Summary
The decision to consume or reject a food based on the degree of acidity is critical for animal survival. However, the gustatory receptors that detect sour compounds and influence feeding behavior have been elusive. Here, using the fly, Drosophila melanogaster, we reveal that a member of the Ionotropic Receptor family, IR7a, is essential for rejecting foods laced with high levels of acetic acid. IR7a was dispensable for repulsion of other acidic compounds, indicating that the gustatory sensation of acids occurs through a repertoire rather than a single receptor. The fly’s main taste organ, the labellum, is decorated with bristles that house dendrites of gustatory receptor neurons (GRNs). IR7a was expressed in a subset of bitter GRNs, rather than GRNs dedicated to sour taste. Our findings indicate that flies taste acids through a repertoire of receptors, enabling them to discriminate foods on the basis of acid composition, rather than just pH.
Keywords: Sour, sourness, acid taste, proton, labeled line, feeding, Drosophila
Introduction
Sour taste allows for the detection of hydrogen ions and organic acids. It is one of the five basic tastes, and along with other chemical and textural features, allows animals from flies to humans to discriminate between foods that are safe and appealing from other options that are dangerous. While low levels of certain acids are attractive, high levels are repulsive. The aversive makes sense, as foods that are very acidic may be spoiled due to excessive microbial growth, and if consumed, can lead to adverse effects.
Despite the critical role of sour detection for food selection, the behavioral and molecular mechanisms controlling the behavioral aversion to feeding on acid-containing foods are poorly understood. This contrasts with sugar, bitter and umami taste modalities, which in mammals such as mice and humans, are detected through G-protein coupled receptors (Liman et al., 2014). In the fly, Drosophila melanogaster, sugar, bitter and amino acid taste occurs primarily through ionotropic receptors, which serve both as receptors and cation channels (Liman et al., 2014). These include a family called “Gustatory Receptors” (GRs) (Clyne et al., 2000; Scott et al., 2001; Kim and Carlson, 2002), another referred to as “Ionotropic Receptors” (IRs) (Benton et al.,2009), as well as TRP channels (Al-Anzi et al., 2006; Kang et al., 2010; Kim et al., 2010). Fly GRs and IRs are distinct from mammalian proteins, although IRs are very distantly related to mammalian ionotropic glutamate receptors. Nevertheless, both mice and flies use ionotropic receptors (ENaCs and IRs) for the gustatory detection of low salt (Chandrashekar et al., 2010; Zhang et al., 2013; Jaeger et al., 2018). In addition, Drosophila use IRs for tasting another mineral, Ca2+ (Lee et al., 2018).
Over the last few years, many candidate sour taste receptors have been implicated in mammals; however, they remain elusive (Liman et al., 2014). In the mouse, the prime candidate is Otopetrin1, which is a H+-selective channel expressed in type III taste receptor cells (Tu et al., 2018). Currently, it remains unclear whether or not loss of this channel impacts on the behavioral detection of acidic foods (Montell, 2018; Tu et al., 2018).
Drosophila also detects sour in foods, and does so through both taste and smell (Ai et al., 2010; Abuin et al., 2011; Ai et al., 2013; Charlu et al., 2013). As in humans and mice, some acidic compounds are attractive at low levels, and aversive at higher concentrations (Charlu et al., 2013). In the Drosophila taste system, the repulsive behavioral responses to acidic stimuli, such as carboxylic acids, are mediated through dual cellular mechanisms—activation of a subset of deterrent gustatory receptor neurons (GRNs) that also respond to bitter compounds, and inhibition of sugar-activated GRNs (Charlu et al., 2013; Jeong et al., 2013). The olfactory detection of acids, as well as the ability to navigate to acid-containing zones for laying eggs is mediated by members of the IR family (Joseph et al., 2009; Ai et al., 2010; Ai et al., 2013; Chen and Amrein, 2017).
Currently, the receptors required in taste systems for inhibiting consumption of sour foods are not known. To reveal the identity of a taste receptor that controls this behavior, we focused on acetic acid taste in Drosophila. We demonstrate that a member of the Ir family, Ir7a, is expressed in a subset of bitter GRNs, and is required for rejection of food containing high levels of acetic acid. Surprisingly, the receptor was narrowly tuned, thereby indicating that different acids, such as carboxylic acids, are sensed through distinct receptors.
Results
Opposing responses to low and high sour taste
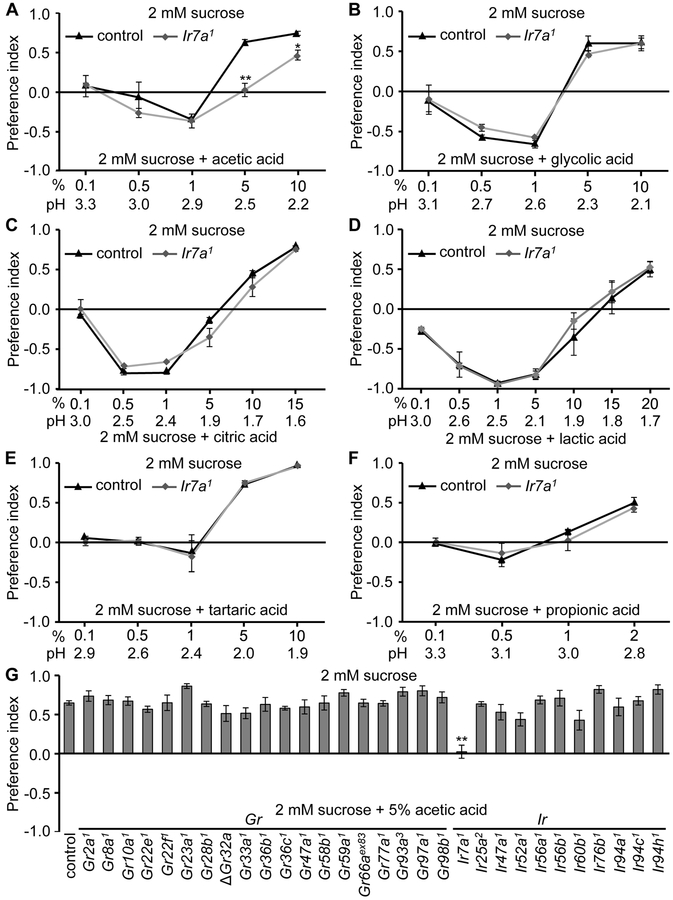
To characterize sour-regulated feeding decisions in flies, we conducted two-way choice assays. We allowed the animals to choose between 2 mM sucrose alone versus 2 mM sucrose combined with different concentrations of carboxylic acids. Control flies showed distinct behavioral responses to sour depending on the specific carboxylic acid and the concentration. In the case of acetic acid, control flies were indifferent to ≤0.5% acetic acid, they mildly preferred 1.0% acetic acid, and strongly avoided 5% and 10% acetic acid (Figure 1A). The responses to several other carboxylic acids showed much greater bivalent responses. 0.5—1% glycolic acid or citric acid were strongly attractive, while either ≥5% glycolic acid, or ≥10% citric acid were highly aversive (Figures 1B and 1C) . The response to lactic acid, a major metabolite of the beneficial bacterium, Lactobacillus, was the most appealing, and elicited robust attraction even at a concentration of 5% (Figure 1D). 10% lactic acid was still slightly preferred, and 15% caused only mild repulsion, which was not significantly different from a PI of 0 (Figure 1D). Not until the concentration increased to 20% was lactic acid strongly aversive (Figure 1D). The flies were neutral to low levels of several other carboxylic acids and to HCl, but avoided high concentrations of these acids (Figures 1E, 1F, S1A and S1B).
Figure 1. Binary food choice assays to test responses of flies to low and high concentration of acids.
(A—F) Two-way choice feeding assays showing preferences of control (w1118) and Ir7a1 flies to 2 mM sucrose versus 2 mM sucrose and the indicated concentrations of organic acids. n=4-6.
(A) Acetic acid.
(B) Glycolic acid.
(C) Citric acid.
(D) Lactic acid.
(E) Tartaric acid.
(F) Propionic acid.
(G) Screening of candidate Gr and Ir mutants for acetic acid avoidance. The control flies were w1118. n=4-6.
The error bars indicate SEMs. The asterisks indicate significant differences from the controls using ANOVA tests with Scheffe’s post hoc analyses between control and Ir7a1 flies. *p < 0.05, **p < 0.01.
The attraction to some sour compounds could be an intrinsic feature of the acids, or depend on the presence of sugar. Therefore, we tested the gustatory responses in the absence of sugar by allowing the flies to choose between agarose alone and agarose plus different concentrations of acetic acid. Under these conditions, the modest preference for 1.0% acetic acid was eliminated in control flies (Figure S1C), indicating that in the absence of sugar, the competition between activation of avoidance GRNs and attraction GRNs cancel out. In Ir7a1 flies, the minor avoidance to 1% acetic acid is wiped out, leaving a very slight attraction (Figure S1C), suggesting the presence of another but minor attractive pathway. However, the strong attractions to glycolic acid, citric acid, and lactic acid were similar with and without sugar (Figures 1B—1D and S1D—S1F). In addition, the robust avoidance of 5% and 10% acetic acid, 15% citric acid, 5%, 10% glycolic acid, and 20% lactic acid still occurred (Figures S1C—S1F).
Ir7a is narrowly tuned for tasting aversive levels of acetic acid
To identify a receptor required for sensing the aversion to sour taste, we focused on acetic acid. A candidate sour taste receptor should be expressed in bitter-sensing GRNs, since these GRNs are activated by carboxylic acids (Charlu et al., 2013). Many members of the “Gustatory Receptor” (GR) family are expressed in bitter GRNs, including GR32a, GR33a and GR66a, which are required broadly for the detection of many bitter tastants (Moon et al., 2006; Lee et al., 2009; Moon et al., 2009; Lee et al., 2010). Therefore, we surveyed the effects of mutations disrupting these three as well as 16 additional Grs for defects in the repulsion to 5% acetic acid. To conduct the behavioral screen we performed two-way choice feeding assays using 2 mM sucrose versus 2 mM sucrose and 5% acetic acid. None of the 19 Gr mutants exhibited a significance difference from the control flies (Figure 1G).
Variant IRs are another class of chemosensory receptors, many of which are expressed in GRNs (Benton et al., 2009; Croset et al., 2010; Zhang et al., 2013; Koh et al., 2014). IRs might function in regulating feeding on the basis of sour taste since two (IR25a, and IR76b) are required in female legs for selecting oviposition sites on the basis of pH (Chen and Amrein, 2017). However, mutation of Ir25a, Ir76b and two other Ir genes (Ir56b, Ir60b) had no significant impact on the rejection of food with 5% acetic acid (Figure 1G). We created knockouts of seven additional Ir genes that are expressed in taste organs (Ir7a, Ir47a, Ir52a, Ir56a, Ir94a, Ir94c, and Ir94h; Figure S1G) (Croset et al., 2010; Koh et al., 2014). Six of these latter mutations also had no impact (Figure 1G).
Of significance here, we found that the Ir7a1 null mutation eliminated the ability to avoid 5% acetic acid mixed with sucrose (Figure 1G), and reduced avoidance to 10% acetic acid (Figure 1A). When we placed the Ir7a1 mutation in trans with a deficiency (Df) that spans Ir7a, we recapitulated the severe defect in repulsion to the 5% acetic acid (Figure S2A). In contrast, Ir7a1 flies showed no significant difference from control flies in attraction to 1% acetic acid (Figure 1A). When the sucrose was eliminated from the assay, and the flies were given a choice between agarose alone versus agarose plus either 5% or 10% acetic acid, their aversion to even 10% acetic acid was virtually eliminated (Figure S1C). The aversion of control flies to feeding on acetic acid, and the indifference exhibited by Ir7a1 flies were indistinguishable in males and females (Figure S2B).
To test whether IR7a is required generally for sensing sour compounds, we surveyed the behavioral responses of Ir7a1 flies to other carboxylic acids and HCl over a range of concentrations. We found that Ir7a1 flies showed normal feeding aversion in response to each of the other acids tested, including glycolic acid, citric acid, lactic acid, tartaric acid, propionic acid, butyric acid and HCl (Figures 1B—1F, S1A and S1B). Mutations affecting 9 other Ir genes also had no impact on repulsion to high levels of these acids (Figure S3).
Action potentials induced by high levels of acetic acid depend on Ir7a
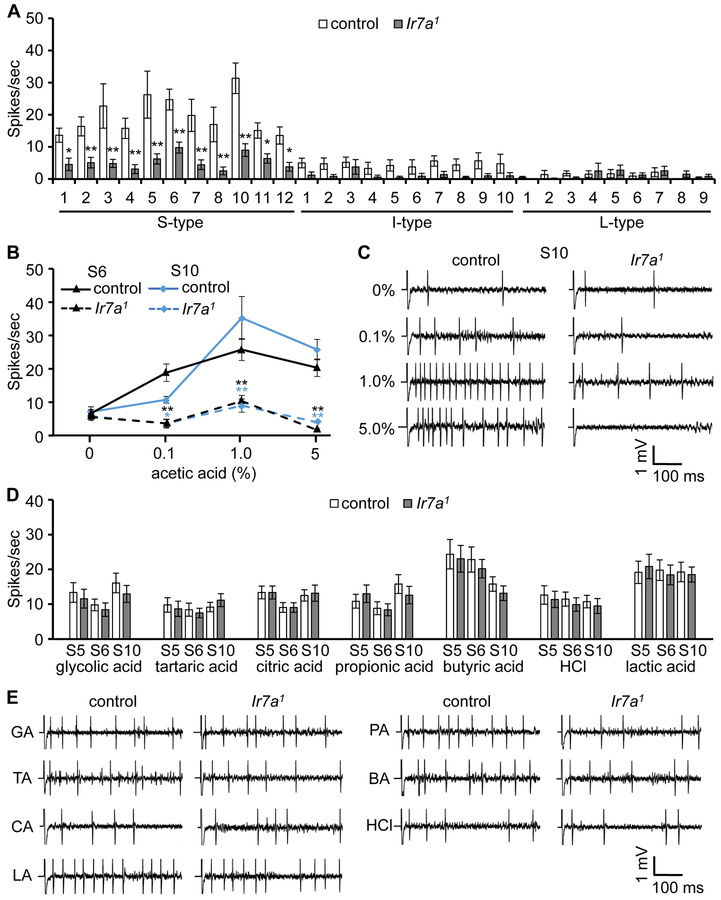
To address a requirement for IR7a for production of acetic acid-induced action potentials, we performed tip recordings. The main taste organ consists of two bilaterally symmetrical labella, each of which is decorated with 31 sensilla (bristles) that fall into three size classes: short-(S), intermediate- (I), and long- (L) sensilla. The L- and S-type sensilla harbor four GRNs while the I-type house two (Hiroi et al., 2002; Meunier et al., 2003; Hiroi et al., 2004). In control flies, all S-type sensilla responded to 1% acetic acid, while the I- and L-type sensilla produced few action potentials (Figure 2A). S3, S5, S6, and S10 elicited the highest neuronal activities. We obtained similar results after subjecting the flies to nutrient deprivation for 24 hours (Figures S2E), indicating that the responses to acetic acid were not sensitive to starvation. We performed a dose response analysis using two of the high responding sensilla (S6 and S10) and found that the action potential frequencies peaked at 1% acetic acid (Figures 2B and 2C). Of significance, the Ir7a1 mutation greatly reduced acetic acid-induced action potentials in S-type sensilla (Figures 2A and 2B). The remaining activity in Ir7a1 flies was highest in response to 1% acetic acid (Figure 2B). When we placed Ir7a1 in trans with a deficiency that uncovered Ir7a, we observed a similar impairment in action potentials provoked by 1% acetic acid. (Figure S2C).
Figure 2. Action potentials induced by acetic acid depend on Ir7a.
Tip recordings performed on gustatory sensilla from labella of control (w1118) and Ir7a1 flies.
(A)A verage spikes produced by GRNs in S, I and L type sensilla in response to 1% acetic acid. n=15-18.
(B) Responses of S6 and S10 sensilla to the indicated concentrations of acetic acid. n=15-16.
(C) Representative traces obtained from S10 in response to the indicated concentrations of acetic acid.
(D) Frequencies of action potentials elicited from S5, S6, and S10 sensilla using 1% glycolic acid, 1% tartaric acid, 5% citric acid, 1% propionic acid, 1% butyric acid, 10 mM HCl, and 20% lactic acid. n=20.
(E) Representative traces on S10 sensilla in response to 1% glycolic acid (GA), 1% tartaric acid (TA), 5% citric acid (CA), 1% propionic acid (PA), 1% butyric acid (BA), 10 mM HCl, and 20% lactic acid (LA).
The error bars represent SEMs. The asterisks indicate significant differences from the controls (*p < 0.05, **p<0.01). Each frequency of the same sensillum between control and Ir7a1 mutant was compared using unpaired Student t-test.
Ir25a and Ir76b are proposed to form heteromultimers with other narrowly tuned IRs (Benton et al., 2009; Rimal and Lee, 2018). However, in contrast to Ir7a1 flies, the Ir25a2 and Ir76b1 mutants displayed high levels of neuronal firing upon application of 1% acetic acid (Figure S2D). While there were reductions in the action potential frequencies relative to the control, the differences were not significant, consistent with the normal acetic acid rejection exhibited by the Ir25a2 and Ir76b1 mutants (Figure 1G).
To address whether IR7a is required for neuronal firing in response to other sour compounds, we tested several organic acids, and the inorganic acid, HCl. We focused on S5, S6 and S10 sensilla, which had relatively high responses to acetic acid (Figure 2A). The Ir7a1 responses to 1% glycolic acid, 1% tartaric acid, 5% citric acid, 1% propionic acid, 1% butyric acid, and 10 mM HCl were similar to the controls (Figures 2D and 2E). We screened the 10 available Ir mutants for roles in sensing other sour compounds, focusing on S6 and S10 sensilla, which respond to these acids. The neuronal firing rates in the mutant backgrounds were not significantly different from the control flies (Figure S4).
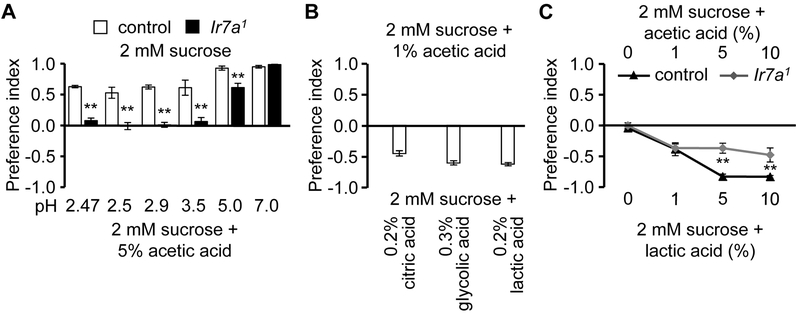
The electrophysiological and behavioral data indicate that IR7a is narrowly tuned. IR7a could be responding to the carbon backbone of acetic acid, to the pH or both. To test whether the carbon backbone is sufficient for IR7a-induced action potentials, we performed tip recordings using other compounds with the same carbon backbone as acetic acid, but are not acids. We found that the responses to ethanol and acetaldehyde were indistinguishable between control and Ir7a1 flies (Figure S5A—S5D). Because 5% acetic acid is repulsive, while 1% is slightly attractive (Figure 1A), we adjusted the pH of 5% acetic acid from 2.5 to 2.9 (the pH of 1% acetic acid) and performed two-way choice feeding assay. Control flies still avoid 5% acetic acid at pH2.9, while Ir7a1 flies do not (Figure 3A). Ir7a was also required for the repulsion to 5% acetic acid at pH3.5. When we adjusted the pH so that it was only slightly sour (5.0), the rejection of acetic acid was only partially dependent on IR7a (Figure 3A). When the pH of acetic acid was neutral (7.0), control flies still reject acetic acid. Of significance, the distaste for acetic acid at pH7.0 was the same as in control flies (Figure 3A). These data demonstrate that IR7a does not function simply by detecting the 3D chemical structure of acetic acid, but that the recognition of acetic acid by IR7a is also pH-dependent.
Figure 3. Both pH and chemical structure of acids affect behavior.
(A) Two-way choice feeding assays showing preferences to 2 mM sucrose versus either 2 mM sucrose plus 5% acetic acid (pH 2.47), or 2 mM sucrose plus 5% acetic acid, with the pH adjusted to 2.5 to 7. n=5-7.
(B) Two-way choice feeding assays using 2 mM sucrose plus an attractive level of acetic acid (1%, pH 2.9), versus 2 mM sucrose plus concentrations of other acids at the identical pH (2.9). n=4-6.
(C) Two-way choice feeding assays using 2 mM sucrose plus the indicated concentrations of acetic acid, versus 2 mM sucrose plus lactic acid at indicated identical concentrations. n=4-6.
The error bars represent SEMs. The asterisks indicate significant differences from the controls (**p<0.01) using ANOVA with Scheffe’s analysis as a post hoc test.
1% acetic acid plus sucrose is only slightly attractive, while 1% glycolic acid, citric acid or lactic acid and sucrose are strongly appealing to the flies. To address whether the different preferences to the same concentration of acid were functions of the pH or the carbon backbone, we allowed the flies to choose between 1% acetic acid plus sucrose, versus sucrose mixed with other acids at the same pH of 2.9 (0.2% citric acid, 0.3% glycolic acid, and 0.2% lactic acid). We found that the control flies preferred each of these other acids over acetic acid at the same pH of 2.9 (Figure 3B). This indicates that sour attraction is also a behavior influenced by the chemical structure of the carboxylic acid, rather than just pH. Consistent with the higher intrinsic appeal of some carboxylic acids over others, we found that control flies prefer lactic acid over the identical concentration of acetic acid (Figure 3C). The preference for 5% and 10% lactic acid is reduced in the Ir7a1 mutant (Figure 3C), consistent with the observation that repulsion to these levels of acetic acid is reduced in the Ir7a1 flies.
Carboxylic acids such as acetic acid suppress feeding through dual effects on two types of GRNs. They activate GRNs that respond to aversive compounds and suppress sugar-activated GRNs (Charlu et al., 2013). To test whether IR7a was required for suppression of the sugar response, we used tip recordings to assay the effects of different concentrations of acetic acid on sucrose-induced action potentials elicited by L4 and L6 sensilla. In control flies, sucrose stimulates a high level of neuronal firing, which is mildly suppressed by increasing concentrations of acetic acid (Figure S5E). At 1% acetic acid, ~80% of the sucrose-induced activity remained, potentially accounting for the attraction of the sucrose/1% acetic acid mixture (Figure 1A). The attenuation of the sugar-induced activity was Ir7a-independent, as it was identical in the Ir7a1 mutant (Figure S5E).
Ir7a functions in acetic acid detection in bitter-sensing GRNs
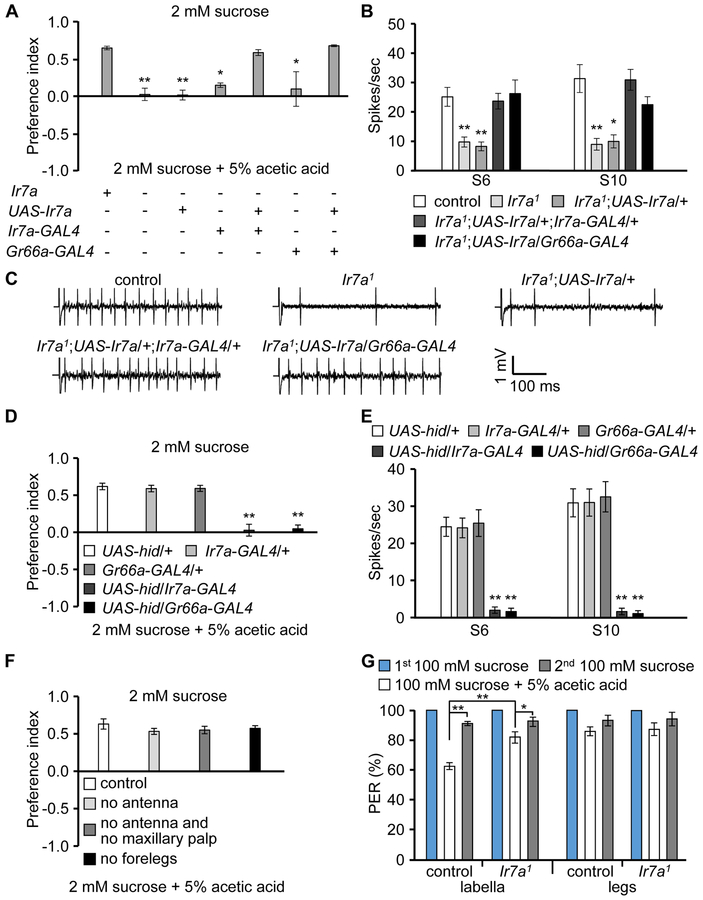
Bitter-sensing GRNs are required for acid sensation (Charlu et al., 2013). To test whether Ir7a is required in these GRNs, we performed phenotypic rescue experiments using the Gr66a-GAL4, which is expressed in bitter GRNs. We generated UAS-Ir7a transgenic flies, which we expressed in the Ir7a1 background under the control of either the Ir7a-GAL4 or the Gr66a-GAL4. We found that either GAL4 in combination with UAS-Ir7a rescued the repulsion to 5% acetic acid (Figure 4A), and restored high frequencies of acetic acid-induced action potentials (Figures 4B and 4C). We also expressed the proapoptotic transgene, UAS-hid, under control of either the Ir7a-GAL4 or Gr66a-GAL4, and performed binary food choice assays and tip recordings. The Ir7a-GAL4/UAS-hid and Gr66a-GAL4/UAS-hid flies exhibited greatly diminished avoidance and action potentials in response to acetic acid (Figures 4D and 4E). The requirement for Ir7a in bitter-responsive GRNs was not dependent on the presence of sugar, since we observed similar rescue of the acetic acid rejection when we assayed repulsion to 5% acetic acid in the absence of sugar (Figure S6A). When we removed sugar from the two-way choice assays, we also observed a similar loss of acetic acid avoidance due to expression of UAS-hid under control of the Ir7a-GAL4 (Figure S6B).
Figure 4. Ir7a is necessary for acetic acid detection in bitter-sensing GRNs in the labellum.
(A) Two-way choice feeding assays showing rescue of the avoidance defect in Ir7a1 in response to acetic acid. n=4-6.
(B) Rescue of the Ir7a1 defect in neuronal firing in response to 1% acetic acid by expression of Ir7a in either Ir7a- or Gr66a-positive GRNs. n=10-12.
(C) Representative traces obtained from S10 sensilla in response to 1% acetic acid.
(D) Two-way food choice assays after expressing the cell death gene, hid (UAS-hid), under control of the Ir7a-GAL4 or the Gr66a-GAL4. n=6.
(E) Average frequency of action potentials elicited after expressing UAS-hid under control of the Ir7a-GAL4 or the Gr66a-GAL4. n=11-13.
(F) Two-way choice feeding assays after removing the indicated organs. n=4-6.
(G) Average percentages of flies extending their proboscis after applying the indicated stimuli to labella or legs. The 1st and 2nd 100 mM sucrose refers to application of sucrose before and after application of 100 mM sucrose + 5% acetic acid. n=4-6.
To test whether chemosensory organs other than the labellum were critical for the gustatory avoidance of acetic acid, we performed surgical dissections. The two main olfactory organs are the antenna and maxillary palp. We found that flies missing these olfactory tissues displayed a similar repulsion to acetic acid as control animals (Figure 4F). In addition to the labellum, the legs contain taste sensilla. After removing the forelegs, the flies rejected acetic acid to a similar extent as control flies (Figure 4F). However, it was not possible to remove the remaining midlegs and hindlegs without greatly impairing movement.
To interrogate the responses of both the labellum and legs to the gustatory rejection of acetic acid, we performed proboscis extension response (PER) assays. 100% of starved flies extend their proboscis when we briefly touch either the labellum or legs with a probe with a 1st offering of 100 mM sucrose (Figure 4G). Upon addition of 5% acetic acid to the sucrose, the PER diminished 37.8 ±2.1% when we applied the food to the labellum (PER=62.2 ±2.0%), and 16.9 ±2.2% (PER=83.1 ±2.4%) when we touched the legs (Figure 4G). Upon offering the flies sucrose only again (2nd sucrose offering) the PER returned to >90% (Figure 4G; % PER: labellum, 90.7 ±1.0; legs, 92.6 ±3.1). , The slight reduction from the 100% response to 1st sucrose only offering was possibly due to residual effects of the exposure to acetic acid. We found that the Ir7a1 mutant flies displayed significantly less distaste for acetic acid-containing sucrose than control flies, but only when we applied the probe to the labellum (Figure 4G; % PER: labellum, 82.1 ±3.2; legs, 86.1 ±3.7). Thus, while both the labellum and legs sense acetic acid, the labellum appears to be more important in suppressing feeding in response to acetic acid, and Ir7a contributes to this behavior.
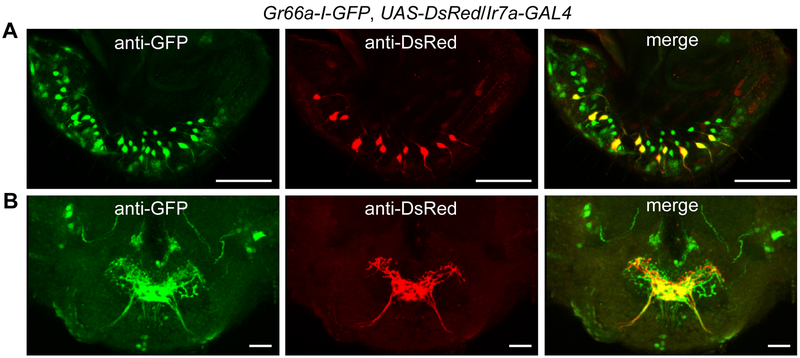
To address whether Ir7a was expressed in bitter-sensing GRNs, we performed co-labeling experiments. We combined a transgene expressing GFP under direct control of the Gr66a promoter (Gr66a-I-GFP) with the Ir7a-GAL4 and UAS-DsRed. The UAS-DsRed driven by Ir7a-GAL4 was expressed in GRNs in 11 sensilla, all of which were S-type and were co-labelled with Gr66a-I-GFP (Figures 5A and S7A). The Ir7a and Gr66a GRNs also sent their projections to the same medial area in SEZ (Figure 5B). We additionally found that Ir7a is expressed in two neurons in the prothoracic tarsi and one neuron in the pharynx (Figures S7B and S7C). Collectively, our behavioral, electrophysiological, rescue, cell ablation and immunolocalization data demonstrate that the same GRNs that sense sour (acetic acid) are bitter-sensing GRNs in S-type sensilla decorating the labellum. However, Ir7a was not required for the aversive responses to any of the other bitter chemicals tested (Figures S5F and S5G).
Figure 5. Ir7a is expressed in a subset of bitter-sensing GRNs.
Relative spatial distributions of the Gr66a (green; anti-GFP) and Ir7a (red; anti-DsRed) reporters in (A) labella, and (B) brains of Gr66a-I-GFP,Ir7a-GAL4/UAS-DsRed flies. The images were acquired by confocal microscopy. Shown are 3-D reconstructions generated by maximum transparency. The scale bars represent 25 μm.
IR7a is a sensor for acetic acid taste
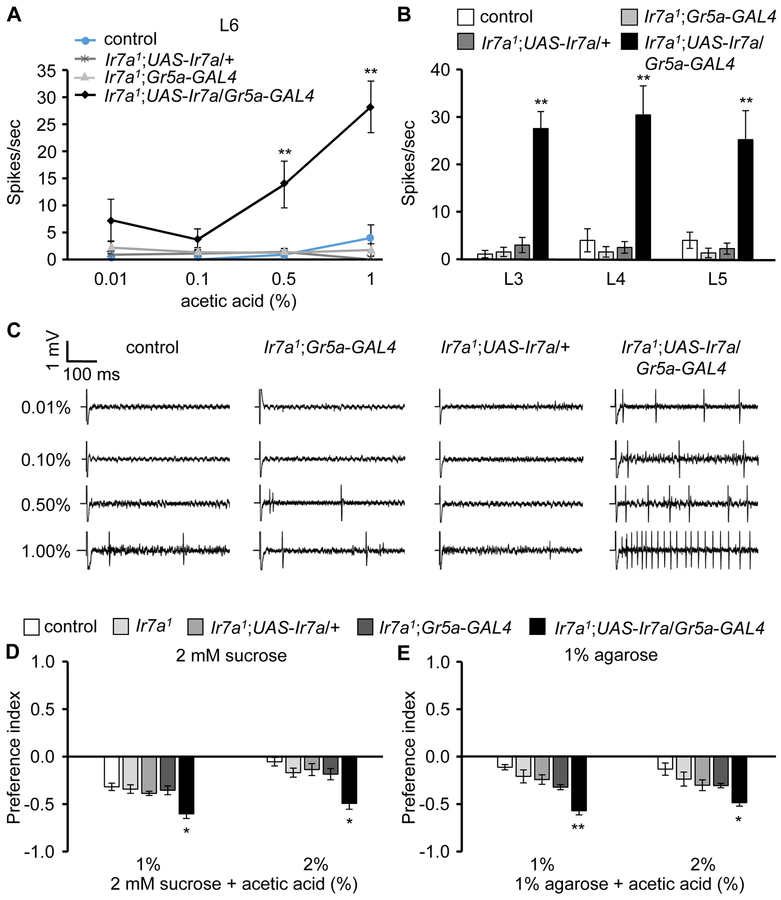
To test whether IR7a is sufficient to confer acetic acid-induced action potentials to cells that do not normally respond this acid, we ectopically expressed Ir7a in sugarsensing GRNs. We introduced UAS-Ir7a in sweet GRNs using the Gr5a-GAL4 and performed tip recordings. The GRNs in L-type sensilla, such as L3, L4, L5 and L6, are normally not stimulated by acetic acid (Figures 6A and 6B). Flies harboring just the UAS-Ir7a or the Gr5a-GAL4 transgene are also insensitive to acetic acid. In contrast, we found that sweet GRNs expressing Ir7a (Ir7a1;UAS-Ir7a/Gr5a-GAL4) elicited robust neuronal firing upon presentation of 1% acetic acid (Figures 6A—6C). There was also a trend towards increased acetic acid attraction, either in the presence or absence of sucrose (Figures 6D and 6E). Thus, activation of IR7a in sugar GRNs by acetic acid was sufficient to overcome the modest inhibition of the sugar-activated action potentials by acetic acid (Figure S5E). Furthermore, sucrose plus either 1% or 2% acetic acid were equally attractive (Figure 6D). This suggests that the slight difference in inhibition of sugar GRNs by 1% and 2% acetic acid (inhibition L4: 23.3 and ~24.7% for 1% and 2% acetic acid, respectively; inhibition L6: 21.4 and ~23.8% for 1% and 2% acetic acid respectively; Figure S5E), is counterbalanced by a slight increase in activation of sugar-activated GRNs by 2% acetic acid, relative to 1% acetic acid.
Figure 6. Misexpression of Ir7a confers acetic acid sensitivity to sugar GRNs.
UAS-Ir7a was expressed in sugar GRNs in Ir7a1 flies under control of the Gr5a-GAL4.
(A—C) Tip recordings to monitor acetic acid-induced action potentials from labellar sensilla from the indicated flies.
(A) Average frequencies of action potentials elicited from L6 sensilla. n=15-18.
(B) Average frequencies of neuronal firing from L3, L4, and L5 sensilla using 1% acetic acid. n=15-18.
(C) Representative traces from L6 sensilla.
(D and E) Two-way food choice assay to test attraction to acetic acid due to ectopic expression of Ir7a in sweet GRNs: (D) with sucrose (n=8-10) and (E) without sucrose (n=4-6).
The error bars represent SEMs. The asterisks indicate significant differences (**p < 0.01, *p < 0.05.) using single factor ANOVA with Scheffe’s analysis as a post hoc test to compare two sets of data.
Discussion
Sour sensing of different carboxylic acids through distinct receptors
A major but unresolved question in the taste field is whether protons and protonated organic acids are sensed through the same or discrete molecular mechanisms. Flowever, molecular and genetic evidence in favor of this concept has been lacking. Our study provides strong support for the model that protons and organic acids are sensed through distinct mechanisms, since mutation of Ir7a greatly impairs the gustatory avoidance of an organic acid, but has no impact on the taste aversion to HCl.
Rather than being required broadly for organic acids, IR7a was narrowly tuned. Avoidance of most organic acids other than acetic acid was either normal or showed only moderate effects. Loss of IR7a greatly impaired avoidance of acetic acid, but had no behavioral impact on the repulsion to any of the organic acids tested. These findings indicate that there must be distinct receptors for different carboxylic acids. However, none of the 10 other IRs tested significantly impaired the aversion to feeding on sugar mixed with HCl or any of the organic acids tested. These include Ir25a and Ir76b, which function in the selection of acid-containing oviposition sites (Chen and Amrein, 2017). Additional candidate sour taste receptors include other IRs, TRP channels, and ENaC channel family members (PPK). One or more of the three Otopetrin channels may also function in sensing either mineral or organic acids, since mammalian Otopetrin1 is a H+ channel, which is expressed in taste receptor cells (Tu et al., 2018). However, the contribution of Otopetrin 1 to taste behavior remains to be defined.
A feature of sour taste, which is reminiscent of Na+ taste, is that low levels of some organic acids are very attractive while high levels are aversive. Notable examples are citric acid and glycolic acid, which are rich in various types of fruit. In contrast to citric acid, glycolic acid and lactic acid, which are highly attractive at lower levels, acetic acid is only slightly appealing to flies at lower concentrations (1%). The attraction to 1% acetic acid is so subtle that it is essentially eliminated if flies are given a choice between agarose alone and agarose plus 1% acetic acid, indicating that a combination of the acetic acid and sugar is necessary for the mild attraction. Because the attraction to lower levels of citric acid, glycolic acid and lactic acid are robust, these acids retain their appeal in the absence of sugar. The attraction to low concentrations of acids is not dependent on Ir7a, and may require one or more IRs, TRPs, PPKs or Otopetrin channels. Nevertheless, the observation that IR7a is narrowly tuned, and only functions in the repulsion rather than the attraction to acetic acid, highlights that the potential set of receptors that function in sour taste is complex.
A question concerns the full repertoire of subunits required for acetic acid sensation. Our finding that ectopic expression of IR7a in sugar-activated GRNs is sufficient to enable these cells to be activated by acetic acid suggests that IR7a is the only subunit required to generate an acetic acid receptor. IRs are distantly related to mammalian ionotropic glutamate receptors (Rimal and Lee, 2018), and appear to be cation channels. However, we found that introduction of IR7a in Drosophila S2 cells was insufficient to confer an acetic acid-induced conductance. We suggest that IR7a interacts in vivo with an obligatory subunit that is missing from S2 cells, or that the unusual ionic composition of the endolymph is necessary to induce activation of IR7a.
Chemical basis for taste discrimination of acetic acid from other organic acids
Our data strongly argue that IR7a is required for recognition of a combination of the sour pH and structure of acetic acid. The sensation of acetic acid by IR7a was not simply due to recognition of just the carbon backbone, since ethanol and acetaldehyde induce the same number of action potentials in control and Ir7a flies. Rather, acidic pH is critical for the IR7a-dependent recognition of acetic acid. The repulsion of 5% acetic acid is strongly dependent on IR7a only if the pH is ≤3.5. At these low pH levels, Ir7a1 flies do not discriminate between sucrose only versus sucrose plus 5% acetic acid. However, control and Ir7a1 flies avoid 5% acetic acid to the same extend if the pH is neutral. Thus, the rejection of acetic acid at a neutral pH is through a receptor other than IR7a.
While the Ir7a-dependent rejection of 5% acetic acid depends on its acidic pH, IR7a is not simply sensing low pH either. Sugar plus lactic acid at pH 2.1 is very attractive to the flies, while sugar plus acetic acid at pH 2.5 is highly aversive. Moreover, when we allowed the flies to choose between 5% acetic acid, versus the same concentration of lactic acid, the animals stronger preferred the lactic acid, even though the pH of the lactic acid was lower. Thus, the discrimination of acetic acid from other acids involves a combination of the carbon backbone and the acid quality of the chemical.
Potential ethological advantage for recognizing different carboxylic acids
An intriguing question is why flies have a receptor that is narrowly tuned to acetic acid. Acetic acid is a fermentation product of a subset of bacteria, such as Acetobacter. We propose that the detection of this carboxylic acid by a narrowly-tuned receptor may provide the animals with the ability to differentiate between foods that have an acidic pH due to growth of acetic acid-producing bacteria, versus foods that are sour due to fermentation by other microorganisms such as fungi or Lactobacillus that generate sour compounds such as citric acid or lactic acid. Indeed, lactic acid is very attractive even at relatively high concentrations such as 5%. Thus, the fly’s capacity to discern between different acid metabolites might endow the animals with the ability to recognize and react to the particular microbial flora in their foods.
A “valence labeled line” mechanism for avoidance of high acetic acid levels
A cellular taste coding mechanism, called the labeled line model, is concerned with instances in which distinct taste qualities that are sensed by different receptor cells which uninterruptedly transfer the information to the brain. However, the distributive model has taste receptor cells that react to multiple types of taste qualities to varying amounts. The brain then deciphers the taste quality according to the intensity of the repertoire of the cellular activities.
Consistent with a previous study showing that acid sensing GRNs comprise a subset of bitter GRNs (Charlu et al., 2013), we found that acetic acid-sensing Ir7a GRNs are not dedicated specifically to sour taste repulsion. Rather, the Ir7a-positive GRNs constitute a subset of GRNs that express a key bitter taste receptor, Gr66a. Detection of high levels of acetic acid in food, through Ir7a, or the sensation of bitter compounds, both lead to a negative behavioral valence—gustatory repulsion. Thus, our findings support the model that avoidance of high concentrations of acetic acid in particular, and possibly sour taste avoidance in general, conforms to a valence labeled line model. According to this latter model, a single GRN responds to more than one taste quality, but activation of the GRN leads to the same behavioral output (Liman et al., 2014). In a similar fashion, sour-sensing type taste receptor cells in mammals may also respond to other taste qualities with the same valence, such as high salt (Montell, 2018).
Another feature common between bitter compounds and aversive levels of acids is that they both inhibit feeding in flies through dual mechanisms: activation of sour/bitter GRNs and inhibition of sugar GRNs. However, acid-induced inhibition of sugar GRNs is not dependent on Ir7a, since this effect is not attenuated in Ir7a1 flies.
1% acetic acid is slightly attractive, and higher concentrations are aversive. At 5% acetic acid, there appears to be a balance between acetic acid attraction, and acetic acid inhibition of the sugar response, since Ir7a mutants are indifferent to sugar alone versus sugar plus 5% acetic acid. Furthermore, this indicates that the activation of bitter GRNs is the primary mode for avoidance of ≥5% acetic acid. At still higher levels of acetic acid, inhibition of sugar GRNs by acetic acid appears to come into play since the mutant animals prefer sugar only, versus sugar plus acetic acid.
The mechanism through which high levels of acetic acid and other sour compounds inhibits sugar GRNs remains to be determined. In the case of bitter compounds, they inhibit sugar GRNs through two mechanisms. According to one, stimulation of bitter GRNs activate GABAergic interneurons, which then inhibit sweet GRNs (Chu et al., 2014). A second mechanism involves binding of bitter compounds to an odorant binding protein in the endolymph, which in turn inhibits sugar-activated gustatory receptors (Jeong et al., 2013). It is intriguing to speculate that organic acids such as acetic acid inhibit sugar attraction through similar strategies.
STAR Methods
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The control strain used in this study was w1118. We obtained the following lines from the Bloomington Drosophila Stock Center (https://bdsc.indiana.edu/): Gr2a1, Gr10a1, Gr22e1, Gr23a1, Gr22f1, Gr28b1, Gr36b1, Gr36c1, Gr58b1, Gr59a1, Gr77a1, Gr97a1, Ir56b1, Ir7a deficiency line (Bloomington Stock #6698), UAS-DsRed, UAS-hid and Ir7a-GAL4. We previously generated the following null mutant lines, which we deposited at the BDSC: Ir76b1 (Zhang et al., 2013), Gr33aGAL4 (Moon et al., 2009), Gr33a1 (Moon et al., 2009), Gr8a1 (Lee et al., 2012), Gr98b1 (Shim et al., 2015), Gr93a3 (Lee et al., 2009). H. Amrein provided the ΔGr32a, Gr66a-GAL4 and Gr5a-GAL4, K. Scott provided Gr66a-I-GFP, and L. Vosshall provided Ir25a2. The Ir7a1, Ir47a1, Ir52a1, Ir56a1, Ir94a1, Ir94c1, and Ir94h1 null mutants, and the UAS-Ir7a transgenic line were generated as described below.
METHOD DETAILS
Generation of IR mutant lines
The Ir7a1, Ir47a1, Ir52a1, Ir56a1, Ir94a1, Ir94c1, and Ir94h1 mutations were generated by ends-out homologous recombination (Gong and Golic, 2003). To create each construct for injections, we amplified two 3-kb genomic fragments by PCR, and subcloned the DNAs into the pw35 vector (Moon et al., 2009). Using the “A” of the “ATG” start codon as +1, the deleted regions were as follows: 1) Ir7a: −32 to +1300, 2) Ir47a1: −62 to +560, 3) Ir52a1: −12 to +586, 4) Ir56a1; +4 to +795, 5) Ir94a1: −201 to +655, 6) Ir94c1: −427 to +982, and 7) Ir94h1: −117 to +527. The constructs were injected into w1118 embryos by either BestGene Inc. (Ir7a, Ir47a, and Ir56a), or the KAIST Injection Facility (Ir52a, Ir94a1, Ir94c1, and Ir94h). We outcrossed all the mutants with w1118 for 5 generations.
To obtain the UAS-Ir7a transgenic line, we amplified the full-length Ir7a cDNA by RT-PCR using proboscis mRNA and subcloned the cDNA into the pUAST vector (Brand and Perrimon, 1993) between the Xbal and Not1 sites. We verified the cDNA by DNA sequencing. The transformation vector was injected into w1118 embryos (BestGene Inc.).
Chemical sources
Sucrose, sulforhodamine B, acetic acid, propionic acid, butyric acid, glycolic acid, tartaric acid, citric acid and tricholine citrate were purchased from Sigma-Aldrich Co. Brilliant blue FCF was purchased from Wako Pure Chemical Industry Ltd. HCl was purchased from Samchun Pure Chemical.
Binary food choice assay
We performed two choice food assays as previously described (Meunier et al., 2003; Moon et al., 2006). 50–70 flies (3–6 days old; mixed sexes, unless indicated otherwise) were starved for 18 h in a humidified chamber. We conducted two types of binary food assays. In one type of assay, we allowed the flies to choose between 1% agarose and 2 mM sucrose versus 1% agarose and 2 mM sucrose mixed with different concentrations of acids. In the second assay, we left sucrose out of the food, and allowed the flies to select either 1% agarose alone or 1% agarose and the indicated concentrations of acids. In one set of experiments (Figure 3A), we adjusted the pH of 2 mM sucrose and 5% acetic acid from 2.5 to 2.9 with KOH. The two foods in each binary test were mixed with either blue food coloring (brilliant blue FCF, 0.125 mg/mL) or red food coloring (sulforhodamine B, 0.1 mg/mL). The two mixtures were distributed in alternating wells of a 72-well microtiter dish (Thermo Fisher Scientific, cat. no. 438733). Within 30 min of preparation, we introduced the 50–70 starved flies into the dish, which we kept in a dark, humidified chamber, and allowed the flies to feed for 90 min at room temperature. To sacrifice the flies, we transferred them to −20°C and then analyzed the color of their abdomens under a stereomicroscope. Blue (NB), red (NR), or purple (NP) flies were tabulated. The preference index (P.I.) was calculated according to the following equation: (NB-NR)/(NR+NB+NP) or (NR-NB)/(NR+NB+NP), depending on the dye/tastant combinations. P.I.s = 1.0 and −1 indicated complete preferences for one of the other food. A P.I. = 0 indicates no bias between the two food choices.
Proboscis Extension Response (PER) assay
We performed PER assays as described previously (Poudel and Lee, 2016). Briefly, 5-10 day-old flies were starved in an empty vial for 18—24 h with a piece of Kimwipe soaked with water. To apply stimuli to the labella, we anesthetized the flies on ice and fixed them in small Pipetman tips to expose their heads. To stimulate the legs, we anesthetized the flies on ice and fixed them on a glass slides by applying glue to their wings. The flies were then incubated in a humid chamber for 1 h to allow them to recover from the procedure. To exclude a water-associated response, before stimulating a fly with a tastant, we applied a Kimwipe wick with water to the labella or legs. Once the fly no longer responded to water alone, we used a Kimwipe wick to offer 100 mM sucrose to either the labella or legs. Flies that did not response to the sucrose (1st 100 mM sucrose offering) were discarded. Mixtures of 100 mM sucrose and 5% acetic acid mixture were subsequently applied to the labella or legs. Extension of the proboscis was scored as a PER. Finally, 100 mM sucrose (2nd 100 mM sucrose offering) to confirm that an absence of a PER to the sucrose/acetic acid mixture was not to an unresponsive fly. ≥10 flies were used for each test. Each experiment was carried out ≥4 times.
Electrophysiology
We performed tip recordings assay as previously described (Moon et al., 2006). We immobilized freshly eclosed flies by keeping them on ice, and then inserted a reference glass electrode filled with Ringer’s solution into the thorax of the fly, extending the electrode towards their proboscis. We stimulated the sensilla with tastants introduced in the buffer solution of the recording pipet (10–20 μm tip diameter). We used 1 mM KCl (S-type and I-type sensilla) or 30 mM tricholine citrate (L-type sensilla) as the electrolyte for the recordings. The recording electrode was connected to a preamplifier (Taste PROBE, Syntech, Hilversum, The Netherlands; http://www.ockenfels-syntech.com/), and the signals were collected and amplified 10x, using a signal connection interface box (Syntech) in conjunction with a 100–3000 Hz band-pass filter. Recordings of action potentials were acquired using a 12-kHz sampling rate, and analyzed using Autospike 3.1 software (Syntech).
Immunohistochemistry
The labella of Gr66a-I-GFP,UAS-DsRed/Ir7a-GAL4 flies were dissected and fixed using 4% paraformaldehyde (Electron Microscopy Sciences, cat. no. 15710) in phosphate buffered saline and 0.2% Triton X-100 (PBS-T) for 15 min at room temperature. The tissue were then washed three times with PBS-T, and cut in half with a razor blade. The samples were then incubated for 30 min at room temperature in blocking buffer: 0.5% goat serum in 1× PBS-T. The following primary antibodies (1:1000 dilution) were added to fresh blocking buffer and incubated with the labellum overnight at 4°C: mouse anti-GFP (Molecular Probes, Cat No A11120) and rabbit anti-DsRed (Clontech, Cat No 632496). The samples were washed three times with PBS-T at 4°C and incubated with the following secondary antibodies (1:200 dilution in the blocking buffer) for 4 h at 4°C: goat anti-mouse Alexa488 (Cat No A11029) and goat anti-rabbit Alexa568 (Cat No A11036). The tissues were washed three times with PBS-T and placed in 1.25x PDA mounting buffer (37.5% glycerol, 187.5 mM NaCl, 62.5 mM Tris pH 8.8), and viewed using a Carl Zeiss LSM700 confocal microscope.
Immunostaining of adult brains were performed as previously described (Lee et al., 2009). Briefly, we dissected the brains of Gr66a-I-GFP,UAS-DsRed/Ir7a-GAL4 flies, and fixed them on ice for 30-45 min by inserting them in 24-well cell culture cluster plates (SPL life sciences, Cat No 30024) containing 940 μL of Fix Buffer (0.1 M Pipes pH 6.9, 1 mM EGTA, 1% TritonX-100, 2 mM MgSO4, 150 mM NaCl) and 60 μL of 37% formaldehyde. The tissues were washed three times in PBS and 0.2% saponin (PBS-S) and blocked overnight at 4°C in blocking buffer (1 mL 1x PBS, 0.1% saponin, 5 mg/mL BSA). The tissues were incubated for 16 h at 4°C with the primary antibodies (mouse anti-GFP and rabbit anti-DsRed) diluted 1:1000 in the freshly prepared blocking buffer. The samples were washed three times for 20 min each and incubated for 4 h at 4°C with the secondary antibodies (Alexa 488 and Alexa 568; Invitrogen-Molecular Probes) diluted 1:200 in the blocking buffer, and washed three times. The tissues were transferred into 1.25x PDA Solution and viewed by confocal microscopy (Carl Zeiss LSM700.
QUANTIFICATION AND STATISTICAL ANALYSIS
All error bars represent SEMs. Single factor ANOVA with Scheffe’s analysis was used as a post hoc test to compare multiple sets of data. Asterisks indicate statistical significance (*p < 0.05, **p < 0.01).
Supplementary Material
Acknowledgments
We thank H. Amrein, L. Vosshall, and the Bloomington Stock Center for fly stocks. S.R. and S.P. were supported by the Global Scholarship Program for Foreign Graduate Students at Kookmin University in Korea. This work is supported by grants to Y.L. from the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1A2B6004202), and to C.M. from the National Institute on Deafness and other Communication Disorders (DC007864).
References
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, and Benton R (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai M, Blais S, Park JY, Min S, Neubert TA, and Suh GS (2013). Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J Neurosci 33, 10741–10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, and Suh GS (2010). Acid sensing by the Drosophila olfactory system. Nature 468, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Anzi B, Tracey WD Jr., and Benzer S (2006). Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr Biol 16, 1034–1040. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, and Vosshall LB (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJ, and Zuker CS (2010). The cells and peripheral representation of sodium taste in mice. Nature 464, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlu S, Wisotsky Z, Medina A, and Dahanukar A (2013). Acid sensing by sweet and bitter taste neurons in Drosophila melanogaster. Nat Commun 4, 2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, and Amrein H (2017). Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr Biol 27, 2741–2750 e2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B, Chui V, Mann K, and Gordon MD (2014). Presynaptic gain control drives sweet and bitter taste integration in Drosophila. Curr Biol 24, 1978–1984. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, and Carlson JR (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, and Benton R (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6, e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WJ, and Golic KG (2003). Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA 100, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi M, Marion-Poll F, and Tanimura T (2002). Differentiated response to sugars among labellar chemosensilla in Drosophila. Zoolog Sci 19, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Hiroi M, Meunier N, Marion-Poll F, and Tanimura T (2004). Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila. J Neurobiol 61, 333–342. [DOI] [PubMed] [Google Scholar]
- Jaeger AH, Stanley M, Weiss ZF, Musso PY, Chan RC, Zhang H, Feldman-Kiss D, and Gordon MD (2018). A complex peripheral code for salt taste in Drosophila. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YT, Shim J, Oh SR, Yoon HI, Kim CH, Moon SJ, and Montell C (2013). An Odorant-Binding Protein required for suppression of sweet taste by bitter chemicals. Neuron 79, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RM, Devineni AV, King IF, and Heberlein U (2009). Oviposition preference for and positional avoidance of acetic acid provide a model for competing behavioral drives in Drosophila. Proc Natl Acad Sci USA 106, 11352–11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL, and Garrity PA (2010). Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464, 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, and Carlson JR (2002). Gene discovery by e-genetics: Drosophila odor and taste receptors. J Cell Sci 115, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lee Y, Akitake B, Woodward OM, Guggino WB, and Montell C (2010). Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc Natl Acad Sci USA 107, 8440–8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, and Carlson JR (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kang MJ, Shim J, Cheong CU, Moon SJ, and Montell C (2012). Gustatory receptors required for avoiding the insecticide L-canavanine. J Neurosci 32, 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim SH, and Montell C (2010). Avoiding DEET through insect gustatory receptors. Neuron 67, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Moon SJ, and Montell C (2009). Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci USA 106, 4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Poudel S, Kim Y, Thakur D, and Montell C (2018). Calcium taste avoidance in Drosophila. Neuron 97, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Zhang YV, and Montell C (2014). Peripheral coding of taste. Neuron 81, 984–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier N, Marion-Poll F, Rospars JP, and Tanimura T (2003). Peripheral coding of bitter taste in Drosophila. J Neurobiol 56, 139–152. [DOI] [PubMed] [Google Scholar]
- Montell C (2018). pHirst sour taste channels pHound? Science 359, 991–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SJ, Köttgen M, Jiao Y, Xu H, and Montell C (2006). A taste receptor required for the caffeine response in vivo. Curr Biol 16, 1812–1817. [DOI] [PubMed] [Google Scholar]
- Moon SJ, Lee Y, Jiao Y, and Montell C (2009). A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol 19, 1623–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S, and Lee Y (2016). Gustatory Receptors Required for Avoiding the Toxic Compound Coumarin in Drosophila melanogaster. Mol Cells 39, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimal S, and Lee Y (2018). The multidimensional ionotropic receptors of Drosophila melanogaster. Insect Mol Biol 27, 1–7. [DOI] [PubMed] [Google Scholar]
- Scott K, Brady R Jr., Cravchik A, Morozov P, Rzhetsky A, Zuker C, and Axel R (2001). A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673. [DOI] [PubMed] [Google Scholar]
- Shim J, Lee Y, Jeong YT, Kim Y, Gee MG, Montell C, and Moon SJ (2015). The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat Commun 6, 8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu YH, Cooper AJ, T. B, Chang RB, Artiga DJ, Turner HN, Mulhall EM, Ye W, Smith AD, and Liman ER (2018). An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Ni J, and Montell C (2013). The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.