Abstract
CD133 is a cellular surface protein, which has been reported to be a cancer stem cell marker, and thus is considered a potential target for cancer treatment. Metformin, one of the biguanides used for the treatment of diabetes, is also known to reduce the risk of cancer development and cancer stem-like cells (CSCs), including the expression of CD133. However, the mechanism underlying the reduction of the expression of CD133 by metformin is not yet understood. This study shows that metformin suppressed CD133 expression mainly by affecting the CD133 P1 promoter via adenosine monophosphate (AMP)-activated protein kinase (AMPK) signaling but not the mammalian target of rapamycin (mTOR). AMPK inhibition rescued the reduction of CD133 by metformin. Further experiments demonstrated that CCAAT/enhancer-binding protein beta (CEBPβ) was upregulated by metformin and that two isoforms of CEBPβ reciprocally regulated the expression of CD133. Specifically, the liver-enriched activator protein (LAP) isoform increased the expression of CD133 by directly binding to the P1 promoter region, whereas the liver-enriched inhibitory protein (LIP) isoform suppressed the expression of CD133. Consistent with these findings, a three dimensional (3D) culture assay and drug sensitivity assay demonstrated that LAP-overexpressing cells formed large spheroids and were more resistant to 5-fluorouracil (5-FU) treatment, whereas LIP-overexpressing cells were more sensitive to 5-FU and showed combined effects with metformin. Our results indicated that metformin-AMPK-CEBPβ signaling plays a crucial role in regulating the gene expression of CD133. Additionally, regulating the ratio of LAP/LIP may be a novel strategy for targeting CSCs for the treatment of cancer.
Abbreviations: ACC, Acetyl-CoA-carboxylase; AMPK, AMP-activated protein kinase; CSC, Cancer stem-like cells; CEBPβ, CCAAT/enhancer-binding protein beta; DMEM, Dulbecco's modified Eagle's medium; FACS, Fluorescence activated cell sorting; H&E, Hematoxylin and eosin; LAP, Liver-enriched activator protein; LIP, Liver-enriched inhibitory protein; SDS, Sodium dodecyl sulfate; TBS, Tris-buffered saline
Introduction
During previous decades, drug repositioning, which describes the attempt to use a known drug to for the treatment of new diseases, has been a feature of investigations throughout the world. Metformin, which is universally accepted to treat type 2 diabetes, is also considered to be one of these drugs and has recently been shown to have tumor-suppressive effects by inhibiting cancer cell proliferation and regulating immune cells, including regulatory T cells, in the tumor microenvironment [1], [2].
Cancer stem-like cells (CSCs) are described as a small population in cancer tissue that plays a key role in tumor growth and maintenance. Therefore, CSCs are considered to be one of factors of high importance in relation to drug resistance, recurrence, metastasis, and poor prognosis [3], [4]. To date, there are no drugs targeting CSCs approved for clinical application, and the development of these drugs is of great interest for cancer therapy.
CD133, also known as PROM1, is one of the most widely used markers for identifying CSCs in tumors of several types of cancer, including liver, colon, brain, prostate, and pancreatic cancer [5], [6], [7], [8], [9]. CD133 is a five-transmembrane glycoprotein consisting of an N-terminal extracellular domain, transmembrane domain, and C-terminal intracellular domain. In terms of posttranscriptional modification, with the exception of glycosylation, acetylation and phosphorylation are involved in affecting several intracellular signaling pathways. However, no specific extracellular ligand has been identified so far [10]. CD133 is not only a CSC marker but also plays an important role in CSC maintenance by regulating several cell signaling pathways [11], and the suppression of CD133 expression appears to be one strategy for cancer treatment.
Metformin (1,1-dimethylbiguanide) is a low-molecular-weight compound belonging to one of the guanidine derivatives and is approved for the treatment of type 2 diabetes. Metformin improves insulin resistance and reduces blood glucose concentration by suppressing gluconeogenesis in hepatocytes [12]. Although several mechanisms of action have been shown to explain the effects of metformin, including inhibiting mitochondrial respiratory chain complex I resulting in an increase in the AMP/ATP ratio, several questions remain in explaining all the effects of metformin [13], [14]. In addition, with respect to the anticancer effect of metformin, various reports have shown that metformin selectively affects CSCs by decreasing CD133+ cells and by increasing their sensitivity to anticancer drugs [15], [16], [17]. However, the detailed mechanism underlying the reduction of CD133 remains largely unknown.
CCAAT/enhancer-binding protein beta (CEBPβ) is a transcription factor, belonging to the CEBP family (α, β, δ, ε, γ, and ζ). The members of this family commonly contain a basic leucine zipper DNA binding domain and regulate the transcription of several target genes by binding as a homodimer and/or heterodimer [18]. It has been shown that CEBPβ is involved in several cell biological properties, including cell proliferation, differentiation, immune response, and adipogenesis [18], [19]. Furthermore, in certain tumors, CEBPβ also functions as an oncogene and tumor suppresser via the regulation of the cell cycle, autophagy, and senescence in a context-dependent manner [20], [21], [22].
In the present study, we investigated which factors are essential for reducing the expression of CD133 by metformin in vitro and revealed that metformin-AMPK-CEBPβ signaling plays a crucial role in regulating the gene expression of CD133.
Materials and Methods
Cells and Reagents
HepG2 cells and Huh1cells (provided by RIKEN RBC, Tsukuba, Japan in December 2015) were cultured in Dulbecco's modified Eagle's medium (DMEM, Nacalai Tesque, Kyoto, Japan). JHH2–7 cells (provided by JCRB Cell Bank, Osaka, Japan in April 2017) were cultured in William's Medium E + GlutaMAX (Thermo Fisher Scientific, Waltham, MA, USA). All mediums were supplemented with 10% fetal bovine serum (Moregate Biotec, Bulimba, Australia), 100 U/ml penicillin, and 100 μg/ml streptomycin (WaKo Pure Chemical Industries, Osaka, Japan), and cells were incubated at 37 °C under 20% O2 and 5% CO2 conditions. The cells were authenticated by short-tandem repeat-PCR method at the cell bank. In our laboratory, the cells were cultured on a massive scale after shipping and were stored at −80 °C. Stocked cell aliquots were used within short term passages. Metformin hydrochloride was purchased from Cayman Chemical (Ann Arbor, MI, USA) and dissolved in water. Compound C, rapamycin, and C188–9 were purchased from Merck Millipore, (Burlington, MA, USA) and dissolved in dimethyl sulfoxide (DMSO).
Plasmids and Small Interfering RNAs (siRNAs)
Complementary DNAs (cDNAs) of CEBPβ, including LAP* (345 amino acids (aa)), LAP (322 aa), and LIP (147 aa), were obtained by reverse transcription-polymerase chain reaction (RT-PCR), and the resulting fragments were cloned into the pCMV-3 × FLAG vector, which was constructed by inserting oligonucleotides encoding 3× FLAG into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). For retrovirus-mediated gene transfer, CEBPβ fragments were cloned into the pBABE-puro vector. The CD133 clones containing the promoter fragments P1-Luc were provided by Professor S. Tanaka of Hokkaido University (Sapporo, Japan) [23]. siRNAs for AMP-activated protein kinase alpha (AMPKα1) and scramble (control siRNA) were purchased from Qiagen (Hilden, Germany). siRNA transfection was performed with Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer's instruction. The final concentration of siRNAs was 5 nM.
Transient Transfection and Reporter Gene Assay
The cells were plated at a density of 1 × 105 cells in 24-well plates with 500 μl culture medium. Following incubation for 24 h at 37 °C, the cells were transfected with 100 ng luciferase plasmid DNA and 10 ng Renilla pGL4.75 (hRluc-CMV) vector (Promega, Madison, WI, USA) as an internal control by using lipofectamine LTX (Invitrogen), and the culture medium was replaced with/without metformin. A reporter gene assay was performed 24 h following transfection by using the Dual-Glo Luciferase reporter assay system (Promega), and the luminescence intensity was measured using GLOMAX 20/20 luminometer (Promega), according to the manufacturer's protocol. The transcription activity was normalized according to Renilla luciferase activity. Experiments were performed in triplicate.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from the cells using an RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. A total of 500 ng total RNA was reverse transcribed into cDNA by using Prime Script RT Master Mix (Takara, Kusatsu, Japan). PCR was performed in a 20-μl volume containing 9 μl cDNA (15 ng/μl) and 10 μl SYBR Premix Ex Taq (Takara). Following an initial denaturation at 95 °C for 20 s, a two-step cycle procedure was used (denaturation at 95 °C for 1 s and annealing and extension at 60 °C for 20 s) for 40 cycles in a Step One real-time PCR system (Applied Biosystems, Foster City, CA, USA). Gene expression levels were determined using the ddCt method with β-actin as an endogenous control. The data were analyzed with Step One software version 2.2.2 (Applied Biosystems). Primers used for qRT-PCR are listed in Table 1.
Table 1.
Primer sequences used for qRT-PCR
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| AMPKα1 | CGGAGCCTTGATGTGGTAGG | TTCATCCAGCCTTCCATTCTT |
| AMPKα2 | ATCGTTCTGTCGCCACTCTC | ACACACTTCTTTCACAGCCTCA |
| β-actin | TTGCCGACAGGATGCAGAA | GCCGATCCACACGGAGTACT |
| CD133 (total) | TGGCCACCGCTCTAGATACT | GCTTTTCCTATGCCAAACCA |
| CD133 (P1) | ACCTGGCCATGCTCTCAGCT | TCACGCGGCTGTACCACATAGA |
| CD133 (P2) | GTGACTAGGGCGGGAGCAG | TCACGCGGCTGTACCACATAGA |
| CD133 (P3) | GTGCCCGTCCAACCCAAACTT | TCACGCGGCTGTACCACATAGA |
| CD133 (P4) | CGCCAAAAGCACTCCAGATGAC | TCACGCGGCTGTACCACATAGA |
| CD133 (P5) | TGGATCTGGACCCCAGGAGTT | TCACGCGGCTGTACCACATAGA |
| CEBPβ | CCCGCCCGTGGTGTTATTTA | GCATCAACTTCGAAACCGGC |
Western Blot Analysis
The cells were washed with ice-cold Tris-buffered saline (TBS) (−) and lysed in 1× sodium dodecyl sulfate (SDS) sample buffer containing 2% 2-mercaptoethanol. The samples were heated at 95 °C for 5 min and then subjected to SDS-polyacrylamide gel electrophoresis. The separated proteins were transferred onto Immobilon-P polyvinylidene fluoride (PVDF) membranes (Merck Millipore), which were subsequently incubated with TBS with 0.05% Tween 20 containing 5% dried nonfat milk for 30 min at room temperature. The membranes were probed with primary antibodies for AMPKα, phosphorylated (p-)AMPKα (T172), acetyl-CoA-carboxylase (ACC), p-ACC (S79), β-actin, CD133, p-CEBPβ (T235), p70-S6K, p-p70-S6K (T389), signal transducer and activator of transcription (STAT)3, p-STAT3 (Y705) (1:1000, Cell Signaling Technology, Inc., Danvers, MA, USA), and CEBPβ (1:200, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), and bound antibodies were detected with peroxidase-labeled rabbit or mouse antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and visualized by Immobilon western HRP Substrate detection reagents (Merck Millipore).
DNA Microarray
The DNA microarray was performed by using the GeneChip Human Gene 2.0 ST array (Applied Biosystems) according to the manufacturer's instructions. In brief, the HepG2 cells were treated with or without metformin (5 mM) for 48 h. Total RNA was extracted from cells by using an RNeasy Mini kit according to the manufacturer's instructions. The experiments following RNA extraction was outsourced to GeneticLab, Co., Ltd. (Sapporo, Japan). Data analysis was performed with GeneSpring GX version 12.6 (Agilent Technologies, Santa Clara, CA, USA).
Retrovirus-Mediated Gene Transfer
Using pBABE-puro-vectors (Empty, LAP, and LIP), replication-incompetent retrovirus expressing Empty, LAP, and LIP were produced and transduced as described previously [24], [25]. Stable cell lines were established by selection with 1 μg/ml puromycin (Thermo Fisher Scientific) for 2 weeks.
Chromatin Immunoprecipitation (ChIP) Assay
The stable cell lines (HepG2-Empty, LAP, and LIP) were plated in a 10-cm dish with 10 ml culture medium. Following incubation for 48 h at 37 °C, the cells were fixed with 1% formaldehyde, and chromatin was subsequently prepared using an Enzymatic Chromatrap Premium Pro G 24 column shearing kit (Porvair Sciences, Norfolk, UK). All manipulations were performed according to the manufacturer's instruction. In brief, immunoprecipitation (IP) was performed using 2 μg sheared chromatin and 4 μg anti-CEBPβ (Santa Cruz Biotechnology, Inc.) or anti-normal mouse immunoglobulin G (IgG; Santa Cruz Biotechnology, Inc.). The DNA collected by ChIP was amplified by qRT-PCR. The target primer was designed for the location of P1 around the CEBPβ binding site (5′-TGTAGCTTGTGCATCCATCC-3′ and 5′-GGTGTCTAGCATACAGCAGG-3′).
The accumulation of LAP and/or LIP for the P1 promoter (around the CEBPβ binding site) was calculated from the ratio of IP anti-CEBPβ sample data/IP anti-IgG sample data.
Flow Cytometry and Fluorescence Activated Cell Sorting (FACS)
FACSCantoII (BD Biosciences, Franklin Lakes, NJ, USA) was used for flow cytometry. The cells were suspended in Hank's balanced salt solution (Invitrogen) containing 0.5% bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) and stained with PE-anti-CD133 (Miltenyi Biotec, Bergisch Gladbach, Germany) at 1:40 on ice for 30 min. Dead cells were eliminated using 7-AAD (BD Biosciences) according to the manufacturer's instruction. To fractionate CD133High (CD133H) and CD133Low (CD133L), the cells were sorted by FACSAriaIII (BD Biosciences).
3D Culture of HepG2 Cells
3D culture was performed as described previously with modification [26], [27]. In brief, the stable cell lines (HepG2-Empty, LAP, and LIP) were seeded at 4,000 cells/well in a 96-well plate in 50 μl Matrigel (Corning, Corning, NY, USA). Following solidification, 500 μl of advanced DMEM/F12 (Thermo Fisher Scientific) supplemented with 1× Glutamax and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (15 mM) was added and incubated at 37 °C for 2 weeks. The growth medium was replenished every other day. The 3D spheroids were recovered by digesting the Matrigel with DispaseI (1,000 PU/ml, Wako Pure Chemical Industries), fixed overnight in 4% paraformaldehyde (Wako Pure Chemical Industries), and then embedded in 2% bacto-agar and 2.5% gelatin and subjected to histologic analyses. Hematoxylin and eosin (H&E) staining was performed as described previously [28]. Data analysis was performed with NanoZumer Digital Pathology View 2 (Hamamatsu Photonics, Hamamatsu, Japan).
Drug Sensitivity (WST-8 assay)
A total of 10,000 cells (HepG2-Empty, LAP, and LIP) were seeded per well in a 96-well plate. After 24 h, drugs were added at the following concentrations: Metformin, 5 mM; and 5-FU, 20 μM. After 24 h, the WST-8 assay was performed using a cell counting kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. The absorbance (450–750 nm) was measured using the GloMax Multi+ Detection System (Promega), and the survival rate was determined.
Statistical Analysis
Data are expressed as the mean ± standard deviation. Comparisons of parameters between two groups were performed with the (two-tailed) Student's t-test. Comparisons of parameters among groups were performed by one-way analysis of variance, followed by Tukey's multiple comparison test. Differences were considered significant at P < .05.
Results
Metformin Decreases the Expression of CD133 at the Transcription Level
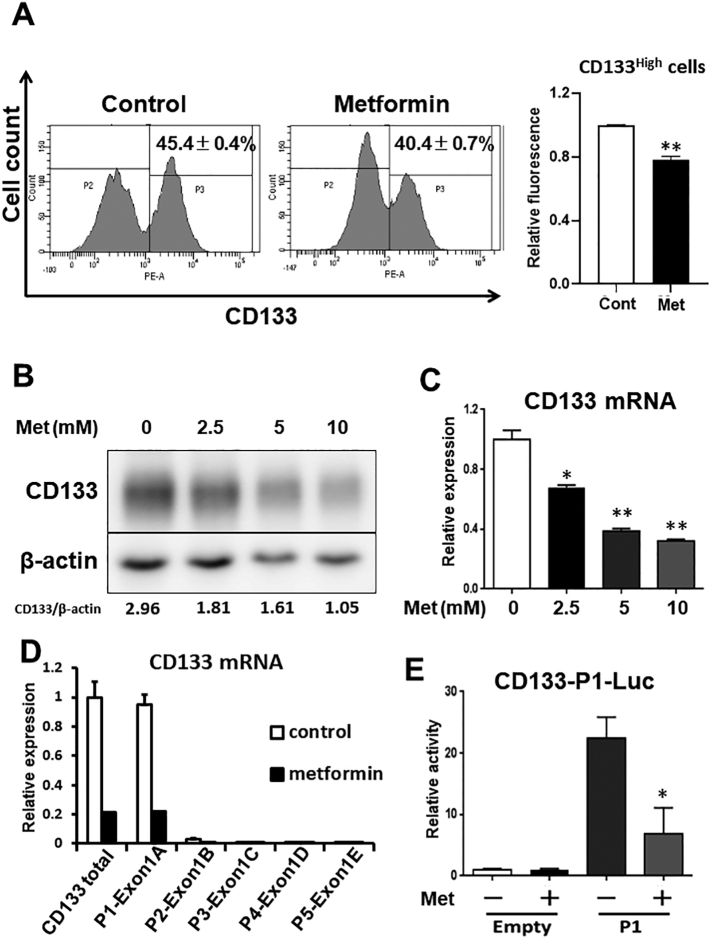
We first examined the protein expression of CD133 in HepG2 cells. The FACS and western blot analysis showed that the protein expression of CD133 was decreased by metformin (Figure 1, A and B, respectively). The dose-dependent decrease in CD133 was also confirmed at the protein and mRNA levels (Figure 1, B and C, respectively). Consistent with the results of HepG2 cells, the expression of CD133 was decreased by metformin in other hepatocellular carcinoma (HCC) cell lines (JHH6, JHH7, and Huh1) (Figure S1, A and B).
Figure 1.
Metformin decreases the expression of CD133 at the transcription level.
(A) HepG2 cells were treated with or without metformin (5 mM) for 48 h. The cells were then stained with PE-anti-CD133 antibody and analyzed by FACS (left). Fluorescence intensity was measured for quantitation (right). ⁎⁎P < .01, vs. Control treatment (n = 3). (B and C) HepG2 cells were treated with different doses of metformin for 48 h, and the expression of CD133 was examined by western blot and qRT-PCR analyses. ⁎P < .05, vs. Met (0 mM), ⁎⁎P < .01, vs. Met (0 mM). (D) mRNA expression of CD133 from each promoter was detected using exon-specific primers. (E) Promoter activity of P1 in HepG2 cells. Cells were treated with or without metformin (5 mM) for 24 h following plasmid transfection. Luciferase activity was then measured. ⁎P < .05, vs. P1-Met (−). Met; metformin.
It has been demonstrated that CD133 gene transcription is modulated by five alternative promoters in the 5′-untranslated region of the CD133 gene, P1, P2, P3, P4, and P5, resulting in CD133 transcription variants [29]. Therefore, to clarify which promoter is crucial for the effect of metformin, we performed qRT-PCR by using specific primer pairs for each variant. Although the expression levels of all five mRNAs were downregulated by metformin, mRNA driven from P1 predominantly contributed to the reduction in CD133 in the HepG2 cells (Figure 1D). Consistent with the qRT-PCR experiment, P1 promoter activity was also reduced by metformin (Figure 1E). These results suggest that metformin mainly affects the CD133-P1 promoter region and reduces the expression of CD133 at the transcription level.
Metformin Decreases the Expression of CD133 Through AMPK Signaling
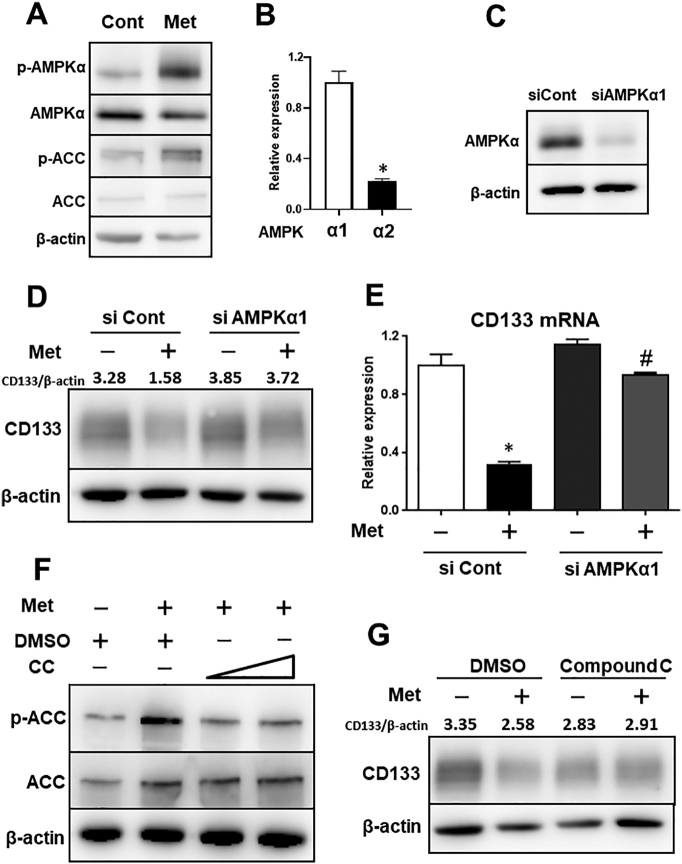
It is widely accepted that metformin activates AMPK by inhibiting mitochondrial respiratory chain complex I, resulting in a reduction in the ATP/AMP ratio [30], [31]. Therefore, we examined whether the activation of AMPK is essential for regulating the expression of CD133. Metformin certainly induced the activation of AMPK and phosphorylation of its substrate ACC (Figure 2A). In addition, since the catalytic subunit of AMPK is encoded by two distinct genes (AMPKα1 and AMPKα2), we confirmed the expression of AMPKα1 and AMPKα2 by qRT-PCR in HepG2 cells. In this cell line, the expression of AMPKα1 was higher compared with that of AMPKα2 and therefore, we focused on AMPKα1 (Figure 2B). We then knocked down AMPKα1 by using siRNA. Knockdown efficiency was confirmed by western blotting (Figure 2C); the knockdown of AMPKα1 partially abrogated the reduction of CD133 by metformin at the protein and mRNA levels (Figure 2, D and E, respectively).
Figure 2.
Metformin decreases the expression of CD133 through AMPK signaling.
(A) HepG2 cells were treated with or without metformin (5 mM) for 48 h. Levels of phosphorylated and total AMPKα and ACC were examined by western blotting. (B) mRNA expression of AMPKα1 and AMPKα2 were examined by qRT-PCR in HepG2 cells. (C) AMPKα1 was knocked down by using siRNA in HepG2 cells at a concentration of 5 nM, and knockdown efficiency was confirmed by western blotting. (D and E) HepG2 cells were transfected with siRNA (control or AMPKα1). Following transfection for 24 h, cells were treated with or without metformin for 48 h, and expression levels of CD133 were examined by western blot and quantitative polymerase chain reaction analyses. ⁎P < .05, vs. siCont-Met (−), #P < .05, vs. siCont-Met (+). (F) HepG2 cells were pretreated with or without Compound C (5, 10 μM) for 30 min and then treated with or without metformin (5 mM) for 2 h. The effects of Compound C were confirmed by western blotting (p-ACC levels). (G) HepG2 cells were pretreated with or without Compound C (5 μM) for 30 min and then treated with or without metformin (5 mM) for 48 h; expression of CD133 was examined by western blotting. Met; metformin, CC; Compound C, Cont; control, ACC; acetyl-CoA-carboxylase, p-; phosphorylated.
Consistent with the results of AMPK silencing, the inhibition of AMPK by compound C, an AMPK inhibitor, also eliminated the effects of metformin (Figure 2, F and G). The effects of compound C were confirmed by ACC phosphorylation (Figure 2F). Furthermore, AICAR, an AMPK activator, also suppressed CD133 expression as metformin treatment (Figure S1, C and D). These results suggest that the expression of CD133 is regulated, at least in part, by AMPK signaling.
Janus Kinase (JAK)/STAT3 Pathway is Partially Involved in the Metformin/AMPK-Induced Suppression of CD133
AMPK phosphorylates a variety of substrates and regulates various signaling pathways. Among those pathways, it is well known that the mTOR pathway is suppressed by AMPK activators, including metformin [32]. The JAK/STAT3 pathway, which is essential for the immune response, is also suppressed by AMPK via the phosphorylation of JAK (Figure S2A) [33]. Moreover, it has been reported that interleukin (IL)-6 increases the expression of CD133 through the activation of STAT3 [34]. Therefore, we investigated whether mTOR and/or the JAK/STAT pathway were involved in the metformin-induced suppression of CD133. Metformin suppressed the phosphorylation of p70-S6 kinase, a representative mTOR substrate, and suppressed the phosphorylation of STAT3 (Figure S2B). However, rapamycin, an mTOR inhibitor, did not affect the expression of CD133 (Figure S2, C and D), whereas C188–9, a STAT3 inhibitor, significantly decreased the expression of CD133 (Figure S2, E and F). Meanwhile, contrary to our expectation, the P1 promoter activity was not reduced by the inhibition of STAT3 (Figure S2G). These results suggest that other novel factors are involved in P1 promoter suppression by metformin.
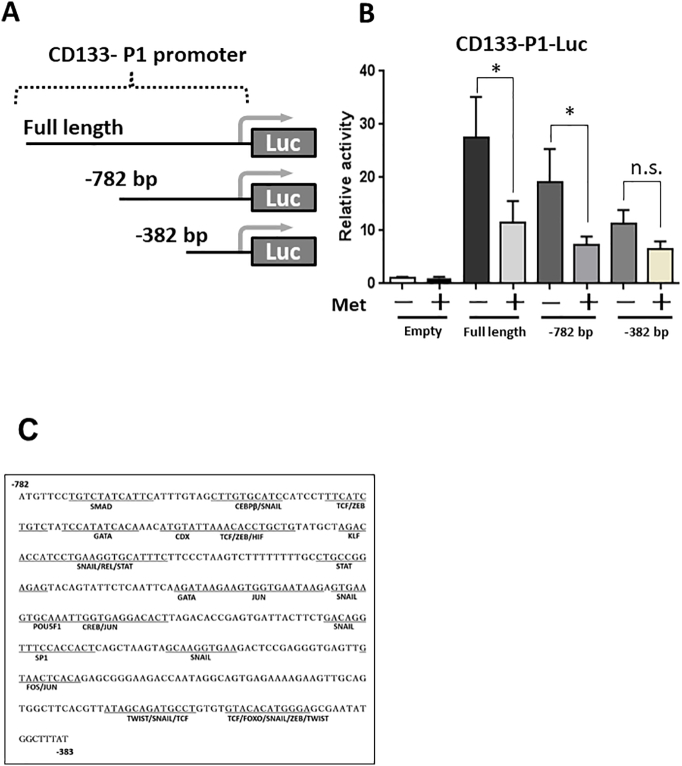
The Region Between −782 and −382 bp is Essential for the Regulation of CD133 P1 Promoter Activity by Metformin
To identify factors required for the metformin-induced suppression, we first focused on the P1 promoter sequence. To elucidate the crucial region in the P1 promoter, we used a series of deletion mutants of the P1 promoter (pGL3enh-P1–782 bp, −382 bp; Figure 3A), and the reporter gene assay was performed. The promoter activities of the P1 full length and the −782 bp deletion mutant were significantly decreased by metformin, whereas the activity of the −382 bp deletion mutant was not affected (Figure 3B). These results indicated that the region between −782 and− 382 bp is essential for the metformin-induced suppression of CD133. We then analyzed P1 promoter region between −782 and −382 bp by using an open-access database transcription factor binding profile search (JASPAR2018) [34]. The results showed that 18 transcription factors have probability to bind to the region (Figure 3C).
Figure 3.
Metformin affects the region between −782 and− 382 bp of the CD133-P1 promoter.
(A) Schematic representation of luciferase reporter constructs containing the deleted sequence of the P1 promoter. (B) Promoter activity of P1-deleted constructs in HepG2 cells. Cells were treated with or without metformin (5 mM) for 24 h following plasmid transfection and luciferase activity was measured. ⁎P < .05, vs. P1-Met (−). (C) CD133-P1 promoter sequence between −782 and− 382 bp containing CEBPβ binding site.
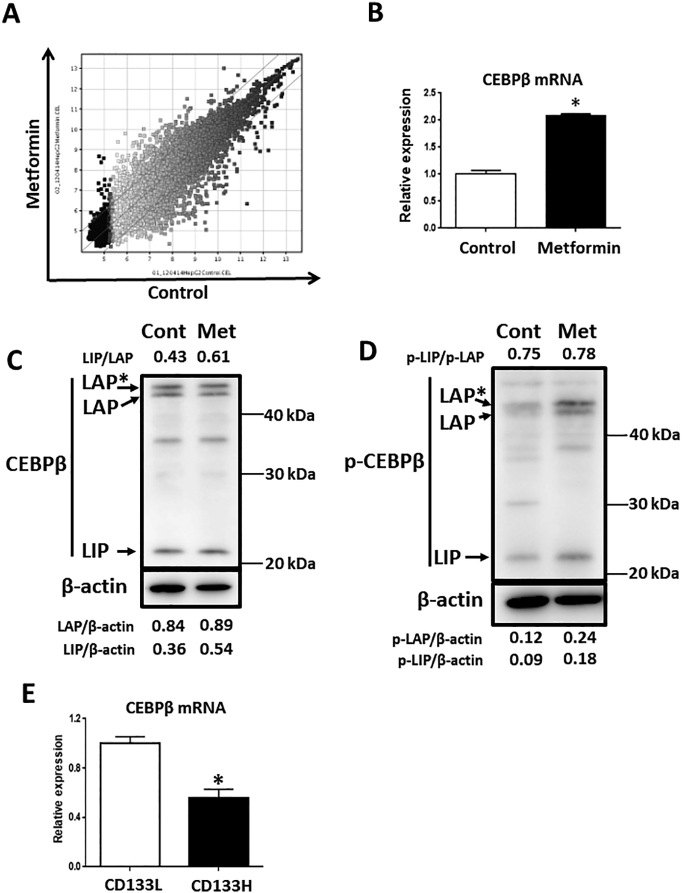
CEBPβ is Up-Regulated by Metformin and in the CD133Low fraction of HepG2 cells
As metformin appeared to not be able to bind to the P1 promoter directly, the involvement of other downstream factors, including transcriptional factors, was considered. Therefore, to elucidate pivotal factors that regulate the CD133 P1 promoter region by metformin, we performed DNA microarray analysis and compared gene expression profiles following metformin treatment in HepG2 cells. Among 36,725 transcript clusters detected, 676 clusters were increased over 2.0-fold and 592 clusters were decreased less than 2.0-fold (Figure 4A). We then selected transcription-related genes and confirmed mRNA expression levels by qRT-PCR for validation. Finally, we evaluated 13 candidate genes by comparing sorted HepG2-CD133H cells with HepG2- CD133L cells. Following a series of experiments, and because transcription factor binding site is also included in the region between −782 and− 382 bp of the P1 promoter, we focused on the CEBPβ gene. Metformin increased the expression of CEBPβ (Figure 4, B and C) and induced the phosphorylation of CEBPβ (Figure 4D). In addition, when the HepG2 cells were fractionated to CD133H and CD133L cells (Figure S3, A–C), the expression of CEBPβ was increased in the CD133L cells (Figure 4E).
Figure 4.
Metformin increases the gene expression of CEBPβ.
(A) Scatter plot of DNA microarray results in HepG2 cells (control treatment vs. metformin treatment). (B-D) HepG2 cells were treated with or without metformin (5 mM) for 48 h, and expression levels of CEBPβ were examined by qRT-PCR and western blot analyses. ⁎P < .05, vs. Control. Met; metformin, p-; phosphorylated. (E) Sorted CD133H and CD133L cells were cultured for 48 h, and the expression levels of CEBPβ were examined by qRT-PCR analyses. *P < .05, vs. CD133L.
Two isoforms of CEBPβ reciprocally regulate the expression of CD133
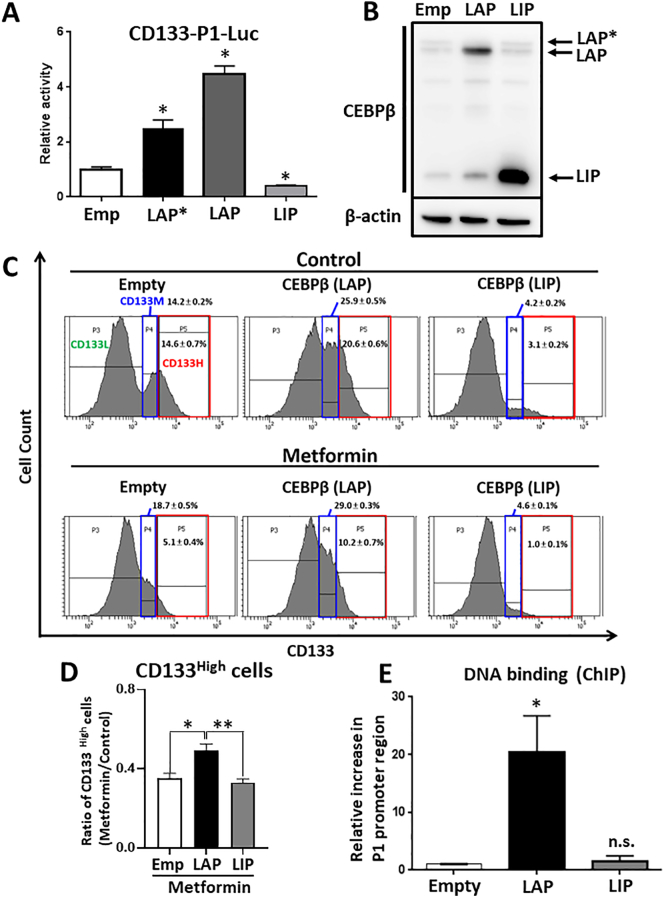
CEBPβ is comprised of three isoforms (LAP*, LAP, and LIP) [35]. In general, LAP* and LAP are regarded as transcriptional activators, as they contain a transactivation domain [36]. By contrast, the LIP isoform does not have a transactivation domain and acts as a transcriptional inhibitor [37]. The expression of all three isoforms was detected in the HepG2 cells (Figure 4C). The LIP isoform, but not the LAP isoform, was upregulated by metformin treatment. Meanwhile, with respect to the phosphorylation of Thr235, all isoforms were upregulated by metformin treatment (Figure 4D). To investigate the involvement of CEBPβ isoforms in the transcription of CD133, we constructed the plasmids of the three isoforms and analyzed P1 promoter activities following the transient overexpression of the isoforms. As expected, the P1 promoter activity was increased by the overexpression of both LAP* and LAP but not by the overexpression of LIP (Figure 5A). We next generated stable cells ectopically expressing CEBPβ (LAP and LIP) in HepG2 cells. The expression levels of LAP and LIP were confirmed by western blotting (Figure 5B). As expected, in the LAP-overexpressing cells, the expression of CD133 was increased and not affected by metformin compared with mock and LIP-overexpressing cells. On the other hand, in the LIP-overexpressing cells, the expression of CD133 was decreased (Figure 5, C and D). Furthermore, the ChIP assay showed that the LAP isoform strongly bound to the CEBPβ binding site of the P1 promoter region (Figure 5E). By contrast, the LIP isoform did not bind to the P1 promoter region. These results indicated that the two isoforms of CEBPβ reciprocally regulate the expression of CD133, that is, the LAP isoform increases the expression of CD133 by directly binding to P1 promoter region, whereas the LIP isoform suppresses the expression of CD133, most likely via an indirect promoter binding mechanism.
Figure 5.
Two isoforms of CEBPβ reciprocally regulate the expression of CD133.
(A) Promoter activity of P1 in HepG2 cells transiently transfected with CEBPβ isoforms (LAP*, LAP, or LIP). ⁎P < .05, vs. Empty. (B) CEBPβ isoform (LAP or LIP) transduction was performed by retrovirus-mediated gene transfer into HepG2 cells. Expression levels of LAP and LIP were examined by western blotting. (C) Stable cells (overexpressing LAP or LIP) were treated with or without metformin (5 mM) for 48 h. The cells were then stained with PE-anti-CD133 antibody and analyzed by FACS. CD133-positive cells were divided into two population (CD133M and CD133H) for analyzing the change of CD133 expression in detail. Reference range are defined by Empty-control cells (CD133M; 14.2%, CD133H; 14.6%). (D) Ratio of CD133H cells (Control treatment/Metformin treatment). ⁎P < .05, ⁎⁎P < .01. (E) ChIP assay showing the binding of LAP to the CD133 P1 promoter around CEBPβ binding sites. ⁎P < .05, vs. Empty. Met; metformin, n.s.; not-significant.
Two Isoforms of CEBPβ are Involved in the Characteristic Feature of CSCs
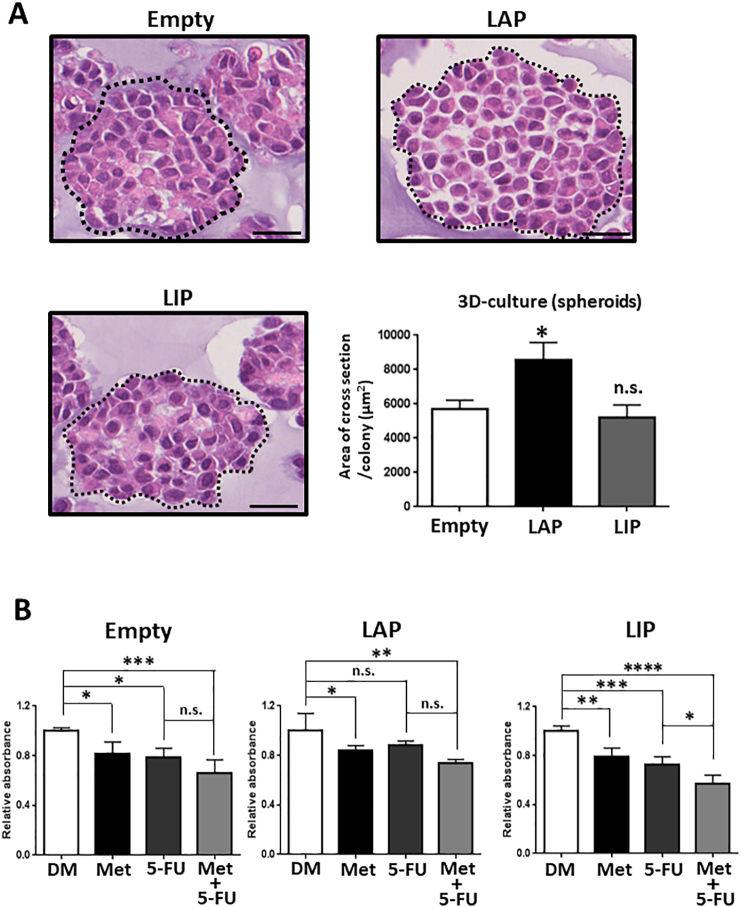
To reveal whether or not CEBPβ is involved in the characteristic feature of CSCs, we performed a 3D culture assay by using CEBPβ-stable cells, as the formation of 3D spheroids is considered to represent tumorigenicity in vitro [26]. The spheroids derived from the LAP-overexpressing cells, but not the LIP-overexpressing cells, were significantly larger than those from the control cells (Figure 6A). In addition, the LAP-overexpressing cells were more resistant to 5-FU, a chemotherapeutic agent often used to treat patients with hepatocellular carcinoma (HCC) [38], than the Empty cells (Figure 6B). By contrast, the LIP-overexpressing cells were more sensitive to 5-FU and showed combined effects with metformin. These results indicated that CEBPβ, particularly the LIP isoform, is important for the suppression of CSC characteristics by metformin, whereas the LAP isoform supports CSC maintenance.
Figure 6.
Two isoforms of CEBPβ are involved in the characteristic features of cancer stem-like cells.
(A) 3D spheroids were generated with stable cells (Mock, LAP, and LIP) and analyzed by H&E staining (n = 3). Scale bars = 25 μm. ⁎P < .05, vs. Empty. (B) Stable cells (Mock, LAP, and LIP) were treated with or without metformin (5 mM) and 5-FU (20 μM) for 24 h. Cell viability was measured using the WST-8 assay (n = 4). ⁎P < .05, ⁎⁎P < .01, ⁎⁎⁎P < .001, ⁎⁎⁎⁎P < .0001. Met; metformin, DM; DMSO, n.s.; not-significant.
Discussion
In the present study, we focused on the regulation of CD133 expression by metformin and demonstrated that 1) metformin suppresses the expression of CD133 via AMPK signaling; 2) metformin regulates the activity of the CD133 P1 promoter through the region between −782 and− 382 bp; 3) metformin suppresses CD133 P1 promoter activity through upregulating the expression of CEBPβ, particularly the LIP isoform; and 4) two isoforms of CEBPβ reciprocally regulate the expression of CD133 and are involved in the characteristic feature of CSCs.
It has been demonstrated that CD133 is not only a CSC marker but also plays an important role in CSC maintenance by regulating several cell signaling pathways [11]. CD133 physically interacts with HDAC6 and stabilizes β-catenin, resulting in increasing cell proliferation and tumorigenesis [39]. Phosphoinositide-3-kinase/AKT and p38-mitogen-activated protein kinase signaling were upregulated in CSCs through epidermal growth factor receptor stabilization by CD133 and/or the phosphorylation of CD133 by Src and Fyn kinases [11], [40]. Moreover, xCT, a cystine transporter, is increased in CD133+ CSCs and promotes glutathione synthesis, resulting in the reduction of reactive oxygen species [41]. Therefore, as CD133 has multiple functions in CSC maintenance, targeting the expression of CD133 appears to be a promising strategy for cancer treatment.
CD133 has five promoters functioning in a tissue-dependent manner [29]. For example, P1 is active in several tissues, including the prostate, kidney, and lungs, whereas P2 is active in the brain, and P4 is active in the testis. In the present study, we used an HCC cell line and examined the expression of all five promoter-derived mRNAs and found that P1 promoter was predominant in the HepG2 cells (Figure 1D). This result was consistent with previous reports demonstrating that P1 is a main promoter in liver tissues, including in HCC [29], [42].
It has been reported that metformin reduces the expression of CD133 [15], [16], [17]. However, the detailed mechanism underlying the reduction of CD133 has remained largely unknown. In the present study, we showed for the first time, to the best of our knowledge, that metformin suppressed the expression of CD133 via AMPK but not mTOR (Figure 2, D–F, Figure S2, C and D). In addition, it has been reported that STAT3 is suppressed by AMPK and increases the expression of CD133 [33], [34]. Consistent with a previous study, our results showed that metformin reduced the phosphorylation of STAT3 and the STAT3 inhibitor (C188–9) decreased the expression of CD133 (Figure S2, B, E, and F). By contrast, the P1 promoter activity was not suppressed by C188–9 (Figure S2G). Therefore, the results suggested that metformin suppresses the expression of CD133 by at least two means; by reducing P1 promoter activity and inactivating STAT3. Although the mechanism underlying the reduction in the expression of CD133 by the metformin/STAT3 pathway is unclear, it is possible that STAT3 regulates CD133 mRNA stabilization and/or another promoter. However, as the P1 promoter mentioned above was predominant in the HepG2 cells (Figure 1D), our observations suggest that P1 is the main promoter responsible for the suppression of CD133.
There have been several studies demonstrating the regulation of CD133 transcription other than STAT3. We previously reported that the expression of CD133 was regulated at the P5 promoter by hypoxia-inducible factors through Ets transcription factors in colon cancer cells [23]. Octamer-binding transcription factor 4 and SRY-box 2, known as stemness-related transcription factors, promote the expression of CD133 in hypoxia [43]. By contrast, IKaros, a transcription factor containing a zinc finger domain, represses the expression of CD133 through directly binding to the P1 promoter [42]. In addition, two reports exist on p53 regulating the expression of CD133. Chen et al. revealed that p53 promoted the expression of CD133 [44], whereas Park et al. showed that p53 repressed the expression of CD133 [45]. As AMPK is known to activate p53 activity via direct phosphorylation at Ser15 of p53 [46], we examined whether or not metformin induces the phosphorylation of p53. However, its phosphorylation status was unchanged by metformin (data not shown). Therefore, we performed a DNA microarray to identify downstream factors of the metformin/AMPK pathway regulating the CD133-P1 promoter, with subsequent focus on the CEBPβ gene.
In the present study, it was shown that metformin increased the expression of the CEBPβ LIP isoform but not the LAP isoform (Figure 4C). Yang et al. reported that 5-aminoimidazole-4-carboxamide ribonucleotide, an AMPK activator, increased the expression of CEBPβ via AMPK/peroxisome proliferator-activated receptor gamma co-activator-alpha signaling in monocytes [47]. Therefore, this pathway may explain our results; however, the mechanism underlying this action, particularly the increase in only the LIP isoform, remains to be elucidated, and further studies are required.
As shown in Figure 4D, metformin also increased the phosphorylation of CEBPβ at Thr235. It has been reported that three kinases (MAPK, CDK2, and RPS6KA1) phosphorylate CEBPβ and promote its transcriptional activity [48], [49], [50]. However, with respect to AMPK, there is no evidence that CEBPβ is one of the substrates for AMPK. In the present study, two isoforms of CEBPβ reciprocally regulated the expression of CD133. The LAP isoform increased CD133-P1 promoter activity by direct binding, whereas the LIP isoform decreased its activity (Figure 5A). As both LAP and LIP were similarly phosphorylated by metformin (Figure 4E), it appears that the LAP/LIP ratio, rather than phosphorylation, is important for the regulation of CD133.
Conclusion
Metformin decreases the expression of CD133 mainly by affecting the CD133 P1 promoter via the AMPK/CEBPβ pathway. Furthermore, the present study showed for the first time, to the best of our knowledge, that the LAP/LIP ratio is important for regulating the expression of CD133. Our results appear to demonstrate a novel mechanism for explaining the antitumor effects of metformin. Therefore, regulating the ratio of LAP/LIP may be a novel strategy for targeting CSCs in cancer treatment.
The following are the supplementary data related to this article.
Metformin and AICAR suppress the expression of CD133. (A) HCC cell lines were cultured for 48 h, and expression levels of CD133 were confirmed by qRT-PCR analysis. (B) JHH6, JHH7, and Huh1 cells were treated with or without metformin (5 mM) for 48 h, and the expression levels of CD133 were examined by qRT-PCR analysis. *P < .05, vs. Cont. (C, D) Sorted HepG2 cells were treated with or without AICAR (1 mM) for 48 h, and the expression levels of CD133 were examined by FACS and qRT-PCR analysis. *P < .05, vs. Cont. Cont, control; Met, metformin.
The JAK/STAT3 pathway is partially involved in the metformin/AMPK-induced suppression of CD133. (A) Schematic of representative metformin/AMPK signaling. Metformin increases cellular AMP levels and activates AMPK and consequently suppresses JAK and mTOR activities. (B) HepG2 cells were treated with or without metformin (5 mM) for 48 h. Levels of phosphorylated and total STAT3 and p70S6K were examined by western blotting. (C and D) HepG2 cells were treated with or without rapamycin (10 nM) for 48 h, and expression levels of CD133 were examined by western blot and qRT-PCR analyses. (E and F) HepG2 cells were treated with or without C188-9 (20 μM) for 48 h, and expression levels of CD133 were examined by western blot and qRT-PCR analyses. ⁎P < .05, vs. DMSO. (G) Promoter activity of P1 in HepG2 cells. Cells were treated with or without C188-9 (20 μM) for 24 h following plasmid transfection, and luciferase activity was then measured. Met; metformin, Rapa; rapamycin, Cont; control, n.s.; not-significant, p-; phosphorylated.
(A, B and C) Sorted CD133H and CD133L cells were cultured for 48 h, and expression levels of CD133 were confirmed by FACS, western blotting, and qRT-PCR analyses. *P < .05, vs. CD133L.
Acknowledgments
Not applicable.
Footnotes
Funding: This work was supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS, 21592397), and a Grant-in-Aid for the JSPS Research Fellow (OM, 17J00434).
References
- 1.Cai X, Hu X, Cai B, Wang Q, Li Y, Tan X, Hu H, Chen X, Huang J, Cheng J. Metformin suppresses hepatocellular carcinoma cell growth through induction of cell cycle G1/G0 phase arrest and p21CIP and p27KIP expression and downregulation of cyclin D1 in vitro and in vivo. Oncol Rep. 2013;30:2449–2457. doi: 10.3892/or.2013.2718. [DOI] [PubMed] [Google Scholar]
- 2.Kunisada Y, Eikawa S, Tomonobu N, Domae S, Uehara T, Hori S, Furusawa Y, Hase K, Sasaki A, Udono H. Attenuation of CD4(+)CD25(+) regulatory T cells in the tumor microenvironment by metformin, a type 2 diabetes drug. EBioMedicine. 2017;25:154–164. doi: 10.1016/j.ebiom.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Yin S, Li J, Hu C, Chen X, Yao M, Yan M, Jiang G, Ge C, Xie H, Wan D. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120:1444–1450. doi: 10.1002/ijc.22476. [DOI] [PubMed] [Google Scholar]
- 6.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 7.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 8.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 9.Hermann P, Huber S, Herrler T, Aicher A, Ellwart J, Guba M, Bruns C, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Jang JW, Song Y, Kim SH, Kim J, Seo HR. Potential mechanisms of CD133 in cancer stem cells. Life Sci. 2017;184:25–29. doi: 10.1016/j.lfs.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Glumac PM, LeBeau AM. The role of CD133 in cancer: a concise review. Clin Transl Med. 2018;7:18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Li X, Zhang H, Lu Y. Molecular mechanisms of metformin for diabetes and cancer treatment. Front Physiol. 2018;9:1039. doi: 10.3389/fphys.2018.01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales DR, Morris AD. Metformin in cancer treatment and prevention. Annu Rev Med. 2015;66:17–29. doi: 10.1146/annurev-med-062613-093128. [DOI] [PubMed] [Google Scholar]
- 15.Saito T, Chiba T, Yuki K, Zen Y, Oshima M, Koide S, Motoyama T, Ogasawara S, Suzuki E, Ooka Y. Metformin, a diabetes drug, eliminates tumor-initiating hepatocellular carcinoma cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Yuan W, Tong D, Liu G, Lan W, Zhang D, Xiao H, Zhang Y, Huang Z, Yang J. Low concentrations of metformin sensitively inhibit CD133+ cell proliferation in pancreatic cancer and have anticancer action. PLoS One. 2016;8 doi: 10.1371/journal.pone.0063969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai X, Chu H, Yang X, Meng Y, Shi P, Gou S. Metformin increases sensitivity of pancreatic cancer cells to gemcitabine by reducing CD133+ cell populations and suppressing ERK/P70S6K signaling. Sci Rep. 2015;5 doi: 10.1038/srep14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 19.Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Tanikawa T, Kryczek I, Xia H, Li G, Wu K, Wei S, Zhao L, Vatan L, Wen B. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab. 2018;28:87–103 e6. doi: 10.1016/j.cmet.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salotti J, Sakchaisri K, Tourtellotte WG, Johnson PF. An Arf-Egr-C/EBPbeta pathway linked to ras-induced senescence and cancer. Mol Cell Biol. 2015;35:866–883. doi: 10.1128/MCB.01489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abreu MM, Sealy L. The C/EBPbeta isoform, liver-inhibitory protein (LIP), induces autophagy in breast cancer cell lines. Exp Cell Res. 2010;316:3227–3238. doi: 10.1016/j.yexcr.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohnishi S, Maehara O, Nakagawa K, Kameya A, Otaki K, Fujita H, Higashi R, Takagi K, Asaka M, Sakamoto N. Hypoxia-inducible factors activate CD133 promoter through ETS family transcription factors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natsuizaka M, Ohashi S, Wong GS, Ahmadi A, Kalman RA, Budo D, Klein-Szanto AJ, Herlyn M, Diehl JA, Nakagawa H. Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{beta}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells. Carcinogenesis. 2010;31:1344–1353. doi: 10.1093/carcin/bgq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maehara O, Sato F, Natsuizaka M, Asano A, Kubota Y, Itoh J, Tsunematsu S, Terashita K, Tsukuda Y, Nakai M. A pivotal role of Kruppel-like factor 5 in regulation of cancer stem-like cells in hepatocellular carcinoma. Cancer Biol Ther. 2015;16:1453–1461. doi: 10.1080/15384047.2015.1070992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natsuizaka M, Whelan KA, Kagawa S, Tanaka K, Giroux V, Chandramouleeswaran PM, Long A, Sahu V, Darling DS, Que J. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat Commun. 2017;8:1758. doi: 10.1038/s41467-017-01500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasagi Y, Chandramouleeswaran PM, Whelan KA, Tanaka K, Giroux V, Sharma M, Wang J, Benitez AJ, DeMarshall M, Tobias JW. The esophageal organoid system reveals functional interplay between notch and cytokines in reactive epithelial changes. Cell Mol Gastroenterol Hepatol. 2018;5:333–352. doi: 10.1016/j.jcmgh.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maehara O, Suda G, Natsuizaka M, Ohnishi S, Komatsu Y, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K. Fibroblast growth factor-2-mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis. 2017;38:1073–1083. doi: 10.1093/carcin/bgx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shmelkov SV, Jun L, St Clair R, McGarrigle D, Derderian CA, Usenko JK, Costa C, Zhang F, Guo X, Rafii S. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein AC133. Blood. 2004;103:2055–2061. doi: 10.1182/blood-2003-06-1881. [DOI] [PubMed] [Google Scholar]
- 30.El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 31.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- 32.Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- 33.Rutherford C, Speirs C, Williams JJ, Ewart MA, Mancini SJ, Hawley SA, Delles C, Viollet B, Costa-Pereira AP, Baillie GS. Phosphorylation of Janus kinase 1 (JAK1) by AMP-activated protein kinase (AMPK) links energy sensing to anti-inflammatory signaling. Sci Signal. 2016;9:ra109. doi: 10.1126/scisignal.aaf8566. [DOI] [PubMed] [Google Scholar]
- 34.Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Cheneby J, Kulkarni SR, Tan G. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Won C, Kim BH, Yi EH, Choi KJ, Kim EK, Jeong JM, Lee JH, Jang JJ, Yoon JH, Jeong WI. Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology. 2015;62:1160–1173. doi: 10.1002/hep.27968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Descombes P, Chojkier M, Lichtsteiner S, Falvey E, Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990;4:1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita T, Arai K, Sunagozaka H, Ueda T, Terashima T, Yamashita T, Mizukoshi E, Sakai A, Nakamoto Y, Honda M. Randomized, phase II study comparing interferon combined with hepatic arterial infusion of fluorouracil plus cisplatin and fluorouracil alone in patients with advanced hepatocellular carcinoma. Oncology. 2011;81:281–290. doi: 10.1159/000334439. [DOI] [PubMed] [Google Scholar]
- 39.Mak AB, Nixon AM, Kittanakom S, Stewart JM, Chen GI, Curak J, Gingras AC, Mazitschek R, Neel BG, Stagljar I. Regulation of CD133 by HDAC6 promotes beta-catenin signaling to suppress cancer cell differentiation. Cell Rep. 2012;2:951–963. doi: 10.1016/j.celrep.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng CC, Kuo KK, Su HT, Hsiao PJ, Chen YW, Wu DC, Hung WC, Cheng KH. Pancreatic tumor progression associated with CD133 overexpression: Involvement of increased TERT expression and epidermal growth factor receptor-dependent Akt activation. Pancreas. 2016;45:443–457. doi: 10.1097/MPA.0000000000000460. [DOI] [PubMed] [Google Scholar]
- 41.Song Y, Jang J, Shin TH, Bae SM, Kim JS, Kim KM, Myung SJ, Choi EK, Seo HR. Sulfasalazine attenuates evading anticancer response of CD133-positive hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2017;36:38. doi: 10.1186/s13046-017-0511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Li H, Ge C, Li M, Zhao FY, Hou HL, Zhu MX, Tian H, Zhang LX, Chen TY. Inhibitory effects of transcription factor Ikaros on the expression of liver cancer stem cell marker CD133 in hepatocellular carcinoma. Oncotarget. 2014;5:10621–10635. doi: 10.18632/oncotarget.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iida H, Suzuki M, Goitsuka R, Ueno H. Hypoxia induces CD133 expression in human lung cancer cells by up-regulation of OCT3/4 and SOX2. Int J Oncol. 2012;40:71–79. doi: 10.3892/ijo.2011.1207. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Guan H, Liu XD, Xie DF, Wang Y, Ma T, Huang B, Zhou PK. p53 positively regulates the expression of cancer stem cell marker CD133 in HCT116 colon cancer cells. Oncol Lett. 2018;16:431–438. doi: 10.3892/ol.2018.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park EK, Lee JC, Park JW, Bang SY, Yi SA, Kim BK, Park JH, Kwon SH, You JS, Nam SW. Transcriptional repression of cancer stem cell marker CD133 by tumor suppressor p53. Cell Death Dis. 2015;6:e1964. doi: 10.1038/cddis.2015.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imamura K, Ogura T, Kishimoto A, Kaminishi M, Esumi H. Cell cycle regulation via p53 phosphorylation by a 5'-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2001;287:562–567. doi: 10.1006/bbrc.2001.5627. [DOI] [PubMed] [Google Scholar]
- 47.Yang CS, Kim JJ, Lee HM, Jin HS, Lee SH, Park JH, Kim SJ, Kim JM, Han YM, Lee MS. The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy. 2014;10:785–802. doi: 10.4161/auto.28072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Kim JW, Gronborg M, Urlaub H, Lane MD, Tang QQ. Role of cdk2 in the sequential phosphorylation/activation of C/EBPbeta during adipocyte differentiation. Proc Natl Acad Sci U S A. 2007;104:11597–11602. doi: 10.1073/pnas.0703771104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buck M, Poli V, Hunter T, Chojkier M. C/EBPbeta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol Cell. 2001;8:807–816. doi: 10.1016/s1097-2765(01)00374-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metformin and AICAR suppress the expression of CD133. (A) HCC cell lines were cultured for 48 h, and expression levels of CD133 were confirmed by qRT-PCR analysis. (B) JHH6, JHH7, and Huh1 cells were treated with or without metformin (5 mM) for 48 h, and the expression levels of CD133 were examined by qRT-PCR analysis. *P < .05, vs. Cont. (C, D) Sorted HepG2 cells were treated with or without AICAR (1 mM) for 48 h, and the expression levels of CD133 were examined by FACS and qRT-PCR analysis. *P < .05, vs. Cont. Cont, control; Met, metformin.
The JAK/STAT3 pathway is partially involved in the metformin/AMPK-induced suppression of CD133. (A) Schematic of representative metformin/AMPK signaling. Metformin increases cellular AMP levels and activates AMPK and consequently suppresses JAK and mTOR activities. (B) HepG2 cells were treated with or without metformin (5 mM) for 48 h. Levels of phosphorylated and total STAT3 and p70S6K were examined by western blotting. (C and D) HepG2 cells were treated with or without rapamycin (10 nM) for 48 h, and expression levels of CD133 were examined by western blot and qRT-PCR analyses. (E and F) HepG2 cells were treated with or without C188-9 (20 μM) for 48 h, and expression levels of CD133 were examined by western blot and qRT-PCR analyses. ⁎P < .05, vs. DMSO. (G) Promoter activity of P1 in HepG2 cells. Cells were treated with or without C188-9 (20 μM) for 24 h following plasmid transfection, and luciferase activity was then measured. Met; metformin, Rapa; rapamycin, Cont; control, n.s.; not-significant, p-; phosphorylated.
(A, B and C) Sorted CD133H and CD133L cells were cultured for 48 h, and expression levels of CD133 were confirmed by FACS, western blotting, and qRT-PCR analyses. *P < .05, vs. CD133L.