with the Truseq Nano DNA kit (Illumina). The three libraries were sequenced in a single lane on an Illumina Hiseq2000 instrument with 2×101-bp paired-ends reads (200-cycle kit v3) and a single 7 bp index read.
Identification of a linked genomic interval and identification of candidate mutations within this interval were done by a custom-made mapping pipeline similar to SHOREmap (Schneeberger et al., 2009). Briefly, paired FASTQ reads were aligned to the Arabidopsis thaliana reference genome (TAIR10) by Bowtie 2 (Langmead et al., 2012). Single nucleotide polymorphisms (SNPs) were called by comparison with the reference genome using BCFtools and VCFtools (Danecek et al., 2011). Only EMS type mutations, which are C to T and G to A mutations, were included. SNPs of rda2 that were also found in the Col-0 pRAB18::GFP parental line or rda1 were excluded. Ratios of rda2-specific SNPs over the reference-type base (TAIR 10) were analyzed for every base position by R-values which were quantified by dividing the number of reads with a non-reference base by [the number of reads with a reference base multiplied by the total read number at each position] (Schneeberger et al., 2009). A moving average was calculated to display a candidate interval. Within this interval, mutations were prioritized according to their anticipated effect using the SnpEff tool (Cingolani et al., 2012).
Stomatal movement analyses
Fully expanded leaves of 4-5 week-old plants were detached and floated in stomatal assay buffer (5 mM KCl, 50 μM CaCl2, and 10 mM MES-Tris, pH 5.6) for 2.5 h. Epidermal peels were then prepared using a perforated-tape epidermal detachment method (Ibata et al., 2013). Images of guard cells from the abaxial side of leaves were taken using an inverted light microscope as “Control”. The same epidermal peels were then treated with 30 μM DFPM or solvent, and after 30 min ABA was added to a final concentration of 10 μM. 1.5 h after ABA addition, the same guard cells observed in “Control” images were identified and taken as “ABA” or “DFPM/ABA” images. Stomatal apertures were measured using Image J software independently by two different persons in a blind manner. Statistical analyses were performed with Prism (Graphpad).
Whole leaf time-resolved gas exchange analyses
Gas exchange analyses were performed as published previously (Hauser et al., 2018) with mild modification. Briefly, fully expanded rosette leaves of 6-8 week-old plants were cut at the end of the petiole with a fresh razor blade and immediately dipped in water. Another cut was made in the middle of the petiole to avoid clogging of the transpiration stream and petioles were immediately dipped in microcentrifuge tubes with 30 μM DFPM solution. In experiments without DFPM, petioles of detached leaves were dipped in a 0.02% DMSO solution as a solvent control. Stomatal conductance was evaluated using portable gas exchange analyses systems, LI-6800 or LI-6400XT (LI-COR). Detached leaves were clamped with a plant chamber of LI-6800 or LI-6400XT and equilibrated for 40-60 min at a relative humidity of 75%, airflow of 200 rpm and CO2 concentration of 400 ppm. Once stabilized, steady-state stomatal conductance was measured for 10 min. ABA was then added to a final concentration of 2 μM to the petiole-exposed solution while not touching clamped leaves. Stomatal conductance (mmol m−2min−1) was measured for additional 40-60 min. Stomatal conductance is normalized by averages of 10 min steady-state stomatal conductance before ABA addition. Maximum rates of stomatal conductance reduction were analyzed by calculating maximum slopes of each graph and were averaged.
Quantitative gene expression analyses
Two week-old Arabidopsis seedlings were treated with 10 μM DFPM for 1 hour, then ABA was added to a final concentration of 10 μM. DMSO was used as a solvent control for DFPM and EtOH was used as a solvent control for ABA. After the indicated time periods, total RNA was extracted using Spectrum™ Plant Total RNA Kit (Sigma-Aldrich), treated with TURBO™ DNase (Thermo-Fisher) and reverse-transcribed using First-Strand cDNA Synthesis Kit (GE Healthcare). qPCR analyses were performed on a BioRad CFX96 qPCR System using SYBR® Green JumpStart (Sigma) with gene specific primers (Table S2). Levels of transcript were normalized against the housekeeping gene PDF2. Statistical analyses were performed with Prism (Graphpad).
Measurement of phosphorylated MAP kinases
Ten to fourteen day-old seedlings were incubated in ½ MS liquid media with 1% sucrose containing 30 μM DFPM, 0.06% DMSO (solvent control) or 1 μM flg22 (GenScript) for the indicated time periods. For experiments with kinase inhibitors, plants were pre-treated with 10 μM of GW5074 (Sigma), Sorafenib (Sigma) or PD098059 (Sigma) 30 min before DFPM treatment. Total proteins were extracted in extraction buffer (25 mM Tris-HCl (pH 7.8), 75 mM NaCl, 10 mM MgCl2, 15 mM EGTA, 1 mM DTT, 1 mM NaF, 0.5 mM NaVO3, 15 mM β-glycerolphosphate, 15 mM p-nitrophenylphosphate, 0.1% Tween 20, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin). 20 μg of total proteins for each genotype and treatment were subjected to SDS-PAGE. Western blotting was performed with the anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) monoclonal antibody (9101, Cell Signaling Technology) and the secondary anti-rabbit antibody (BioRad) by the manufacturer’s protocol. The intensity of phosphorylated MAP kinase bands was quantified using Image J software. The band intensities were presented as ratios to the DFPM control without inhibitor treatment. Blots were stained with Coomassie blue solution to visualize the amount of loaded proteins.
Recombinant protein purification and in vitro kinase assay
The coding sequence of the RDA2 intracellular domain was PCR amplified using RDA2-pGEX-USER-F and RDA2-pGEX-USER-R primers (Table S2), and cloned into the modified pGEX-6P1 vector with a C-terminal GST tag and an additional N-terminal Strep II tag (Brandt et al., 2012) using USER® enzyme (New England Biolabs) resulting in pGEX-6P1-RDA2_intracellular-StrepII. The lysine 552 to glutamic acid mutation (K552E) was introduced to pGEX-6P1-RDA2_intracellular-StrepII by PCR with K552E-F and K552E-R primers (Table S2), resulting in pGEX-6P1-RDA2_intracellular-K552E-StrepII.
Escherichia coli strain BL21 (DE3) was transformed with the plasmids and grown to OD600 ~0.6. Protein expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and growth overnight at room temperature. E. coli cells were harvested by centrifugation at 4000 × g for 15 min at 4 °C and resuspended in buffer W (100 mM Tris, 150 mM NaCl, pH 8.0) supplemented with 1 mg/mL lysozyme and protease inhibitor cocktail (Roche). After sonicating E. coli cells by three 30-s pulses, insoluble cell debris was removed by centrifugation at 25000 × g for 30 min at 4 °C. RDA2-intracellular domain and RDA2-intracellular-K552E domain proteins were purified with Strep-Tactin® Macroprep® (IBA) following the manufacturer’s protocol.
10 ng/μL of recombinant RDA2 intracellular domain was incubated in the phosphorylation buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM EGTA, 0.1% Triton X-100, 1 mM DTT) containing 50 ng/μL MyBP together with 50, 100 or 200 μM DFPM or DMSO (control) for 30 min at room temperature. For experiments with kinase inhibitors K252a (Calbiochem) and staurosporine (Sigma), protein samples were pre-incubated with 20 μM of the inhibitors for 30 min before phosphorylation reactions. 200 μM ATP and 0.05 μCi/μL [γ−32P]-ATP (PerkinElmer) were then added to start phosphorylation reactions. Phosphorylation reactions were stopped by addition of SDS-PAGE sample buffer after 30 min incubation at room temperature. Phosphorylated proteins were detected by autoradiography after SDS-PAGE.
ROS production measurements
Leaf discs from 3-4 week-old Arabidopsis plants were floated on sterile water in white 96-well plates overnight at room temperature under light. At least 12 plants were used per genotype per condition. Each leaf disc was placed in one well of 96-well plates with its adaxial side up. After 20-24 hours, water in 96-well plates was replaced with 50 μL of fresh sterile water and leaf discs were incubated for additional 1 hour. 50 μL of the 2× elicitation solution (100 μg/mL Horseradish Peroxidase (Sigma, P6782), 4 μM L-012 (Wako chemical, 120-04891)) was added to each well. Luminescence was measured with a Mithras LB940 micro plate reader (Berthold). Flg22, PEP1 or DFPM were added to individual wells with a built-in injector in the plate reader to a final concentration of 100 nM flg22, 100 nM PEP1 and 30 μM DFPM. Water control was injected to examine mechanical-induced ROS production. DMSO (0.06%) was used as a solvent control for DFPM.
Pathogen tolerance assays
Pseudomonas syringae pv. tomato DC3000 (Pst) culture was grown overnight at 28°C in liquid low salt LB media containing 50 μg/mL rifampicin to the OD600 ranging from 0.6 to 1.0. The bacterial cells were collected by centrifugation at 2600 × g for 10 min, room temperature, and resuspended in 10 mM MgCl2 to the OD600 of 0.2. The bacterial solution was diluted 1000-fold (105 colony forming units per mL) in 10 mM MgCl2 and then infiltrated into the abaxial side of Arabidopsis leaves with a needleless syringe. Four week-old Arabidopsis plants were used, and the leaves were either first infiltrated with 1 μM flg22 or H2O one day prior infiltration with Pst, or infiltrated with Pst supplemented with 30 μM DFPM or 0.02% v/v DMSO (mock treatment). The inoculated plants were covered with lids to maintain a high humidity. Two 6 mm-diameter leaf disks from eight different plants per treatment were collected and homogenized in 10 mM MgCl2. Serial dilutions were plated on low salt LB agar media containing 50 μg/mL rifampicin and 12.5 μg/mL cycloheximide. The plates were incubated for 2 days at 28°C and the colonies were counted manually.
Supplementary Material
Table S1. List of SNP mutations in the mapping region of rda2.
Table S2. Primers used in the study.
Figure S1. Screening for rda mutants.
Figure S2. Three rda mutants were less sensitive to DFPM in inhibiting ABA-mediated reporter pRAB18::GFP expression than the wild-type Col-0.
Figure S3. Mutants of EDS1, PAD4, RAR1, SGT1b, VICTR and VICTL1 exhibited wild-type levels of DFPM sensitivity in inhibiting ABA-mediated pRAB18::GFP reporter expression in leaf epidermal pavement cells.
Figure S4. Allelism test between rda2 and T-DNA insertion mutants of genes that harbor rda2-specific SNPs.
Figure S5. DFPM-mediated activation of MAP kinases remained high between 15 and 60 min of DFPM in some experimental conditions.
Figure S6. MAP kinase activation in response to flg22 in rda2 mutants, and ROS production in response to flg22, PEP1 and DFPM.
Figure S7. rda2 mutants showed a wild-type response to DFPM inhibition of root growth.
Acknowledgements
We thank Prof. Shuqun Zhang (University of Missouri) for providing seeds of mpk3-1 (SALK_151594), mpk6-2 (SALK_073907) and mkk4-18 mkk5-18; Prof. Teruaki Taji (Tokyo University of Agriculture, Japan) for seeds of NIL-Col-0 and NIL-Bu-5. We thank Drs. Detlef Weigel and Christa Lanz (MPI for Developmental Biology, Germany) for sequencing support. We thank Drs. Detlef Weigel (MPI for Developmental Biology, Germany) and Sebastian Schulze (UC San Diego) for critical reading. This research was funded by the National Institutes of Health (GM060396-ES010337) to J.I.S., by Hellman Fellowship to A. H., and partly by the Next-Generation BioGreen 21 Program (SSAC grant PJ01335001), and the Rural Development Administration, Republic of Korea to T.-H. K. J.P. was funded by Human Frontier Science Program Long-term postdoctoral fellowship and K.D. was funded by Ciencias sem Fronteiras/CNPq fellowship (200260/2015-4).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Ariga H, Katori T, Tsuchimatsu T, Hirase T, Tajima Y, Parker JE, Alcazar R, Koornneef M, Hoekenga O, Lipka AE, Gore MA, Sakakibara H, Kojima M, Kobayashi Y, Iuchi S, Kobayashi M, Shinozaki K, Sakata Y, Hayashi T, Saijo Y and Taji T (2017) NLR locus-mediated trade-off between abiotic and biotic stress adaptation in Arabidopsis. Nat Plants, 3, 17072 10.1038/nplants.2017.72 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM and Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–83. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Asselbergh B, Achuo AE, Hofte M and Van Gijsegem F (2008a) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol Plant Pathol, 9, 11–24. 10.1111/j.1364-3703.2007.00437.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D and Hofte M (2008b) Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact, 21, 709–19. 10.1094/MPMI-21-6-0709 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Jain R and Urwin PE (2015) The Response of Plants to Simultaneous Biotic and Abiotic Stress In Combined Stresses in Plants. (Mahalingam R, ed). Switzerland: Springer International Publishing; pp. 181–201. [Google Scholar]
- Atkinson NJ and Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot, 63, 3523–43. 10.1093/jxb/ers100 [DOI] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB and Hofte MM (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol, 128, 491–501. 10.1104/pp.010605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S and Boller T (2015) Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J Exp Bot, 66, 5183–93. 10.1093/jxb/erv180 [DOI] [PubMed] [Google Scholar]
- Bellande K, Bono JJ, Savelli B, Jamet E and Canut H (2017) Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside? Int J Mol Sci, 18 10.3390/ijms18061164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH and Gassmann W (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science, 334, 1405–8. 10.1126/science.1211592 [DOI] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S and Zhou JM (2018) Receptor-like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-activated Protein Kinase Cascades in Arabidopsis. Plant Cell. 10.1105/tpc.17.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM, Pye MF and Roubtsova TV (2014) Predisposition in plant disease: exploiting the nexus in abiotic and biotic stress perception and response. Annu Rev Phytopathol, 52, 517–49. 10.1146/annurev-phyto-081211-172902 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, Mccormack M, Lee H, Shan L, He P, Bush J, Cheng SH and Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature, 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjarvi J, Ghassemian M, Stephan AB, Hu H and Schroeder JI (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci U S A, 109, 10593–8. 10.1073/pnas.1116590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA (2010) Tackling the threat to food security caused by crop pests in the new millennium. Food Security, 2, 133–141. 10.1007/s12571-010-0061-8 [DOI] [Google Scholar]
- Buhrow LM, Cram D, Tulpan D, Foroud NA and Loewen MC (2016) Exogenous Abscisic Acid and Gibberellic Acid Elicit Opposing Effects on Fusarium graminearum Infection in Wheat. Phytopathology, 106, 986–96. 10.1094/PHYTO-01-16-0033-R [DOI] [PubMed] [Google Scholar]
- Chen D, Cao Y, Li H, Kim D, Ahsan N, Thelen J and Stacey G (2017) Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun, 8, 2265 10.1038/s41467-017-02340-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, Ma B, Wang Y, Zhao X, Li S and Zhu L (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J, 46, 794–804. 10.1111/j.1365-313X.2006.02739.x [DOI] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY and Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science, 343, 290–4. 10.1126/science.343.6168.290 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ and Raikhel NV (1991) Lectins, lectin genes, and their role in plant defense. Plant Cell, 3, 1–9. 10.1105/tpc.3.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X and Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin), 6, 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D and Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol, 16, 537–52. 10.1038/nri.2016.77 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR and Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol, 61, 651–79. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, Mcvean G, Durbin R and Genomes Project Analysis, G. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–8. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michele R, Mcfarlane HE, Parsons HT, Meents MJ, Lao J, Gonzalez Fernandez-Nino SM, Petzold CJ, Frommer WB, Samuels AL and Heazlewood JL (2016) Free-Flow Electrophoresis of Plasma Membrane Vesicles Enriched by Two-Phase Partitioning Enhances the Quality of the Proteome from Arabidopsis Seedlings. J Proteome Res, 15, 900–13. 10.1021/acs.jproteome.5b00876 [DOI] [PubMed] [Google Scholar]
- Deutsch CA, Tewksbury JJ, Tigchelaar M, Battisti DS, Merrill SC, Huey RB and Naylor RL (2018) Increase in crop losses to insect pests in a warming climate. Science, 361, 916–919. 10.1126/science.aat3466 [DOI] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B and Coaker G (2012) Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics, 11, M111 014555 10.1074/mcp.M111.014555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hill L, Crooks C, Doerner P and Lamb C (2009) Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol, 150, 1750–61. 10.1104/pp.109.137943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R (2013) Abscisic Acid synthesis and response. Arabidopsis Book, 11, e0166 10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Byerlee D and Edmeades G (2009) Can technology deliver on the yield challenge to 2050? How to feed the World in 2050. Proceedings of a technical meeting of experts, Rome, Italy, 24-26 June 2009, 2009 Food and Agriculture Organization of the United Nations (FAO), pp. 1–48. [Google Scholar]
- Frei Dit Frey N, Garcia AV, Bigeard J, Zaag R, Bueso E, Garmier M, Pateyron S, De Tauzia-Moreau ML, Brunaud V, Balzergue S, Colcombet J, Aubourg S, Martin-Magniette ML and Hirt H (2014) Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol, 15, R87 10.1186/gb-2014-15-6-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K and Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol, 9, 436–42. 10.1016/j.pbi.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Hatfield JL and Walthall CL (2015) Meeting Global Food Needs: Realizing the Potential via Genetics x Environment x Management Interactions. Agronomy Journal, 107, 1215–1226. 10.2134/agronj15.0076 [DOI] [Google Scholar]
- Hauser F, Ceciliato PHO, Lin Y-C, Guo D, Gregerson JD, Abbasi N, Youhanna D, Park J, Dubeaux G, Shani E, Poomchongkho N and Schroeder JI (2018) A seed resource for screening functionally redundant genes and isolation of new mutants impaired in CO2 and ABA responses. Journal of Experimental Botany. 10.1093/jxb/ery363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Li Z, Waadt R and Schroeder JI (2017) SnapShot: Abscisic Acid Signaling. Cell, 171, 1708–1708 e0 10.1016/j.cell.2017.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat Q, Hayat S, Irfan M and Ahmad A (2010) Effect of exogenous salicylic acid under changing environment: A review. Environmental and Experimental Botany, 68, 14–25. 10.1016/j.envexpbot.2009.08.005 [DOI] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L and Parker JE (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science, 334, 1401–4. 10.1126/science.1211641 [DOI] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N and Millar AH (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res, 45, D1064–D1074. 10.1093/nar/gkw1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PK, Takahashi Y, Munemasa S, Merilo E, Laanemets K, Waadt R, Pater D, Kollist H and Schroeder JI (2018) Abscisic acid-independent stomatal CO2 signal transduction pathway and convergence of CO2 and ABA signaling downstream of OST1 kinase. Proc Natl Acad Sci U S A, 115, E9971–E9980. 10.1073/pnas.1809204115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata H, Nagatani A and Mochizuki N (2013) Perforated-tape Epidermal Detachment (PED): A simple and rapid method for isolating epidermal peels from specific areas of Arabidopsis leaves. Plant Biotechnology, 30, 497–U112. 10.5511/plantbiotechnology.13.0903b [DOI] [Google Scholar]
- Jurca ME, Bottka S and Feher A (2008) Characterization of a family of Arabidopsis receptor-like cytoplasmic kinases (RLCK class VI). Plant Cell Rep, 27, 739–48. 10.1007/s00299-007-0494-5 [DOI] [PubMed] [Google Scholar]
- Khan MI, Fatma M, Per TS, Anjum NA and Khan NA (2015) Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front Plant Sci, 6, 462 10.3389/fpls.2015.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Hauser F, Ha T, Xue S, Bohmer M, Nishimura N, Munemasa S, Hubbard K, Peine N, Lee BH, Lee S, Robert N, Parker JE and Schroeder JI (2011) Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol, 21, 990–7. 10.1016/j.cub.2011.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kunz HH, Bhattacharjee S, Hauser F, Park J, Engineer C, Liu A, Ha T, Parker JE, Gassmann W and Schroeder JI (2012) Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell, 24, 5177–92. 10.1105/tpc.112.107235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, Yamane H, Miyao A, Takatsuji H, Takahashi A and Hirochika H (2010) A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J, 63, 599–612. 10.1111/j.1365-313X.2010.04264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzai Y, Mochizuki S, Nakajima K, Desaki Y, Hayafune M, Miyazaki H, Yokotani N, Ozawa K, Minami E, Kaku H, Shibuya N and Nishizawa Y (2014) Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol Plant Microbe Interact, 27, 975–82. 10.1094/MPMI-03-14-0068-R [DOI] [PubMed] [Google Scholar]
- Kunz HH, Park J, Mevers E, Garcia AV, Highhouse S, Gerwick WH, Parker JE and Schroeder JI (2016) Small Molecule DFPM Derivative-Activated Plant Resistance Protein Signaling in Roots Is Unaffected by EDS1 Subcellular Targeting Signal and Chemical Genetic Isolation of victr R-Protein Mutants. PLoS One, 11, e0155937 10.1371/journal.pone.0155937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD and Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J, 22, 2623–33. 10.1093/emboj/cdg277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr W and Raschke K (1988) Abscisic-acid contents and concentrations in protoplasts from guard cells and mesophyll cells ofVicia faba L. Planta, 173, 528–531. 10.1007/bf00958966 [DOI] [PubMed] [Google Scholar]
- Langmead B and Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods, 9, 357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannoo N and Van Damme EJM (2014) Lectin domains at the frontiers of plant defense. Frontiers in Plant Science, 5 10.3389/fpls.2014.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Wang G, Zhao JL, Zhang LQ, Ai LF, Han YF, Sun DY, Zhang SW and Sun Y (2014) The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell, 26, 2538–2553. 10.1105/tpc.114.125187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, He Y, Wang J and Wang HB (2012) Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell, 24, 3406–19. 10.1105/tpc.112.102475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y and Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell, 16, 3386–99. 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B and Mauch F (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol, 8, 409–14. 10.1016/j.pbi.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Maxmen A (2013) Crop pests: Under attack. Nature, 501, S15–7. 10.1038/501S15a [DOI] [PubMed] [Google Scholar]
- Melillo JM, Richmond TTC and Yohe GW (2014) Climate Change Impacts in the United States: The Third National Climate Assessment. U.S. Global Change Research Program, U.S. National Climate Assessment; 10.7930/J0Z31WJ2 [DOI] [Google Scholar]
- Meng X and Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol, 51, 245–66. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Mine A, Seyfferth C, Kracher B, Berens ML, Becker D and Tsuda K (2018) The Defense Phytohormone Signaling Network Enables Rapid, High-amplitude Transcriptional Reprogramming During Effector-Triggered Immunity. Plant Cell. 10.1105/tpc.17.00970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R and Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol, 61, 443–62. 10.1146/annurev-arplant-042809-112116 [DOI] [PubMed] [Google Scholar]
- Mohr PG and Cahill DM (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Functional Plant Biology, 30, 461–469. 10.1071/Fp02231 [DOI] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E and Yoshioka K (2010) The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol, 152, 1901–13. 10.1104/pp.109.152603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh M, Janda T, Horváth E, Páldi E and Szalai G (2002) Exogenous salicylic acid increases polyamine content but may decrease drought tolerance in maize. Plant Science, 162, 569–574. 10.1016/s0168-9452(01)00593-3 [DOI] [Google Scholar]
- Oerke EC (2005) Crop losses to pests. The Journal of Agricultural Science, 144, 31 10.1017/s0021859605005708 [DOI] [Google Scholar]
- Olson AJ, Pataky JK, Darcy CJ and Ford RE (1990) Effects of Drought Stress and Infection by Maize-Dwarf Mosaic-Virus on Sweet Corn. Plant Disease, 74, 147–151. 10.1094/Pd-74-0147 [DOI] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, Mccourt P, Zhu JK, Schroeder JI, Volkman BF and Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–71. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch CM and Sonnewald U (2013) Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol, 162, 1849–66. 10.1104/pp.113.221044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramegowda V and Senthil-Kumar M (2015) The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J Plant Physiol, 176, 47–54. 10.1016/j.jplph.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schaffer M, Illig T, Westphal L, Knirel YA, Sanchez-Carballo PM, Zahringer U, Huckelhoven R, Lee J and Scheel D (2015) A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol, 16, 426–33. 10.1038/ni.3124 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D and Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods, 6, 550–1. 10.1038/nmeth0809-550 [DOI] [PubMed] [Google Scholar]
- Singh P and Zimmerli L (2013) Lectin receptor kinases in plant innate immunity. Front Plant Sci, 4, 124 10.3389/fpls.2013.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E and Mittler R (2014) Abiotic and biotic stress combinations. New Phytol, 203, 32–43. 10.1111/nph.12797 [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M and Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol, 14, 519–29. 10.1016/j.pbi.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V and Mauch-Mani B (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci, 14, 310–7. 10.1016/j.tplants.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Toth R and Van Der Hoorn RA (2010) Emerging principles in plant chemical genetics. Trends Plant Sci, 15, 81–8. 10.1016/j.tplants.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Ulferts S, Delventhal R, Splivallo R, Karlovsky P and Schaffrath U (2015) Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol, 15, 7 10.1186/s12870-014-0409-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid N, Macovei A and Tuteja N (2013) Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol Plant, 6, 1405–18. 10.1093/mp/sst033 [DOI] [PubMed] [Google Scholar]
- Van Holle S and Van Damme EJM (2018) Signaling through plant lectins: modulation of plant immunity and beyond. Biochem Soc Trans, 46, 217–233. 10.1042/BST20170371 [DOI] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED and Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife, 3, e01739 10.7554/eLife.01739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hsu PK and Schroeder JI (2015) Abscisic acid and other plant hormones: Methods to visualize distribution and signaling. BioEssays, 37, 1338–1349. 10.1002/bies.201500115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y and Mou Z (2017) A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. Elife, 6 10.7554/eLife.25474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC and Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell, 19, 63–73. 10.1105/tpc.106.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B and Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta, 218, 1–14. 10.1007/s00425-003-1105-5 [DOI] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV, Jehle AK, Gotz F, Kulik A, Molinaro A, Lipka V, Gust AA and Nurnberger T (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci U S A, 108, 19824–9. 10.1073/pnas.1112862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J and Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci, 20, 56–64. 10.1016/j.tplants.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, Ochiai H, Tada Y, Shimamoto K, Yoshioka H and Kawasaki T (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe, 13, 347–57. 10.1016/j.chom.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y and Huffaker A (2011) Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol, 14, 351–7. 10.1016/j.pbi.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S and Nakashita H (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell, 20, 1678–1692. 10.1105/tpc.107.054296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Su J, Zhang Y, Xu J and Zhang S (2018) Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol, 45, 1–10. 10.1016/j.pbi.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Zhao C, Nie H, Shen Q, Zhang S, Lukowitz W and Tang D (2014) EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet, 10, e1004389 10.1371/journal.pgen.1004389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G and Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of SNP mutations in the mapping region of rda2.
Table S2. Primers used in the study.
Figure S1. Screening for rda mutants.
Figure S2. Three rda mutants were less sensitive to DFPM in inhibiting ABA-mediated reporter pRAB18::GFP expression than the wild-type Col-0.
Figure S3. Mutants of EDS1, PAD4, RAR1, SGT1b, VICTR and VICTL1 exhibited wild-type levels of DFPM sensitivity in inhibiting ABA-mediated pRAB18::GFP reporter expression in leaf epidermal pavement cells.
Figure S4. Allelism test between rda2 and T-DNA insertion mutants of genes that harbor rda2-specific SNPs.
Figure S5. DFPM-mediated activation of MAP kinases remained high between 15 and 60 min of DFPM in some experimental conditions.
Figure S6. MAP kinase activation in response to flg22 in rda2 mutants, and ROS production in response to flg22, PEP1 and DFPM.
Figure S7. rda2 mutants showed a wild-type response to DFPM inhibition of root growth.


