Abstract
The unicellular green macroalga Acetabularia acetabulum L. Silva is an excellent system for studying regional differentiation within a single cell. In late adults, physiologically mediated extracellular alkalinity varies along the long axis of the alga with extracellular pH more alkaline along the apical and middle regions of the stalk than at and near the rhizoid. Respiration also varies with greater respiration at and near the rhizoid than along the stalk. We hypothesized that the apical and middle regions of the stalk require greater carbonic anhydrase (CA) activity to facilitate inorganic carbon uptake for photosynthesis. Treatment of algae with the CA inhibitors acetazolamide and ethoxyzolamide decreased photosynthetic oxygen evolution along the stalk but not at the rhizoid, indicating that CA facilitates inorganic carbon uptake in the apical portions of the alga. To examine the distribution of enzymatic activity within the alga, individuals were dissected into apical, middle, and basal tissue pools and assayed for both total and external CA activity. CA activity was greatest in the apical portions. We cloned two CA genes (AaCA1 and AaCA2). Northern analysis demonstrated that both genes are expressed throughout much of the life cycle of A. acetabulum. AaCA1 mRNA first appears in early adults. AaCA2 mRNA appears in juveniles. The AaCA1 and AaCA2 mRNAs are distributed asymmetrically in late adults with highest levels of each in the apical portion of the alga. mRNA localization and enzyme activity patterns correlate for AaCA1 and AaCA2, indicating that mRNA localization is one mechanism underlying regional differentiation in A. acetabulum.
Plant cells require differentiation and maintenance of subcellular regions to carry out many of their functions. Polar transport of molecules and tip growth are just two examples of how differentiation within a cell plays an important role in plant growth and development. Subcellular differentiation has been studied extensively in several animal systems, often through genetic, biochemical, and molecular characterization of large, single cells like frog oocytes and Drosophila blastoderms. Acetabularia acetabulum L. Silva is a unicellular plant system with potential to contribute to our understanding of subcellular differentiation in plants (Berger et al., 1987; Menzel, 1994; Mandoli, 1998a, 1998b).
A. acetabulum is a classic system for studies of morphogenesis, development, and interactions between the nucleus and cytoplasm (Hämmerling, 1934, 1936). It remains unicellular and uninucleate throughout the majority of its life cycle even while it undergoes elaborate morphological changes and passes through several phases of development. A. acetabulum grows siphonously with new whorls of hairs initiated at the stalk apex. For this unicell to differentiate poles and initiate whorls, it must create subcellular domains with specific functions. Recent advances have made A. acetabulum more amenable to molecular analysis of cellular processes (Hunt and Mandoli, 1992, 1996; Mandoli and Larsen, 1993; Mandoli, 1998b). We have chosen to examine the process of regional differentiation in this unicellular green alga.
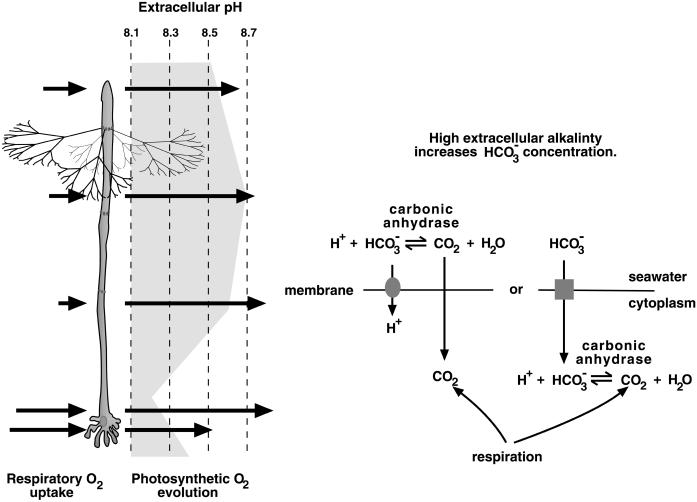
Several overlapping metabolic and physiological subcellular patterns exist in A. acetabulum (Serikawa et al., 2000). Three overlapping patterns involve extracellular pH along the cell, oxygen evolution through photosynthesis, and oxygen uptake through respiration. Along the apical two-thirds of the stalk, physiological processes lead to a higher extracellular alkalinity than at and near the rhizoid. Meanwhile, photosynthetic oxygen evolution occurs at a high level along all of the stalk and to a lesser extent at the rhizoid, and oxygen uptake for respiration occurs at a higher level near and at the rhizoid than elsewhere in the alga (Fig. 1). Wishing to understand how these patterns are created and maintained in this unicell, we began by examining some of the implications these patterns have for photosynthetic inorganic carbon uptake.
Figure 1.
CA and inorganic carbon uptake. The shading next to the diagram of a late adult A. acetabulum (left) indicates extracellular alkalinity along the stalk (measurements made in artificial seawater at pH 8.1). The alga is drawn with only one whorl of hairs for clarity. The arrows on the left of the alga indicate oxygen uptake in the dark due to respiration and the arrows on the right indicate the pattern of photosynthetic oxygen evolution along the stalk (Serikawa et al., 2000). These measurements of extracellular pH and oxygen flux were made using self-referencing ion-specific micro-electrodes and represent the mean values measured from 13 individuals. The lengths of the arrows are proportional to the amount of oxygen flux. The external pH along most of the plant is alkaline relative to the surrounding medium, whereas the region at and near the rhizoid shows external acidity in nearly half of the individuals measured. The diagram (right) illustrates two potential roles for CA in inorganic carbon uptake when the extracellular medium is alkaline. In alkaline media, the equilibrium among forms of inorganic carbon shifts toward bicarbonate, leaving only a small pool of carbon dioxide available for diffusion across the plasma membrane into the cytoplasm (Badger and Price, 1994; Raven, 1995). Without CA activity, interconversion between bicarbonate and carbon dioxide occurs slowly and the carbon dioxide pool is quickly depleted. With extracellular CA activity, the interconversion is faster so that the pool of external carbon dioxide available for photosynthesis is continually replenished. If an alga actively takes up bicarbonate, internal CA activity can then convert that bicarbonate into carbon dioxide for use in photosynthesis. In addition, respiration can also provide inorganic carbon for photosynthesis.
In the ocean, which is alkaline, much of the dissolved inorganic carbon is in the form of bicarbonate. Around the apical portion of the stalk, the pH outside of the cell wall can be even more alkaline. As a consequence, more of the available inorganic carbon near the apical portion of the alga than near the rhizoid is in the form of bicarbonate, which cannot freely diffuse across the cell membrane. In addition, the greater rate of respiration at and near the base of the stalk provides more internal carbon dioxide near the base than at the apex. These two physiological differences along the stalk indicate that inorganic carbon may be less available for the apical versus the basal portions of the alga. Because photosynthesis occurs at high levels all along the stalk (Serikawa et al., 2000), it seems likely that some mechanism exists to facilitate inorganic carbon uptake in the apical portions of the stalk.
Carbonic anhydrase (CA) catalyzes the interconversion of CO2 and HCO3−, and many algae use CA to facilitate inorganic carbon (Badger and Price, 1994; Raven, 1995; Mercado et al., 1998). In some algae, CA activity outside the cell increases the rate of HCO3− to CO2 conversion as CO2 diffuses into the organism, thus increasing the availability of CO2. Other algae actively transport HCO3− into the cell, where intracellular CA converts that HCO3− into CO2 (Fig. 1). Virtually nothing is known about which forms of inorganic carbon are taken up by A. acetabulum. It seems possible that more CA activity might be needed in the apical and middle portions of the stalk. If so, CA might be a useful molecular marker for subcellular localization and regional differentiation in A. acetabulum. We decided to test the role of CA in photosynthesis, examine if enzyme activity is localized to the apical portion of the stalk, and, if so, clone CA as a marker for subcellular differentiation.
RESULTS
CA Activity and Photosynthetic Oxygen Evolution
We hypothesized that the asymmetric distribution of extracellular alkalinity and respiration along the algal body (Fig. 1) might necessitate increased CA activity in the apical portion to facilitate inorganic carbon uptake for photosynthesis. To test whether CA facilitates photosynthesis we examined the effects of the CA inhibitors acetazolamide (AZ) and ethoxyzolamide (EZ) on photosynthetic oxygen evolution. These inhibitors have been used to differentiate between the contribution of extracellular and intracellular CAs to photosynthesis with AZ presumably unable to pass cell membranes, whereas EZ can enter cells and inhibit both internal and external CAs (Mercado et al., 1997, 1998). A caveat, however, is that at least in Chlamydomonas it appears that AZ can enter the cell (Williams and Turpin, 1987), indicating that use of these inhibitors does not provide definitive proof of the site of CA action. We performed this experiment using artificial seawater made at either pH 7.6 or 8.3 to further test the necessity of CA in photosynthesis. If CA facilitates inorganic carbon uptake in A. acetabulum through conversion of HCO3− to CO2 under conditions of high extracellular alkalinity, lowering the pH of the medium should reduce or abolish any effects of CA inhibition.
Both inhibitors decreased photosynthetic oxygen evolution by populations of whole algae growing in pH 8.3 but not in pH 7.6 when assayed with a Clark-type polarigraphic oxygen electrode (data not shown). We next used the self-referencing oxygen microelectrode technique to examine where and to what degree oxygen evolution was affected by the inhibitors (Porterfield and Smith, 2001). This technique allows real-time measurements of oxygen flux at the surface of individual, intact cells (Porterfield and Smith, 2001; Serikawa et al., 2000). Algae were measured at the apex, one-third from the apex, one-third from the rhizoid, and at the rhizoid (Fig. 2). Our hypothesis about the role of CA activity in photosynthetic inorganic carbon uptake predicts that addition of the inhibitors would affect photosynthetic oxygen efflux more in the apical than the basal regions of the alga.
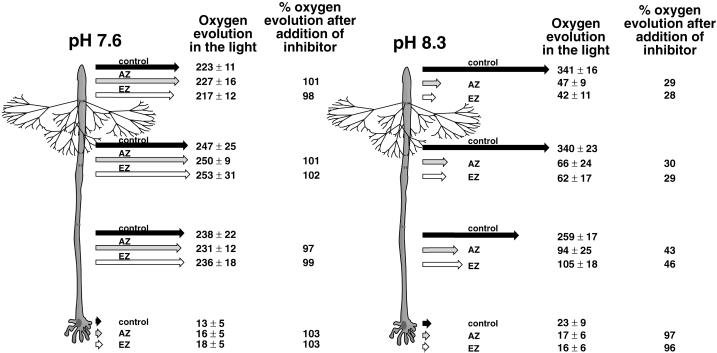
Figure 2.
Effects of AZ and EZ on photosynthetic oxygen evolution in media with different pHs. Oxygen evolution along the stalk of late adult algae growing at pH 7.5 or 8.3 was measured using self-referencing oxygen microelectrodes in the light. Measurements were made at the apex, one-third from the apex, one-third from the rhizoid, and at the rhizoid before and after addition of either AZ or EZ. Ten algae were measured for each combination of pH and inhibitor. Black arrows represent oxygen flux before inhibitor addition. Gray arrows represent flux after AZ addition. White arrows represent flux after EZ addition. Next to each arrow is the flux value ±sd. Next to the flux values for AZ and EZ are the percentages for oxygen evolution seen after inhibitor addition relative to before. For these calculations, gross photosynthetic oxygen evolution was taken as the sum of oxygen efflux in the light plus the magnitude of oxygen influx in the dark (dark values taken from Serikawa et al., 2000). This method, although not highly rigorous, does provide an internally consistent relative baseline.
Inhibitor addition decreased the rate of oxygen evolution at pH 8.3 but did not affect oxygen evolution at pH 7.6 (Fig. 2). The lack of inhibition at pH 7.6 is consistent with CA acting to convert HCO3− to CO2 for use in photosynthesis. The magnitude of the decrease in oxygen evolution in the light at pH 8.3 was roughly equivalent for AZ and EZ, consistent with the main contribution of CAs to photosynthesis being outside the plasma membrane (in the periplasmic space and/or cell wall). When oxygen evolution along the stalk after inhibitor addition is calculated as a percentage of oxygen evolution before inhibitor addition, apical regions of the alga (Fig. 2, positions 1 and 2) appear more affected than basal regions (Fig. 2, positions 3 and 4). The rhizoid showed no apparent decrease in photosynthetic oxygen evolution after inhibitor addition, indicating that this region did not require CA activity for photosynthesis. The magnitude of oxygen evolution in the apical portions of the stalk was approximately 50% higher at pH 8.3 than at pH 7.6 (Fig. 2), indicating that under alkaline conditions photosynthesis might actually be more efficient than at lower pHs, even though proportionally more inorganic carbon was theoretically present in the form of HCO3−.
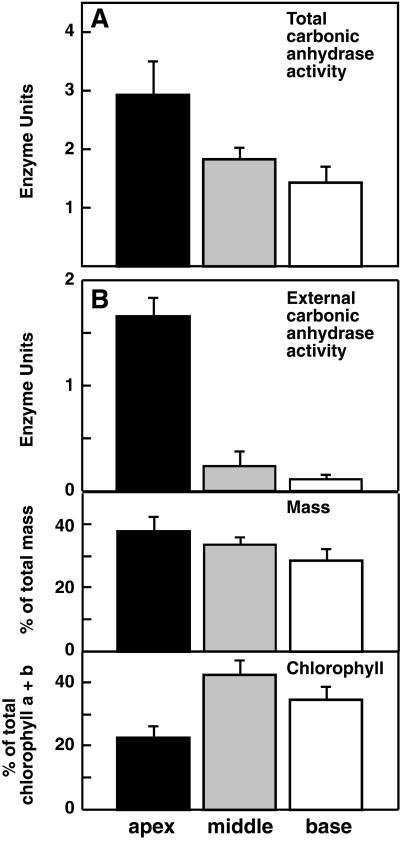
Having established that CAs do play a role in photosynthesis, we examined the distribution of CA activity along the stalk. Populations of algae were dissected into apical, middle, or basal portions. Each subpopulation was homogenized and assayed for CA activity. Enzyme activity was distributed in an apical to basal gradient with the highest levels of activity in the apical portions (Fig. 3A). It should be noted that our grinding buffer contained the sulfhydryl reagent β-mercaptoethanol, which might affect the activity of the CA enzyme through the disruption of disulfide bonds. Our activity values, therefore, are primarily qualitative and relative measures of enzyme activity and may not accurately reflect full CA activity.
Figure 3.
Spatial distribution of CA activity along the stalk of late adults. A, Enzyme assays of CA were performed using tissue extracts from pools of apical, middle, or basal portions of dissected late adults. The means of four trials are graphed with se bars. B, Enzyme assays of intact pools of apical, middle, or basal enucleate fragments were also performed. For these samples the fresh weight (middle) and chlorophyll A + B content (bottom) were also measured to provide a control for equal dissections. The means of three trials are graphed with se bars.
Given that our experiments with AZ and EZ were consistent with the primary role for CA in the extracellular domain, we also assayed for external CA activity alone. We dissected plants into three enucleate portions (apical, middle, and basal) and, after an interval to allow wound healing, assayed populations of these intact, living cell fragments for extracellular CA activity (Fig. 3B). These populations were then weighed and ground for measurements of chlorophyll A and B content, thus allowing activity to be compared with both mass and chlorophyll content. For both of these measures it appeared that our tissue pools contained approximately equal amounts of tissue (Fig. 3B, bottom panels) with the differences seen being too small to account for the dramatic differences in external CA activity in the different tissue pools.
Overall, the distribution of both total and external CA activity agrees with our expectations based on our hypothesis. An interesting finding, given our inhibitor work suggesting that CA acts extracellularly to facilitate photosynthesis (Fig. 2), is that protein extracts contain more CA activity than intact fragments (compare Fig. 3, A and B, top panel). This indicates the existence of cytoplasmic CAs with possible roles outside of photosynthesis.
Cloning of CA Genes
To examine the molecular and cellular mechanisms underlying the asymmetric distribution of CA activity, we used degenerate PCR to clone CA genes from A. acetabulum. Our screens resulted in the isolation of two different CA genes, AaCA1 and AaCA2 (Figs. 4 and 5). Both fall into the α family of CAs based on sequence homology (Hewett-Emmett and Tashian, 1996). In addition, the genomic region surrounding AaCA1 was isolated and sequenced.
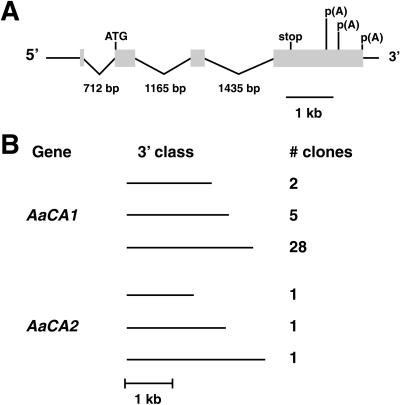
Figure 4.
Genomic organization of AaCA1 and cDNA classes identified for AaCA1 and AaCA2. A, AaCA1 consists of four exons (shaded boxes). Start of translation is indicated with an ATG and the end of translation is labeled “stop.” The positions of the three polyadenylation sites are shown along the 3′ end of the fourth exon. B, Both AaCA1 and AaCA2 can be polyadenylated at alternative sites. Different cDNA size classes (short, medium, and long) and number of clones identified for each class are given.
Figure 5.
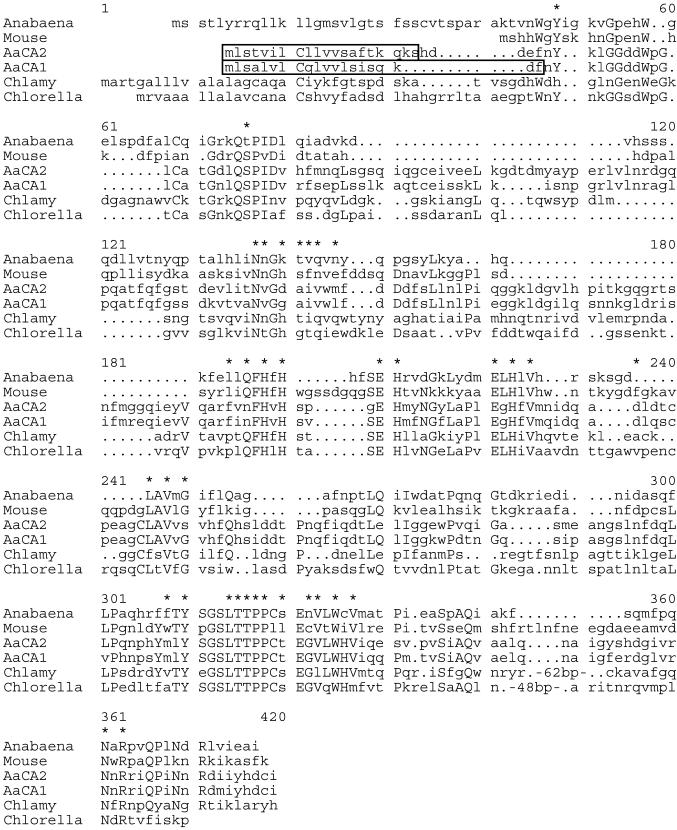
Comparison of AaCA1 and AaCA2 with other α-CAs. Residues located within the active site of CAs (Eriksson and Liljas, 1992; Hewett-Emmett and Tashian, 1996) are indicated with asterisks. Residues conserved among at least four of the six sequences are capitalized. Sequences used are from Anabaena (Soltes et al., 1997), mouse (Curtis et al., 1983), Chlamydomonas (abbreviated as Chlamy in the figure; Fukuzawa et al., 1990b), and Chlorella (Satoh et al., 1998). Sequences were aligned using the pileup program of the GCG suite of sequence analysis programs. The large proportion of hydrophobic amino acids within the first 20 amino acids of AaCA1 and AaCA2 (boxed regions) suggests the presence of an endoplasmic reticulum signal sequence (von Heijne, 1983; Fukuzawa et al., 1990b). Periods represent gaps introduced within sequences to maximize alignments.
AaCA1 has four exons (Fig. 4A) with the 3′-untranslated region (UTR) transcribed as a single long unit without any introns. The 3′ UTRs for some A. acetabulum genes are unusually long (> 1 kb, for AaCA1 and 2; Serikawa and Mandoli, 1999).
Sequencing indicated that AaCA1 and AaCA2 both use at least three different polyadenylation sites (Fig. 4B). However, for AaCA1 these sites were not equally used. PCR screening of 35 plaques from our initial library screen revealed that, for AaCA1, the site leading to the longest 3′ UTR was used 80% of the time, whereas the site leading to a 3′ UTR of medium length was used 14% of the time, and the site leading to the shortest UTR was only used for 6% of clones screened. Because we isolated only three cDNAs for AaCA2, it is not known whether a similar bias exists for polyadenylation of AaCA2. However, northern analyses with AaCA1 or AaCA2 fails to detect the medium or short forms (data not shown).
Regions of homology were evident both within and between the 3′ UTRs of AaCA1 and AaCA2 (data not shown). The AaCA1 3′ UTR contains a repeated element, separated by 25 bases, which shows conservation of 90 of 109 bases. The AaCA2 3′ UTR contains a repeated element showing conservation of 38 of 51 bp with the repeats spaced 366 bp apart. In addition, the 3′ UTRs of AaCA1 and AaCA2 share a region of homology showing conservation of 84 of 100 bases. Both 3′ UTRs also contain several copies of the six-nucleotide sequence AGCATY (five copies in AaCA1; eight copies in AaCA2). The functional significance of these conserved and repeated regions is unknown, but they may play a role in the post-transcriptional regulation of these two genes (Lipshitz and Smibert, 2000).
Comparison of AaCA1 and AaCA2 with CAs from other species reveals considerable sequence divergence (Fig. 5). Several key regions and residues are conserved, however, especially residues necessary to maintain metal binding activity and enzymatic function. AaCA1 and AaCA2 are much more similar to each other than to CAs from other species, including those from other green algae (75% identity between AaCA1 and AaCA2 as compared, for example, with only 29% identity between AaCA1 and CAH1 from Chlamydomonas). This indicates that duplication of CA genes may have occurred recently in A. acetabulum.
Given that CA is both intra- and extracellular (Figs. 2 and 3), we examined the N-terminal regions of AaCA1 and AaCA2 for evidence of an endoplasmic reticulum signal sequence. Targeting to the endoplasmic reticulum would be the first step in secretion of CA into the periplasmic space. The first several amino acids of both predicted proteins have characteristics of signal sequences (von Heijne, 1983; Fukuzawa et al., 1990a), most notably a high proportion of hydrophobic amino acids such as Leu and Val within the first 20 amino acids (nine valines, iso-Leus, or leucines in AaCA1 and eight in AaCA2) (Fig. 5, boxed region). This hydrophobic region in the algal proteins precedes the start of significant homology, which is also consistent with this region being a signal sequence that is cleaved from the mature protein. In contrast, the mouse protein, which is not secreted, lacks a putative signal sequence.
The C-terminal regions of AaCA1 and AaCA2 do not contain the same long extension seen in the Chlamydomonas and Chlorella proteins (Fig. 5). This particular CA in Chlamydomonas is cleaved within that extension, yielding two polypeptides that combine to form a heterotetrameric complex (Fukuzawa et al., 1990a; Kamo et al., 1990). The absence of this region in AaCA1 and AaCA2 suggested that these two CAs are not cleaved post-translationally in the same way as the Chlamydomonas protein.
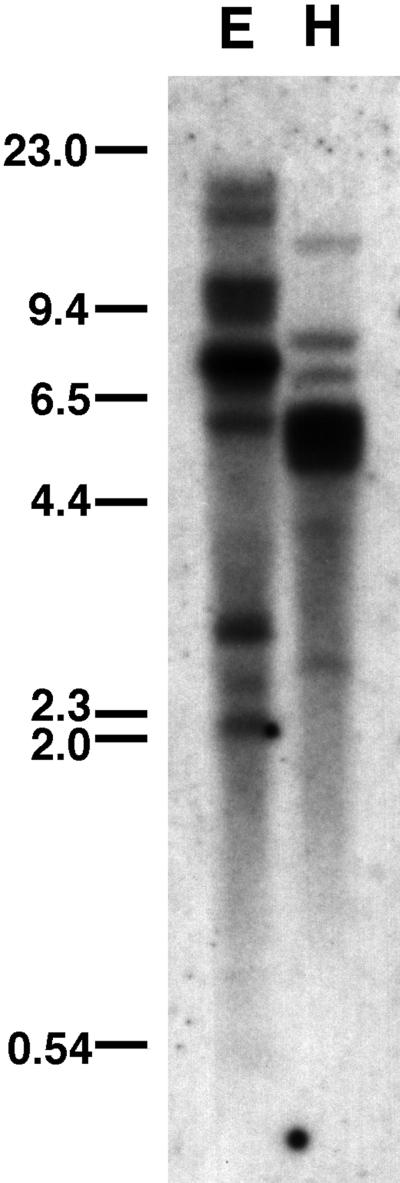
A low stringency Southern analysis of AaCA1 indicated that the α CAs are encoded by a multigene family in A. acetabulum (Fig. 6) as they are in Chlamydomonas (Fukuzawa et al., 1990a; Funke et al., 1997; Karlsson et al., 1998).
Figure 6.
Southern analysis of AaCA1. DNA (10 μg) from strain Aa0006 was digested with either EcoRI (E) or HindIII (H) and probed at low stringency with the longest AaCA1 cDNA. Sizes are in kilobase pairs.
Expression Patterns of AaCA1 and AaCA2
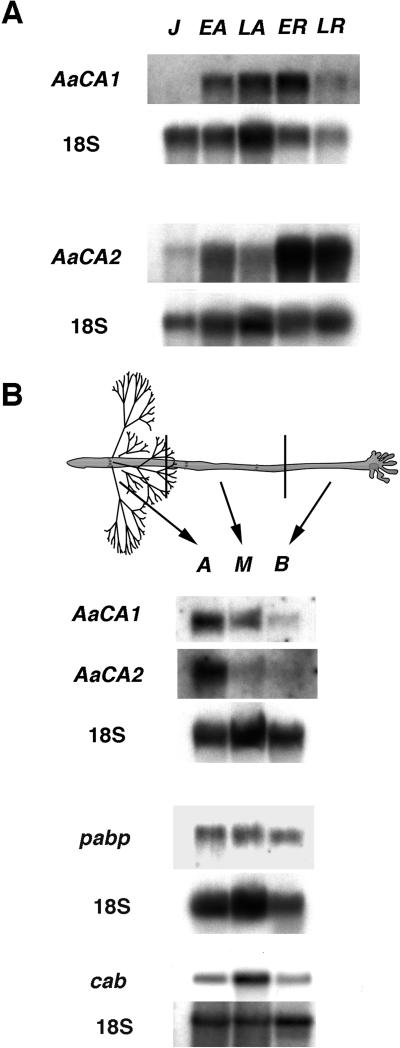
AaCA1 and AaCA2 mRNAs had distinct expression profiles during several phases of development (Fig. 7A). AaCA1 mRNA first appeared in vegetative early adults and was most abundant early in reproductive development. AaCA2 mRNA first appeared in juvenile algae, declined slightly in late adults, and then reached much higher levels during reproduction. The shorter polyadenylated forms were not detected even in longer exposures of our blots (data not shown). This corroborates our PCR survey of 3′-UTR lengths for AaCA1 cDNAs, which indicated that the shorter forms were much lower in abundance than the longest form.
Figure 7.
Northern analysis of AaCA1 and AaCA2. A, Total RNA isolated from several developmental phases was probed with either AaCA1 or AaCA2. J, Juvenile; EA, early adult; LA, late adult; ER, early reproductive; LR, late reproductive. In this nomenclature, adults are vegetative (Mandoli, 1998a). 18S RNA was probed as a loading control. B, Total RNA was isolated from pools of tissue derived from dissected late adults. Probing with the CA cDNAs revealed that AaCA1 and AaCA2 mRNA are present in gradients with the highest concentration of mRNA in the apical portion of the body plan. Probing with two other A. acetabulum genes, pabp, a putative poly(A) binding protein clone, or cab, a chlorophyll a/b binding clone (GenBank accession no. AF093617) revealed that these mRNAs are more evenly distributed. The gels probed with AaCA1, AaCA2; pabp; and cab were loaded with 5, 10, and 1 μg of total RNA per lane, respectively. 18S RNA was probed as a loading control. A, Apical portion; M, middle portion; B, basal portion.
To examine whether the spatial distribution patterns of AaCA1 and AaCA2 mRNAs correlated with measured enzyme activity (Fig. 3), we dissected vegetative late adults into apical, middle, and basal portions and isolated total RNA from each tissue pool. Probing these RNA samples with either AaCA1 or AaCA2 revealed that both mRNAs are asymmetrically localized within the body plan (Fig. 7B). Both mRNAs occurred in a gradient with the highest amounts of mRNA located in the apical portion of the plant, the region with the highest level of CA activity (Fig. 3). In contrast, the mRNAs for both a putative poly(A) binding protein and a chlorophyll a/b binding protein show a lesser degree of localization (Fig. 7B).
DISCUSSION
CA Activity and Inorganic Carbon Use
CA activity displayed a nonuniform distribution along the body plan of A. acetabulum and the mRNA localization of two CA genes paralleled the distribution of enzyme activity. These data are consistent with inorganic carbon uptake being facilitated by CA in the apical portion of the alga and support the hypothesis that A. acetabulum creates and maintains subcellular regions through mRNA localization.
Obtaining inorganic carbon for photosynthesis is a fundamental problem for marine plants. The ocean is generally alkaline, which means that most of the available carbon is in the form of bicarbonate and carbonate (Korb et al., 1997; Nimer et al., 1997; Mercado et al., 1998). These negatively charged molecules do not freely diffuse across the plasma membrane, necessitating cellular mechanisms that either actively transport these molecules into the alga or increase the abundance of carbon dioxide outside the cell wall. An additional complexity is that respiration also provides carbon dioxide for use in photosynthesis. In A. acetabulum, respiration is asymmetrically distributed, which results in the rhizoid and base of the stalk having more carbon dioxide available internally than does the apical portion of the stalk.
We suggest that A. acetabulum uses more than one mechanism to facilitate carbon uptake: reduction of alkalinity of the medium near the base and increased external CA activity in the middle and apical portions of the stalk. In this model, proton efflux at the base of the stalk decreases the local pH of the surrounding seawater and thus increases the proportion of carbon dioxide among inorganic carbon species. Combined with respiration, this provides the inorganic carbon pool for photosynthesis. In contrast, in the apical portion of the alga, the combination of a higher external pH (the result of proton influx, [Serikawa et al., 2000]) and less respiration (Fig. 1) necessitates additional strategies to acquire inorganic carbon. We believe that the effects of external alkalinity are ameliorated by the activity of external CAs that hasten the interconversion of bicarbonate and carbon dioxide. Thus, as carbon dioxide is taken up by the alga, CA activity re-establishes the equilibrium between carbon dioxide and bicarbonate more rapidly than would occur otherwise, thereby increasing the effective availability of carbon dioxide.
Our results are consistent with this model for inorganic carbon uptake in A. acetabulum. First, inhibition of CA with either AZ or EZ decreased but did not abolish oxygen evolution (Fig. 2), indicating that CA enhances photosynthetic oxygen evolution but is not absolutely required. This inhibition shows a polar distribution with more inhibition occurring in the apical portion of the alga than at the base, indicating that CA facilitates photosynthesis primarily in the apical portion of the alga. Algal species rely to different degrees upon CA activity. Some algae are completely or partially inhibited in photosynthetic oxygen evolution without CA activity whereas others show little or no effect when CA is inhibited (Mercado et al., 1997, 1998; Nimer et al., 1997; Sukenik et al., 1997). A. acetabulum falls in the middle of that range with AZ and EZ inhibiting but not preventing photosynthetic oxygen evolution.
AZ and EZ had approximately equal effects on oxygen evolution, consistent with the role of CA in photosynthesis being located outside of the plasma membrane, although this is not definitive proof given the possibility that AZ may cross membranes (Williams and Turpin, 1987).
The lack of inhibition by AZ and EZ under conditions of decreased extracellular alkalinity (Fig. 2, pH 7.6 versus pH 8.3) is also consistent with a role for CA in facilitating inorganic carbon uptake through inter-conversion of bicarbonate and carbon dioxide. When the external pH is lowered, which leads to a higher concentration of carbon dioxide, AZ and EZ have no inhibitory effect on photosynthetic oxygen evolution.
The distributions for total and extracellular CA activity support our model of two, spatially separated mechanisms for inorganic carbon uptake and correlate with the results of our inhibitor studies. As hypothesized based on the distribution of extracellular alkalinity along the stalk (Fig. 1; Serikawa et al., 2000), we measured more CA activity in the apical portions of the alga (Fig. 3). The amount of CA activity measured in cellular extracts shows a less dramatic localization to the apical region and is greater overall than that measured externally (compare Fig. 3, A and B). In addition to a possible role in photosynthesis, either intra- or extracellular, this overall pool may have other physiological functions (Raven, 1995). As more CAs of the α and other classes (β and γ) are cloned and studied, these roles should become clearer.
Our studies do not preclude facilitated uptake of bicarbonate as a third mechanism for inorganic carbon uptake. In this process, bicarbonate is actively taken into the cytoplasm and later converted to carbon dioxide for use by Rubisco (Lucas, 1983; Badger and Price, 1994). Our experiments did not address the significance of this third mechanism for inorganic carbon uptake for photosynthesis. In this mechanism, internal CA might facilitate the dehydration of bicarbonate to carbon dioxide.
Whereas our experiments indicate that external CA activity is important for photosynthesis, several questions remain. Our experiments were performed under low CO2 conditions (ambient CO2). In Chlamydomonas, the levels of CA activity are reduced under high CO2 conditions as compared with ambient CO2 (Sültemeyer, 1998). A similar phenomenon may occur in A. acetabulum. How closely mRNA levels parallel protein distribution and how CAs reach the extracellular environment also remain unknown.
mRNA Localization as a Mechanism for Regional Differentiation in A. acetabulum
Morphogenetic gradients have been hypothesized in A. acetabulum ever since Hämmerling first described the different developmental potentials of dissected portions of this alga (Hämmerling, 1932, 1936). His experiments showed that when an alga is dissected into three enucleate portions (apical, middle, and basal), the apical portion has the most developmental potential (it can form new whorls and a cap), the middle less (it can make few whorls), and the base the least (sometimes new whorls). Over time the hypothesis has developed that these morphogenetic gradients arise from localized mRNA populations (Mandoli, 1998a).
The correlation between localization patterns of AaCA1 and AaCA2 mRNAs and the distribution of CA activity along the stalk indicates that A. acetabulum uses mRNA localization to establish a gradient of enzyme activity. mRNA localization is an important developmental mechanism in a variety of organisms (Mowry and Melton, 1992; Glotzer and Ephrussi, 1996; Hesketh, 1996; Bashirullah et al., 1998) but thus far has not been extensively studied in plant systems (Bouget et al., 1996; Serikawa and Mandoli, 1999). mRNA gradients can lead to either a gradient of activity or can result in the formation of sharp boundaries within a cell, delineating subcellular regions and domains (Bashirullah et al., 1998).
The possibility that mRNA localization directly controls the distribution of CA activity in A. acetabulum must be viewed with caution because of limitations of the enzyme assay. Our Southern analyses indicate that additional CAs exist in A. acetabulum. We therefore cannot make a direct connection between AaCA1 and AaCA2 mRNA distribution and overall enzyme activity because the enzyme assay does not differentiate between different CA isoforms. Nevertheless, the parallels between mRNA localization and enzyme activity are intriguing because, as reported here and elsewhere (Serikawa and Mandoli, 1999), mRNA localization is not a general pattern displayed by all genes in late adults of this species.
Results reported here and elsewhere (Vogel, 1998; Serikawa and Mandoli, 1999) indicate that gradients of specific mRNAs exist in A. acetabulum in both the apical and basal directions. With AaCA1 and AaCA2 we now have tools to dissect how gradients are formed and what relationship gradients have to the physiology and phenotype of the organism. mRNA localization in A. acetabulum is particularly interesting because this species uses post-transcriptional mechanisms to control several aspects of its development (Shoeman et al., 1983; Li-Weber and Schweiger, 1985). For example, if the nucleus-containing rhizoid is removed from a vegetative alga, the remaining, enucleate portion can continue development for days to weeks and, depending upon the age of the alga upon enucleation, can even undergo the transition from vegetative to reproductive phase (Hämmerling, 1934). Reliance on post-transcriptional mechanisms makes sense given that the apex, where morphogenesis occurs, can be 2 to 3 cm away from the nucleus. Using post-transcriptional mechanisms such as mRNA localization to regulate development may allow quicker responses to both external and internal cues.
MATERIALS AND METHODS
Strains and Culture Conditions
All experiments used heterogeneous wild-type strains of Acetabularia acetabulum L. Silva, either Aa0006 (Ladenburg no. 5) or Aa0005 (Ladenburg no. 17). The growth conditions have been detailed elsewhere (Serikawa and Mandoli, 1999; Serikawa et al., 2000).
O2 Evolution and Inhibitors of CA
Measuring oxygen evolution using the self-referencing micro-electrode approach has been detailed elsewhere (Porterfield and Smith, 2001). This work was done using a modified version of a commercially available DC-coupled amplifier (model IPA-2, Applicable Electronics, Forestdale, MA) using ASET software (Applicable Electronics, East Falmouth, MA). For these experiments, late adult algae were grown for a week in artificial seawater adjusted to pH 7.6 or 8.3. For measurements of individual algae, one alga was placed in a small plastic dish in the appropriate pH and, to provide a control level of oxygen flux in the light, measured at four positions along the long axis: at the apex; one-third of stalk length from the apex; one-third stalk length from the rhizoid; and at the rhizoid. Either AZ or EZ (Sigma, St. Louis) was then added to a final concentration of 100 μm. After a few minutes to allow for equilibration, oxygen evolution was again measured at the four locations. Ten individual algae were measured for each pH/inhibitor concentration, resulting in four independent data sets.
Enzyme Activity Assays
Assays of CA activity were performed using a variation of the method of Wilbur and Anderson (Wilbur and Anderson, 1948). Either protein extracts or intact cell fragments were assayed. To make protein extracts, pools of apical, middle, and basal tissue were gathered and frozen. Each pool consisted of 30 apices, middles, or bases and was kept on dry ice until homogenization. Each pool was homogenized on ice in 0.5 mL of grinding buffer (10 mm Tris-HCl, pH 8.3, 2 mm EDTA, and 10 mm 2-mercaptoethanol) using a Tissue Tearor (Whatman, Clifton, NJ) for approximately 10 s. Assays were prepared by adding 0.4 mL of tissue homogenate to 1.8 mL of 25 mm Veronal buffer, pH 8.3. Once the pH stabilized, 0.6 mL of ice-cold, CO2-saturated water was added to the veronal buffer/tissue homogenate mix and the time for the pH to drop 1 unit was measured. As a baseline, we measured the time for the pH to drop 1 unit when CO2 saturated water was added to veronal buffer/grinding buffer without tissue homogenates. Enzyme units were defined as (Tc/Ta) − 1, where Tc is the time without tissue homogenates and Ta is the time with tissue homogenates. Assays were carried out at 1°C to 2°C, and experiments were done four times.
Measurements of external CA activity were performed in a similar fashion except that the fragments were not ground prior to the enzyme assays. Live late adult individual algae were dissected into three approximately equal, enucleate portions through pressure wounding and cutting with a scalpel (Kratz et al., 1998). These fragments were kept in the dark at 20°C to 22°C for 2 d to permit healing. They were then exposed to normal light conditions for 24 h and assayed in groups of 40 intact fragments as described above except that the Veronal buffer was also made up to 500 mm NaCl. After measuring the external enzyme activities, the fragment pools were dried by pipetting off excess buffer and weighed. Next, each tissue pool was homogenized in 80% (v/v) acetone, and absorbance was measured at 663 and 646 nm to estimate chlorophyll content (Lichtenthaler, 1987). Three different groups of fragments were assayed, and the results were averaged.
Cloning of CA Genes
The BLOCKS WWW server (http://blocks.fhcrc.org/blocks/) at the Fred Hutchinson Cancer Research Center (Seattle) was used to create sequence alignments among α CAs from several organisms. Alignment blocks were run through the CODEHOPS algorithm (http://blocks.fhcrc.org/blocks/codehop.html) to design degenerate primers for PCR (Rose et al., 1998). The two primers designed were CAa1 (forward primer sequence 5′-GTG CCA CAG GCA ACA AGC ARW SNC CNA T-3′) and CAa2 (reverse primer sequence 5′-CAC TGT ACT CCT TCT GAA CAG GGN GGN GTN GT-3′). CAa1 corresponds to the amino acid sequence CATGNKQSPI and CAa2 corresponds to the sequence TTPPCSEGVKW. Amplification using these primers is expected to yield PCR fragments of 600 to 700 bp. To make template for PCR, 1 μg of poly(A) mRNA isolated from vegetative late adult plants was reverse transcribed using random hexamers and avian myeloblastoma virus reverse transcriptase (Life Technologies/Gibco-BRL, Cleveland) according to the manufacturers instructions. The final concentrations in the 50-μL PCR reactions were 1× PCR buffer (Life Technologies/Gibco-BRL), 2.5 mm MgCl2, 200 nm nucleotides, and 1 mm of each primer. One-hundredth of the reverse transcription reaction was added as template. The cycling program was a variant of touchdown PCR (Don et al., 1991). Samples were heated to 94°C, and 0.5 μL of Taq DNA polymerase (2.5 units, Life Technologies/Gibco-BRL) was added. The first five cycles consisted of decreasing annealing temperatures (63°C, 62°C, 61°C, 60°C, and 59°C), and the next 30 cycles were at 58°C. Times for all cycles were 1-min melting at 94°C, 30-s annealing, and 1-min extension at 72°C. At the end of these cycles samples were treated with an additional 5-min extension time at 72°C.
PCR products were purified on agarose gels, cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and then used as probes to screen approximately 3 × 105 plaques from a ZAP Express cDNA library (Stratagene, La Jolla, CA). The library had been constructed from mRNA isolated from algae of mixed ages. Prehybridization (2 h) and hybridization (12–14 h) were carried out in 50% (v/v) formamide, 0.25 m NaCl, 7% (w/v) SDS, and 0.2 m Na2PO4 at 37°C. Filters were washed twice for 30 min at 37°C in 0.2× SSC, 0.1% (w/v) SDS. Whereas over 50 clones of varying intensity were seen in the initial screen, only 10 were selected at random for additional purification. Of these, seven isolates were of AaCA1 and three were of AaCA2.
Approximately 3 × 105 plaques from a Lambda DASH II library (Stratagene) constructed from genomic DNA from Aa0006 were screened at 42°C with the longest AaCA1 cDNA. Six positive clones were isolated, subcloned into Bluescript SK− (Stratagene), ordered through Southern analysis with various regions of the AaCA1 cDNA, and sequenced (GenBank accession nos. are as follows: AaCA1 mRNA, AF170174; AaCA1 genomic clone, AF170175; and AaCA2 mRNA, AF170173).
Southern Blots
DNA was isolated from gametangia of A. acetabulum with DNAzol extra strength (Molecular Research Center, Cincinnati) according to the manufacturer's instructions. Genomic DNA (10 μg) was digested in large volumes (200 μL) overnight, ethanol precipitated, fractionated on 0.8% (w/v) agarose gels and transferred to nylon membranes (Hybond N+, Amersham, Buckinghamshire, UK). Membranes were hybridized under the same conditions as library screens except that high stringency blots were hybridized and washed at 42°C.
Northern Blots
For developmental phase blots, RNA was isolated from whole algae at several stages of development using the method of Chang et al. (1993). For subcellular localization blots, RNA was isolated from pooled portions of late adults and early reproductive algae using TRIzol Reagent (Life Technologies/Gibco-BRL) according to the manufacturer's instructions. To gather tissue from subcellular portions, a glass Petri plate was laid on a bed of crushed dry ice. Individual plants were laid on the Petri plate in parallel and cut with a razor blade into thirds (apical, middle, and basal portions). The tissue was transferred to 15-mL conical tubes using chilled forceps.
RNA samples were denatured and run on a 1.2% (w/v) denaturing formaldehyde gel in MOPS [3-(N-morpholino)propanesulfonic acid] buffer. The gel was rinsed for 1 h in several changes of water, stained for 20 min in 0.5 μg/mL ethidium bromide, destained for 2 to 3 h in water, and photographed. RNA was then transferred to filters (Hybond N+, Amersham) and hybridized overnight at 65°C in 5× SSC, 3× Denhardt's reagent, 50 μg mL−1 salmon sperm DNA, 50 μg/mL tRNA, and 1% (w/v) SDS. Blots were washed twice for 30 min at 65°C in 0.2× SSC, 0.1% (w/v) SDS.
ACKNOWLEDGMENTS
We thank Dr. Robert Cleland for laboratory space and advice, Dr. Luca Comai for materials and advice, Kari Stiles, Dr. Rainer Stahlberg, and Dr. Liz VanVolkenburgh for equipment and physiology expertise, Richard Ivey for technical suggestions, and Chris Higgens, Christina Richmond, and Lori Krueger for comments and suggestions on the manuscript.
Footnotes
This work was supported by the U.S. National Science Foundation (postdoctoral grant to K.A.S. and grant no. IBN–9630618 to D.F.M.).
LITERATURE CITED
- Badger MR, Price GD. The role of carbonic anhydrase in photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:369–392. [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Berger S, de Groot EJ, Neuhaus G, Schweiger M. Acetabularia: a giant single cell organism with valuable advantages for cell biology. Euro J Cell Biol. 1987;44:349–370. [Google Scholar]
- Bouget F-Y, Gerttula S, Shaw SL, Quatrano RS. Localization of actin mRNA during the establishment of cell polarity and early cell division in Fucus embryos. Plant Cell. 1996;8:189–201. doi: 10.1105/tpc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Curtis PJ, Withers E, Demuth D, Wyatt R, Venta PJ, Tashian RE. The nucleotide sequence and derived amino acid sequence of cDNA coding for mouse carbonic anhydrase II. Gene. 1983;25:325–332. doi: 10.1016/0378-1119(83)90237-8. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson E, Liljas A. X-ray crystallographic studies of carbonic anhydrase isozymes I, II and III. In: Dodgson SJ, Tashian RE, Gros G, Carter NS, editors. The Carbonic Anhydrases: Cellular Physiology and Molecular Genetics. New York: Plenum Press; 1992. pp. 33–48. [Google Scholar]
- Fukuzawa H, Fujiwara S, Tachiki A, Miyachi S. Nucleotide sequences of two genes CAH1 and CAH2 which encode carbonic anhydrase polypeptides in Chlamydomonas reinhardtii. Nucleic Acids Res. 1990a;18:6441–6442. doi: 10.1093/nar/18.21.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S. cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci USA. 1990b;87:4383–4387. doi: 10.1073/pnas.87.11.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke RP, Kovar JL, Weeks DP. Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2. Plant Physiol. 1997;114:237–244. doi: 10.1104/pp.114.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer JB, Ephrussi A. mRNA localization and the cytoskeleton. Semin Cell Dev Biol. 1996;7:357–365. [Google Scholar]
- Hämmerling J. Entwicklung und regeneration bei Acetabularia mediterranea. Z Indukt Abstammungs- Vererbungsl. 1932;62:92–93. [Google Scholar]
- Hämmerling J. Regenerationsversuche an kernhaltigen und kernlosen Zellteilen von Acetabularia wettsteinii. Biol Z. 1934;54:650–655. [Google Scholar]
- Hämmerling J. Studien zum Polaritatsproblem I-III. Zool Jahrb Abt Allg Zool Physiol Tiere. 1936;56:439–486. [Google Scholar]
- Hesketh JE. Sorting of messenger RNAs in the cytoplasm: mRNA localization and the cytoskeleton. Exp Cell Res. 1996;225:219–236. doi: 10.1006/excr.1996.0172. [DOI] [PubMed] [Google Scholar]
- Hewett-Emmett D, Tashian RE. Functional diversity, conservation, and convergence in the evolution of the α, β and γ-carbonic anhydrase gene families. Mol Phylogenet Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- Hunt BE, Mandoli DF. Axenic cultures of Acetabularia (Chlorophyta): a decontamination protocol with potential application to other algae. J Phycol. 1992;28:407–414. [Google Scholar]
- Hunt BE, Mandoli DF. A new artificial seawater that facilitates growth of large numbers of cells of Acetabularia acetabulum (Chlorophyta) and reduces the labor inherent in cell culture. J Phycol. 1996;32:483–495. [Google Scholar]
- Kamo T, Shimogawara K, Fukuzawa H, Muto S, Miyachi S. Subunit constitution of carbonic anhydrase from Chlamydomonas reinhardtii. Eur J Biochem. 1990;192:557–562. doi: 10.1111/j.1432-1033.1990.tb19261.x. [DOI] [PubMed] [Google Scholar]
- Karlsson J, Clarke AK, Chen Z-Y, Hugghins SY, Park Y-I, Husic HD, Moroney JV, Samuelsson G. A novel α-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 1998;17:1208–1216. doi: 10.1093/emboj/17.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb RE, Saville PJ, Johnston AM, Raven JA. Sources of inorganic carbon for photosynthesis by three species of marine diatom. J Phycol. 1997;33:433–440. [Google Scholar]
- Kratz RF, Young PA, Mandoli DF. Timing and light regulation of apical morphogenesis during reproductive development in wild-type populations of Acetabularia acetabulum (Chlorophyceae) J Phycol. 1998;34:138–146. [Google Scholar]
- Lichtenthaler HK. Chlorophylls and cartenoids: pigments of photosynthetic membranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Lipshitz HD, Smibert CA. Mechanisms of RNA localization and translational regulation. Curr Opin Genet Dev. 2000;10:476–488. doi: 10.1016/s0959-437x(00)00116-7. [DOI] [PubMed] [Google Scholar]
- Li-Weber M, Schweiger H-G. Evidence for and mechanism of translational control during cell differentiation in Acetabularia. Eur J Cell Biol. 1985;38:73–78. [Google Scholar]
- Lucas WJ. Photosynthetic assimilation of exogenous HCO3− by aquatic plants. Annu Rev Plant Physiol. 1983;34:71–104. [Google Scholar]
- Mandoli DF. Elaboration of body plan and phase change during development of Acetabularia: how is the complex architecture of a giant unicell built? Annu Rev Plant Physiol Plant Mol Biol. 1998a;49:173–198. doi: 10.1146/annurev.arplant.49.1.173. [DOI] [PubMed] [Google Scholar]
- Mandoli DF. What ever happened to Acetabularia: bringing a once-classic model system into the age of molecular genetics. Int Rev Cytol. 1998b;182:1–67. [Google Scholar]
- Mandoli DF, Larsen T. Improved mating efficiency in Acetabularia acetabulum: recovery of 48–100% of the expected zygotes. Protoplasma. 1993;176:53–63. [Google Scholar]
- Menzel D. Tansley review no. 77 cell differentiation and the cytoskelton in Acetabularia. New Phytol. 1994;128:369–393. doi: 10.1111/j.1469-8137.1994.tb02984.x. [DOI] [PubMed] [Google Scholar]
- Mercado J, Gordillo FJL, Figueroa FL, Niell FX. External carbonic anhydrase and affinity for inorganic carbon in intertidal macroalgae. J Exp Mar Biol Ecol. 1998;221:209–220. [Google Scholar]
- Mercado JM, Figueroa FL, Niell FX. A new method for estimating external carbonic anhydrase activity in macroalgae. J Phycol. 1997;33:999–1006. [Google Scholar]
- Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Nimer NA, Iglesias-Rodrigues MD, Merrett MJ. Bicarbonate utilization by marine phytoplankton species. J Phycol. 1997;33:625–631. [Google Scholar]
- Porterfield DM, Smith PJS. Single-cell, real-time measurements of extracellular oxygen and proton fluxes from Spirogyra grevilleana. Protoplasma. 2000;212:80–88. [Google Scholar]
- Raven JA. Photosynthetic and non-photosynthetic roles of carbonic anhydrase in algae and cyanobacteria. Phycologia. 1995;34:93–101. [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Sietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly-related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Iwasaki T, Odani S, Shiraiwa Y. Purification, characterization and cDNA cloning of soluble carbonic anhydrase from Chlorella sorokiniana grown under ordinary air. Planta. 1998;206:657–665. doi: 10.1007/s004250050444. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Mandoli DF. Aaknox1, a kn1-like homeobox gene in Acetabularia acetabulum, undergoes developmentally regulated subcellular localization. Plant Mol Biol. 1999;41:785–793. doi: 10.1023/a:1006387107071. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Porterfield DM, Smith PJ, Mandoli DF. Calcification and measurements of proton and oxygen flux reveal subcellular domains in Acetabularia acetabulum 1. Planta. 2000;211:474–483. doi: 10.1007/s004250000318. [DOI] [PubMed] [Google Scholar]
- Shoeman RL, Neuhaus G, Schweiger H-G. Gene expression in Acetabularia: III. Comparison of stained cytosolic proteins and in vivo and in vitro translation products. J Cell Sci. 1983;60:1–12. doi: 10.1242/jcs.60.1.1. [DOI] [PubMed] [Google Scholar]
- Soltes RE, Mulligan ME, Coleman JR. Identification and characterization of a gene encoding a vertebrate-type carbonic anhydrase in cyanobacteria. J Bacteriol. 1997;179:769–774. doi: 10.1128/jb.179.3.769-774.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukenik A, Tchernov D, Kaplan A, Huertas E, Lubian LM, Livne A. Uptake, efflux and photosynthetic utilization of inorganic carbon by the marine eustigmatophyte Nannochloropis sp. J Phycol. 1997;33:969–974. [Google Scholar]
- Sültemeyer D. Carbonic anhydrases in eukaryotic algae: characterization, regulation, and possible function during photosynthesis. Can J Bot. 1998;76:962–972. [Google Scholar]
- Vogel H. Untersuchungen zu spezifischen mRNA-Gradienten in der einzelligen, siphonalen Grünalge Acetabularia acetabulum (L.) Silva. PhD thesis. Giessen, Germany: Universität Giessen; 1998. [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- Wilbur KM, Anderson NG. Electrometric and colorimetric determination of carbonic anhydrase. J Biol Chem. 1948;176:147–154. [PubMed] [Google Scholar]
- Williams TG, Turpin DH. The role of external carbonic anhydrase in inorganic carbon acquisition by Chlamydomonas reinhardii at alkaline pH. Plant Physiol. 1987;83:92–96. doi: 10.1104/pp.83.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]