Abstract
Background
Approximately 25% of elite controllers (ECs) lose their virological control by mechanisms that are only partially known. Recently, immunovirological and proteomic factors have been associated to the loss of spontaneous control. Our aim was to perform a metabolomic approach to identify the underlying mechanistic pathways and potential biomarkers associated with this loss of control.
Methods
Plasma samples from EC who spontaneously lost virological control (Transient Controllers, TC, n = 8), at two and one year before the loss of control, were compared with a control group of EC who persistently maintained virological control during the same follow-up period (Persistent Controllers, PC, n = 8). The determination of metabolites and plasma lipids was performed by GC-qTOF and LC-qTOF using targeted and untargeted approaches. Metabolite levels were associated with the polyfunctionality of HIV-specific CD8+T-cell response.
Findings
Our data suggest that, before the loss of control, TCs showed a specific circulating metabolomic profile characterized by aerobic glycolytic metabolism, deregulated mitochondrial function, oxidative stress and increased immunological activation. In addition, CD8+ T-cell polyfunctionality was strongly associated with metabolite levels. Finally, valine was the main differentiating factor between TCs and PCs.
Interpretation
All these metabolomic differences should be considered not only as potential biomarkers but also as therapeutic targets in HIV infection.
Fund
This work was supported by grants from Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Fondos FEDER; Red de Investigación en Sida, Gilead Fellowship program, Spanish Ministry of Education and Spanish Ministry of Economy and Competitiveness.
Keywords: Elite controllers, Energy metabolism, HIV-1, Immunometabolism, Loss of control, Metabolomic profile
Research in context.
Evidence before this study
Elite controllers are those HIV-infected patients who maintain an undetectable viral load without the use of combined antiretroviral treatment. This definition makes them the closest model to the functional HIV cure. However, some studies have elucidated that this group of patient is not homogeneous. Actually, there is a subgroup of elite controllers who eventually loses viral control. The mechanisms that dictate whether an elite controller is going to lose the virological control are still unknown.
Added value of this study
Previous studies reported that differences in metabolite plasma levels are relate to changes in the metabolic flux of T-cells due to its effector functions need significant energetic and biosynthetic requirements. However, these metabolic alterations have not been described in relation to HIV spontaneous control, neither have been associated to immune parameters associated with persistent HIV control. We found for the first time a metabolic reprogramming associated with persistent HIV spontaneous control consisting in increased glycolyis in subjects who were going to transiently control HIV. Importantly this metabolic profiling was associated with HIV-specific CD8+ T-cell response, one of the main factors associated to HIV spontaneous control to date.
Implications of all the available evidence
The current study envisages a specific metabolomic signature associated with the spontaneous loss of virological control in elite controllers. This metabolic profile was characterized by immunometabolism deregulation. All these observed metabolic differences can not only be used as potential biomarkers for a rapid screening of the loss of spontaneous virological control but can also be suggested as susceptible targets for the design of immunotherapeutic strategies in order to achieve the long-term HIV remission without treatment.
Alt-text: Unlabelled Box
1. Introduction
Elite controllers (ECs) are rare individuals who are able to naturally control viral load (VL) below the detection limit in the absence of combined antiretroviral therapy (cART) [1]. It has been suggested that ECs hold the key to how a functional HIV cure can be reached [2]. In-depth characterization of these patients has identified multiple immunological mechanisms involved in the controller phenomenon including host genetic factors, such as HLA-B*57 prevalence [3], a distinct and effective HIV-1-specific T-cell response mediated by high cytokine and chemokine production [4,5] and a remarkably broad array of HLA class II molecules [6].
Several observations have confirmed that ECs are heterogeneous from a clinical point of view and with regard to both immunological [[7], [8], [9]] and virological features, including a variable proportion of ECs who lose HIV control over time [10,11]. That heterogeneity also reflects that no single mechanism is responsible for controlling viral replication. In fact, recent findings have identified persistent controllers (PCs), compared to subjects who eventually lose viral control, as subjects with low viral diversity, low HIV-DNA levels, decreased immune activation and proinflammatory cytokine levels, efficient high Gag-specific T-cell polyfunctionality and a proteomic profile characterized by lower levels of inflammation, transendothelial migration and coagulation [[12], [13], [14], [15]]. PCs may help to define the right model of functional cure and may be a good example for the design of eradication strategies and HIV vaccine development.
New studies of immune system metabolism (“immunometabolism”) have tried to explore the role of metabolic pathways within immune cells and how this interplay regulates immune response outcomes. It is becoming apparent that changes in the metabolomic profile of cells upon viral infection are important for viral replication and importantly for the seeding of HIV reservoir in CD4+ T-cells [[16], [17], [18]]. The measurement of specific metabolic products in plasma has revealed differences in the phenotypes of HIV-infected individuals and identified biomarkers associated with HIV natural evolution [[19], [20], [21]] that may participate in viral containment together with distinct immune features. While not much is known in HIV-1 controllers, a distinct tryptophan catabolic feature may explain the persistence of the functional T-cell response [22,23]. However, to date, no longitudinal study determining the metabolomic profile associated with the loss of spontaneous HIV-1 elite control has been performed, and there is no evidence of an interplay between the metabolic signature and immune outcomes in these patients.
Given the importance of immunometabolism in HIV-1 infection and the lack of metabolomic information in this heterogeneous group of HIV-infected patients, the performance of metabolomic approaches could reveal important metabolic differences between persistent and transient elite controllers, which could lead to new insights into future designs of effective vaccines and cure strategies using persistent spontaneous control as a model. Therefore, the purpose of this study was to perform targeted and untargeted metabolomic strategies in order to study the underlying mechanisms associated with the loss of spontaneous virological control in ECs and to identify potential predictive biomarkers that could explain the inefficient HIV-specific T-cell response observed in transient controllers.
2. Methods
2.1. Patients and study design
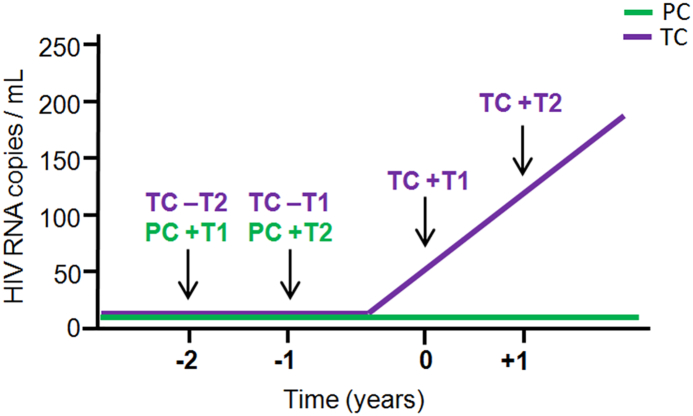
ECs were defined as subjects with VL determinations below the detection limit (<50 HIV-1-RNA copies/mL) in the absence of cART for at least 12 months. A total of 16 subjects were included in the study based on frozen plasma and peripheral blood mononuclear cell (PBMC) sample availability according to a similar study design that was previously described [12,13] (Fig. 1). The samples were received, processed and stored in the Spanish HIV HGM BioBank belonging to the AIDS Research Network (RIS) [24], and data were registered in the RIS cohort of HIV Controllers Study Group (ECRIS) (Annex I, Supplementary Material). A detailed description of the cohort's characteristics has been previously published [11,25]. The 16 subjects were analysed; eight ECs who experienced loss of spontaneous virological HIV-1 control (at least two consecutive measurements of VL above the detection limit in 12 months) were called transient controllers “TCs”, and another group of eight ECs who persistently maintained virological control during the same follow-up period were called persistent controllers “PCs” (see the study design in Fig. 1). Up to four determinations were assessed in TCs: at two years and one year before the loss of virological control, “pre-loss of control period”, (−T2 and –T1 respectively), and two determinations in the “post-loss of control period”, including the closest time point and one year after the loss of virological control (+T1 and + T2, respectively). In PCs, up to two determinations were assessed at one-year intervals (+T1 and +T2).
Fig. 1.
Schematic representation of the study design and follow-up time points determined in transient controllers (TCs) and in persistent controllers (PCs).
None of the subjects took drugs with known metabolic effects (such as lipid-lowering agents or antidiabetics agents, among others). As an example, it has been well described the immunomudulatory and anti-inflammatory effects of statins in infectious disease [26].Therefore, its use may bias immune response and lipidomics results. For this reason, its use was considered within the exclusion criteria. All subjects included in the study gave their informed consent, and protocols were approved by the institutional ethical committees.
Laboratory evaluations were performed at the Laboratory of Joan XXIII University Hospital in Tarragona, IISPV, Rovira i Virgili University (Spain) and at the Laboratory of Immunovirology, Institute of Biomedicine of Seville (IBiS), Virgen del Rocío University Hospital in Seville (Spain).
2.2. Laboratory determinations
Absolute counts of CD4+ and CD8+ T-cells were determined in fresh whole blood by using an Epic XL-MCL flow cytometer (Beckman-Coulter, Brea, California) according to the manufacturers' instructions. Plasma HIV-1 RNA concentration was measured by quantitative polymerase chain reaction (COBAS Ampliprep/COBAS Taqman HIV-1 test, Roche Molecular Systems, Basel, Switzerland) according to the manufacturer's protocol. The detection limit for this assay was 50 HIV-1-RNA copies/mL. Hepatitis C virus (HCV) RNA was determined using an available PCR procedure Kit (COBAS Amplicor, Roche Diagnosis, Barcelona, Spain) with a detection limit of 10 IU/mL.
Total DNA was extracted from peripheral blood mononuclear cells (PBMCs) using the MagNa Pure LC DNA isolation kit. HLA-B group alleles were genotyped using a reverse sequence-specific oligonucleotide bound to a fluorescently coded microsphere system (LABType SSO, RSSO1B, One Lambda, Canoga Park, CA), following manufacturer's instructions.
2.3. Determination of the Krebs cycle metabolites and energy metabolism by GC-(EI)qTOF/MS
Metabolite extraction from plasma samples was performed by a protein precipitation method with methanol/water 8:1 (v/v) containing an internal standard mixture. Supernatants were collected and evaporated to dryness under N2 stream and freeze dried before derivatization with methoxyamine hydrochloride and MSTFA +1% TMCS. Samples were analysed on a 7200 GC-QTOF Gas chromatograph-quadrupole time of flight mass spectrometer (GC-qTOF) from Agilent Technologies (Sta. Clara, CA, USA) using a HP5-MS chromatographic column from J&W Scientific (30 m × 0·25 mm i.d., 0·25 μm film) and helium (99·999% purity) as a carrier gas. Ionization was performed by electronic impact (EI), with an electron energy of 70 eV, and the mass analyzer was operated in Full Scan mode, recording data in a range between 35 and 700 m/z at a scan rate of 5 spec/s [27,28].
Absolute quantification of the Krebs cycle metabolites was performed with an internal standard calibration curve using the corresponding analytical standard for each determined metabolite and a deuterated internal standard depending on the family of metabolite. In addition, an untargeted approach was performed by deconvoluting the acquired raw data by Unknown Analysis software from Agilent and using Fiehn RT library to identify by EI-MS spectra and library retention time the compounds present in the sample. For these compounds, their peak areas were used for relative quantification between samples.
2.4. Liposcale test
We measured triglyceride and cholesterol concentrations, the size and particle numbers for very-low-density lipoprotein (VLDL), low-density lipoprotein (LDL) and high-density lipoprotein (HDL) classes, and the particle numbers of nine subclasses: large, medium and small VLDLs, LDLs and HDLs of frozen samples using the Liposcale test as described elsewhere [[29], [30], [31]].
2.5. Lipidomic analysis by LC-qTOF/MS
Lipid extraction from plasma samples was carried out by a liquid-liquid extraction using methyl-tert-butyl ether/methanol/water. Then, the upper organic layer was collected, evaporated to dryness in a SpeedVac concentrator and reconstituted with methanol:toluene (9:1, v,v) for analysis by UHPLC-(ESI)qTOF.
Samples were analysed on an Agilent 1290 Infinity UHPLC coupled to an Agilent 6550 qTOF mass spectrometer. The chromatographic column used was a Kinetex C18-EVO (2·1 × 100 mm, 2 · 7 μm) from Phenomenex, and the mobile phase was 2-propanol/methanol/water (5:4:1) and 2-propanol/water (99:1), both with 5 mM NH4OAc and 0·1% acetic acid. The mass spectrometry analysis was performed both in positive and negative ionization in separated runs operating in full scan mode between 300 and 1200 m/z at 2 spectra/s. Additionally, tandem mass analysis (MS/MS) was performed at a fixed collision energy of 20 eV for a data dependence analysis (DDA) using the 10 most abundant precursor ions with a dynamic exclusion of 30 s for lipid identification.
Data analysis was performed using Mass Profinder software from Agilent Technologies for raw data signal deconvolution using recursive feature extraction algorithm for these features previously identified with SimLipid software (PREMIER Biosoft), ((http://www.lipidmaps.org/). The generated features were aligned across all samples to create a compound matrix to perform statistical analyses.
2.6. Cell stimulation and intracellular cytokine staining
The stimulation assay and intracellular cytokine staining analysis were evaluated previously in a study with a different objective [12].
Briefly, PBMCs were thawed, washed and in vitro stimulated with 2 μg/mL of overlapped HIV (Gag)-specific peptide pool (NIH AIDS Research and Referenced Reagent Program (https:// www.aidsreagent.org/index.cfm) and stained with conjugated monoclonal anti-CD107a-BV786 (clone H4A3; BD Biosciences, Franklin Lakes, NJ) at the beginning of incubation as previously described [4,12].
Stimulated PBMCs were washed and stained with LIVE/DEAD fixable Violet Dead Cell Stain (Life Technologies, CA, USA). Then, cells were surface stained with anti-CD14-PB, anti-CD19-PB, anti-CD56-PB (Biolegend, San Diego, CA), anti-CD8+-PerCP-Cy5.5, anti-CD45RA-FITC, anti-CD27-BV605 and anti-CD57-PE-CF595 (BD Biosciences). Cells were then stained intracellularly for 30 min with 100 μl of PBS with anti-CD3-APC-H7, anti-IFN-γ-PCy7, anti-TNF-α-Alexa700, anti-IL-2-APC and anti-Perforin-PE (BD Biosciences) and then washed twice and fixed in PBS containing 4% paraformaldehyde. Unstimulated condition and cell stimulation with staphylococcal enterotoxin B (SEB) condition as a positive control were included in each experiment. Alive lymphocytes were defined as low forward/side scatter, no violet dead cell staining and expressed CD3, and/or no CD8+ but not CD19, CD14, and CD56 (Supplementary Fig. 1). PBMCs were analysed by using an LSR Fortessa Cell Analyzer (BD Biosciences, Spain). A minimum of 1,500,000 total events were recorded for each panel and condition.
2.7. Statistical analysis
All variables were considered non-parametric due to the sample size. Differences between categorical values were determined by the Chi-square test. Differences between unpaired groups were determined by univariate comparisons through the non-parametric Mann-Whitney U test. Correlations between variables were assessed using the Spearman rank test. p values <0·05 were considered statistically significant. Additionally, the fold change (FC) of each variable was calculated as ‘A/B', where ‘A' was the median value of the TC group pre-loss time points, and ‘B' was median value of the PC group. Alternatively, ‘A' was the median value of the post-loss time points in TCs, and ‘B‘was the median value of pre-loss time points in TCs. The results were represented with heat maps and hierarchical clustering analysis (HCA).
In PCs, a PCA analysis and Wilcoxon signed-rank test were applied in paired samples to assess metabolite changes as an effect of time. As shown in Supplementary Fig. 2, no changes were observed over time in PCs. Thus, we simplified the following analysis to a single variable, which was expressed as the mean value of the two consecutive longitudinal determinations in PCs. We then focused the analysis on pre-loss of control time points from TCs compared to PCs in order to identify potential biomarkers for the loss of spontaneous HIV-1 control. To simplify, for all the comparisons, we confirmed that no differences were observed between –T1 and –T2, and +T1 and +T2 (data not shown), and the mean values of the –T1 and –T2 time points (pre-loss time point) and +T1 and +T2 time points (post-loss time point) were calculated.
Multivariate statistics were also used to improve the refining and distilling of all the metabolomics data and for pattern recognition purposes. Thus, principal component analysis (PCA) was performed. Moreover, random forest analysis receiver operating characteristic (ROC) curves were also generated to identify the variables that make the largest contributions to the discrimination between groups.
Polyfunctionality was defined as the percentage of lymphocytes producing multiple cytokines. Polyfunctionality was quantified with the polyfunctionality index algorithm [32] employing the 0.1.2 beta version of the “FunkyCells Boolean Dataminer” software (www.FunkyCells.com) provided by Dr. Martin Larson (INSERM U1135, Paris, France).
The statistical software used included the programme ‘R', version 3.4.4 (http://cran.r-project.org) and the SPSS 23.0 package (IBM, Madrid, Spain). Graphs were generated with Prism, version 5.0 (GraphPad Software, Inc.).
3. Results
3.1. Characteristics of the studied subjects
Subjects were recruited based on samples availability based on the study design shown in Fig. 1. The clinical and demographic characteristics of the subjects at baseline (first follow-up time point) were defined. As shown in Table 1, no differences were observed in age, sex, transmission route, HCV coinfection, CD4+ and CD8+ T-cell counts and the CD4:CD8 ratio between TCs and PCs. The TC group presented a shorter time since diagnosis than the PC group (median [interquartile range] of 3 [2–8] vs 13 [10–17] years; p = 0·005). The VL evolution from TCs after the loss of control was 539 [295–1120] at +T1 and 2740 [985–22,250] HIV-RNA copies/mL at +T2. No differences were found in HLA-B57, 27 and 35 frequencies between groups (p >0.05).
Table 1.
Patients' characteristics.
| Transient controllers |
Persistent controllers |
p-Value | |
|---|---|---|---|
| TC (n = 8) | PC (n = 8) | ||
| Age (years) | 41 [34–60] | 44 [40–46] | 0·635 |
| Male sex, n (%) | 5 (62.5) | 4 (50) | 0·614 |
| IDU, n (%) | 3 (37.5) | 4 (50) | 0·198 |
| Time since diagnosis (years) | 3 [2–8] | 13 [10–17] | 0·005 |
| HCV RNA detected, n (%) | 3 (37·5) | 3 (37·5) | 0·999 |
| CD4+ T-cells (cell/μL) | 676 [623–963] | 724 [609–985] | 0·817 |
| CD8+ T-cells (cell/μL) | 787 [553–1162] | 636 [432–1026] | 0·482 |
| CD4:CD8 Ratio | 0.86 [0·53–1·55] | 1.08 [0·93–1·47] | 0·406 |
Values from TC are taken from –T2 and values from PC are taken from the first follow-up time point. Values are given as percentage for categorical variables or median and interquartile range for continous variables. The Mann-Whitney U and Chi-squared tests were used. All p values <0·05 were considered significant and are highlighted in bold. IDU, Intravenous drug users.
3.2. A specific metabolomic signature precedes the loss of spontaneous HIV-1 control in transient controllers
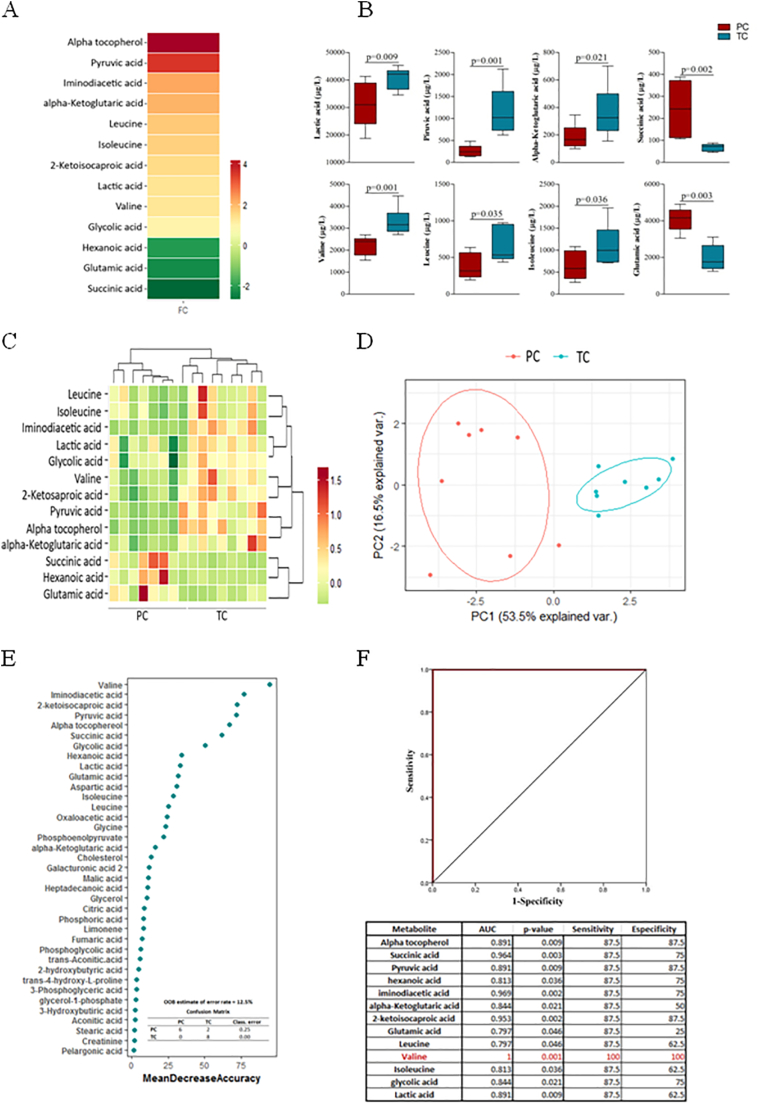
A total of 70 metabolites were identified and quantified by GC-QTOF/MS in plasma samples. When we focused the analysis on pre-loss of control time from TCs compared to PCs in order to identify the metabolomic signature associated with the loss of spontaneous HIV-1 control, only the plasma levels of thirteen metabolites were significantly different (Fig. 2A). Fig. 2B shows the data distribution of some of these 13 significantly metabolites. In addition, these 13 metabolites showed good clustering and reliable differentiation between the two studied groups (Fig. 2C–D).
Fig. 2.
Metabolomic analysis comparing PCs and TCs before the loss of natural HIV-1 control. Heat map representation of the fold change of each statistically significant metabolite, the scale from green (low abundance) to red (high abundance) represents the normalized abundance in arbitrary unit (A). Box-plots graph of some of the 13 significantly metabolites Hierarchical combined tree showing the clusterization of metabolites (C) and Principal Component Analysis (PCA) showing that these 13 metabolites allow the differentiation between the studied groups (D), TCs (blue, n = 8) and PCs (red, n = 8). Random Forest analysis showed metabolites by importance of classification (E). Logistic regression and receiver operator characteristic (ROC) curves elucidated the amino acid valine as the main differentiator between PCs and TCs before the loss of control (F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A Random Forest analysis (out-of-bag (OOB) error = 12·5%) was then performed in order to select the best parameter for group discrimination. As shown in Fig. 2E, valine, iminodiacetic acid, 2-ketoisocaproic acid, pyruvic acid, alpha tocopherol, succinic acid and glycolic acid showed higher classification power (mean decreased accuracy (MDA) > 50). Finally, receiver operating characteristic (ROC) curves demonstrated that, among the 13 significant metabolites, valine was the only metabolite that could discriminate TCs from PCs with 100% sensitivity and specificity (Fig. 2F).
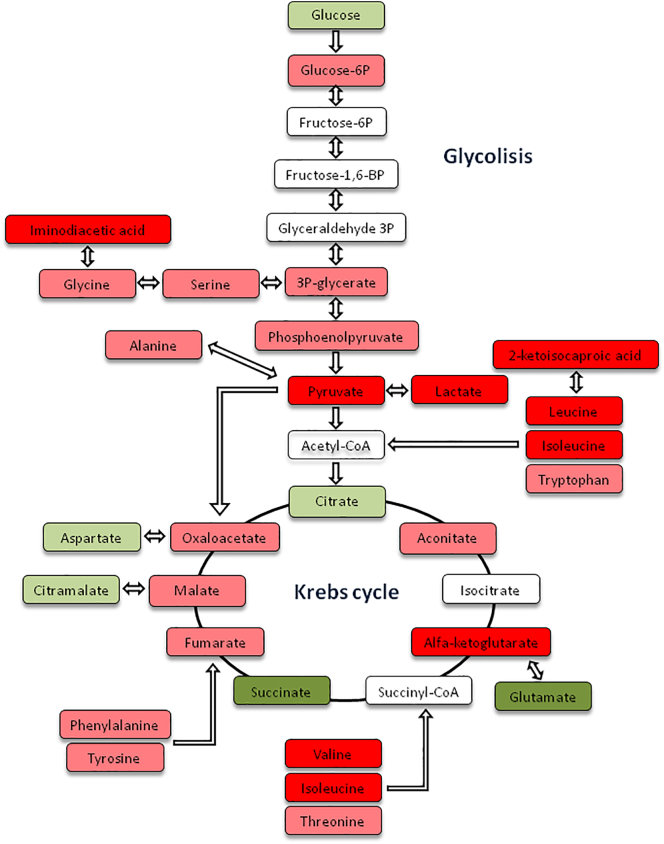
A metabolic pathway analysis revealed that the most significant deregulated metabolites were mainly involved in glycolysis, the Krebs cycle and amino acid metabolism. A schematic representation including the statistically significant metabolites between groups and the rest of metabolites involved in the pathway are shown in Fig. 3.
Fig. 3.
A schematic representation of the relevant deregulated metabolic pathways involved in the loss of virological control. Metabolites observed at lower levels in TCs compared with PC are shown in green (dark green p < 0·05, light green p > 0·05); increased levels of metabolites observed in TCs are shown in red (dark red p < 0·05, light red p > 0·05); and undetected metabolites are shown in white. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Finally, regarding changes in TC metabolites as an effect of detectable viremia, only monostearin levels were different between TCs at pre-loss and post-loss time points, being significantly lower after the loss of control (Supplementary Fig. 3).
3.3. Gag-specific CD8+ T-cells response is associated with metabolite levels
In a previous work [12], we showed that PCs presented higher levels of Gag-specific CD4+ and CD8+ T-cells than TCs before the loss of control, as determined by the intracellular production of IFN-γ, TNF-α and IL-2 by T cells. Polyfunctionality, which is understood as simultaneous multiple production of these cytokines by T cells, was studied only in subjects categorized as responders and calculated by the index of polyfunctionality (pINDEX). T-cell response was defined as the frequency of cells (>0·05%) with detectable IFN-γ, TNF-α and/or IL-2 intracellular cytokine production after stimulation. Due to the low number of Gag-specific CD4+ T-cell responders among TCs, polyfunctionality was only assessed in responders with CD8+ T-cells.
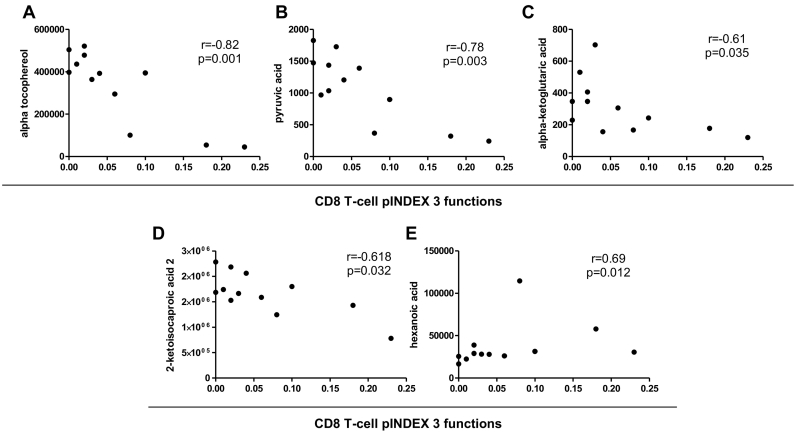
To associate immunological changes with the most significant metabolic results, three-function CD8+ T-cell polyfunctionality (calculated by the pINDEX) from PCs and TCs before the loss of control was strongly inversely correlated with alpha tocopherol, pyruvic acid, alpha-ketoglutaric acid, and 2-ketoisocaproic acid plasma levels (r = −0·82, p = 0·001; r = −0·78, p = 0·003; r = −0·61, p = 0·035; and r = −0·618, p = 0·032, respectively) and directly associated with hexanoic acid (r = 0·69, p = 0·012) (Fig. 4A–E). The same associations with those metabolites remained significant with 4- and 5-function total CD8+ T-cell polyfunctionality (cytokine combinations plus CD107a and plus perforin, respectively; data not shown).
Fig. 4.
Correlation of plasma levels of metabolites with CD8+ T-cell polyfunctionality. Correlations between alpha tocopherol (A), pyruvic acid (B), alpha-ketoglutaric acid (C), 2-ketoisocaproic acid 2 (D), hexanoic acid (E) levels and Gag-specific total CD8+ T-cells expressing combinations of IFN-γ, TNF-α, IL-2 (pINDEX 3-functions) from PCs and TCs before the loss of virological control (n = 12 in each analysis). The Spearman ρ correlation coefficient test was used.
3.4. Plasma lipid analysis comparing persistent controllers and transient controllers and changes in transient controllers due to the loss of control
Using the Liposcale test we found no significant differences in triglycerides, cholesterol and lipoproteins comparing PCs and TCs before the loss of control. Regarding paired samples from TCs, LDL-cholesterol, the number of medium LDL particles and non-HDL particles and the ratio of LDL particles/HDL particles were increased after the loss of control (p = 0·039, p = 0·007, p = 0·039 and p = 0·015 respectively).
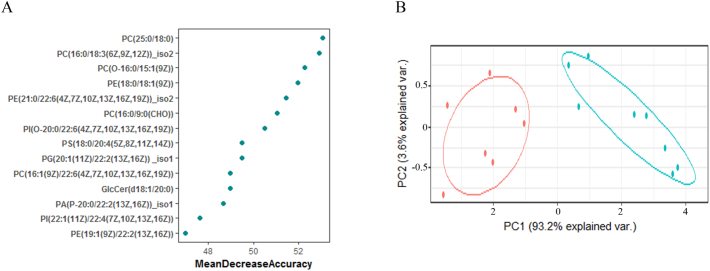
On the other hand, similar to the metabolomic approach described before, we also analysed differences in the plasma lipidome from PCs and TCs before the loss of control. A total of 334 lipids were detected in plasma, but only 93 were significantly different between the two groups (Supplementary Table 1). We then ordered these significant lipids according to their classification power using a Random Forest analysis (OOB error = 6·25%). The results demonstrated that only 7 lipids performed with an MDA higher than 50 (Fig. 5A). These plasma lipid levels were significantly lower in TCs before the loss of control than in PCs, but, more importantly, as shown by the PCA analysis (Fig. 5B), these lipids had great discriminatory potential between the studied groups.
Fig. 5.
Lipidomic analysis comparing TCs before the loss of HIV-1 control and PCs. Random Forest analysis showed lipid levels by importance of classification (A). Principal Component Analysis (PCA) showing that lipids with a mean decrease in accuracy higher than 50 allowed a great differentiation between the studied groups (B). Transient controllers (TCs, blue, n = 8) and persistent controllers, (PCs, red, n = 8). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Regarding changes in TC plasma lipids as an effect of viral loss of control, 43 lipids were significantly different after the loss of control compared with pre-loss time points in TCs; most of them decreased after the loss of virological control (Supplementary Fig. 4).
3.5. Lipid levels are associated with Gag-specific CD8+ T-cell response and metabolite plasma levels
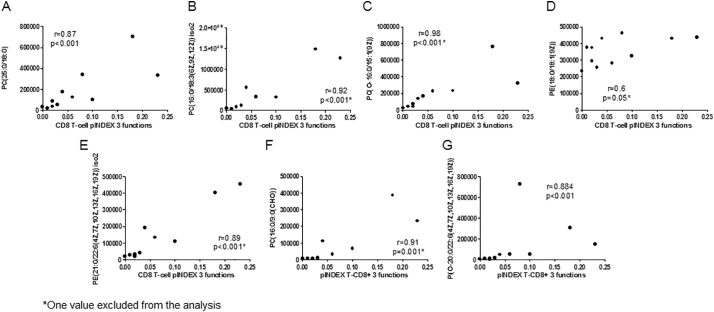
To identify the associations between lipids and Gag-specific CD8+ T-cell response, we focused on the most significant lipids associated with the loss of control mentioned before. All of these plasma lipid levels were higher in PCs than in TCs before the loss of control, and interestingly, these levels were strongly and directly associated with the three-function CD8+ T-cell pINDEX (n = 11) (p < 0·001 for all associations) (Fig. 6A–G). The same associations with these lipids remained significant with 4- and 5-function total CD8+ T-cell polyfunctionality (plus CD107a and plus perforin, respectively) (Supplementary Fig. 5 A–H).
Fig. 6.
PC(25:0/18:0), PC(16:0/18:3(6Z,9Z,12Z)) iso2, PC(O-16:0/15:1(9Z)) and PE(21:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) iso2 plasma levels correlations with Gag-specific CD8+ T-cell pINDEX 3-function from PCs and TCs before the loss of control (n = 11). An outlier (*) excluded from the analysis (n = 10). The Spearman ρ correlation coefficient test was used.
Correlations between the most relevant lipids and metabolites associated with the loss of control are represented in Supplementary Fig. 6. Lipid and metabolite levels were strongly associated in PCs and TCs before the loss of control (n = 15).
4. Discussion
The current study elucidated a metabolomic signature associated with the spontaneous persistent elite control of HIV-1 infection and that distinguish the transient controllers from persistent controllers. There were strong interrelations among metabolites, lipids levels and an important immune function parameter associated with spontaneous control of HIV as is HIV-specific T-cell response. Moreover, the amino acid valine stood out as the main differentiating factor between the studied groups.
Among the observed metabolomic changes, the most significant differences were related to metabolic pathways as important as glycolysis, the Krebs cycle, and amino acid catabolism, mostly that of branched chain amino acids. Previous studies have demonstrated that all these pathways are targeted during viral infection [16,33], but so far these metabolic disturbances in energy metabolism have not been described in HIV-1 elite controllers.
Immunometabolism is now an important field in immunology. Microbial infections results in important changes to the physiology and metabolism of innate immunity host cells, such as macrophages and T-cells. In this sense, differences in metabolite plasma levels may reflect changes in cellular metabolic flux and could be also related to the outcome of the infectious process [34,35].
T-cell activation, T-cell mediated antiviral responses and other T-cell effector functions require significant energetic and biosynthetic requirements from the cell [36,37]. During HIV infection, energy demand increases dramatically, which is translated as a decrease in circulating glucose levels (the main cellular energy source) and activation of the glycolysis pathway [37,38]. This is exactly what we observed in TCs before the loss of control: a lower glucose concentration and a significant increase in glycolytic intermediates, such as 3P-glycerate, phosphoenolpyruvate and pyruvate, compared with PCs. Some studies have proposed that the accumulation of glycolytic intermediates is used in activated T-cells for biosynthetic processes [39]. Moreover, our previous results demonstrated that TCs are characterized by high immune activation and a distinct inflammatory profile [12,13], which can be explained by the observed metabolomic differences. Recent studies have also highlighted the role of glycolytic enzymes and intermediates in regulating T-cell cytokine production, especially IFN-γ, which reinforces our results [40,41]. Moreover, regarding HIV-1 infection, glucose metabolism provides precursors for HIV biosynthesis and intermediate enzymes that may disengage glycolysis and regulate HIV reactivation [42]. Importantly, a recent study has highlighted the important link between increased glycolysis and HIV-1 reservoir seeding in CD4+ T-cells [17]. This is in accordance with our data in TC who also showed higher proviral DNA levels [12].
Furthermore, activated T cells undergo a metabolic reprogramming; instead of relying on mitochondrial oxidative phosphorylation to generate the energy needed for cellular process, they enhance aerobic glycolysis, a phenomenon commonly known as the Warburg effect [43]. As a result, activated lymphocytes increase lactate production even in the presence of oxygen [44,45]. This is exactly what we observed in TCs at pre-loss time points.
Other studies have proposed lactic acidemia or hyperlactatemia as an indicator of mitochondrial oxidative deregulation [[46], [47], [48]]. In this sense, our results showed a general deregulation of the Krebs cycle metabolites in TCs before the loss of control. Actually, only the increased Krebs cycle intermediates in TCs before the loss of control compared to PCs are those that can be supplied by anaplerotic reactions. The most remarkable example is α-ketoglutarate concentration, which was increased in TCs. This could be directly related to the significant decrease in glutamate levels. When energy levels are reduced, glutamate can be converted into α-ketoglutarate and can enter the Krebs cycle for energy production, trying to fully compensate for the lack of oxidative Krebs cycle activity. Moreover, another indicator of defective mitochondrial function was the significant decrease in succinate levels in TCs [[49], [50], [51]], reinforcing the key role of mitochondria in the progression of HIV infection [52,53].
As mentioned before, our results suggested that valine is the main differentiating factor between PCs and TCs. Valine is one of a group of the three essential amino acids with crucial roles in metabolism, called branched-chain amino acids (BCAAs: leucine, isoleucine and valine) [54]. Our results demonstrated a significant increase in this type of amino acids and in some of their catabolic intermediates, such as α-ketoisocaproic acid, in TCs before the loss of control compared to PCs. In this sense, some previous studies have linked high concentrations of BCAAs with deficiencies in its catabolism [55]. Thus, since the catabolic products of BCAAs could enter the Krebs cycle for energy generation, the accumulation of these products may also be related to activated T-cell reprogramming into glycolytic metabolism, reducing the oxidative phosphorylation pathways [43,56,57].
High concentrations of BCAAs also promote oxidative stress and, therefore, mitochondrial damage and dysfunction [58]. α-tocopherol, commonly known as vitamin E, plays an important role as an antioxidant agent and has been used in HIV-infected patients as a supplement to reduce oxidative stress [59]. Our results demonstrated that TCs have higher levels of vitamin E that may mirror a compensatory mechanism.
The changes in mitochondrial dynamics in TCs presented herein also explain the differences observed in lipid levels. Mitochondria have an important role in the maintenance of lipid homeostasis by orchestrating the synthesis of key membrane phospholipids [60]. Moreover, phospholipids are the main building blocks of cell membranes; thus, changes in phospholipid composition could affect mitochondrial function, structure and biogenesis and rely on the metabolism of phospholipids themselves [61]. Therefore, it is not surprising that phospholipid alterations have been associated with several diseases, including HIV infection [61,62]. During infection, innate immune cells suffer from significant remodelling of their lipid compartments, including the plasma membrane [63]. Moreover, lipid synthesis is critical in effector T-cell differentiation [64] and secondary responses of memory CD8+ T-cells [65]. Interestingly, in Pernas et al., we observed that the effective Gag-specific response from PCs was mediated mainly by the central memory CD8+ T-cell subset [12]. This may explain the associations between lipid plasma levels and Gag-specific response that we have shown in this work. The specific role of each lipid subclasses on immune cells is uncertain and may vary according to lipid subtypes, but new evidence is emerging in the scenario of HIV infection [[66], [67], [68]]. Modulation of fatty acid synthesis and oxidation pathways could be used as strategies to prevent inflammatory T-cell differentiation.
The main limitation of our study is the small sample size of our study population. However, as mentioned in previous our published works using this cohort [12,13], these types of patients are rare, and it is difficult to perform a long follow-up with stored available samples. For this reason and to try to overcome this constraint, we have used a highly sensitive mass spectrometry approach. Moreover, the shorter time for diagnosis observed in TCs may be considered an inherent characteristic of this group because of the faster loss of EC status.
In conclusion, this study envisages a specific metabolomic profile associated with the spontaneous loss of virological control in ECs. This profile was characterized by T-cell metabolic reprogramming to the aerobic glycolytic pathway and by a decrease in mitochondrial function and increased oxidative stress and immunological activation. Metabolite and lipid plasma levels were also strongly correlated with immunological parameters. Moreover, valine could be considered a potential biomarker for the prediction of virological progression in elite controllers. Therefore, all these observed metabolic differences can not only be used as biomarkers for rapid screening of future loss of spontaneous control but can also be suggested as therapeutic targets in HIV-1 infection. New strategies focused on improving metabolic and inflammatory conditions in HIV-1 patients are needed in order to enhance protective immunity. In this sense, the use of statin and aspirin are being use in clinical trials (NCT02081638), and in addition antioxidant vitamins, amino acids or other dietary supplements have started to be widespread in the HIV-infected community [18]. Based on our results, metformin and thiazolidinediones, widely used to treat type 2 diabetes, could be potential therapies given that it has been demonstrated that they improve glucose metabolism [69]. Another therapeutic option could be the use of antioxidants, such us vitamin E mentioned before [59]. Moreover, some studies are evaluating the potential benefit of niacin or vitamin B3 in reducing immune activation, another important point during HIV-1 infection [70]. However, further external validation studies with larger cohorts are needed to consolidate our results in order to identify the persistent controller phenotype and to pinpoint susceptible targets for the design of immunotherapeutic strategies with the aim of achieving long-term HIV remission in the absence of ART.
Acknowledgments
Acknowledgements
Authors would like to thank Y. Milanés-Guisado for the help on graphic design and statistical analysis.
Funding sources
This work was supported by grants from the Fondo de Investigación Sanitaria, Instituto de Salud CarlosIII, Fondos Europeos para el Desarrollo Regional, FEDER (grant numbers PI10/02635, PI13/00796, and PI16/00503 to FV and PI12/02283, PI16/00684, and CPII014/00025 to ER.-M.; FI14/00431 to LT-D.; FI17/00186 to MR-JL); Programa de Suport als Grups de Recerca (grant numbers 2017SGR948 and 2014SGR250); the Gilead Fellowship Program (grant numbers GLD14/293 and GLD17/00299); the Red de Investigación en Sida (grant numbers RD12/0017/0005, RD16/0025/0006, RD12/0017/0029, RD16/0025/0020). ER-M. is supported by Consejería de Salud y Bienestar Social of Junta de Andalucía through the Nicolás Monardes Program (C-0032/17). FV is supported by a grant from the Programa de Intensificación de Investigadores, Instituto de Salud Carlos III (grant numbers INT11/240, INT12/282, and INT15/226). AR is supported by a grant from the Acció Instrumental d'incorporació de científics i tecnòlegs (PERIS SLT002/16/00101), Departament de Salut, Generalitat de Catalunya. The funders had no role in study design, data collection, data analysis, interpretation, writing of the report.
Declaration of interests
The authors declare no competing interests.
Author contributions
FV and ER-M conceived and coordinated the study development. LT-D, ER-G and AR designed and performed experiments, analysed and interpreted the data, designed the figures and wrote the manuscript.VA and PH contributed in sample preparation and in mass spectrometry analysis. MR-JL participated in the methodology and data analysis. JP, CV, IP and ML collaborated with patient's characterization and sample collection. ER-M and FV participated in critical revisions for intellectual content. All authors critically reviewed, edited and approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.004.
Contributor Information
Ezequiel Ruiz-Mateos, Email: eruizmateos-ibis@us.es.
Francesc Vidal, Email: fvidalmarsal.hj23.ics@gencat.cat.
Appendix A. Supplementary data
Supplementary material
References
- 1.Deeks S.G., Walker B.D. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27(3):406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Shasha D., Walker B.D. Lessons to be learned from natural control of HIV - future directions, therapeutic, and preventive implications. Front Immunol. 2013;4:1–8. doi: 10.3389/fimmu.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Migueles S.A., Sabbaghian M.S., Shupert W.L., Bettinotti M.P., Marincola F.M., Martino L. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci. 2000;97(6):2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrando-Martinez S., Casazza J.P., Leal M., Machmach K., Munoz-Fernandez M.A., Viciana P. Differential gag-specific polyfunctional T cell maturation patterns in HIV-1 elite controllers. J Virol. 2012;86(7):3667–3674. doi: 10.1128/JVI.07034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez-Cirion A., Sinet M., Shin S.Y., Urrutia A., Versmisse P., Lacabaratz C. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with gag-specific CD8 T cell responses. J Immunol. 2009;182(12):7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 6.Galperin M., Farenc C., Mukhopadhyay M., Jayasinghe D., Decroos A., Benati D. CD4+ T cell-mediated HLA class II cross-restriction in HIV controllers. Sci Immunol. 2018;3(24):1–13. doi: 10.1126/sciimmunol.aat0687. [DOI] [PubMed] [Google Scholar]
- 7.Pereyra F., Addo M.M., Kaufmann D.E., Liu Y., Miura T., Rathod A. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197(4):563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 8.Boufassa F., Saez-Cirion A., Lechenadec J., Zucman D., Avettand-Fenoel V., Venet A. CD4 dynamics over a 15 year-period among HIV controllers enrolled in the ANRS French Observatory. PLoS One. 2011;6(4):1–7. doi: 10.1371/journal.pone.0018726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benito J.M., Ortiz M.C., León A., Sarabia L.A., Ligos J.M., Montoya M. Class-modeling analysis reveals T-cell homeostasis disturbances involved in loss of immune control in elite controllers. BMC Med. 2018;16(1):30. doi: 10.1186/s12916-018-1026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noel N., Lerolle N., Lécuroux C., Goujard C., Venet A., Saez-Cirion A. Immunologic and virologic progression in HIV controllers: the role of viral “blips” and immune activation in the ANRS CO21 CODEX study. PLoS One. 2015;10(7):1–11. doi: 10.1371/journal.pone.0131922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leon a Perez I., Ruiz-Mateos E., Benito J.M., Leal M., Lopez-Galindez C. Rate and predictors of progression in elite and viremic HIV-1 controllers. AIDS. 2016;5:1209–1220. doi: 10.1097/QAD.0000000000001050. [DOI] [PubMed] [Google Scholar]
- 12.Pernas M., Tarancon-Diez L., Rodriguez-Gallego E., Gomez J., Prado J.G., Casado C. Factors leading to the loss of natural elite control of HIV-1 infection. J Virol. 2018;1 doi: 10.1128/JVI.01805-17. [e01805-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Gallego E., Tarancón-Diez L., García F., del Romero J., Benito J.M., Alba V. Proteomic profile associated with loss of spontaneous human immunodeficiency virus type 1 elite control. J Infect Dis. 2019;219(6):867–876. doi: 10.1093/infdis/jiy599. [DOI] [PubMed] [Google Scholar]
- 14.El-Far M., Kouassi P., Sylla M., Zhang Y., Fouda A., Fabre T. Proinflammatory isoforms of IL-32 as novel and robust biomarkers for control failure in HIV-infected slow progressors. Sci Rep. 2016;6 doi: 10.1038/srep22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avettand-Fenoel V., Bayan T., Gardiennet E., Boufassa F., Lopez P., Lecuroux C. Dynamics in HIV-DNA levels over time in HIV controllers. J Int AIDS Soc. 2019;22(1) doi: 10.1002/jia2.25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollenbaugh J.A., Munger J., Kim B. Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC-MS/MS analysis. Virology. 2011;415(2):153–159. doi: 10.1016/j.virol.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valle-Casuso J.C., Angin M., Volant S., Passaes C., Monceaux V., Mikhailova A. Cellular metabolism is a major determinant of HIV-1 reservoir seeding in CD4+ T cells and offers an opportunity to tackle infection. Cell Metab. 2018;29:1–16. doi: 10.1016/j.cmet.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Dagenais-Lussier X., Mouna A., Routy J.P., Tremblay C., Sekaly R.P., El-Far M. Current topics in HIV-1 pathogenesis: the emergence of deregulated immuno-metabolism in HIV-infected subjects. Cytokine Growth Factor Rev. 2015;26(6):603–613. doi: 10.1016/j.cytogfr.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Scarpelini B., Zanoni M., Sucupira M.C.A., Truong H.H.M., Janini L.M.R., Segurado I.D.C. Plasma metabolomics biosignature according to HIV stage of infection, pace of disease progression, viremia level and immunological response to treatment. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0161920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghannoum M.A., Mukherjee P.K., Jurevic R.J., Retuerto M., Brown R.E., Sikaroodi M. Metabolomics reveals differential levels of oral metabolites in HIV-infected patients: toward novel diagnostic targets. OMICS. 2013;17(1):5–15. doi: 10.1089/omi.2011.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F., Zhang S., Jeon R., Vuckovic I., Jiang X., Lerman A. Interferon gamma induces reversible metabolic reprogramming of M1 macrophages to sustain cell viability and pro-inflammatory activity. EBioMedicine. 2018;30:303–316. doi: 10.1016/j.ebiom.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenabian M.A., Patel M., Kema I., Kanagaratham C., Radzioch D., Thébault P. Distinct tryptophan catabolism and Th17/Treg balance in HIV progressors and elite controllers. PLoS One. 2013;8(10):1–13. doi: 10.1371/journal.pone.0078146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dagenais-Lussier X., Aounallah M., Mehraj V., El-Far M., Tremblay C., Sekaly R.-P. Kynurenine reduces memory CD4 T-cell survival by interfering with IL-2 signaling early during HIV-1 infection. J Virol. 2016;90 doi: 10.1128/JVI.00994-16. [JVI.00994-16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Merino I., de las Cuevas N., Jiménez J.L., Gallego J., Gómez C., Prieto C. The Spanish HIV BioBank: a model of cooperative HIV research. Retrovirology. 2009;6(27):1–5. doi: 10.1186/1742-4690-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Molina B., Leon A., Rodriguez C., Benito J.M., Lopez-Galindez C., Garcia F. Analysis of non-AIDS-defining events in HIV controllers. Clin Infect Dis. 2016;62(10):1304–1309. doi: 10.1093/cid/ciw120. [DOI] [PubMed] [Google Scholar]
- 26.Parihar S.P., Guler R., Brombacher F. Statins: a viable candidate for host-directed therapy against infectious diseases. Nat Rev Immunol. 2019;19(2):104–117. doi: 10.1038/s41577-018-0094-3. [DOI] [PubMed] [Google Scholar]
- 27.Riera-Borrull M., Rodríguez-Gallego E., Hernández-Aguilera A., Luciano F., Ras R., Cuyàs E. Exploring the process of energy generation in pathophysiology by targeted metabolomics: performance of a simple and quantitative method. J Am Soc Mass Spectrom. 2016;27(1):168–177. doi: 10.1007/s13361-015-1262-3. [DOI] [PubMed] [Google Scholar]
- 28.Kind T., Wohlgemuth G., Lee D.Y., Lu Y., Palazoglu M., Shahbaz S. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallol R., Amigó N., Rodríguez M.A., Heras M., Vinaixa M., Plana N. Liposcale: a novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J Lipid Res. 2015;56(3):737–746. doi: 10.1194/jlr.D050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Gallego E., Gómez J., Pacheco Y.M., Peraire J., Viladés C., Beltrán-Debón R. A baseline metabolomic signature is associated with immunological CD4+ T-cell recovery after 36 months of ART in HIV-infected patients. AIDS. 2017;32(5):565–573. doi: 10.1097/QAD.0000000000001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Gallego E., Gómez J., Domingo P., Ferrando-Martínez S., Peraire J., Viladés C. Circulating metabolomic profile can predict dyslipidemia in HIV patients undergoing antiretroviral therapy. Atherosclerosis. 2018;273:28–36. doi: 10.1016/j.atherosclerosis.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Larsen M., Sauce D., Arnaud L., Fastenackels S., Appay V., Gorochov G. Evaluating cellular polyfunctionality with a novel polyfunctionality index. PLoS One. 2012;7(7):1–10. doi: 10.1371/journal.pone.0042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Zhu W., Zhang L., Lei H., Wu X., Guo L. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci Rep. 2015;5:8421. doi: 10.1038/srep08421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell D.G., Huang L., VanderVen B.C. Immunometabolism at the interface between macrophages and pathogens. Nat Rev Immunol. 2019 doi: 10.1038/s41577-019-0124-9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neill L.A.J., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fox C.J., Hammerman P.S., Thompson C.B. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5(11):844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 37.Palmer C.S., Ostrowski M., Gouillou M., Tsai L., Yu D., Zhou J. Increased glucose metabolic activity is associated with CD4+T-cell activation and depletion during chronic HIV infection. AIDS. 2014;28(3):297–309. doi: 10.1097/QAD.0000000000000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marko A.J., Miller R.A., Kelman A., Frauwirth K.A. Induction of glucose metabolism in stimulated T lymphocytes is regulated by mitogen-activated protein kinase signaling. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacIver N.J., Michalek R.D., Rathmell J.C. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang C.-H., Curtis J.D., Maggi L.B.J., Faubert B., Villarino A.V., O'Sullivan D. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poznanski S.M., Barra N.G., Ashkar A.A., Schertzer J.D. Immunometabolism of T cells and NK cells: metabolic control of effector and regulatory function. Inflamm Res. 2018;67:813–828. doi: 10.1007/s00011-018-1174-3. [DOI] [PubMed] [Google Scholar]
- 42.Palmer C.S., Cherry C.L., Sada-Ovalle I., Singh A., Crowe S.M. Glucose metabolism in T cells and monocytes: new perspectives in HIV pathogenesis. EBioMedicine. 2016;6:31–41. doi: 10.1016/j.ebiom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vander Heiden M., Cantley L., Thompson C. Understanding the Warburg effect: the metabolic Requiremetns of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinhouse S., Warburg O., Burk D., Schade A.L. On respiratory impairment in cancer cells. Science. 1956;124 (267–217) [PubMed] [Google Scholar]
- 45.Donnelly R.P., Finlay D.K. Glucose, glycolysis and lymphocyte responses. Mol Immunol. 2015;68:513–519. doi: 10.1016/j.molimm.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 46.Robinson B.H. Lactic acidemia and mitochondrial disease. Mol Genet Metab. 2006;89:3–13. doi: 10.1016/j.ymgme.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Garrabou G., Morén C., Gallego-Escuredo J.M., Milinkovic A., Villarroya F., Negredo E. Genetic and functional mitochondrial assessment of hiv-infected patients developing HAART-related hyperlactatemia. J Acquir Immune Defic Syndr. 2009;52:443–451. doi: 10.1097/QAI.0b013e3181bd564c. [DOI] [PubMed] [Google Scholar]
- 48.Pérez-Matute P., Pérez-Martínez L., Blanco J.R., Oteo J.A. Role of mitochondria in HIV infection and associated metabolic disorders: focus on nonalcoholic fatty liver disease and lipodystrophy syndrome. Oxid Med Cell Longev. 2013;2013:493413. doi: 10.1155/2013/493413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giorgi-Coll S., Amaral A.I., Hutchinson P.J.A., Kotter M.R., Carpenter K.L.H. Succinate supplementation improves metabolic performance of mixed glial cell cultures with mitochondrial dysfunction. Sci Rep. 2017;7:1003. doi: 10.1038/s41598-017-01149-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nowak G., Clifton G.L., Bakajsova D. Succinate ameliorates energy deficits and prevents dysfunction of complex I in injured renal proximal tubular cells. J Pharmacol Exp Ther. 2008;324:1155–1162. doi: 10.1124/jpet.107.130872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tretter L., Patocs A., Chinopoulos C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim Biophys Acta Bioenerg. 2016;1857:1086–1101. doi: 10.1016/j.bbabio.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Younes S.-A., Talla A., Pereira Ribeiro S., Saidakova E.V., Korolevskaya L.B., Shmagel K.V. Cycling CD4+ T cells in HIV-infected immune nonresponders have mitochondrial dysfunction. J Clin Invest. 2018;128(11):5083–5094. doi: 10.1172/JCI120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villeneuve L.M., Purnell P.R., Stauch K.L., Callen S.E., Buch S.J., Fox H.S. HIV-1 transgenic rats display mitochondrial abnormalities consistent with abnormal energy generation and distribution. J Neurovirol. 2016;22:564–574. doi: 10.1007/s13365-016-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holeček M. Branched-chain amino acids in health and disease: metabolism, alterations in blood plasma, and as supplements. Nutr Metab. 2018;15:33. doi: 10.1186/s12986-018-0271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manoli I., Venditti C.P. Metabolic diseases: Foundations of clinical management, genetics, and pathology. 2017. Disorders of branched chain amino acid metabolism. [Google Scholar]
- 56.Shao D., Villet O., Zhang Z., Choi S.W., Yan J., Ritterhoff J. Glucose promotes cell growth by suppressing branched-chain amino acid degradation. Nat Commun. 2018;9:2935. doi: 10.1038/s41467-018-05362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li P., Yin Y.-L., Li D., Woo Kim S., Wu G. Amino acids and immune function. Br J Nutr. 2007;98(02):237. doi: 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- 58.Zhenyukh O., Civantos E., Ruiz-Ortega M., Sánchez M.S., Vázquez C., Peiró C. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 activation. Free Radic Biol Med. 2017;104:165–177. doi: 10.1016/j.freeradbiomed.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Allard J.P., Aghdassi E., Chau J., Tam C., Kovacs C.M., Salit I.E. Effects of vitamin E and C supplementation on oxidative stress and viral load in HIV-infected subjects. AIDS. 1998;12 doi: 10.1097/00002030-199813000-00013. (1653-1559) [DOI] [PubMed] [Google Scholar]
- 60.Mesmin B. Mitochondrial lipid transport and biosynthesis: a complex balance. J Cell Biol. 2016;214:9–11. doi: 10.1083/jcb.201606069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schenkel L.C., Bakovic M. Formation and regulation of mitochondrial membranes. Int J Cell Biol. 2014;2014 doi: 10.1155/2014/709828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y.W., Claypool S.M. Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front Genet. 2015;6:3. doi: 10.3389/fgene.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Donnell V.B., Murphy R.C. New families of bioactive oxidized phospholipids generated by immune cells: identification and signaling actions. Blood. 2012;120(10):1985–1992. doi: 10.1182/blood-2012-04-402826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20(11) doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 65.Lochner M., Berod L., Sparwasser T. Fatty acid metabolism in the regulation of T cell function. Trends Immunol. 2015;36(2):81–91. doi: 10.1016/j.it.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 66.Yeoh H.L., Cheng A.C., Cherry C.L., Weir J.M., Meikle P.J., Hoy J.F. Immunometabolic and Lipidomic markers associated with the frailty index and quality of life in aging HIV+ men on antiretroviral therapy. EBioMedicine. 2017;22:112–121. doi: 10.1016/j.ebiom.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kabagambe E.K., Ezeamama A.E., Guwatudde D., Campos H., Fawzi W. Plasma n6-fatty acid levels are associated with CD4 cell counts, hospitalization and mortality in HIV-infected patients Edmond. J Acquir Immune Defic Syndr. 2016;15(5):598–605. doi: 10.1097/QAI.0000000000001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong G., Trevillyan J.M., Fatou B., Cinel M., Weir J.M., Hoy J.F. Plasma lipidomic profiling of treated HIV-positive individuals and the implications for cardiovascular risk prediction. PLoS One. 2014;9(4):1–7. doi: 10.1371/journal.pone.0094810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdelgadir E., Ali R., Rashid F., Bashier A. Effect of metformin on different non-diabetes related conditions, a special focus on malignant conditions: review of literature. J Clin Med Res. 2017;9(5):388–395. doi: 10.14740/jocmr2922e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lebouché B., Jenabian M.A., Singer J., Graziani G.M., Engler K., Trottier B. The role of extended-release niacin on immune activation and neurocognition in HIV-infected patients treated with antiretroviral therapy - CTN PT006: study protocol for a randomized controlled trial. Trials. 2014;15:390. doi: 10.1186/1745-6215-15-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material