Abstract
Keloids are wounding-induced fibroproliferative human tumor–like skin scars of complex genetic makeup and poorly defined pathogenesis. To reveal dynamic epigenetic and transcriptome changes of keloid fibroblasts, we performed RNA-seq and ATAC-seq analysis on an early passage keloid fibroblast cell strain and its paired normal control fibroblasts. This keloid strain produced keloid-like scars in a plasma clot-based skin equivalent humanized keloid animal model. RNA-seq analysis reveals gene ontology terms including Hepatic fibrosis, Wnt–βcatenin, TGF-β, regulation of Epithelial Mesenchymal Transition (EMT), STAT3, and adherens junction. ATAC-seq analysis suggests STAT3 signaling is the most significantly enriched gene ontology term in keloid fibroblasts, followed by Wnt signaling (Wnt5) and regulation of the EMT pathway. Immunohistochemistry confirms that STAT3 (Tyr705 phospho-STAT3) is activated and β−catenin is up-regulated in the dermis of keloid clinical specimens and keloid skin equivalent implants from the humanized mouse model. A non-linear dose response of Cucurbitacin I, a selective JAK2/STAT3 inhibitor, in collagen type I expression of keloid-derived plasma clot-based skin equivalents implicates a likely role of STAT3 signaling in keloid pathogenesis. This work also demonstrates the utility of the recently established humanized keloid mouse model in exploring the mechanism of keloid formation.
Keywords: fibroblasts, artificial skin, animal model
1. BACKGROUND
Keloids are wounding induced fibroproliferative human tumor-like skin scars of ill-defined pathogenesis and with a higher incidence in dark-skinned individuals indicating a genetic predisposition.[1–5] Due to high recurrence following surgical excision and the lack of effective medical interventions as well as the accompanied psychological distress in affected individuals, keloids have become a burden on health-related quality of life in society.[6,7]
Wound repair is the inherent ability of an organism to protect itself against injuries and may result in either organ regeneration or scarring.[8] Keloid formation is preceded by excessive inflammation, pruritis, and pain. Scars expand beyond the confines of wounds and progress with a proliferative edge forming a raised scar and a collapsed center.[9–12] Fibroblasts are the principal mediator of fibroproliferative disorders.[13, 14] Recent lineage tracing and single-cell transcriptional profiling studies reveal that skin fibroblasts consist of distinct subpopulations arising from different lineages with different roles in determining dermal architecture in skin development and repair.[15–17] A report combining site-specific in situ microdissection and gene expression profiling of keloid tissues, found distinct gene signatures of clear keloid regions highlighting morphological heterogeneity within the keloid scar.[18] The study reveals novel keloid dermis gene signatures and supports the roles of dermal fibroblasts in the deposition of extracellular matrix (ECM) and in all phases of wound healing.
2. QUESTIONS ASKED
What significant gene ontology terms identified through an unbiased omics study can distinguish keloid fibroblasts from normal fibroblasts?
3. EXPERIMENTAL DESIGN
Here we conducted a vertical study revealing epigenetic and transcriptomic changes of keloid fibroblasts followed by in vivo candidate molecule expression confirmation and subsequent functional testing in an organ culture. Early passages of a freshly isolated keloid fibroblast cell strain that produces keloid-like scars in the plasma clot-based skin equivalent humanized keloid animal model were compared to its paired normal control.[12] These cell strains were chosen to avoid inconsistent results due to clinical specimen scarcity and variability, representing late but diverse stages of lesion maturation and inherent biological variability between and within keloid lesions, (i.e., raised border, flattened center, reddish inflammatory/expanding edge)[12] which complicate data interpretation.
4. RESULTS AND DISCUSSION
4.1. RNA-seq and Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) analyses identify differential transcriptome and epigenome profiling in keloid fibroblasts
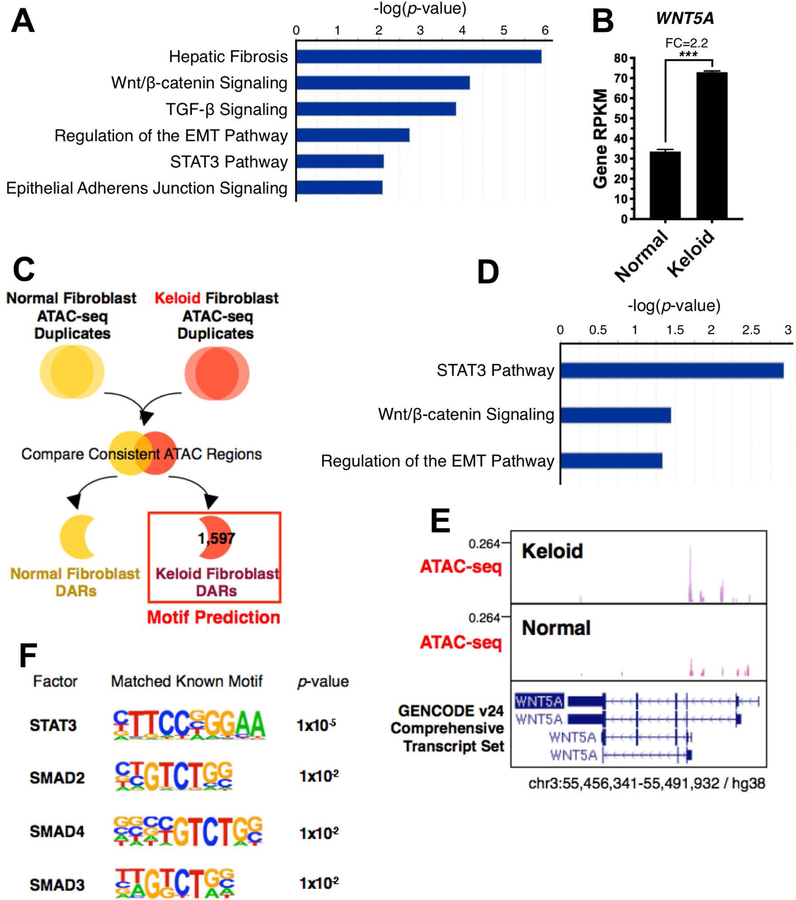
To gain insight into differential gene expression that might underlie keloid fibrosis, total RNA was collected from 3 technical replicates of passage 5 fibroblasts derived from one control and one keloid patient specimen and sequenced by the USC Molecular Genomic Core facility. Bioinformatics analysis revealed that hepatic fibrosis, Wnt, TGF-β, STAT3 and epithelial-mesenchymal transition (EMT) pathways were the most significantly enriched gene ontology terms for keloid fibroblasts (Fig. 1A; S1, S2). Keloid specific differentially expressed genes involved in these pathways are listed (Table S1). Since STAT3 was implicated in keloids, it is interesting to note that STAT3 phosphorylation may impact the keloid fibrotic phenotype because targeting STAT3 reduced fibroblast collagen production, cell proliferation, and migration in in vitro monolayer culture of keloid fibroblasts and fibroblast – keratinocytes co-cultures.[19–23] Furthermore, RNA-seq data showed that Wnt5a was increased 2.2-fold in keloids compared to normal fibroblasts (Fig. 1B), consistent with a recent keloid microarray study.[18] In addition to Wnt5a, Wnt2B, Wnt5B and Wnt6 are also elevated in keloid fibroblasts. Wnt5a mRNA and protein levels and β-catenin levels are elevated in keloid fibroblasts compared to normal fibroblasts.[24] Furthermore, treating normal and keloid fibroblasts with recombinant Wnt5a peptide increased total and phosphorylated forms of β-catenin,[24] suggesting that the Wnt/β-catenin canonical signaling pathway may be triggered by Wnt5a in keloid fibroblasts.
Figure 1. Significantly enriched pathways highlighted by RNA-seq and ATAC-seq analysis of keloid versus normal fibroblasts.
Fig. 1A. Significantly enriched pathways of differentially expressed genes from RNA-seq using human keloid fibroblasts compared to normal fibroblasts.
Fig. 1B. RNA-seq analysis that Wnt5a is upregulated 2.2-fold in keloid fibroblasts compared to normal fibroblasts (p<0.001).
Fig. 1C. Schematic of ATAC-seq analysis strategy to obtain keloid-specific Differentially Accessible Regions (DARs) and significantly enriched transcription factor binding motifs.
Fig. 1D. Significantly enriched canonical pathways of proximal promoters of keloid-specific DARs.
Fig. 1E. ATAC-seq analysis of open chromatin shows the Wnt5a promoter is more accessible in keloids than normal fibroblasts.
Fig. 1F. Significantly enriched transcription factor binding motifs at keloid-specific DARs.
We performed ATAC-seq to examine regions of transposase accessible open chromatin in keloid compared to control fibroblasts to further identify upstream networks. After confirming the samples were of acceptable quality, we identified keloid-specific differentially accessible regions (DARs) (Fig. 1C; Fig. S3 and S4) located in promoters, 5’ UTRs and non-coding RNA (Fig. S5), indicating that a global gene activation network acts in keloids. Gene network members were identified by focusing on open chromatin regions lying near promoters; these genes mostly controlled the STAT3, Wnt signaling and regulation of the EMT pathways (Fig. 1D, Table S2), echoing the RNA-seq result. For example, we showed that Wnt5a has increased accessibility in this assay (Fig. 1E). We further identified the master regulator that controls these regions by performing transcription factor binding motif prediction. STAT3 and SMAD family member motifs were significantly enriched in keloid-specific open regions (Fig. 1F). Our systemic results demonstrate that STAT3 functions in a feed-forward regulatory loop to control downstream target genes involved in keloids. STAT3 may induce Wnt5a in embryonic stem cells, adult normal tissues, and cancer.[25–27]
4.2. STAT3 and Wnt/β-catenin pathways are enriched in keloid dermis versus control clinical specimens
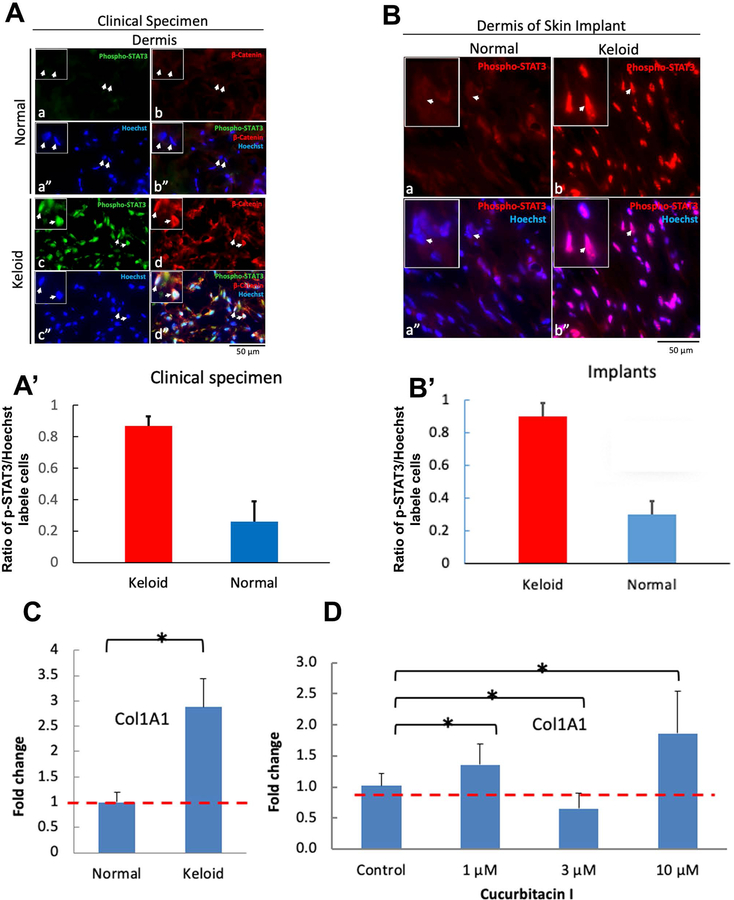
The omics data shows that STAT3 and Wnt pathways were enriched and predicts that STAT3 family members function as master regulators in keloid fibroblasts. Hence, we evaluated STAT3 pathway activation in clinical specimens by immunostaining for p-STAT3 (Tyr705), an activated form of STAT3. We found that keloid dermis ubiquitously expressed high levels of nuclear p-STAT3 (Fig. 2A, panel c), compared with low expression levels in normal dermis (Fig. 2A, panel a). Using immunostaining we confirmed that β-catenin was up-regulated in keloids versus control specimens (Fig. 2A, compare panels b and d). Intriguingly, β-catenin expression was identified in the p-STAT3 positive cell subpopulation, sometimes showing nuclear localization (Fig. 2A, panel d”). Results from studies on clinical specimens suggest STAT3 and Wnt/β-catenin are involved in keloid pathogenesis. These data are quantified and show a 4-fold increase in STAT3 expression in keloid clinical specimens (Fig 2A’).
Figure 2. Immunohistochemistry demonstrates that STAT3, i.e., Tyr705 p-STAT3 is activated and/or β−catenin is up-regulated in dermis of keloid clinical specimens as well as in 18 wk- old fibrin clot-based skin equivalent humanized mouse model keloid implants compared to normal controls.
Fig. 2A. Activation of the STAT3 pathway is up-regulated in keloid clinical specimens. Normal or keloid skin tissue sections were double immunostained with rabbit anti-p-STAT3(Tyr705) polyclonal antibody (green) and mouse anti-β-catenin monoclonal antibody (red). a-b. Both p-STAT3 (a) and β-catenin (b) expression in normal skin dermis appears low or not detectable. a”. Dermal cell nuclei were revealed by Hoechst staining (blue). b”. Merged image. c, d, and d”. Nuclear p-STAT3 expression was ubiquitously detected in the keloid skin dermis (arrows) (c). Note that the p-STAT3 positive cell subpopulation also express β-catenin (arrows in c and d). d”. Merged image. Bar, 50µm.
Fig. 2A’. p-STAT3 levels are significantly increased in keloid compared to control clinical specimens.
Fig. 2B. STAT3 pathway activation is up-regulated in the humanized keloid mouse model. Immunostaining with rabbit polyclonal anti-p-STAT3 (Tyr705) antibodies. A-B. p-STAT3 expression was hardly detectable in N102 implant dermal fibroblasts (A and A”), but was upregulated in K119 implant fibroblasts (B and B”). Bar, 50µm.
Fig. 2B’. p-STAT3 levels are significantly increased in keloid compared to control clinical specimens mimicking what is seen in clinical specimens.
Fig. 2C. Cucurbitacin I modulates collagen type 1 expression in plasma-clot – based artificial skin constructs. Collagen type 1 (a marker of fibrosis) expression levels were determined from constructs harvested at day 7 by qPCR analysis. Data expressed as the mean ± s.d. (n = 6) are representative of three independent experiments. *, p-value < 0.05. A. K119 skin constructs show significantly elevated collagen type 1 mRNA expression compared to N102 skin constructs.
Fig. 2D. Cucurbitacin I, a selective JAK2/STAT3 inhibitor, affects collagen type 1 expression. In the tested concentration range, the expression of collagen type 1 was increased at 1 µM and 10 µM of Cucurbitacin I, respectively and was decreased at 3 µM.
4.3. Activation of the STAT3 pathway was observed in the plasma clot-based skin equivalent humanized keloid animal model as predicted by RNA-seq and ATAC-seq
A long-standing challenge facing keloid research is the lack of an animal model enabling pathogenesis and translational study. From the perspective of disease development, an ideal model would reveal the initiation and propagation dynamics seen during keloid formation. Recently, the plasma clot-based skin equivalent humanized keloid animal model was established by implanting a porous polyethylene ring-supported plasma/fibrin-based epidermal–dermal skin constructs on the dorsum of athymic NU/J mice.[12] The implants were stable up to 18 weeks, contained abundant human cells, and keloid cell–derived implants remodeled to yield scar architecture characteristic of keloid fibrosis compared with normal implants and clinical specimens.[12] To investigate whether STAT3 pathway activation is associated with keloid fibrosis as predicted by the RNA-seq and ATAC-seq data, 18-week-old implants in the keloid mouse model were immunostained with an anti-p-STAT3 (Tyr705) antibody. p-STAT3 was widely expressed in the K119 keloid implant dermal fibroblasts (Fig. 2B, panels b and b”), but was hardly detectable in N102 normal skin implant fibroblasts (Fig. 2B, panels a and a”). These results indicate that the humanized keloid mouse model not only recapitulates keloid phenotypes but also the activated STAT3 pathway, similar to the clinical specimens.
Pharmacological inhibition of the STAT3 pathway modulates keloid fibrosis in the artificial skin construct-based assay model
Skin wound healing is a complex process that involves a cascade of events beginning with homeostasis, followed by collagen production and scar formation. To evaluate whether STAT3 signaling is involved in collagen synthesis and can be targeted to control keloid fibrosis, N102 and K119 keratinocytes and fibroblasts were used to establish the plasma-based artificial skin constructs as an in vitro assay model.[12] The results show that collagen type 1 mRNA was significantly increased in the K119 keloid skin constructs compared to N102 normal control (Fig. 2C). Cucurbitacin I, a natural selective inhibitor of the JAK2/STAT3 pathway[28] was used to assess the effect of STAT3 signaling on collagen type 1 expression. The result shows that Cucurbitacin I affects collagen type 1 synthesis in a dose dependent manner, stimulating collagen type 1 mRNA synthesis at 1 µM and 10 µM but reducing it at 3 µM (Fig. 2D). Validation of our findings in clinical specimens and the humanized keloid mouse model demonstrates that our system can retain intrinsic keloid characteristics and establish that the keloid-like animal model is capable of discovering biomarkers to reveal the dynamics of cellular and molecular events.
5. CONCLUSION
Here, we show that the plasma clot-based artificial skin construct model, which mimics the early wound healing phase, can be used for preliminary in vitro drug screening as seen in the testing of Cucurbitacin I. Results from this assay would facilitate therapeutic target identification and provide a guideline to determine therapeutic dosage and pharmacokinetic/pharmacodynamic conditions needed for future testing in an animal model.
By investigating diverse samples collected from different locations within keloids and from different stages of keloid formation with the humanized animal model in future studies, this method could define key pathways and networks associated with a particular keloid disease stage. Furthermore, it could facilitate the identification of biomarkers reflecting the entire keloid spectrum. This pilot study provides proof of principle that this type of approach will be fruitful. In the future, we will apply this approach to study additional keloid and control cell strains and more patient specimens for more comprehensive studies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank the USC Molecular Genomics Center for conducting Illumina transcriptome sequencing. RBW, CMC, and research reported in this publication was supported by grants from the NIH NIAMS under Award Numbers AR 47364 and AR 60306 and NIGMS Award Number GM125322, by the Pasadena Guild Endowment for Developmental Biology and Regenerative Medicine and by Bridge Funding awarded to TLT through The Saban Research Institute of Children’s Hospital Los Angeles, Los Angeles, CA, USA. YSL and YCL are partially supported by Drug Development Center grants from the Higher Education Sprout Project, Taiwan through China Medical University, Taiwan.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
Bibliography
- [1].Huang C, Ogawa R, Expert Opin. Pharmacother 2013, 14, 2087. [DOI] [PubMed] [Google Scholar]
- [2].Shih B, Garside E, McGrouther DA, Bayat A, Wound Repair Regen 2010, 18, 139. [DOI] [PubMed] [Google Scholar]
- [3].Tuan TL, Nichter LS, Mol. Med. Today 1998, 4, 19. [DOI] [PubMed] [Google Scholar]
- [4].Velez Edwards DR, Tsosie KS, Williams SM, Edwards TL, Russell SB, Hum. Genet 2014, 133, 1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Glass DA, J. Investig. Dermatol. Symp. Proc 2017, 18, S50. [DOI] [PubMed] [Google Scholar]
- [6].Bijlard E, Kouwenberg CAE, Timman R, Hovius SER, Busschbach JJV, Mureau MAM, Acta Derm. Venereol 2017, 97, 225. [DOI] [PubMed] [Google Scholar]
- [7].Mari W, Alsabri SG, Tabal N, Younes S, Sherif A, Simman R, J. Am. Coll. Clin. Wound Spec 2015, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee Y-S, Wysocki A, Warburton D, Tuan T-L, Birth Defects Res. C Embryo Today 2012, 96, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bagabir R, Byers RJ, Chaudhry IH, Müller W, Paus R, Bayat RA, Br. J. Dermatol 2012, 167, 1053. [DOI] [PubMed] [Google Scholar]
- [10].Bux S, Madaree A, Cells Tissues Organs 2010, 191, 213. [DOI] [PubMed] [Google Scholar]
- [11].Jumper N, Paus R, Bayat A, Histol Histopathol 2015, 30, 1033–1057. [DOI] [PubMed] [Google Scholar]
- [12].Lee Y-S, Hsu T, Chiu W-C, Sarkozy H, Kulber DA, Choi A, Kim EW, Benya PD, Tuan T-L, Wound Repair Regen 2016, 24, 302. [DOI] [PubMed] [Google Scholar]
- [13].Hsu C-K, Lin H-H, Harn HI-C, Hughes MW, Tang M-J, Yang C-C, J. Dermatol. Sci 2018, 90, 232. [DOI] [PubMed] [Google Scholar]
- [14].Wynn TA, Ramalingam TR, Nat. Med 2012, 18, 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, Watt FM, Nature 2013, 504, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, H.P., Weissman IL, Longaker MT Science 2015, 348, aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Philippeos C, Telerman SB, Oulès B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, M.D., Watt FM, J. Invest. Dermatol 2018, 138, 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jumper N, Hodgkinson R, Paus R, Bayat A, PLoS One 2017, 12, e0172955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Canady J, Arndt S, Karrer S, Bosserhoff AK, J. Invest. Dermatol 2013, 133, 647. [DOI] [PubMed] [Google Scholar]
- [20].Ghazizadeh M, Tosa M, Shimizu H, Hyakusoku H, Kawanami O, J. Invest. Dermatol 2007, 127, 98. [DOI] [PubMed] [Google Scholar]
- [21].Lim CP, Phan TT, Lim IJ, Cao X, X. Oncogene 2016, 25, 5416–5425. [DOI] [PubMed] [Google Scholar]
- [22].Lim CP, Phan TT, Lim IJ, Cao X, Invest XJ Dermatol 2019, 129, 851. [DOI] [PubMed] [Google Scholar]
- [23].Park G, Yoon BS, Moon JH, Kim B, Jun EK, Oh S, Kim H, Song HJ, Noh JY, Oh C, You S, J. Invest. Dermatol 2008, 128, 2429. [DOI] [PubMed] [Google Scholar]
- [24].Igota S, Tosa M, Murakami M, Egawa S, Shimizu H, Hyakusoku H, M. Ghazizadeh. Int. J. Med. Sci 2013, 10, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Banerjee K, Resat H, Int. J. Cancer 2016, 138, 2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, Miranda-Saavedra D, D. Nucleic Acids Res 2013, 41, 2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Katoh M, Katoh M, Int. J. Mol. Med 2017, 40, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM, Cancer Res 2003, 63, 1270. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.