Abstract
Multidrug-resistant organisms are increasing in healthcare settings, and there are few antimicrobials available to treat infections from these bacteria. Pseudomonas aeruginosa is an opportunistic pathogen in burn patients and individuals with cystic fibrosis (CF), and a leading cause of nosocomial infections. P. aeruginosa is inherently resistant to many antibiotics and can develop resistance to others, limiting treatment options. P. aeruginosa has multiple sigma factors to regulate transcription. The alternative sigma factor, RpoN (σ54), regulates many virulence genes and is linked to antibiotic resistance. Recently, we described a cis-acting peptide, RpoN*, which is a “molecular roadblock”, binding consensus promoters at the -24 site, blocking transcription. RpoN* reduces virulence of P. aeruginosa laboratory strains, but its effects in clinical isolates was unknown. We investigated the effects of RpoN* on phenotypically varied P. aeruginosa strains isolated from CF patients. RpoN* expression reduced motility, biofilm formation, and pathogenesis in a P. aeruginosa-C. elegans infection model. Furthermore, we investigated RpoN* effects on antibiotic susceptibility in a laboratory strain. RpoN* expression increased susceptibility to several beta-lactam-based antibiotics in strain P. aeruginosa PA19660 Xen5. We show that using a cis-acting peptide to block RpoN consensus promoters has potential clinical implications in reducing virulence and improving antibiotic susceptibility.
Subject terms: Bacteriology, Bacterial pathogenesis, Pathogens, Biofilms
Introduction
Multidrug-resistant organisms (MDROs) are an increasing problem in the healthcare setting. Both Gram-negative and Gram-positive MDROs are prevalent globally1. There are few or no antimicrobial agents available for treatment of infections caused by these bacteria2. Pseudomonas aeruginosa, a Gram-negative, opportunistic pathogen is a leading cause of nosocomial infections and is associated with infections in burn patients3,4. P. aeruginosa is also responsible for colonizing the respiratory tract and causing chronic infections in individuals with cystic fibrosis (CF)5. It is the most common pathogen isolated from individuals with CF and is a major source of morbidity and mortality6,7.
In CF patients, P. aeruginosa undergoes a transformation from a non-mucoid form upon initial colonization of the lungs to a mucoid form as the disease progresses. This results in a chronic debilitating pulmonary infection characterized by the overexpression of alginate. Mucoid strains synthesize large quantities of alginate, enhancing biofilm formation and protecting P. aeruginosa from antibiotics or the immune response8, possibly through formation of microcolonies9,10. While aggressive prevention regimens have led to a decline in P. aeruginosa prevalence in CF patients, multidrug resistant strains are still prevalent and occurred in 19.4% of CF infections in 201511. P. aeruginosa is inherently resistant to a number of antibiotics12,13. It can also acquire resistance through exogenous resistance genes via horizontal gene transfer or mutations14, limiting treatment options. Antimicrobial development is directed toward alternative treatments and novel targets. Promising strategies include enhancing activity of currently available antibiotics and decreasing virulence of the bacteria once an infection occurs15,16.
P. aeruginosa virulence is caused by many factors, including toxins, proteases, phospholipases, the presence of pili and flagella, and biofilm formation17. Virulence is regulated by a network of transcription factors, such as sigma factors RpoS and RpoN, and quorum sensing regulators18. The alternative sigma factor, σ54 or RpoN, regulates nitrogen assimilation, quorum sensing, motility, biofilm formation and other virulence factors19–27. RpoN regulation was recently linked to P. aeruginosa tolerance to several antibiotics28–30. RpoN binds to specific promoters with conserved −24, −12 sequences upstream of RpoN-regulated genes throughout the genome and is a key virulence regulator31. The specific and conserved nature through which RpoN controls its regulon led us to develop the RpoN molecular roadblock, RpoN*. RpoN* is a cis-acting peptide that specifically binds the −24 site of RpoN consensus promoters, blocking transcription by RpoN and other factors32. The RpoN* peptide sequence includes the identical amino acids to A. aeolicus RpoN Region III that bind with high affinity to the −24 site33. We previously described engineering the rpoN* gene in the broad-host range plasmid pBBR1MCS-5 under control of the ITPG-inducible trc promoter32. The pBBR1MCS-5 plasmid contains the GmR cassette34 and is present at about 10 copies per cell35. When RpoN* is expressed in P. aeruginosa laboratory strains, transcription is affected globally and virulence is attenuated32. We showed that more than 700 genes are affected, either directly regulated by RpoN or indirectly by other transcription factors under RpoN control32. Furthermore, in P. aeruginosa, some genes may have promoter binding sites for multiple sigma factors31. Thus, loss of RpoN does not always equate to loss of transcription and gene expression. We showed that RpoN* affects virulence in a RpoN-deletion strain of P. aeruginosa PAO1. This demonstrates the roadblock’s ability to attenuate gene expression by blocking transcription of genes under dual-regulation with RpoN and other sigma factors32. This strategy of blocking multiple promoters throughout the P. aeruginosa genome may be an effective method to combat virulence and evade development of resistance.
P. aeruginosa isolated from CF patients are phenotypically and genetically varied36,37. Many P. aeruginosa clinical isolates have mutations, including deletion or loss of function, in the rpoN gene38,39. It was not known how the cis-acting RpoN* peptide would affect virulence phenotypes in P. aeruginosa clinical isolates, particularly in strains that do not express or have low levels of RpoN. In this study, we describe the effects of RpoN* on in vitro and in vivo virulence of P. aeruginosa isolated from CF patients. We also describe RpoN* effects on antibiotic resistance in a laboratory strain. Expression of RpoN* reduced virulence-associated phenotypes in clinical isolates and improved P. aeruginosa susceptibility to multiple antibiotics. This study demonstrates that RpoN* has potential clinical applications and represents an effective strategy to combat antibiotic resistance and infections with P. aeruginosa in CF patients.
Results
Virulence phenotypes were variable in P. aeruginosa isolates from CF patients
P. aeruginosa isolated from different CF patients or within the same CF patient have varied phenotypes and genotypes36,37. P. aeruginosa adapts over time, leading to mutations and changes in expression of genes related to motility, quorum sensing, and overall virulence38,40. To determine the virulence-related phenotypic profiles of the strains used in this study (Table 1), each P. aeruginosa patient isolate was evaluated for motility and biofilm formation, compared to the virulent positive control strain P. aeruginosa PA19660 Xen5. Several patient isolates were highly motile in the swimming assay (flagella), including SCH0057-7, SCH0256-1, SCH0354-1 and UUH0201, while others were nonmotile (Fig. 1A, Supplemental Fig. S1A–C). Most strains were motile in the twitching assay (pili) and produced moderate biofilms, with SCH0254-118 migrating the furthest (Fig. 1B, Supplemental Fig. S1A’–C’) and forming the most extensive biofilm (Fig. 1C). SCH0254-116, SCH0397-3, and UUH0202 did not form biofilms.
Table 1.
P. aeruginosa strains used in this study.
| Location | Strain | Source | CF mutations | Reference |
|---|---|---|---|---|
| Laboratory | ||||
| PAO1-M | — | n/a | C. Manoil (27) | |
| PAO1-S | — | n/a | D. Haas (15) | |
| ∆rpoN | — | n/a | D. Haas (15) | |
| PA19660 Xen5 | Septicemia | n/a | Perkin Elmer | |
| Clinical Isolate | ||||
| Seattle Children’s Hospital (SCH) | ||||
| SCH0057-7 | unknown | ∆F508/∆F508 | This study | |
| SCH0254-23 | unknown | ∆F508/unknown | This study | |
| SCH0254-116 | unknown | ∆F508/unknown | This study | |
| SCH0254-118 | unknown | ∆F508/unknown | This study | |
| SCH0256-1 | sputum | ∆F508/∆F508 | This study | |
| SCH0338-38 | sputum | unknown/unknown | This study | |
| SCH0354-1 | sputum | ∆F508/G551D | This study | |
| SCH0397-3 | unknown | ∆F508/unknown | This study | |
| SCH03269 | sputum | ∆F508/∆F508 | This study | |
| Upstate University Hospital (UUH) | ||||
| UUH0101 | sputum | ∆F508/unknown | This study | |
| UUH0201 | sputum | ∆F508/∆F508 | This study | |
| UUH0202 | sputum | ∆F508/∆F508 | This study | |
Figure 1.
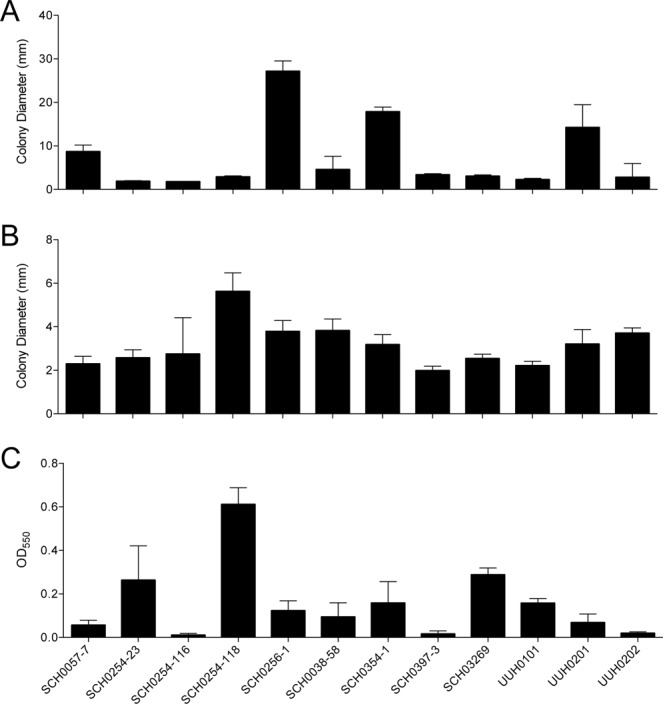
Characterization of virulence phenotypes of P. aeruginosa strains isolated from cystic fibrosis patients. P. aeruginosa CF patient isolates were compared to laboratory strain PA19660 Xen5. All assays were conducted at 37 °C for 24 h. (A) Colony diameter of swimming, or flagellar, motility assay conducted on soft (0.3%) agar. (B) Twitching, or pili, motility assay conducted on semi-hard (1.3%) agar. Colony diameter was measured across point of inoculation to the edges of bacteria colony. (C) Biofilm formation assay was conducted in 96-well microtiter plates. Biofilms were stained with crystal violet (0.1%), solubilized in ethanol (95%), and absorbance measured at OD550. Bars are the mean ± SD; n = 5 to 6 replicates in motility assays and n = 10 in biofilm assay. Each assay was performed at least three separate times and representative results are shown.
The pathogenesis of patient isolates was evaluated in a P. aeruginosa – C. elegans infection model. All patient isolates were compared to E. coli OP50, an avirulent negative control. SCH0057-7 was the most pathogenic in the paralytic killing assay, which is mediated by hydrogen cyanide production41,42 (Fig. 2A). Other strains were moderately pathogenic, including SCH0256-1, SCH0354-1, SCH0397-3, and UUH0202. SCH0057-7, SCH0338-58, and UUH0202 were highly pathogenic in the slow killing assay, which mimics establishment and proliferation of an infection and is mediated by the lasR, gacA, lemA, and ptsP genes43, while UUH0201 was moderately pathogenic (Fig. 2B). As expected, the virulence-associated phenotypes of patient isolates varied widely in vitro and in vivo.
Figure 2.
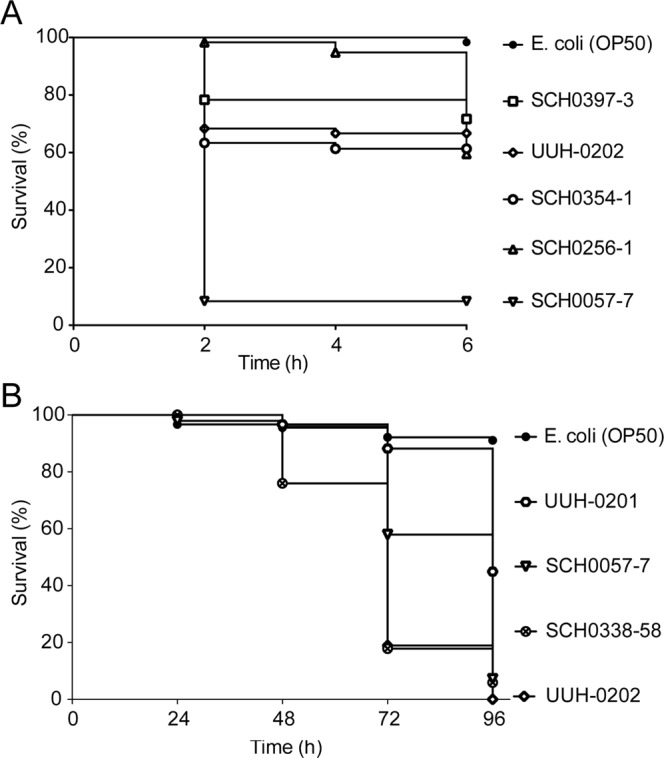
Pathogenesis of P. aeruginosa isolated from cystic fibrosis patients in P. aeruginosa – C. elegans infection model. Kaplan-Meier survival curves for P. aeruginosa – C. elegans infection assays. (A) Paralytic killing assay on BHI agar. Assay was conducted at room temperature and worm status scored every 2 h. (B) Slow killing assay on modified NGM agar (0.35% bactopeptone, 2% bactoagar). Assay was conducted at 20 °C and worm status scored every 24 h. Strains used included CF patient isolates, and E. coli for reference. n = 48 to 90 worms per strain.
RpoN protein levels varied among patient isolates
Others reported that the rpoN gene was mutated or lost in approximately 20% of P. aeruginosa isolates from CF patients38. Loss or mutation in the rpoN gene can result in phenotypes similar to those observed in the patient isolates evaluated here20,22,23. Thus, we evaluated relative protein levels of RpoN in these patient isolates by western blot. RpoN levels were moderately high in the positive control P. aeruginosa PAO1-S, while low or minimal protein levels were detected in the isogenic ΔrpoN mutant negative control (Fig. 3). The faint bands in the ∆rpoN strain and several CF patient isolates are background signals due to non-specific antibody binding to another protein or sigma factor with a similar apparent molecular weight. RpoN levels varied in the CF patient isolates, with high levels in SCH0057-7, SCH0397-3, and UUH0201; intermediate levels in SCH0254-116, SCH0338-58, and UUH0202; and background levels in SCH0254-23, SCH0254-118, SCH0256-1, SCH0354-1, SCH03269, and UUH0101.
Figure 3.
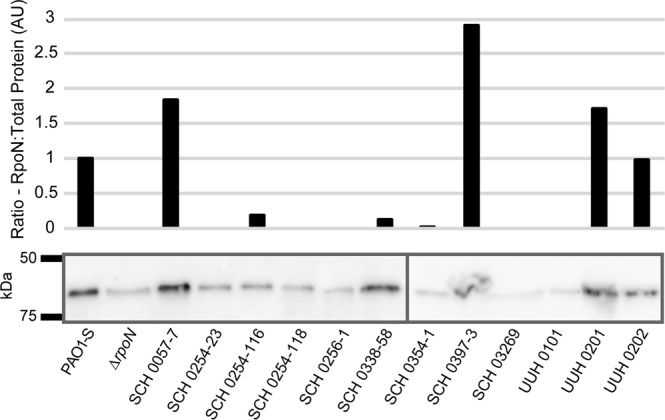
RpoN protein in P. aeruginosa isolates is highly variable. Immunoblot (bottom) and analysis (top) of RpoN expression in P. aeruginosa CF patient isolates and laboratory strains PAO1-S and ΔrpoN. Immunoblots performed on 10% Mini-PROTEAN TGX Stain-Free gels (BioRad). RpoN protein levels were calculated by comparing measured total protein in each lane to the measured RpoN band (presented as arbitrary units (AU)). The background value in ∆rpoN was subtracted from all samples and values were normalized against PAO1-S. The graph and immunoblot are representative of immunoblots from multiple bacterial cultures and western blot analyses. Separate immunoblots are indicated by the dividing line. See Supplemental Fig. S2 for full immunoblots.
RpoN* expressed in CF patient isolates reduced virulence-associated phenotypes in vitro
The effect of RpoN* expression on motility and biofilm formation in patient isolates was not known. Unfortunately, some patient isolates could not be transformed, and so only four isolates were evaluated for the effects of RpoN* expressed from a plasmid. SCH0057-7, SCH0256-1, SCH0338-58, and SCH0354-1 were transformed with a plasmid expressing RpoN* or the empty vector and selected with gentamicin. If RpoN* affected transcription of virulence-related genes in different genetic backgrounds as previously reported32, we expected attenuation of virulence-related phenotypes in P. aeruginosa CF patient isolates. RpoN* significantly reduced colony diameter in all four patient isolates in the swimming motility assay (Student’s t-test, **p ≤ 0.01, ***p ≤ 0.0001) (Fig. 4A, Supplemental Fig. S3 (top two rows)). RpoN* significantly reduced colony diameter in SCH0057-7, SCH0256-1, and SCH0338-58 in the twitching motility assay (Student’s t-test, **p ≤ 0.01, ***p ≤ 0.0001) (Fig. 4B, Supplemental Fig. S3 (bottom two rows)). Colony diameter varied widely in SCH0354-1 when RpoN* was expressed and was always smaller than with empty vector, although the difference was not significant. In the biofilm formation assay, RpoN* significantly reduced biofilm formation by SCH0057-7 and SCH0256-1 (Student’s t-test, p ≤ 0.0001) (Fig. 4C). Thus RpoN* reduced virulence-associated phenotypes of P. aeruginosa isolated from CF patients.
Figure 4.
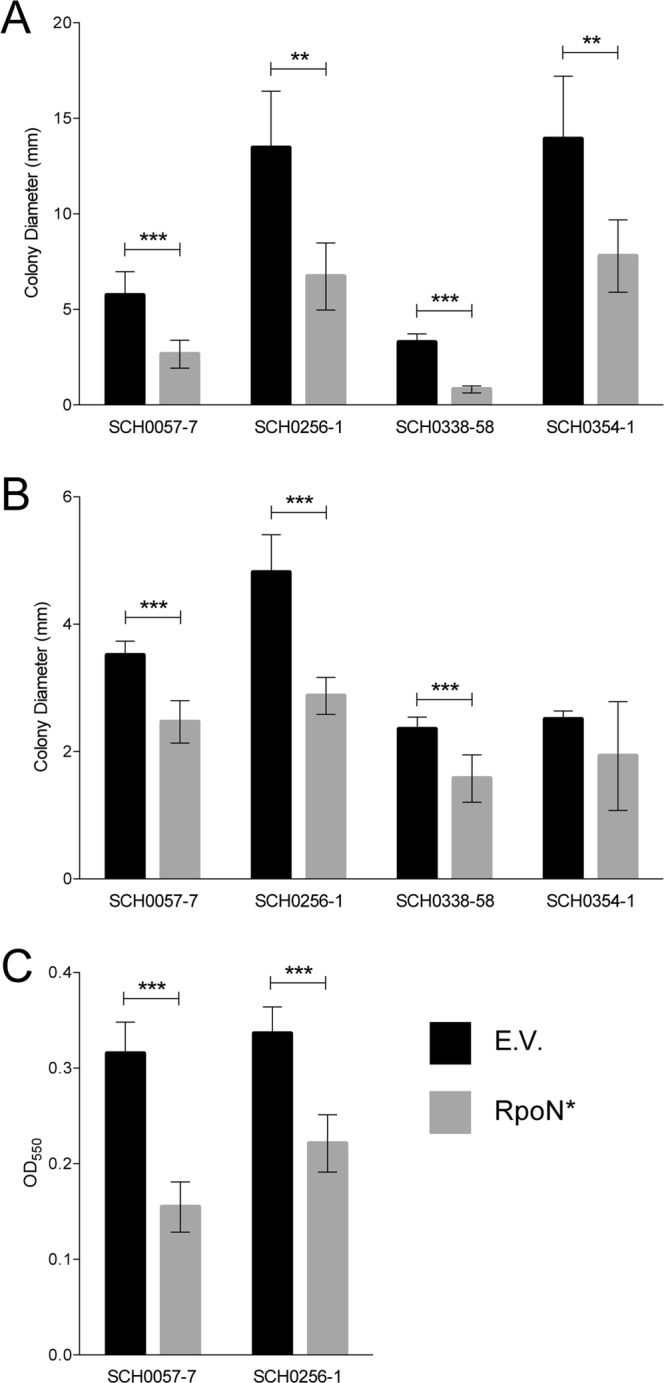
RpoN* expression decreases motility and biofilm formation. P. aeruginosa CF patient isolates with empty vector (E.V., black bars), or with RpoN*-expression vector (gray bars). Media was supplemented with gentamicin (30 mg/L), and IPTG (1 mM) when applicable, and all assays were conducted at 37 °C for 24 h. (A) Colony diameter of swimming, or flagellar, motility assay conducted on soft (0.3%) agar. (B) Colony diameter of twitching, or pili, motility assay conducted on semi-hard (1.3%) agar. (C) Biofilm formation assay conducted in 96-well microtiter plates. Bars are the mean ± SD (Student’s t-test, ***p ≤ 0.0001; **p ≤ 0.01). n = 4 to 5 replicates in motility assays; n = 12 in biofilm assays.
RpoN* expression increased worm survival in P. aeruginosa – C. elegans infection model
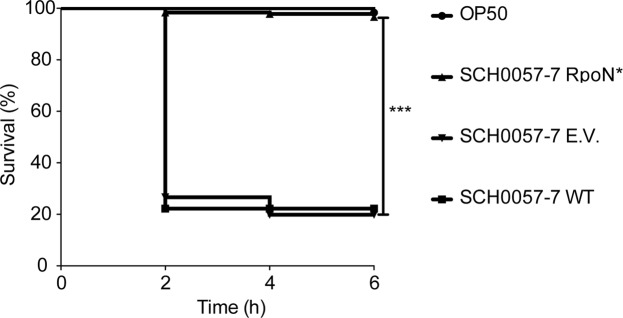
Initial evaluation of patient isolates revealed a single P. aeruginosa strain, SCH0057-7, that was both transformable and pathogenic in the P. aeruginosa – C. elegans infection assay. Therefore, effects of RpoN* on pathogenesis of SCH0057-7 were evaluated using the paralytic killing assay, which is based on P. aeruginosa hydrogen cyanide production and mimics conditions in the CF lung41,42. Wild-type P. aeruginosa SCH0057-7 was the positive, virulent control and E. coli was the negative, avirulent control. The test conditions were P. aeruginosa SCH0057-7 expressing RpoN* or carrying the empty vector plasmid. If RpoN* affected virulence-related phenotypes in P. aeruginosa SCH0057-7, then we expected increased survival of C. elegans. Wild type SCH0057-7 and with the empty vector killed approximately 80% of C. elegans (Fig. 5). In contrast, RpoN* expression significantly increased C. elegans survival (Mantel-Cox Log-Rank Test, p ≤ 0.0001). Thus, RpoN* expression reduced pathogenesis of a patient isolate in a P. aeruginosa – C. elegans infection model.
Figure 5.
RpoN* promotes C. elegans survival in paralytic killing assay. P. aeruginosa CF isolate SCH0057-7 wild type, with empty vector (E.V.), or with RpoN*-expression vector, were compared to avirulent E. coli OP50. Paralytic killing assay was conducted on BHI agar supplemented with gentamicin (30 mg/L) and IPTG (1 mM), when applicable. Assay was conducted at room temperature, and worm status scored every 2 h. Kaplan-Meier survival curves represent combined survival of three separate assays (exception: E. coli). Mantel-Cox log-rank test was used to analyze E.V. and RpoN* curves (***p ≤ 0.0001). n = 180 worms per SCH0057-7 condition, n = 60 worms for E. coli.
RpoN* increased susceptibility to select antibiotics in vitro
Antibiotic resistance is a problem in CF patients with P. aeruginosa infections44–46. We previously reported that RpoN* alters transcription of several genes involved in multidrug efflux pumps that confer natural resistance32. Additionally, RpoN is implicated in tolerance to various classes of antibiotics28–30. We evaluated the effects of RpoN* on antibiotic susceptibility using a MicroScan Neg MIC 43 panel. The test conditions were P. aeruginosa PA19660 Xen5 that was mock-transformed (no vector) or transformed with the empty vector or RpoN* plasmid. We expected that RpoN* would improve antibiotic susceptibility of P. aeruginosa. In PA19660 Xen5 mock-transformed or with the empty vector, antibiotic susceptibility profiles were the same, except for gentamicin, which increased in the empty vector strain due to the GMR selection marker (Table 2). In PA19660 Xen5 expressing RpoN*, susceptibility to five beta-lactam antibiotics was improved at least 2- to 4-fold (Table 2). Results presented in Table 2 are representative of a single experiment, however these results were highly consistent and repeatable over multiple experiments and biological replicates. The antibiotics with improved susceptibility were cefotaxime, cefepime, and ceftazidime (three cephalosporins), piperacillin (a ureidopenicillin), and imipenem (a carbapenem). Susceptibility to some antibiotics was unchanged (data not shown). For piperacillin, there was at least a 2-fold increase in susceptibility. This improvement is clinically relevant as it changed the status from resistant to sensitive (Table 2). For the other drugs, which there was no change in clinical susceptibility status, presence of RpoN* increased the therapeutic potency of the drugs. The results demonstrate that RpoN* expression increased P. aeruginosa susceptibility to several antibiotics.
Table 2.
RpoN* expression increases susceptibility to antibiotics.
| P. aeruginosa PA19660 Xen5 | ||||
|---|---|---|---|---|
| Antibiotic Class and Name: |
Wild Type n = 4 |
Empty Vector n = 6 |
RpoN* n = 6 |
Treatment Guidelines of P. aeruginosa† |
| Minimal Inhibitory Concentration (MIC) | ||||
| Aminoglycosides: | ||||
| Gentamicin (Gm) | 8 μg/mL (I) |
>38 μg/mL (R) |
>38 μg/mL (R) |
On and off-label in combination with other antimicrobials |
| Carbapenems: | ||||
| Imipenem (Imp) | 2 μg/mL (S) |
2 μg/mL (S) |
≤1 μg/mL (S) |
On-label use as an individual drug |
| Cephalosporins: | ||||
| Cefepime (Cpe) [4th generation] |
4 μg/mL (S) |
4 μg/mL (S) |
≤2 μg/mL (S) |
On and off-label use individually or in combination with other antimicrobials |
| Ceftazidime (Caz) [3rd generation] |
4 μg/mL (S) |
4 μg/mL (S) |
≤1 μg/mL (S) |
On and off-label use individually or in combination with other antimicrobials |
| Cefotaxime (Cft) [3rd generation] |
16 μg/mL (R) |
16 μg/mL (R) |
8 μg/mL (R) |
On-label use as an individual drug |
| Penicillins: | ||||
| Piperacillin (Pi) | >64 μg/mL (R) |
>64 μg/mL (R) |
≤16–64 μg/mL (S) |
On and off-label use in combination with tazobactam |
The interpretation as to whether the obtained MIC is resistant (R), intermediate (I), or sensitive (S) is based on therapeutic guidelines for treatment of an infection using a particular antibiotic against a specific organism.
†Treatment information and use for each antibiotic obtained from UpToDate in September 2018.
Discussion
Here, we confirm and expand results of previous studies by showing the ability of RpoN* to abrogate virulence phenotypes in P. aeruginosa isolates from CF patients and to improve susceptibility to several antibiotics. Our working model of the mechanism of action of RpoN* is that it binds the -24 promoter consensus sites, blocking transactivation by RpoN and other sigma factors. By altering the transcriptome, RpoN* reduced virulence in well-characterized laboratory strains32. Thus, the motivation for this study was to understand the clinical relevance of RpoN*. We demonstrated that RpoN* expressed in CF patient isolates reduced motility and biofilm formation in vitro, independently of RpoN protein levels. The RpoN* molecular roadblock protected C. elegans from a highly virulent P. aeruginosa patient isolate in an in vivo infection model. RpoN* also improved P. aeruginosa susceptibility to antibiotics.
P. aeruginosa isolated from CF patients are highly variable36,37, with the rpoN gene often mutated or lost38. The patient isolates evaluated in this study had a broad range of motility, biofilm formation, RpoN protein levels, and virulence in C. elegans. There was no correspondence between most in vitro phenotypes, in vivo pathogenesis, and RpoN levels (Supplemental Fig. S4). The only correlation observed was between twitching, or pili-associated, motility and biofilm formation (Supplemental Fig. S4F, p = 0.0357, R2 = 0.37050). Other studies suggested that in vitro phenotypes of P. aeruginosa isolates can be related to disease status in CF patients47. Patient information and status of P. aeruginosa infections is limited for the isolates described here, so a comparison between phenotypes and patient status is not feasible. Interestingly, two isolates, UUH0201 and UUH0202, were obtained five months apart from the same patient, with UUH0201 collected first. The UUH0202 strain was less motile and RpoN protein levels dropped compared to UUH0201, but virulence increased. This supports the concept that in vitro phenotypes reflect P. aeruginosa infection status in CF patients47. Further work would be needed to fully elucidate such correlations.
The RpoN* molecular roadblock reduced virulence phenotypes in patient isolates with high or low levels of RpoN. Furthermore, there was no substantial difference between CF isolates transformed with the empty vector and in the absence of manipulation (wild type strains). For instance, RpoN protein levels were higher in SCH0057-7 than PAO1-S, and RpoN* reduced flagellar and pili motility, biofilm formation and pathogenesis. In contrast, relative RpoN protein levels were low in SCH0256-1 and SCH0354-1, and yet RpoN* reduced motility. Thus, the roadblock was effective in the presence or absence of the native sigma factor. This is possible since there is a redundancy among P. aeruginosa sigma factors, with multiple sigma factors having consensus promoter sites for the same gene31. Thus, in the absence of RpoN, other sigma factors promote transcription of certain genes. The results here confirm our previous findings, which show that RpoN* reduced virulence in a laboratory strain that was deleted for rpoN32. Unfortunately, barriers to transformation precluded evaluating RpoN* in some of the other clinical isolates. However, the strains that were successfully transformed represented much of the diversity across the patient isolates.
The CF patient isolates demonstrated variable pathogenesis in the C. elegans paralytic killing model that spans 6 hours. Only one pathogenic isolate, SCH0057-7, was transformable and thus possible to evaluate the effects of RpoN* in vivo. This strain and several others were also pathogenic in the slow killing assay. This assay was not used to evaluate RpoN* because of difficulty maintaining the plasmid and RpoN* expression. Gentamicin selection and IPTG induction are not durable, we found32, because the C. elegans cuticle is impermeable and the compounds are poorly absorbed in the intestine48. While it is expected that expressing RpoN* in CF isolates would improve C. elegans survival in the slow killing assay, it is not feasible with the current vector. If issues with maintaining the plasmid and expression of the roadblock were resolved, it would be interesting to evaluate RpoN* in this assay using patient isolates.
The molecular roadblock, RpoN*, binds numerous promoters in bacterial genomes, altering the transcriptome. RpoN* expression in P. aeruginosa greatly reduced transcription of the mex family genes32, which are involved in multidrug efflux pumps49. Increased expression of mex genes is linked to increased resistance to antibiotics14. Therefore, we investigated whether RpoN* alters P. aeruginosa susceptibility to antibiotics. We employed a clinical laboratory assay for testing bacterial susceptibility or resistance to antibiotics and found that RpoN* improved antibiotic susceptibility at least two-fold for five different antibiotics, including imipenem. This agrees with previous studies that showed RpoN is involved in P. aeruginosa tolerance of carbapenems, quinolones, and tobramycin28–30. Unfortunately, the commercial assay uses pre-determined antibiotic concentrations in a 96-well plate, limiting the scope of the roadblock’s effects. Additionally, the P. aeruginosa strain used here is sensitive to quinolones and tobramycin, so RpoN* effects on resistance to these antibiotics was not evaluated. Unfortunately, testing clinical strains was not feasible for this study. However, it will be important to test such strains, particularly those resistant to quinolones, carbapenems, and tobramycin, to determine the effects of RpoN*. Further studies are needed to uncover the full spectrum of RpoN* effects on antibiotic susceptibility.
MRDOs are increasing worldwide, even those with resistance to entire antibiotic classes. Alarmingly, nearly all antibiotics brought to market in the past 30 years are variations on existing drugs50. Research into alternative strategies to treat bacterial infections is a priority, including compounds to enhance activity of existing antibiotics or neutralize virulence factors. The molecular roadblock falls into the latter type. RpoN* binds consensus promoters throughout the P. aeruginosa genome, affecting the transcription of numerous virulence factors. Due to the many binding sites for RpoN*, it is unlikely that resistance to it would develop during treatment. The binding sequence of the RpoN consensus promoter is conserved across gram-negative and gram-positive bacteria (36, 37). The effects of RpoN* on virulence phenotypes of Pseudomonas putida, Burkholderia cepacia, and Escherichia coli have been explored (unpublished data), suggesting that RpoN* may reduce virulence in multiple organisms. More studies are needed to identify the spectrum of RpoN* activity and its resistance frequency. Currently, RpoN* is a tool for antimicrobial development and is not a usable drug. However, a study was recently published using a stapled RpoN-like peptide to reduce transcription of RpoN-related genes in E. coli51. They found that this stapled peptide, which binds to RpoN consensus promoters, could enter P. aeruginosa cells51. Finding a small molecule or stapled-peptide that works in the same cis-acting manner as RpoN* would be an effective, clinically relevant strategy to combat P. aeruginosa virulence and antibiotic resistance.
Materials and Methods
Bacteria and nematodes
P. aeruginosa clinical isolates were provided by Seattle Children’s Hospital (SCH strains) and Upstate University Hospital (UUH strains). P. aeruginosa PAO1-M was provided by C. Manoil41, and P. aeruginosa PAO1-S and ΔrpoN were provided by D. Haas22. P. aeruginosa PA19660 Xen5 was purchased from PerkinElmer. E. coli OP50 was provided by D. Pruyne (SUNY Upstate Medical University). All strains are listed in Table 1. For long-term storage, bacteria were grown overnight in LB broth at 37 °C with shaking, and frozen in 10% glycerol at −80 °C. Caenorhabditis elegans N2 was purchased from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN), and maintained on nematode growth media (NGM) seeded with E. coli OP50 at 20 °C52. Populations were synchronized via egg lay and grown to the young adult stage at 20 °C53.
Plasmids
RpoN* and empty vector plasmids were previously described32. Plasmids were maintained in E. coli INV110 (Invitrogen) with gentamicin selection (30 mg/L). RpoN* expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG, 1 mM).
Transformation
Permissive P. aeruginosa patient isolates and a lab strain were transformed by electroporation prior to all experiments, per standard protocol54. Transformed bacteria were selected on LB agar or BHI agar supplemented with gentamicin (30 mg/L). Individual colonies were picked for each assay.
Western blot analysis
Overnight bacteria cultures were treated with Cell Lytic B Lysis Reagent (Sigma) to generate crude cell lysates. The soluble protein fraction was separated on 10% Mini-PROTEAN TGX Stain-Free protein gels (BioRad), activated for 5 minutes with UV light, imaged and transferred via semi-dry apparatus to a PVDF membrane. Membranes were incubated with primary antibody specific for E. coli RNA σ54 (1:500, BioLegend) overnight, then with secondary antibody HRP goat anti-mouse (1:10,000, Jackson ImmunoResearch). The chemiluminescent signal was generated with the Pierce SuperSignal West Fempto substrate kit (Thermo Scientific) and detected with ChemiDoc MP Imaging System (Bio-Rad Laboratories). Protein bands and total protein per lane were measured with Image Lab (Version 5.2.1; Bio-Rad Laboratories). RpoN bands were compared to corresponding total detected protein in each lane and the background value in ∆rpoN was subtracted from all samples.
Phenotyping assays
Assays to measure swimming, twitching55, and biofilm formation56, were conducted according to standard protocols. Transformed P. aeruginosa clinical isolates were grown in appropriate media supplemented with gentamicin (30 mg/L), and with or without IPTG (1 mM). Motility and biofilm assays were conducted at 37 °C for 24 h. Images of motility assays were obtained with IVIS-50 (Perkin Elmer) and colony diameter was measured with Living Image software (Perkin Elmer). To study biofilm formation, overnight cultures were diluted 1:3 in LB broth with appropriate conditions, grown for 3 h at 37 °C with shaking. Log-phase cultures were diluted 1:50 into M63 minimal media with 0.4% arginine and 1 mM MgSO4 (30 mg/L gentamicin and 1 mM IPTG included for transformed cultures), and 100 µl added per well to a U-bottom 96-well plate and plates incubated at 37 °C for 24 h. Biofilms were stained with 0.1% crystal violet at RT, extracted in 95% ethanol, and absorbance was measured at 550 nm with a μQuant microplate spectrophotometer (BioTek).
P. aeruginosa – C. elegans infection assays
For the paralytic killing assay, laboratory strains, clinical isolates, or transformed P. aeruginosa were spread on Brain Heart Infusion (BHI) agar (Difco) with, when applicable, gentamicin (30 mg/L) and with or without IPTG (1 mM). E. coli was spread on BHI agar. All plates were grown overnight at 37 °C. Bacteria colonies were swabbed onto BHI agar, supplemented with gentamicin and/or IPTG (1 mM) when applicable, and grown at 37 °C for 24 h41. Adult C. elegans were added to plates and the assay was conducted at room temperature, per standard protocol41. For the slow killing assay, laboratory strains or clinical isolates of P. aeruginosa were grown overnight in LB broth at 37 °C with shaking, and cultures were spread on a modified NGM agar (0.35% bactopeptone, 2% bactoagar)57. Plates were incubated at 37 °C for 24 h, then at room temperature for an additional 24 h. The assay was conducted at 20 °C, and worms were scored every 24 h per standard protocol57.
Antibiotic sensitivity testing
Transformed or mock transformed bacteria were grown overnight in LB broth with gentamicin (30 mg/L) and IPTG (1 mM) or only IPTG (1 mM), respectively. MicroScan Neg MIC 43 panels (Beckman Coulter Inc., Brea, CA) were used. Panels were set up per manufacturer’s protocol (MicroScan Gram Negative Procedure Manual, version 09/2016) using the RENOX system (Beckman Coulter Inc., Brea, CA) with a final well concentration of 3–7 × 105 CFU/mL. The following modifications were made to the manufacturer’s protocol: LB broth supplemented with IPTG (1 mM) and with or without gentamicin (30 mg/L) was used in place of saline for whole panel. Plates were incubated at 35 °C for 16–20 h and read using a MicroScan autoSCAN-4 (Beckman Coulter Inc, Brea, CA). MIC values were determined by the MicroScan reader based on optical density. Quality control was performed on the panels per manufacturer’s protocol.
Statistics
Data were analyzed using Excel and GraphPad Prism with a significance of p ≤ 0.05 (Microsoft, Washington; GraphPad Software Inc., California).
Supplementary information
Acknowledgements
We would like to thank the Clinical Microbiology Lab at the Syracuse VA Medical Center in Syracuse, NY for supplying the antibiotic susceptibility testing panels and for use of their facility for reading the results. We would also like to thank Dr. Ran Anbar and Donna Linder at the Golisano Center at Upstate University Hospital and Marcella Blackledge and Dr. Rafael Hernandez at the Seattle Children’s Hospital for supplying the P. aeruginosa CF patient isolates (Seattle Children’s’ Hospital CF patient isolates obtained under NIH P30 DK089507).
Author Contributions
M.G.L. wrote the manuscript. C.T.N. and J.F.M. conceived the study. M.G.L. and J.L.V. conducted the experiments. M.G.L. generated the figures. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43060-6.
References
- 1.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Freire-Moran L, et al. Critical shortage of new antibiotics in development against multidrug-resistant bacteria-Time to react is now. Drug Resist Updat. 2011;14:118–124. doi: 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 4.Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 5.Davis PB, Drumm M, Konstan MW. Cystic fibrosis. Am J Respir Crit Care Med. 1996;154:1229–1256. doi: 10.1164/ajrccm.154.5.8912731. [DOI] [PubMed] [Google Scholar]
- 6.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol. 2002;34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 7.Ahlgren HG, et al. Clinical outcomes associated with Staphylococcus aureus and Pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm Med. 2015;15:67. doi: 10.1186/s12890-015-0062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoiby N. Understanding bacterial biofilms in patients with cystic fibrosis: current and innovative approaches to potential therapies. J Cyst Fibros. 2002;1:249–254. doi: 10.1016/S1569-1993(02)00104-2. [DOI] [PubMed] [Google Scholar]
- 9.Fegan M, Francis P, Hayward AC, Davis GH, Fuerst JA. Phenotypic conversion of Pseudomonas aeruginosa in cystic fibrosis. J Clin Microbiol. 1990;28:1143–1146. doi: 10.1128/jcm.28.6.1143-1146.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam J, Chan R, Lam K, Costerton J. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infection and immunity. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cystic Fibrosis Foundation Patient Registry 2015. Annual Data Report (Cystic Fibrosis Foundation, Bethesda, Maryland, 2016).
- 12.Hancock RE, Speert DP. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat. 2000;3:247–255. doi: 10.1054/drup.2000.0152. [DOI] [PubMed] [Google Scholar]
- 13.Porras-Gómez M, Vega-Baudrit J, Núñez-Corrales S. Overview of multidrug-resistant Pseudomonas aeruginosa and novel therapeutic approaches. Journal of Biomaterials and Nanobiotechnology. 2012;3:519. doi: 10.4236/jbnb.2012.324053. [DOI] [Google Scholar]
- 14.Riou M, et al. Increase of efflux-mediated resistance in Pseudomonas aeruginosa during antibiotic treatment in patients suffering from nosocomial pneumonia. Int J Antimicrob Agents. 2016;47:77–83. doi: 10.1016/j.ijantimicag.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Dickey, S. W., Cheung, G. Y. C. & Otto, M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov, 10.1038/nrd.2017.23 (2017). [DOI] [PubMed]
- 16.Wagner S, et al. Novel Strategies for the Treatment of Pseudomonas aeruginosa Infections. J Med Chem. 2016;59:5929–5969. doi: 10.1021/acs.jmedchem.5b01698. [DOI] [PubMed] [Google Scholar]
- 17.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 18.Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 20.Totten PA, Lara JC, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. Journal of bacteriology. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damron FH, et al. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol. 2012;194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heurlier K, Denervaud V, Pessi G, Reimmann C, Haas D. Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. Journal of bacteriology. 2003;185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson LS, Webb JS, Rice SA, Kjelleberg S. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS microbiology letters. 2003;220:187–195. doi: 10.1016/S0378-1097(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 24.Shao, X. et al. RpoN-Dependent Direct Regulation of Quorum Sensing and the Type VI Secretion System in Pseudomonas aeruginosa PAO1. J Bacteriol200, 10.1128/JB.00205-18 (2018). [DOI] [PMC free article] [PubMed]
- 25.Caiazza NC, O’Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Ahmar, R., Kirby, B. D. & Yu, H. D. Pyrimidine Biosynthesis Regulates the Small-Colony Variant and Mucoidy in Pseudomonas aeruginosa through Sigma Factor Competition. J Bacteriol201, 10.1128/JB.00575-18 (2019). [DOI] [PMC free article] [PubMed]
- 27.Cai Z, et al. RpoN Regulates Virulence Factors of Pseudomonas aeruginosa via Modulating the PqsR Quorum Sensing Regulator. Int J Mol Sci. 2015;16:28311–28319. doi: 10.3390/ijms161226103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. RpoN Promotes Pseudomonas aeruginosa Survival in the Presence of Tobramycin. Front Microbiol. 2017;8:839. doi: 10.3389/fmicb.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. RpoN Modulates Carbapenem Tolerance in Pseudomonas aeruginosa through Pseudomonas Quinolone Signal and PqsE. Antimicrob Agents Chemother. 2016;60:5752–5764. doi: 10.1128/AAC.00260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viducic D, et al. rpoN gene of Pseudomonas aeruginosa alters its susceptibility to quinolones and carbapenems. Antimicrob Agents Chemother. 2007;51:1455–1462. doi: 10.1128/AAC.00348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulz S, et al. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 2015;11:e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd, M. G. et al. Targeting the alternative sigma factor RpoN to combat virulence in Pseudomonas aeruginosa. Scientific Reports7 (2017). [DOI] [PMC free article] [PubMed]
- 33.Doucleff M, Malak LT, Pelton JG, Wemmer DE. The C-terminal RpoN domain of sigma54 forms an unpredicted helix-turn-helix motif similar to domains of sigma70. J Biol Chem. 2005;280:41530–41536. doi: 10.1074/jbc.M509010200. [DOI] [PubMed] [Google Scholar]
- 34.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 35.Elzer PH, Phillips RW, Kovach ME, Peterson KM, Roop RM., II Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect Immun. 1994;62:4135–4139. doi: 10.1128/iai.62.10.4135-4139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark ST, et al. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci Rep. 2015;5:10932. doi: 10.1038/srep10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duong J, et al. Phenotypic and Genotypic Comparison of Epidemic and Non-Epidemic Strains of Pseudomonas aeruginosa from Individuals with Cystic Fibrosis. PLoS One. 2015;10:e0143466. doi: 10.1371/journal.pone.0143466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darby C, Cosma CL, Thomas JH, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallagher LA, Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. Journal of bacteriology. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mustafa MH, et al. Antimicrobial Susceptibility of Pseudomonas aeruginosa Isolated from Cystic Fibrosis Patients in Northern Europe. Antimicrob Agents Chemother. 2016;60:6735–6741. doi: 10.1128/AAC.01046-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill D, et al. Antibiotic susceptabilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol. 2005;43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomas M, et al. Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2010;54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer-Hamblett N, et al. Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am J Respir Crit Care Med. 2014;190:289–297. doi: 10.1164/rccm.201404-0681OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Reilly LP, Luke CJ, Perlmutter DH, Silverman GA, Pak SC. C. elegans in high-throughput drug discovery. Adv Drug Deliv Rev. 2014;69-70:247–253. doi: 10.1016/j.addr.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poole K, Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr Top Med Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 50.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Payne, S. R. et al. Inhibition of Bacterial Gene Transcription with an RpoN-Based Stapled Peptide. Cell Chem Biol, 10.1016/j.chembiol.2018.05.007 (2018). [DOI] [PMC free article] [PubMed]
- 52.Wood, W. B. The Nematode Caenorhabditis elegans: Introduction to C. elegans biology (Cold Spring Harbor Laboratory Press, 1988).
- 53.Lionaki E, Tavernarakis N. Assessing aging and senescent decline in Caenorhabditis elegans: cohort survival analysis. Methods Mol Biol. 2013;965:473–484. doi: 10.1007/978-1-62703-239-1_31. [DOI] [PubMed] [Google Scholar]
- 54.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 55.O’Toole GA, et al. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 56.O’Toole, G. A. Microtiter dish biofilm formation assay. J Vis Exp, 10.3791/2437 (2011). [DOI] [PMC free article] [PubMed]
- 57.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.