Abstract
Background
TP53 mutation is the most common mutation in hepatocellular carcinoma (HCC), and it affects the progression and prognosis of HCC. We investigated how TP53 mutation regulates the HCC immunophenotype and thus affects the prognosis of HCC.
Methods
We investigated TP53 mutation status and RNA expression in different populations and platforms and developed an immune prognostic model (IPM) based on immune-related genes that were differentially expressed between TP53WT and TP53MUT HCC samples. Then, the influence of the IPM on the immune microenvironment in HCC was comprehensively analysed.
Findings
TP53 mutation resulted in the downregulation of the immune response in HCC. Thirty-seven of the 312 immune response-related genes were differentially expressed based on TP53 mutation status. An IPM was established and validated based on 865 patients with HCC to differentiate patients with a low or high risk of poor survival. A nomogram was also established for clinical application. Functional enrichment analysis showed that the humoral immune response and immune system diseases pathway represented the major function and pathway, respectively, related to the IPM genes. Moreover, we found that the patients in the high-risk group had higher fractions of T cells follicular helper, T cells regulatory (Tregs) and macrophages M0 and presented higher expression of CTLA-4, PD-1 and TIM-3 than the low-risk group.
Interpretation
TP53 mutation is strongly related to the immune microenvironment in HCC. Our IPM, which is sensitive to TP53 mutation status, may have important implications for identifying subgroups of HCC patients with low or high risk of unfavourable survival.
Fund
This work was supported by the International Science and Technology Cooperation Projects (2016YFE0107100), the Capital Special Research Project for Health Development (2014-2-4012), the Beijing Natural Science Foundation (L172055 and 7192158), the National Ten Thousand Talent Program, the Fundamental Research Funds for the Central Universities (3332018032), and the CAMS Innovation Fund for Medical Science (CIFMS) (2017-I2M-4-003 and 2018-I2M-3-001).
Keywords: TP53, Mutation, Immune prognostic model, Immune profile, Hepatocellular carcinoma
Research in context.
Evidence before this study
We searched PubMed through Feb 20, 2019, for research articles containing the terms “immune prognostic model AND hepatocellular carcinoma” without language or date restrictions. This search did not find any previous high-throughput studies that had investigated the potential prognostic role of immune prognostic models in hepatocellular carcinoma. In addition, the same search method was used to identify articles containing the terms “immune prognostic model AND TP53”. This search also identified no previous high-throughput studies that had investigated the relationship between immune prognostic models and TP53.
Added value of this study
We found that the immune phenotype was related to TP53 mutation and developed and validated an immune prognostic model for hepatocellular carcinoma that was affected by TP53 mutation status. This model is based on the expression of 2 immune genes that differentiate patients with a low or high risk of poor survival in both the training and validation cohorts. Our study included 865 patients with hepatocellular carcinoma to establish and validate an immune prognostic model, and to our knowledge, it is the largest prognostic model discovery project for hepatocellular carcinoma. Our results suggest that this immune prognostic model is more accurate than clinicopathological risk factors alone. We further developed a nomogram to predict patient prognosis, and it consisted of the immune prognostic model, vascular tumour invasion and hepatitis C status.
Implications of all available evidence
For the first time, we identified and validated an immune prognostic model based on 2 immune genes. This model has independent prognostic significance for patients with hepatocellular carcinoma and directly quantifies mRNA expression; thus, it has considerable potential for use in future clinical trials and could be implemented for determining the prognoses of individual patients in clinical practice. Moreover, the model reflects the intensity of the immune response triggered by TP53 status in the microenvironment of hepatocellular carcinoma. This study is also the first to describe an immune prognostic model associated with TP53 mutations and can be used as a reference for understanding other cancers.
Alt-text: Unlabelled Box
1. Introduction
Hepatocellular carcinoma (HCC) ranks sixth among the most common types of cancer and has one of the highest mortality rates among cancers [1,2]. Currently, there are a number of established treatments for HCC, including chemotherapy with sorafenib, vascular catheterization, radiofrequency ablation, surgical resection, and liver transplantation [3,4]. However, the recurrence rate is high, even for patients who have received treatment in the early stage, and the survival rate of patients with advanced cancer, including those who receive treatment, is poor [4]. Tumour-promoting immune diseases are considered to enable the development of HCC. HCC cells stimulate a significant immune response, which yields the proper microenvironment for their development [5]. Because of the poor prognosis after standard treatment, immunotherapy is being studied in depth as an additional treatment [6]. In addition, a number of immune-related parameters have been reported to predict the prognosis of patients with HCC, further emphasizing the significance of immune status for determining the outcomes of HCC [5,7]. Nevertheless, few studies have systematically investigated the immune phenotype within the HCC microenvironment and its relationship with prognosis.
Sentences similar to “In human cancer, TP53 is the most commonly mutated gene” have appeared in the introductions of thousands of publications dating back to 1990, one year after the first TP53 mutation was described in colorectal and lung cancer [8,9]. This discovery was followed by the identification of hundreds of new cancer genes, although none of them surpassed the importance of the discovery of TP53. Although thousands of cancer genomes have been sequenced, candidates of paramount importance have not been found. TP53 mutation is often observed and is among the five most conspicuous mutations in common human cancers [10,11]. The wild-type TP53 protein plays important roles in apoptosis after DNA damage and in cell cycle regulation [12]. However, in the event of TP53 mutation, cells with DNA damage can escape apoptosis and transform into cancer cells. Furthermore, the mutant TP53 protein loses its wild-type function and accumulates in the nucleus [13]. This accumulation is considered to be a highly specific marker of malignant tumours [13]. A study covering 12 tumour types with a total of 3281 tumours found that the average mutation frequency of TP53 was approximately 42% [11]. The high mutation rate of TP53 makes its genetic alteration a very attractive potential therapeutic target. Gene therapy, targeted tumour vaccines, and anticancer drugs targeting TP53 mutations, including APR-246, MK-1775, ALT-801, and Kevetrin, are in the early stages of clinical trials. TP53 mutation is also the most common mutation in HCC [14]. This gene plays an important role in maintaining genomic stability, and its functional deletion can cause centrosome amplification, aneuploid cell proliferation and chromosomal instability (CIN) [15]. In particular, when TP53 mutations are combined with functional defects in the tumour suppressor pRb or with spindle checkpoint defects, they are more likely to cause high-level CIN and genomic instability [16]. Considerable data have shown that mutant TP53 proteins simultaneously lose their tumour-suppressive functions and obtain new capacities to advance tumourigenesis [17]. In HCC, TP53 alterations are correlated with serum alpha-fetoprotein (AFP) levels, tumour stage, vascular invasion, tumour differentiation and Child-Pugh class [[18], [19], [20], [21]]. Compared with HCC patients with wild-type TP53, those with tumour TP53 mutations have shorter overall survival (OS) and relapse-free survival times [22]. Thus, understanding the exact effects of TP53 on the pathogenesis of HCC and other forms of cancer is critical.
Interestingly, one of the most recent studies suggested that different immune responses are related to TP53 mutational status [23,24]. Therefore, we speculate that the shorter OS of HCC patients with TP53 mutation may be partly caused by the specific influences of these mutations on the cancer-associated immune system. In this study, we conducted a comprehensive analysis of TP53 mutation status and RNA expression to study the relationship between TP53 mutations and immune responses in HCC. The results showed that the immune response of HCC without TP53 mutation (TP53WT) was markedly stronger than that of HCC with TP53 mutation (TP53MUT). Importantly, our immune prognostic model (IPM) including immunological genes whose expression is affected by TP53 mutations can be used as an important prognostic model and has potential for use in patient management, and the included genes can serve as potential therapeutic biomarkers for HCC.
2. Materials and methods
2.1. RNA-sequencing data
The somatic mutation status for 364 HCC samples (workflow type: VarScan2 Variant Aggregation and Masking), and gene expression data and the corresponding clinical datasheets for 374 HCC samples were obtained from the Cancer Genome Atlas (TCGA) website (https://portal.gdc.cancer.gov/repository) (up to September 10, 2018) [14]. Surgical resection samples were collected from patients diagnosed with HCC, and these patients did not receive prior treatment for their disease [14]. Among these HCC samples, 359 HCC samples with RNA-sequencing data and TP53 mutation information were subjected to subsequent analyses. Sequence data were obtained using the Illumina HiSeq_RNA-Seq and Illumina HiSeq_miRNA-Seq platforms. The study reported herein fully satisfies the TCGA publication requirements (http://cancergenome.nih.gov/publications/publicationguidelines). The gene symbols were annotated based on the Homo_sapiens.GRCh38.91.chr.gtf file (http://asia.ensembl.org/index.html). Log2 transformations were performed for all gene expression data. The function of the trimmed mean of M values (TMM) normalization method of the edgeR R package (Version 3.24.3; http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) in R software (Version 3.5.2; https://www.r-project.org/) was applied to normalize the downloaded data [25]. The average RNA expression value was used when duplicate data were found. Genes with an average expression value >1 were retained, and low-abundance RNA-sequencing data were removed.
2.2. Microarray data
The gene expression profile matrix files from GSE54236 based on platform GPL6480 (including 78 HCC samples and 77 adjacent noncancerous samples), GSE76427 based on platform GPL10558 (including 115 HCC samples and 52 adjacent noncancerous samples), and GSE14520 based on platform GPL571 (including 225 HCC samples and 220 adjacent noncancerous samples) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Among these datasets, only gene expression data for GSE76427 were subjected to log2 transformation. The average RNA expression value was taken when duplicate data were found. Genes with an average expression value >1 were retained, and low-abundance RNA-sequencing data were removed. Three datasets (GSE54236 (n = 78), GSE76427 (n = 115), and GSE14520 (n = 221)) with survival information were integrated into the meta-GEO HCC cohort (n = 414) to validate the IPM. The sva package (Version: 3.30.1; http://bioconductor.org/packages/release/bioc/html/sva.html) was used to eliminate batch effects, and the scale method of the limma R package (Version 3.38.3; http://www.bioconductor.org/packages/release/bioc/html/limma.html) was used to normalize the data [26]. The obtained data were used according to the TCGA and GEO data access policies. Both mRNA profile data and clinical feature data for HCC are publicly obtainable and open access. All analyses were carried out based on pertinent guidelines and regulations.
2.3. Patients in the Peking HCC cohort and sample collection
From 2004 to 2015, 101 patients who underwent surgery and were diagnosed with HCC at Peking Union Medical College Hospital (Beijing, China) participated in this study in accordance with the provisions of the Helsinki Declaration (Table S1). These patients did not undergo neoadjuvant therapy before surgery. Two experienced pathologists examined all haematoxylin and eosin (H&E)-stained slides of each tumour sample. All final diagnoses were based on the morphology of the tumour samples after staining with H&E. Informed consent forms were signed by all patients. One-hundred thirty-one formalin-fixed paraffin-embedded HCC samples were collected to examine the protein levels of immune genes.
2.4. Immunohistochemistry (IHC)
Paraffin-embedded HCC samples were serially sectioned at 4-μm intervals and subsequently mounted on glass slides. The slides were then baked in the oven at 60 °C for 1 h, deparaffinized, and rehydrated. Heat-mediated antigen retrieval was conducted in a pressure cooker in 10 mmol/L Tris-citrate buffer (pH: 6.0). Endogenous peroxidase activity was blocked by incubating the sections with 3% hydrogen peroxide at room temperature for 10 min. After washing with phosphate-buffered saline (PBS) and incubation with goat serum at room temperature for 30 min, the slides were incubated with primary antibodies overnight at 4 °C. After washing with PBS, each slide was incubated with the appropriate peroxidase-labelled AffiniPure goat anti-rabbit IgG (H + L) (111–035-0030, 1:200, Jackson) secondary antibody for 30 min. Each section was washed with PBS and then developed with 3.3′-diaminobenzidine (DAB) solution for 5 min. Each section was washed with water before counterstaining with haematoxylin. The results of IHC staining were evaluated and scored by two pathologists. For EXO1 (exonuclease 1) expression analysis, a primary anti-EXO1 antibody (LS-C408381, 1:100; LifeSpan) was used. EXO1 is localized in the nucleus of tumour cells. The proportion of stained tumour cells was counted by two pathologists. Scores for the intensity of staining were determined as follows: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The staining index (SI) for EXO1 was calculated as staining intensity × the proportion of positive tumour cells. For TREM-1 (triggering receptor expressed on myeloid cells-1) expression analysis, a primary anti-TREM-1 antibody (ab225861, 1:200; Abcam) was used, and two pathologists counted the number of TREM-1-positive infiltrating lymphocytes (TILs). Images were obtained using a NanoZoomer S210 C13239-01 scanner.
2.5. Gene set enrichment analysis (GSEA)
To determine how the immunological pathways and corresponding immune genes differ between HCC samples without (n = 249) and with (n = 110) TP53 mutations in the TCGA HCC cohort, GSEA (Version: 3.0; http://software.broadinstitute.org/gsea/index.jsp) was performed [27]. An annotated gene set file (c5.bp.v6.2.symbols.gm) was selected for use as the reference gene set. The threshold was set at P < 0.05.
2.6. Differentially expressed gene (DEG) analysis
We compared 249 HCC samples without TP53 mutations and 110 HCC samples with TP53 mutations to identify DEGs using the edgeR R package, and the thresholds were |log2-fold change (FC)| > 2.0 and FDR < 0.01 [25].
2.7. Construction and validation of an immune-related prognostic model
Among the 359 HCC samples with RNA-sequencing data and TP53 mutation information, 350 HCC samples with survival information were subjected to subsequent analyses. The expression profiles of the DEGs from 350 HCC patients with survival information were analysed via univariate Cox regression analysis. The prognostic value of the DEGs for OS was defined by univariate Cox regression analysis. In this analysis, genes were regarded as significant at P < 0.001. For highly correlated genes, the traditional Cox regression model cannot be used directly; thus, least absolute shrinkage and selection operator (LASSO) with L1-penalty, which is a popular method for determining interpretable prediction rules that can handle the collinearity problem, was used [28]. Among the immune genes that were significant in the univariate Cox regression analysis, key immune genes were selected by the LASSO method. In this approach, a sub-selection of immune genes involved in HCC patient prognosis was determined by shrinkage of the regression coefficient via the imposition of a penalty proportional to their size. Finally, a relatively small number of indicators with a weight of nonzero remained, and most of the potential indicators were shrunk to zero. Therefore, LASSO-penalized Cox regression was implemented to further reduce the number of immune genes. In this analysis, we subsampled the dataset 1000 times and chose the immune genes that were repeated >900 times [29]. LASSO Cox analysis was performed by using the glmnet R package (Version: 2.0–16; https://cran.r-project.org/web/packages/glmnet/index.html). Finally, an immune-related prognostic model was constructed utilizing the regression coefficients derived from multivariate Cox regression analysis to multiply the expression level of each immune gene. X-tile 3.6.1 software (Yale University, New Haven, CT, USA) was applied to determine the best cutoff for HCC patients classified as low risk and high risk. The log-rank test and Kaplan-Meier survival analysis were used to assess the predictive ability of the prognostic model.
2.8. Estimation of immune cell type fractions
CIBERSORT is an approach to characterizing the cell composition of complex tissues based on their gene expression profiles, and it is highly consistent with ground truth estimations in many cancers [30]. A leukocyte gene signature matrix consisting of 547 genes, which was termed LM22, was used to distinguish 22 immune cell types, and these types contained myeloid subsets, natural killer (NK) cells, plasma cells, naive and memory B cells and seven T cell types. We utilized CIBERSORT in combination with the LM22 signature matrix to estimate the fractions of 22 human haematopoietic cell phenotypes between HCC samples with and without TP53 mutations. The sum of all estimated immune cell type fractions is equal to 1 for each sample.
2.9. Functional enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (Version: 6.8; https://david.ncifcrf.gov/) and the KO-Based Annotation System (KOBAS) (Version: 3.0; http://kobas.cbi.pku.edu.cn/) were used to perform functional and pathway enrichment analyses to assess the biological implications of the prognostic model [31,32]. Significant biological processes and pathways were visualized using the GOplot (Version: 1.0.2; https://cran.r-project.org/web/packages/GOplot/index.html) and ggalluvial (Version: 0.9.1; https://cran.r-project.org/web/packages/ggalluvial/index.html) R packages, respectively.
2.10. Independence of the IPM from traditional clinical features
Among 350 HCC samples with survival information, 213 HCC samples with complete clinical information, including AFP, gender, weight, age, pathologic stage, vascular tumour invasion, weight, histologic grade, hepatitis B status, hepatitis C status, alcohol consumption status and non-alcoholic fatty liver disease status, were subjected to subsequent analyses. To validate whether the predictions of the prognostic model were independent of traditional clinical features (including AFP, gender, weight, age, pathologic stage, vascular tumour invasion, weight, histologic grade, hepatitis B status, hepatitis C status, alcohol consumption status and non-alcoholic fatty liver disease status) for patients with HCC, univariate and multivariate Cox regression analyses were conducted.
2.11. Construction and evaluation of the nomogram
To individualize the predicted survival probability for 1 year, 3 years and 5 years, a nomogram was constructed based on the results of the multivariate analysis. The rms R package (Version: 5.1–3; https://cran.r-project.org/web/packages/rms/index.html) was used to generate a nomogram that included significant clinical characteristics and calibration plots. Calibration and discrimination are the most commonly used methods for evaluating the performance of models. In this study, the calibration curves were graphically assessed by mapping the nomogram-predicted probabilities against the observed rates, and the 45° line represented the best predictive values. A concordance index (C-index) was used to determine the discrimination of the nomogram, and it was calculated by a bootstrap approach with 1000 resamples [33]. In addition, the predictive accuracies of the nomogram and separate prognostic factors were compared using the C-index and receiver operating characteristic (ROC) analyses. All statistical tests were two-tailed with a statistical significance level set at 0.05 in this study.
3. Results
3.1. Association between immune phenotype and TP53 mutations in HCC
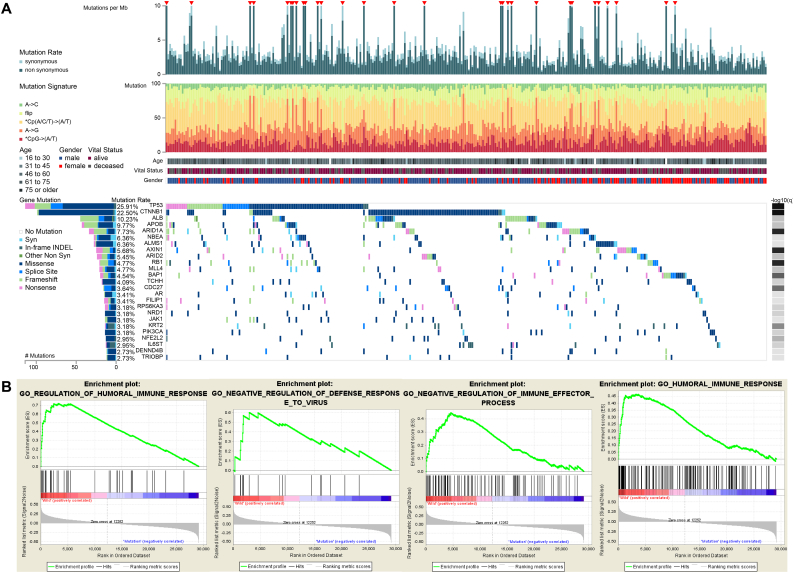
In HCC, TP53 mutation is the most common type of mutation (Fig. 1A). Pioneering investigations demonstrated that TP53 mutation is associated with OS in patients with HCC [22]. Although the pathogenetic role of TP53 mutations in the prognosis of patients with HCC has been well documented, their specific influences on immune profiles in HCC have not been thoroughly investigated. Hence, for the first time, we utilized gene expression data and clinical information on HCC patients in TCGA to find immune-related biological processes linked to TP53 status. GSEA analysis of HCC samples without (n = 249) and with (n = 110) TP53 mutations was performed. The results showed that TP53WT HCCs were significantly enriched in 414 biological processes, and 4 immune-related biological processes were selected: REGULATION_OF_HUMORAL_IMMUNE_RESPONSE (normalized enrichment score, NES = 2.201, size = 47), NEGATIVE_REGULATION_OF_DEFENSE_RESPONSE_TO_VIRUS (NES = 1.722, size = 17), NEGATIVE_REGULATION_OF_IMMUNE_EFFECTOR_PROCESS (NES = 1.681, size = 95), and HUMORAL_IMMUNE_RESPONSE (NES = 1.586, size = 153) (P < 0.05) (Fig. 1B) (Table S2). In contrast, TP53MUT HCCs were not enriched in any immune-related biological processes (Table S3).
Fig. 1.
Gene set enrichment analysis of TP53 in the TCGA dataset. (A) Genomic landscape of HCC and the mutational signatures in the TCGA dataset, which were assayed on the FireBrowse platform. (B) Significant enrichment of the immune-related phenotype in TP53WT HCC patients compared with that in TP53MUT HCC patients.
3.2. Identification of differentially expressed immune-related genes between HCC samples with and without TP53 mutations
To identify the correlations between TP53 status and 4 immune-related processes, 312 immune-related genes were obtained from the 4 immune-related processes. To identify differentially expressed immune-related genes between TP53WT HCC and TP53MUT HCC tissues, we performed differential expression analysis using the edgeR package [25]. Of the 312 immune-related genes investigated, 37 genes were differentially expressed between TP53WT and TP53MUT HCCs (FDR < 0.05 and |log2 FC| > 1) (Table S4).
3.3. Construction of an IPM and evaluation of its predictive ability in the TCGA HCC cohort
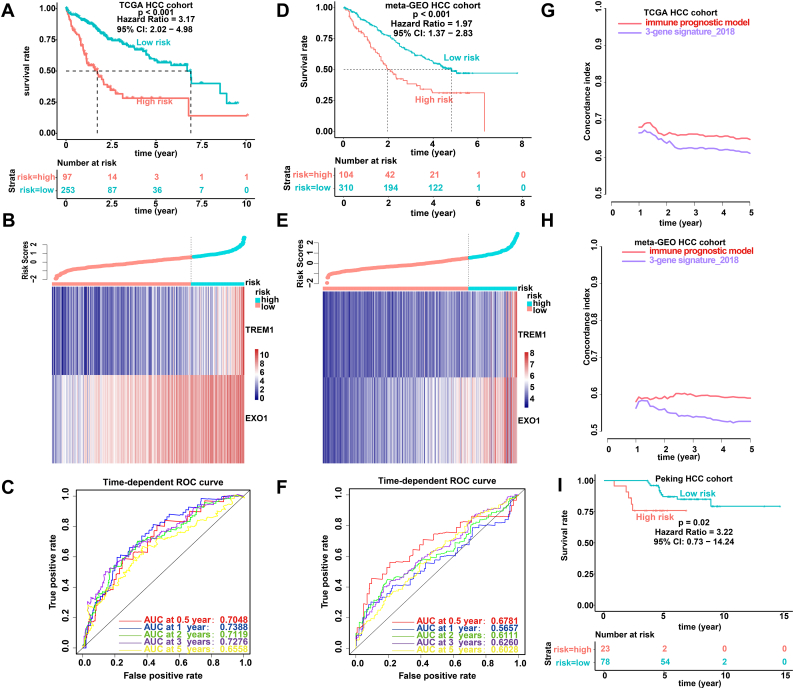
Taking the differences in immune status between TP53WT and TP53MUT HCCs into consideration, we attempted to assess the predictive ability of the DEGs. Univariate Cox regression analysis was performed, and it revealed that 7 of the 37 DEGs were significantly related to OS (Table S5). To find the genes with the greatest prognostic value, we applied Cox-proportional hazards analysis based on the L1-penalized (LASSO) estimation, and two genes (TREM1 and EXO1) that appeared >900 times out of 1000 repetitions were selected [29,34]. We used LASSO because it is suitable for constructing models when there are a large number of correlated covariates [34]. To obtain a uniform cutoff value to stratify the patients into high- and low- risk groups, we conducted normalization of the expression levels of TREM1 and EXO1 in the TCGA, meta-GEO and Peking HCC cohorts with mean value = 0 and standard deviation (SD) = 1 [35]. Then, by weighting the normalized expression level of each immune gene to the regression coefficients of the multivariate Cox regression analysis, we established a risk score model to predict patient survival (risk score = normalized expression level of TREM1 * 0.336 + normalized expression level of EXO1 * 0.392). We calculated the risk score for each patient and categorized the patients into high-risk or low-risk groups according to the optimal cutoff point (1.37) obtained from X-tile software. The cutoff point (1.37) in the TCGA HCC cohort served as the cutoff to assign patients into high- and low- risk groups across all the HCC cohorts. As shown in Fig. 2A, the high-risk patients had a shorter OS than their low-risk counterparts. In addition, the high-risk group showed a 3.17-fold higher risk (95% confidence interval (CI): 2.02–4.98, P < 0.001) than the low-risk group. The risk score distribution and gene expression data are shown in Fig. 2B. Fig. 2C shows the predictive potential of the IPM using time-dependent ROC curves. The area under the ROC curve (AUC) of the prognostic model for OS was 0.7048 at 0.5 years, 0.7388 at 1 year, 0.7119 at 2 years, 0.7276 at 3 years and 0.6558 at 5 years.
Fig. 2.
Prognostic analysis of the IPM. Kaplan-Meier survival, risk score and time-dependent ROC curves of the IPM for the TCGA HCC cohort (A-C) and meta-GEO HCC cohort (D-F). (A and D) OS was significantly higher in the low-risk score group than in the high-risk score group. (B and E) Relationship between the risk score (upper) and the expression of two prognostic immune genes (bottom) is shown. (C and F) Time-dependent ROC curve analysis of the IPM. (G-H) The C-index was used to evaluate prognostic performance for survival prediction. Performance was compared between the IPM and 3-gene signature_2018 by calculating the C-index in the TCGA and meta-GEO HCC cohorts. (I) Kaplan-Meier survival of the IPM for the Peking HCC cohort by using immunohistochemistry.
3.4. Validation and evaluation of the IPM in the meta-GEO HCC cohort and Peking HCC cohort
To determine whether the IPM was robust, the performance of the IPM with the TCGA HCC cohort was assessed in the meta-GEO HCC cohort, which consisted of 414 HCC patients. With the same formula and the same cutoff obtained from the TCGA HCC cohort, the patients in the meta-GEO HCC cohort were divided into a high-risk group and a low-risk group. Consistent with the outcomes of the TCGA HCC cohort, patients who were assigned to the high-risk group had significantly worse OS than those who were assigned to the low-risk group (Fig. 2D). The risk in the high-risk group was 1.97-fold higher than that in the low-risk group (95% CI: 1.37–2.83, P < 0.001), demonstrating the applicability of the developed IPM in different platforms. The risk score distribution and gene expression data are shown in Fig. 2E. Furthermore, the IPM achieved an AUC of 0.6781 at 0.5 years, 0.5657 at 1 year, 0.6111 at 2 years, 0.6260 at 3 years and 0.6028 at 5 years (Fig. 2F). Recently, Yang et al. proposed a prognostic model including 3 genes (secreted phosphoprotein 2 (SPP2); cell division cycle 37-like 1 (CDC37L1); and enoyl-CoA hydratase domain containing 2 (ECHDC2)) to predict the prognosis of patients with HCC [36]. They first integrated 7 HBV-associated HCC datasets to identify DEGs. Second, weighted gene co-expression network analysis (WGCNA) was performed on those DEGs to identify the most significant module. Third, a protein-protein interaction (PPI) network was constructed for the most significant module to identify hub genes. Finally, a three-gene prognostic signature (risk score = expression of SPP2 * - 0.1941 + expression of CDC37L1 * - 0.5466 + expression of ECHDC2 * - 0.4714) for these hub genes was established by univariate and multivariate Cox regression analysis in the GSE14520 dataset. We calculated the C-indexes to compare the prognostic values of their model and our IPM. The C-index is the most commonly used performance measure for survival models; it ranges from 0.5 to 1 and is equal to the AUC [37]. The higher the value of the C-index is, the better the predictability of the model. The C-index of the IPM for 1 to 5-year OS exceeded that of the previous model in both the TCGA and meta-GEO HCC cohorts, suggesting that our IPM had favourable efficacy for predicting both short- and long-term prognosis (Fig. 2G and H ).
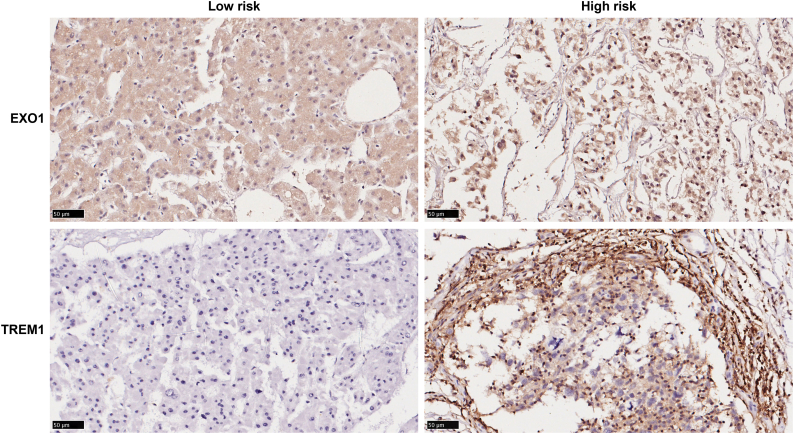
To further examine the robustness and practical application of the IPM, we validated the prognostic power of the IPM using protein values for immune genes and survival information for patients with HCC in our cohort recruited from Peking Union Medical College Hospital. This cohort consisted of 101 HCC patients. We detected the protein levels of two immune genes (TREM1 and EXO1) with IHC. The results revealed that the IPM consisting of these two immune genes at the protein level can differentiate HCC patients with a low or high risk of poor survival based on the same formula and the same cutoff obtained from the TCGA HCC cohort. Representative staining images of TREM1 and EXO1 were demonstrated in Fig. S1. The patients in the high-risk group exhibited poorer OS than the patients in the low-risk group (hazard ratio (HR): 3.22; 95% CI: 0.73–14.24, P = 0.02) (Fig. 2I). Overall, our results demonstrated that the IPM is robust across different molecular levels, platforms and datasets.
Fig. S1.
Representative images of immunostaining at high-risk and low-risk patients.
3.5. Stratification analyses of OS for the IPM according to TP53 status in the TCGA HCC cohort
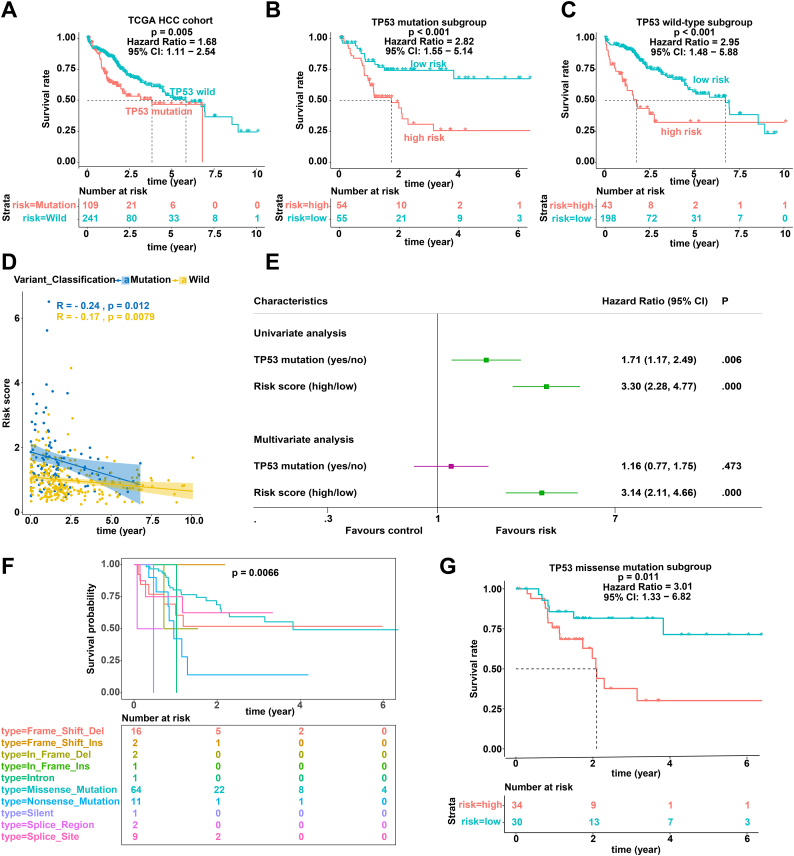
Consistent with the IPM, TP53 status was also significantly related to the prognosis of patients with HCC (Fig. 3A). Stratification analyses were performed to test whether the prognostic value of the IPM was independent of TP53 status. Therefore, patients in the TCGA HCC cohort were divided into two groups according to TP53 status. Stratification analyses suggested that the IPM was significantly related to OS in the TP53WT and TP53MUT TCGA HCC cohorts (Fig. 3B and C). In addition, correlation analyses suggested that the risk score was significantly negatively associated with OS in the TP53WT and TP53MUT TCGA HCC cohorts (Fig. 3D). Furthermore, univariate and multivariate Cox regression analyses showed that the predictive power of the IPM for the OS of patients with HCC is independent of TP53 status (Fig. 3E).
Fig. 3.
Prognostic analysis of TP53 mutation. (A-C) Kaplan-Meier survival of TP53 status (A), TP53 mutation subgroup (B), and TP53 wild-type subgroup (C). (D) Analysis of the correlation between risk score and survival time according to TP53 status. (E) Univariate and multivariate regression analysis of the relation between the IPM and TP53 status regarding prognostic value. Red indicates no statistical significance, and green indicates statistical significance. (F) Kaplan-Meier survival of the different types of TP53 mutations. (G) Kaplan-Meier survival of the TP53 missense mutation subgroup. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Since the TP53 mutation type affects TP53 function, we performed stratification analysis of different TP53 mutation types and found that the TP53 mutation type affects the prognosis of patients with HCC (Fig. 3F) [38,39]. To test whether the prognostic value of the IPM was independent of the TP53 mutation type, we performed prognostic analysis of the TP53 missense mutation subgroup, which has the largest proportion among various TP53 mutation types. As expected, the IPM was able to classify patients into high- and low-risk groups within the TP53 missense mutation subgroup (Fig. 3G).
3.6. Low risk indicated an enhanced local immune phenotype
GSEA was performed between the 253 low-risk and 97 high-risk HCC patients in the TCGA HCC cohort. The GSEA revealed that the low-risk HCC patients were associated with three immune processes: HUMORAL_IMMUNE_RESPONSE (NES = 1.700, size = 153), HUMORAL_IMMUNE_RESPONSE_MEDIATED_BY_CIRCULATING_IMMUNOGLOBULIN (NES = 1.775, size = 63), and REGULATION_OF_HUMORAL_IMMUNE_RESPONSE (NES = 2.157, size = 47) (P < 0.05) (Table S6). In contrast, the high-risk HCCs were related to only one immune process: SOMATIC_DIVERSIFICATION_OF_IMMUNE_RECEPTORS (NES = −0.548, size = 39) (Table S7). Therefore, the local immune signature may confer an intense immune phenotype in the low-risk group and a weakened immune phenotype in the high-risk group.
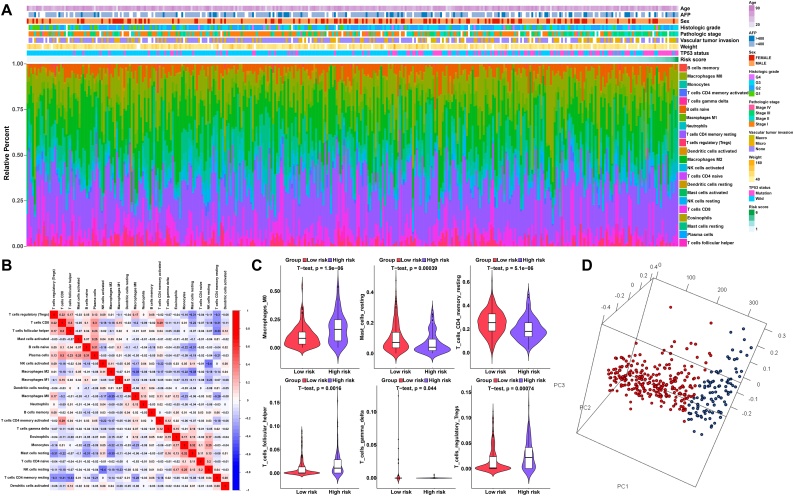
3.7. Immune landscape between the low- and high-risk HCC patients
Using the CIBERSORT method in combination with the LM22 signature matrix, we estimated the differences in the immune infiltration of 22 immune cell types between low- and high-risk HCC patients [30]. Fig. 4A summarizes the results obtained from 350 HCC patients. Within and between groups, the proportion of immune cells in HCC varies (Fig. 4A). Therefore, variations in the proportions of tumour-infiltrating immune cells might represent an intrinsic feature that could characterize individual differences. In addition, the proportions of different subpopulations of tumour-infiltrating immune cells were weakly to moderately correlated (Fig. 4B). The high-risk HCC patients had significantly higher proportions of T cells follicular helper, T cells regulatory (Tregs) and macrophages M0, and significantly lower proportions of T cells CD4 memory resting, T cells gamma delta and mast cells resting than the low-risk HCC patients (P < 0.05) (Fig. 4C). Furthermore, based on the above-identified cell subpopulations, the samples of high-risk HCC patients and low-risk HCC patients were clearly separated into two discrete groups based on principal component analysis (Fig. 4D). Thus, these results suggest that abnormal immune infiltration and the heterogeneity of immune infiltration in HCC may serve as prognostic indicators and targets for immunotherapy and may have significant clinical implications.
Fig. 4.
The landscape of immune infiltration in high- and low-risk HCC patients. (A) Relative proportion of immune infiltration in high- and low-risk patients. (B) Correlation matrix of all 22 immune cell proportions. (C) Violin plots visualizing significantly different immune cells between high-risk and low-risk patients. (D) Principal components analysis performed on HCC patients based on significant differences in immune cells between high-risk and low-risk HCC patients.
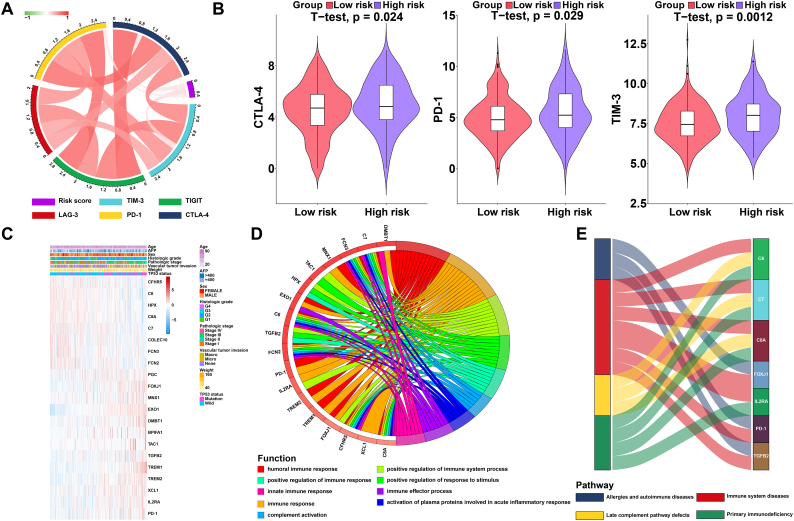
Drugs targeting immune checkpoints have been shown to play antitumour roles by reversing tumour immunosuppressive effects [6]. The expression of immune checkpoints has emerged as a biomarker for the selection of HCC patients for immunotherapy [6]. Therefore, we assessed the correlation between patient risk scores and expression of critical immune checkpoints (CTLA-4, PD-1, TIM-3, LAG-3, and TIGIT) and found that the risk score was significantly related to the expression of CTLA-4, PD-1 and TIM-3 (P < 0.05) (Fig. 5A) (Table S8) [40]. In addition, we investigated the expression of CTLA-4, PD-1 and TIM-3 between the low- and high-risk HCC patients. The expression of CTLA-4, PD-1 and TIM-3 in the high-risk HCC group was significantly higher than that in the low-risk HCC group (P < 0.05), indicating that the poor prognosis of high-risk HCC patients is partly due to the immunosuppressive microenvironment (Fig. 5B).
Fig. 5.
Enrichment analysis of the immune prognostic model. (A) Correlation of the risk score with the expression of several prominent immune checkpoints. (B) Violin plots visualizing significantly different immune checkpoints between high-risk and low-risk patients. (C) Heatmap of immune genes that were differentially expressed in samples from patients with high and low risk scores. (D) Circular plot of the biological processes enriched for the immune genes. (E) Sankey plot of the pathways enriched for the immune genes.
3.8. Altered pathways in high- and low-risk group patients
GO analysis was performed in this study to obtain a novel understanding of the biological effects of the IPM. The immune genes were differentially expressed between the groups at low risk and high risk for HCC (P < 0.05), and genes whose expression correlated with risk scores (absolute Pearson correlation coefficient > 0.2 and P < 0.05) were considered to be risk score-associated genes. Twenty-one immune genes were identified (Fig. 5C) and were subjected to GO and Kyoto Encyclopaedia of Genes and Genomes (KEGG) analyses to identify the potential biological functions (FDR < 0.0001) and pathways (FDR < 0.01) of these genes (Fig. 5D and E) (Table S9, Table S10). According to the results, the genes related to the risk score in the TCGA HCC dataset were mainly enriched in the humoral immune response and immune system diseases pathway (Fig. 5D and E) (Table S9, Table S10).
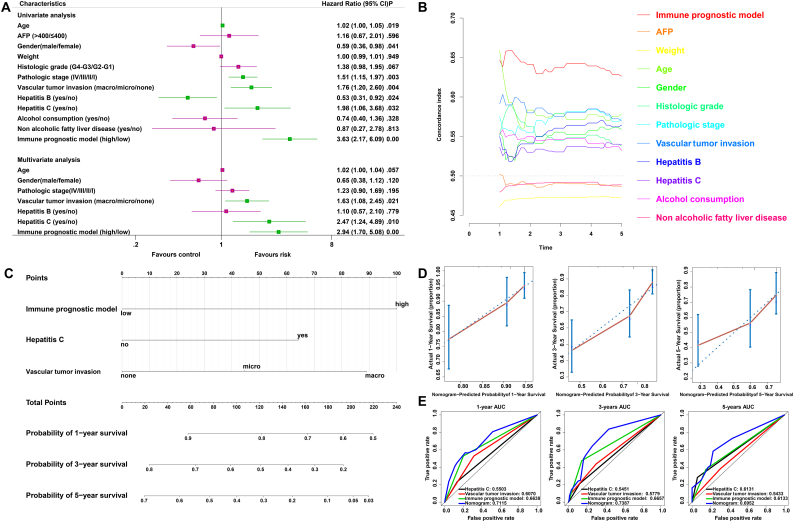
3.9. The IPM is independent of conventional clinical characteristics
Univariate and multivariate Cox regression analyses were conducted to explore whether the prognostic value of the IPM was independent of other clinical factors in the TCGA HCC cohort. After adjusting for clinical characteristics, including gender, age, pathologic stage, vascular tumour invasion, hepatitis B status and hepatitis C status, the IPM remained an independent prognostic factor, thus confirming its robustness for independently predicting HCC prognosis (Fig. 6A). The multivariate Cox regression analysis indicated that the IPM was significantly correlated with the survival information (P < 0.01) and the highest median risk score (HR = 2.94, 95% CI = 1.70–5.08). Furthermore, we compared the C-index between the IPM and conventional clinical characteristics, and of the 11 survival-predictive factors, the IPM had a higher mean C-index (0.6380) than the conventional clinical characteristics (0.5001 to 0.5900) (Fig. 6B). Altogether, these results indicated that the IPM was independent of conventional clinical characteristics and performed better than conventional clinical characteristics in survival prediction.
Fig. 6.
Relationship between the IMP and other clinical information. (A) Univariate and multivariate regression analysis of the relation between the immune prognostic model and clinicopathological features regarding prognostic value. Red indicates no statistical significance, and green indicates statistical significance. (B) The prognostic performance was compared between the IPM and different conventional clinical characteristics by calculating the C-index. (C) Nomogram for predicting the probability of 1-, 3-, and 5-year OS for HCC patients. (D) Calibration plot of the nomogram for predicting the probability of OS at 1, 3, and 5 years. (E) Time-dependent ROC curve analyses of the immune prognostic model, hepatitis C status, vascular tumour invasion, and nomogram. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.10. Construction and validation of a nomogram based on the IPM
To provide clinicians with a quantitative approach to predicting the prognosis of HCC patients, a nomogram that integrated the IPM and independent clinical risk factors (hepatitis C and vascular tumour invasion) was constructed (Fig. 6C). In this nomogram based on multivariate Cox analysis, a point scale was used to assign points to these variables. A straight line was drawn upward to determine the points for the variables, and the sum of the points assigned for each variable was rescaled to a range from 0 to 100. The points of the variables were accumulated and recorded as the total points. The probability of HCC patient survival at 1, 3, and 5 years was determined by drawing a vertical line from the total point axis straight downward to the outcome axis. For example, an HCC patient with high risk (100 points) hepatitis C (64 points), and vascular tumour invasion (micro: 45 points) received a total point score of 209. The probability of 1-year survival was determined by drawing a vertical line from the total point axis at a value of 209 straight downward to the outcome axis, which showed that the probability of 1-year survival was 54%. The IPM was found to contribute the most risk points (ranging from 0 to 100) compared with the other clinical information, which was consistent with our Cox multivariate regression results. The C-index for the nomogram was 0.6969 with 1000 bootstrap replicates (95% CI: 0.6239–0.7698). The bias-corrected line in the calibration plot was found to be close to the ideal curve (the 45-degree line), which indicated good agreement between the prediction and the observation (Fig. 6D). We also compared the predictive accuracy of this nomogram with that of hepatitis C, vascular tumour invasion and the IPM, and the nomogram performance (C-index: 0.6969) was better than the performance of hepatitis C (C-index: 0.5390), vascular tumour invasion (C-index: 0.5867) and the IPM (C-index: 0.6380). The AUC was also the largest for the nomogram (Fig. 6E). In sum, these findings suggest that the nomogram was a better model for predicting short-term or long-term survival in HCC patients than individual prognostic factors.
4. Discussion
In lung adenocarcinoma, TP53 mutation can significantly increase the expression of immune checkpoints, activate effector T cells and increase interferon gamma levels [23]. TP53 mutation can also be used as a predictor of anti-PD-1 immunotherapy in lung cancer [41]. Therefore, it is necessary to further investigate the immune-related effects of TP53 status. However, the mechanism by which TP53 mutation affects the regulation of the HCC immunophenotype and the prognosis of HCC is unknown. In addition, it is important to develop meaningful immune-related prognostic models to determine the immune status of patients because these models represent powerful prognostic biomarkers and can also be used to stratify patients to increase the effectiveness of immunotherapy. In recent years, gene expression signatures representative of tumour immune status have been identified, and their potential clinical relevance in several cancers has been evaluated [42,43]. Several studies have sought to elucidate the immune microenvironment in HCC [44,45]. Rather than employing immune privilege, HCC in fact coordinates a robust immune response involving the innate and adaptive immune systems [44,45]. However, the role of local immune response status in HCC prognosis prediction has not been explored. In the current study, we investigated the role of TP53 mutations in the regulation of immune phenotype in HCC. In GSEA analysis, we found that TP53WT HCCs had a significantly stronger local immunophenotype than TP53MUT HCCs. Then, we profiled an immune-related gene set affected by TP53 mutation and generated a 2-gene-based IPM that could identify patients with HCC who had a high risk of unfavourable prognosis. The results obtained in this study may reveal a feasible therapeutic strategy that involves shaping the immune microenvironment to improve clinical outcomes. The genes (TREM-1 and EXO1) that constitute our IPM could be regarded as individual targets, and they may provide better performance in combination, depending on their immune properties and prognostic significance.
TREM-1 is a cell surface receptor as well as a constituent of the immunoglobulin superfamily, which effectively expands inflammatory responses by secreting proinflammatory mediators [46]. Previous studies have reported that cancer cells can directly upregulate the expression of TREM-1 in patient macrophages, and in patients with non-small cell lung cancer, TREM-1 expression in tumour-associated macrophages is related to poor survival and recurrence [47]. In addition, the expression of TREM-1 by Kupffer cells is a pivotal factor in the evolution and progression of liver cancer [46]. EXO1 is an important nuclease in the mismatch repair system, which helps maintain genomic stability, regulate DNA recombination, and mediate cell cycle arrest [48]. Gene expression profiles in breast tumours show that elevated EXO1 expression is related to unfavourable prognosis, and single-nucleotide polymorphisms (SNPs) of EXO1 are related to hereditary susceptibility to HCC [49,50]. Furthermore, Tanaka et al. performed differential expression analysis between an aggressive recurrence group and a non-aggressive recurrence group of HCC and found that EXO1 was significantly upregulated in the aggressive recurrence group [51]. In our study, for the first time, we discovered that high expression of TREM-1 and EXO1 is linked to unfavourable prognosis in patients with HCC.
Additionally, we demonstrated that the IPM remained an independent prognostic factor after the modification of clinical characteristics. This result suggests that local immune status has the potential to improve the traditional features of accurate prognosis. Therefore, we propose a comprehensive assessment that combines our IPM and other clinical features (hepatitis C, vascular tumour invasion and the IPM). The calibration curve showed satisfactory agreement between the observed values and the predicted values for 1-, 3-, and 5-year OS. The main advantage of this model is that it provides a complementary perspective on individual tumours and develops an individual scoring system for patients; therefore, our nomogram could be a promising tool for clinicians in the future.
During cancer development in immune-competent hosts, to evade antitumour immune responses, less immunogenic cancer cells are selected (immune selection) and immunosuppressive networks are established (immune escape) according to the cancer immunoediting hypothesis [52,53]. Therefore, clinically significant cancers have several immunosuppressive mechanisms, such as increasing various immunosuppressive cells (e.g., Treg cells and tumour-associated macrophages), increasing the expression of various immunosuppressive molecules (e.g., cytotoxic T lymphocyte-associated antigen-4 (CTLA-4)), and decreasing the expression of cancer antigens, which results in the inability of CD8+ T cells to recognize cancer cells [54,55]. By blocking the function of immunosuppressive cells and immunosuppressive mechanisms, potential antitumour immune responses can be released. Here, we investigated the immune mechanisms between patients in the low- and high-risk groups and the possible use of cancer immunotherapy to enhance the antitumour immune response, and the results indicated that the proposed approach has promising clinical efficacy. High-risk HCC patients generally had higher fractions of T cells follicular helper, T cells regulatory (Tregs) and macrophages M0, and lower fractions of T cells CD4 memory resting, T cells gamma delta and mast cells resting than low-risk patients (P < 0.05). In addition, we investigated the expression of immune checkpoints (CTLA-4, PD-1 and TIM-3) between the low- and high-risk groups. The high-risk HCC patients had significantly higher expression of CTLA-4, PD-1 and TIM-3 than the low-risk patients (P < 0.05). Previous research confirmed that T cells CD4 memory resting can be further differentiated and confer various functions, including blocking CD8+ T cell activation and NK cell killing, suppressing harmful immunological reactions to self-antigens and foreign antigens, and aiding CD8+ T cells in tumour rejection [56,57]. Importantly, Tregs also expressed immune checkpoints, such as PD-1 and CTLA-4 [58]. The anti-CTLA-4 antibody ipilimumab inhibits interactions between antigen-presenting cells (APCs) and Tregs [58]. Analyses of anti-CTLA-4 antibodies in mouse models indicated that their antitumour efficacy was based on the depletion of CTLA-4+ Treg cells in tumours through antibody-dependent cellular cytotoxicity (ADCC), since the loss of crystallizable fragment (Fc) function of anti-CTLA-4 mAbs completely eliminates their antitumour effects [[59], [60], [61]]. Therefore, in our model, the risk score was compatible with the ability of tumour-infiltrating immune cells to determine the expression of immune checkpoints, suggesting that the poor prognosis of the high-risk group may be due to the stronger immunosuppressive environment and immune checkpoint expression in this group than in the low-risk group, and these differences promoted HCC growth, progression, invasion, and angiogenesis and resulted in poor prognosis. Furthermore, these results also indicate that high-risk patients will benefit more from immune checkpoint inhibitors than low-risk patients, thereby resulting in a better prognosis.
In the GSEA analysis between the low- and high-risk group patients, the high-risk and low-risk groups had different levels of immune pathway enrichment. The low-risk group was associated with three immune processes, while the high-risk risk group was related to only one immune process; therefore, we speculated that the local immune signature conferred an intense immune phenotype in the low-risk group and a weakened immune phenotype in the high-risk group. In addition, the high-risk patients had significantly higher expression of immunosuppressive molecules (e.g., CTLA-4, PD-1 and TIM-3) and increased levels of various immunosuppressive cells (e.g., Tregs and macrophages) than the low-risk group, suggesting that the weakened immune phenotype in the high-risk group may be due to its stronger immunosuppressive environment and immune checkpoint expression than the low-risk group. Therefore, the IPM may reflect the intensity of the immune response triggered by TP53 status.
Our research provides new insights into the HCC immune microenvironment and immune-related therapies. However, our research is limited because it was retrospective, and our results should thus be further confirmed by prospective studies. In addition, functional and mechanistic studies of the two genes individually and in combination should be conducted to support their clinical application.
In summary, for the first time, we identified and validated an IPM that is based on 2 immune genes, has independent prognostic significance for HCC patients and reflects the overall intensity of the immune response in the HCC microenvironment. This study is also the first to describe an IPM associated with TP53 mutations and can be used as a reference for understanding other cancers. Notably, the IPM provides an immunological perspective to elucidate the mechanisms that determine the clinical outcome of HCC.
The following are the supplementary data related to this article.
Clinicopathological parameters of HCC patients in the Peking HCC cohort.
Gene enrichment in TP53WT HCCs.
Gene enrichment in TP53MUT HCCs.
Differentially expressed immune-related genes between TP53WT and TP53MUT HCCs.
Univariate Cox regression analysis of differentially expressed immune-related genes.
Gene enrichment in low-risk HCCs.
Gene enrichment in high-risk HCCs.
Analysis of correlations between risk score and immune checkpoints.
Functional enrichment analysis of the twenty-one immune genes identified.
Pathway enrichment analysis of the twenty-one immune genes identified.
Funding sources
This work was supported by the International Science and Technology Cooperation Projects (2016YFE0107100), the Capital Special Research Project for Health Development (2014-2-4012), the Beijing Natural Science Foundation (L172055 and 7192158), the National Ten Thousand Talent Program, the Fundamental Research Funds for the Central Universities (3332018032), and the CAMS Innovation Fund for Medical Science (CIFMS) (2017-I2M-4-003 and 2018-I2M-3-001). The funders did not play a role in manuscript design, data collection, data analysis, data interpretation or writing of the manuscript.
Declaration of interests
None.
Author contributions
All authors searched the literature, designed the study, interpreted the findings and revised the manuscript. Junyu Long, Anqiang Wang, and Yi Bai carried out data management and statistical analysis and drafted the manuscript. Jianzhen Lin, Yan Jiang, Xu Yang and Dongxu Wang helped with cohort identification and data management. Xiaobo Yang helped with the statistical analysis. Haitao Zhao performed project administration.
Acknowledgements
None.
References
- 1.Li C., Li R., Zhang W. Progress in non-invasive detection of liver fibrosis. Cancer Biol Med. 2018;15(2):124–136. doi: 10.20892/j.issn.2095-3941.2018.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Heimbach J.K., Kulik L.M., Finn R.S., Sirlin C.B., Abecassis M.M., Roberts L.R. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi: 10.1002/hep.29086. (Baltimore, Md) [DOI] [PubMed] [Google Scholar]
- 5.Huang Y., Wang F.M., Wang T., Wang Y.J., Zhu Z.Y., Gao Y.T. Tumor-infiltrating FoxP3+ Tregs and CD8+ T cells affect the prognosis of hepatocellular carcinoma patients. Digestion. 2012;86(4):329–337. doi: 10.1159/000342801. [DOI] [PubMed] [Google Scholar]
- 6.Long J., Lin J., Wang A., Wu L., Zheng Y., Yang X. PD-1/PD-L blockade in gastrointestinal cancers: lessons learned and the road toward precision immunotherapy. J. Hematol. Oncol. 2017;10(1):146. doi: 10.1186/s13045-017-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Wang F., Wang Y., Zhu Z., Gao Y., Ma Z. Intrahepatic interleukin-17+ T cells and FoxP3+ regulatory T cells cooperate to promote development and affect the prognosis of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2014;29(4):851–859. doi: 10.1111/jgh.12418. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi T., Nau M.M., Chiba I., Birrer M.J., Rosenberg R.K., Vinocour M. p53: a frequent target for genetic abnormalities in lung cancer. Sci (New York, NY) 1989;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 9.Baker S.J., Fearon E.R., Nigro J.M., Hamilton S.R., Preisinger A.C., Jessup J.M. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Sci (New York, NY) 1989;244(4901):217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 10.Stratton M.R. Exploring the genomes of cancer cells: progress and promise. Sci (New York, NY) 2011;331(6024):1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 11.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai P.B., Chi T.Y., Chen G.G. Different levels of p53 induced either apoptosis or cell cycle arrest in a doxycycline-regulated hepatocellular carcinoma cell line in vitro. Apoptosis. 2007;12(2):387–393. doi: 10.1007/s10495-006-0571-1. [DOI] [PubMed] [Google Scholar]
- 13.Dowell S.P., Wilson P.O., Derias N.W., Lane D.P., Hall P.A. Clinical utility of the immunocytochemical detection of p53 protein in cytological specimens. Cancer Res. 1994;54(11):2914–2918. [PubMed] [Google Scholar]
- 14.Comprehensive and integrative genomic characterization of hepatocellular carcinomaCell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. (e23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao C.V., Asch A.S., Yamada H.Y. Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. Carcinogenesis. 2017;38(1):2–11. doi: 10.1093/carcin/bgw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning A.L., Benes C., Dyson N.J. Whole chromosome instability resulting from the synergistic effects of pRB and p53 inactivation. Oncogene. 2014;33(19):2487–2494. doi: 10.1038/onc.2013.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brosh R., Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9(10):701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 18.Park N.H., Chung Y.H., Youn K.H., Song B.C., Yang S.H., Kim J.A. Close correlation of p53 mutation to microvascular invasion in hepatocellular carcinoma. J. Clin. Gastroenterol. 2001;33(5):397–401. doi: 10.1097/00004836-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Terris B., Laurent-Puig P., Belghitti J., Degott C., Henin D., Flejou J.F. Prognostic influence of clinicopathologic features, DNA-ploidy, CD44H and p53 expression in a large series of resected hepatocellular carcinoma in France. Int. J. Cancer. 1997;74(6):614–619. doi: 10.1002/(sici)1097-0215(19971219)74:6<614::aid-ijc10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 20.Yuan R.H., Jeng Y.M., Chen H.L., Lai P.L., Pan H.W., Hsieh F.J. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J. Pathol. 2006;209(4):549–558. doi: 10.1002/path.2011. [DOI] [PubMed] [Google Scholar]
- 21.Atta M.M., el-Masry S.A., Abdel-Hameed M., Baiomy H.A., Ramadan N.E. Value of serum anti-p53 antibodies as a prognostic factor in Egyptian patients with hepatocellular carcinoma. Clin. Biochem. 2008;41(14–15):1131–1139. doi: 10.1016/j.clinbiochem.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Ma Q., Zhang M., Wang X., Zhang D., Li W. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur J Cancer (Oxford, Engl) 2012;48(15):2328–2338. doi: 10.1016/j.ejca.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017;23(12):3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 24.Biton J., Mansuet-Lupo A., Pecuchet N., Alifano M., Ouakrim H., Arrondeau J. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin. Cancer Res. 2018;24(22):5710–5723. doi: 10.1158/1078-0432.CCR-18-0163. [DOI] [PubMed] [Google Scholar]
- 25.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinforma (Oxford, Engl) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gui J., Li H. Penalized. Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinforma (Oxford, Engl) 2005;21(13):3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- 29.Xu R.H., Wei W., Krawczyk M., Wang W., Luo H., Flagg K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017;16(11):1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 30.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 32.Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiran M., Chatrath A., Tang X., Keenan D.M., Dutta A.A. Prognostic signature for lower grade Gliomas based on expression of Long non-coding RNAs. Mol. Neurobiol. 2018;73:273–282. doi: 10.1007/s12035-018-1416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc Series B Stat Methodol. 2011;73(3):273–282. [Google Scholar]
- 35.Shen S., Wang G., Zhang R., Zhao Y., Yu H., Wei Y. Development and validation of an immune gene-set based prognostic signature in ovarian cancer. EBioMedicine. 2018;40:318–326. doi: 10.1016/j.ebiom.2018.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Lu Q., Shao X., Mo B., Nie X., Liu W. Development of a three-gene prognostic signature for hepatitis B virus associated hepatocellular carcinoma based on integrated Transcriptomic analysis. J. Cancer. 2018;9(11):1989–2002. doi: 10.7150/jca.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid M., Wright M.N., Ziegler A. On the use of Harrell's C for clinical risk prediction via random survival forests. Exp Syst Appl. 2016;63:450–459. [Google Scholar]
- 38.Petitjean A., Achatz M.I., Borresen-Dale A.L., Hainaut P., Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 39.Neskey D.M., Osman A.A., Ow T.J., Katsonis P., McDonald T., Hicks S.C. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck Cancer. Cancer Res. 2015;75(7):1527–1536. doi: 10.1158/0008-5472.CAN-14-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalil D.N., Smith E.L., Brentjens R.J., Wolchok J.D. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016;13(5):273–290. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoulidis F., Hellmann M.D., Awad M.M., Rizvi H., Carter B.W., Denning W. STK11/LKB1 co-mutations to predict for de novo resistance to PD-1/PD-L1 axis blockade in KRAS-mutant lung adenocarcinoma. J. Clin. Oncol. 2017;35(15_suppl):9016. [Google Scholar]
- 42.Li Y., Lu Z., Che Y., Wang J., Sun S., Huang J. Immune signature profiling identified predictive and prognostic factors for esophageal squamous cell carcinoma. Oncoimmunology. 2017;6(11) doi: 10.1080/2162402X.2017.1356147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y., Zhang Q., Hu Y., Li T., Yu J., Zhao L. ImmunoScore signature: a prognostic and predictive tool in gastric Cancer. Ann. Surg. 2018;267(3):504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 44.Inada Y., Mizukoshi E., Seike T., Tamai T., Iida N., Kitahara M. Characteristics of immune response to tumor-associated antigens and immune cell profile in hepatocellular carcinoma patients. Hepatology. 2018;69:653–665. doi: 10.1002/hep.30212. (Baltimore, Md) [DOI] [PubMed] [Google Scholar]
- 45.Zhou G., Sprengers D., Boor P.P.C., Doukas M., Schutz H., Mancham S. Antibodies against immune checkpoint molecules restore functions of tumor-infiltrating T cells in hepatocellular carcinomas. Gastroenterology. 2017;153(4):1107–1119. doi: 10.1053/j.gastro.2017.06.017. [e10] [DOI] [PubMed] [Google Scholar]
- 46.Wu J., Li J., Salcedo R., Mivechi N.F., Trinchieri G., Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012;72(16):3977–3986. doi: 10.1158/0008-5472.CAN-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho C.C., Liao W.Y., Wang C.Y., Lu Y.H., Huang H.Y., Chen H.Y. TREM-1 expression in tumor-associated macrophages and clinical outcome in lung cancer. Am. J. Respir. Crit. Care Med. 2008;177(7):763–770. doi: 10.1164/rccm.200704-641OC. [DOI] [PubMed] [Google Scholar]
- 48.Tsai M.H., Tseng H.C., Liu C.S., Chang C.L., Tsai C.W., Tsou Y.A. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral Oncol. 2009;45(9):e90–e94. doi: 10.1016/j.oraloncology.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Erdal E., Haider S., Rehwinkel J., Harris A.L., McHugh P.J. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31(4):353–369. doi: 10.1101/gad.289769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan S., Qin R., Zhu X., Tan C., Song J., Qin L. Associations between single-nucleotide polymorphisms of human exonuclease 1 and the risk of hepatocellular carcinoma. Oncotarget. 2016;7(52):87180–87193. doi: 10.18632/oncotarget.13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka S., Arii S., Yasen M., Mogushi K., Su N.T., Zhao C. Aurora kinase B is a predictive factor for the aggressive recurrence of hepatocellular carcinoma after curative hepatectomy. Br. J. Surg. 2008;95(5):611–619. doi: 10.1002/bjs.6011. [DOI] [PubMed] [Google Scholar]
- 52.Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 53.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Sci (New York, NY) 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 54.Zou W., Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8(6):467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 55.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crouse J., Xu H.C., Lang P.A., Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol. 2015;36(1):49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Rosenberg J., Huang J. CD8(+) T cells and NK cells: parallel and complementary soldiers of immunotherapy. Curr Opin Chem Eng. 2018;19:9–20. doi: 10.1016/j.coche.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romano E., Kusio-Kobialka M., Foukas P.G., Baumgaertner P., Meyer C., Ballabeni P. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc. Natl. Acad. Sci. U. S. A. 2015;112(19):6140–6145. doi: 10.1073/pnas.1417320112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bulliard Y., Jolicoeur R., Windman M., Rue S.M., Ettenberg S., Knee D.A. Activating fc gamma receptors contribute to the antitumor activities of immunoregulatory receptor-targeting antibodies. J. Exp. Med. 2013;210(9):1685–1693. doi: 10.1084/jem.20130573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selby M.J., Engelhardt J.J., Quigley M., Henning K.A., Chen T., Srinivasan M. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. 2013;1(1):32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 61.Simpson T.R., Li F., Montalvo-Ortiz W., Sepulveda M.A., Bergerhoff K., Arce F. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 2013;210(9):1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinicopathological parameters of HCC patients in the Peking HCC cohort.
Gene enrichment in TP53WT HCCs.
Gene enrichment in TP53MUT HCCs.
Differentially expressed immune-related genes between TP53WT and TP53MUT HCCs.
Univariate Cox regression analysis of differentially expressed immune-related genes.
Gene enrichment in low-risk HCCs.
Gene enrichment in high-risk HCCs.
Analysis of correlations between risk score and immune checkpoints.
Functional enrichment analysis of the twenty-one immune genes identified.
Pathway enrichment analysis of the twenty-one immune genes identified.