Abstract
Measuring biological data across time and space is critical for understanding complex biological processes and for various biosurveillance applications. However, such data are often inaccessible or difficult to directly obtain. Less invasive, more robust, and higher-throughput biological recording tools are needed to profile cells and their environments. DNA-based cellular recording is an emerging and powerful framework for tracking intracellular and extracellular biological events over time across living cells and populations. Here, we review and assess DNA recorders that utilize CRISPR nucleases, integrases, and base-editing strategies, as well as recombinase and polymerase-based methods. Quantitative characterization, modelling, and evaluation of these DNA-recording modalities can guide their design and implementation for specific application areas.
Subject categories: Biological sciences / Biological techniques / Genetic techniques, Biological sciences / Biological techniques / Molecular engineering / Synthetic biology, Biological sciences / Biological techniques / Cytological techniques / Lineage tracking, Biological sciences / Biological techniques / Sensors and probes, Biological sciences / Genetics / CRISPR-Cas systems / CRISPR-Cas9 genome editing, Biological sciences / Biological techniques / Genetic engineering
Table of contents blub
In this Review Sheth and Wang describe emerging synthetic biology approaches for using DNA as a memory device for recording cellular events, including the various methodological steps from detecting diverse signals, converting them into DNA alterations, and reading out and interpreting the recorded information. Furthermore, they discuss potential applications as biotechnological and environmental biosensors.
Introduction
Biological life is one of the most complex and dynamic systems in nature. Through evolution and natural selection, vast biochemical and biological diversity has emerged, from complex molecules to multicellular life. These multi-scale biological systems precisely generate and respond to a myriad of biotic signals of varying order and magnitude1. Signals can take the form of ions, metabolites, nucleic acids or proteins, producing biochemical gradients and signalling cascades that propagate across many length and time scales within cells and across populations. The integration of these signals through genetic and epigenetic regulation at the transcriptional, translational and post-translational levels result in robust cellular behaviours2. The spatiotemporal delineation and chronology of these biological signals and cellular states is thus paramount to our understanding of the fundamental organizing principles of biology3.
Tracking multiple biological events simultaneously over time remains a challenge given the sheer number and diversity of signals present within a cell at any given moment. Quantifying these signals and processes in their native cellular and environmental context, which is often inaccessible, poses further practical and technical difficulties. Cellular information can currently be measured by a plethora of methodologies, each with their strengths and weaknesses (Box 1). In the emerging genomic era, where DNA can be readily analyzed and altered, new modalities of DNA-based cellular recording are poised to overcome these traditional limitations in biological information storage and analysis in a variety of settings.
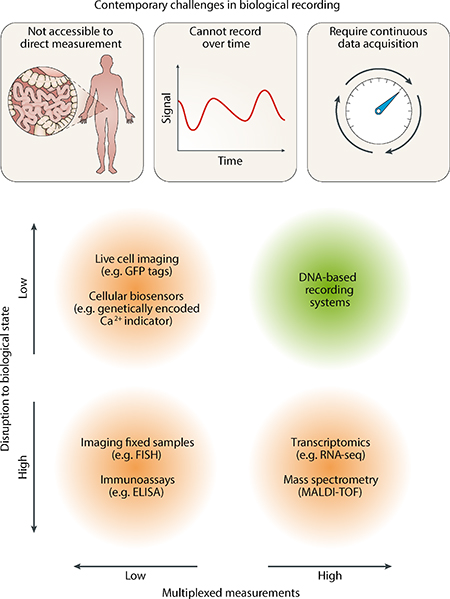
Box 1. Contemporary limits of biological recording.
Different types of biological data can be acquired from cells, including identity, quantity, spatial and temporal information. DNA, RNA, proteins and organic/inorganic metabolites constitute the key classes of molecules that are often measured when analyzing biological processes. In addition to these molecular data, cellular phenotypes that are reflective of more global states, such as growth rate, membrane permeability, electrochemical gradients and oxidative stress can also be quantified. Contemporary approaches to measure these biological data suffer from three key limitations (see the figure). Many environments are difficult to access for direct measurement (e.g. the gut or brain). Methods that require destructive processing steps (e.g. cell or tissue fixation) cannot yield temporal biological data. Methods that allow continuous data acquisition, such as live-cell imaging, require direct access to the biological sample and instruments that cannot be miniaturized down to cellular sizes. Furthermore, the multiplexing capacity of most current methods are either limited or require considerable disruption to the biological state of cells. In theory, DNA-based recording systems can overcome many of these challenges when deployed in live cells to store biological data into a permanent DNA medium over time for analysis at a later point. ELISA, enzyme-linked immunosorbent assay; FISH, fluorescence in situ hybridization; MALDI-TOF, matrix-assisted laser desorption/ionization–time of flight; RNA-seq, RNA sequencing; TFs, transcription factors.
DNA is the fundamental molecule by which information is stored and utilized to produce life. DNA is a high-density storage medium4–6 that can be quickly copied by exponential polymerase chain reaction (PCR) amplification and stably preserved for decades to millennia7. Biological information encoded in DNA can be directly converted into actionable cellular responses through gene regulation and expression. Although DNA is often thought of as a long-term information bearing molecule, there are many examples of biological information storage and access through DNA within a single life cycle of an organism. Examples include phase variation8, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-mediated immunity9, mammalian adaptive immune systems10, diversity-generating retroelements11, and programmed genome rearrangements12,13. Advances in next-generation sequencing (NGS)14 and nucleic acid synthesis15 have ushered in a new era of rapid and cheap DNA reading and writing, which has further elevated the relevance of DNA as a meaningful information storage medium.
In this Review, we discuss recent progress in the emerging field of DNA-based recording technologies in living cells. We highlight key elements of biological information storage, suggest quantitative metrics to assess different recording approaches, and outline technical challenges and knowledge gaps that still need to be addressed. We end by offering possible applications of DNA-based cellular recording and speculate on the future of this exciting area of research and development. Although epigenetic mechanisms, both molecular and cellular, such as protein-based feedback circuits, DNA methylation, chromatin conformation, prion states, and neuronal networks, are clearly interesting and important modes of biological information transmission and storage16–18, they are beyond the scope of this focused Review. For technologies that employ DNA barcodes for lineage tracing applications, we direct the readers to a recent in-depth review19.
Strategies for DNA-based memory in cells
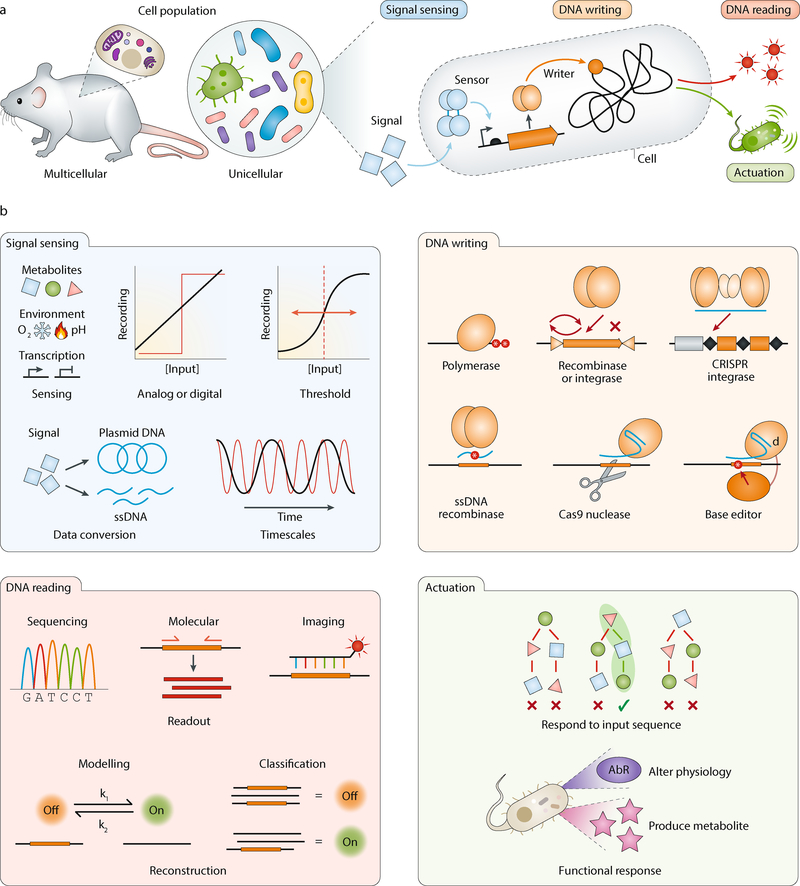
A universal information recording and storage system requires several essential elements: first, transformation of the information of interest into a standardized data format or data stream; second, recording the data into a physical medium; and third, conversion of the stored data back to a desired form that can be interpreted by the user or utilized by another system. A biological DNA-based version of such a memory system must also possess these key capacities (Figure 1). First, information within a cell such as the presence of a metabolite or expression of a gene must be transformed into a format that is compatible with the recording system, for example a biological signal that induces the expression of recording components. Next, this information must be written directly into DNA by alteration, deletion or addition of bases through various DNA modifying enzymes, such as nucleases, integrases or recombinases. Finally, the stored data is read back out from the DNA using a multitude of techniques such as sequencing or imaging. The stored information can be further used to directly actuate or elicit a specific set of biological responses, such as gene expression. Below, we delineate each of these components and their implementation in contemporary DNA-based data recording and storage systems (Table 1).
Figure 1. Components of cellular memory.
a | Cellular recording devices can be engineered into multicellular organisms or unicellular populations, and their general architecture can be broken down into four major components: signal sensing, DNA writing, DNA reading and actuation. b | Properties or examples of each of the four major components. AbR, antibiotic resistance; ssDNA, single-stranded DNA.
Table 1.
Major demonstrated DNA-recording approaches.
| Recombinase | ssDNA editing | Base editing | Cas9 array collapse | Self-targeting gRNA | Polymerase ticker tape | CRISPR array | |
|---|---|---|---|---|---|---|---|
| Device architecture | |||||||
| Detection: input signals | Transcriptional | Transcriptional | Transcriptional | Transcriptional | Transcriptional | Ion concentration | Transcriptional |
| Transformation | Expression of recombinase | Conversion to ssDNA via retron | Expression of base editor and gRNA | Expression of Cas9 | Expression of Cas9 | Alteration of misincorporation rate | Conversion to DNA via copy-inducible plasmid |
| DNA writing: bit | Multiple base pairs | Multiple base pairs | Single base pair | Multiple base pairs | Multiple base pairs | Multiple base pairs | Multiple base pairs |
| DNA writing: address | Fixed; flanked by recognition sites | Flexible; ssDNA targeted | Flexible; dCas9 targeted | Fixed; engineered target array | Fixed; self-targeting gRNA sequence | Fixed; polymerase target | Fixed; CRISPR array |
| DNA writing: write operation | Defined; inversion, excision or integration | Defined; ssDNA and/or recombinasemediated allelic replacement | Defined; C•G-to-T•A base editing | Stochastic; collapse of target array by random indels | Stochastic; sequence evolution of gRNA by random indels | Stochastic; patterns of polymerase misincorporation across target sequence | Directional; integration of CRISPR spacers |
| Reading and actuation | Allele PCR, sequencing, functional response | Sequencing functional response | Sequencing functional response | Sequencing | Sequencing | Sequencing | Sequencing |
| Performance metrics | |||||||
| In vivo temporal resolution | + | + | + | + | + | +++ (theoretical) | + |
| In vivo storage capacity | + | ++ | ++ | + | + | +++ (theoretical) | +++ |
| Demonstrated host range | Prokaryotic and eukaryotic | Escherichia coli | Prokaryotic and eukaryotic | Eukaryotic | Eukaryotic | In vitro | Escherichia coli |
| Representative implementations | |||||||
| Refs | (RSM)55; (BLADE)57 | (SCRIBE)58 | (CAMERA)59 | (GESTALT)68; (MEMOIR)70 | mSCRIBE79, Kalhor et al.78 | Zamft et al.81 | Shipman et al.85,86; (TRACE)31 |
Contemporary recording approaches are classified into seven major approaches, and properties falling under each the recording device component and delineated. In addition, demonstrated performance metrics of each device are qualitatively assessed (+, low performance; +++, high performance). BLADE, Boolean logic and arithmetic through DNA excision; CAMERA, CRISPR- mediated analog multi-event recording apparatus; dCas9, catalytically dead Cas9; GESTALT, genome editing of synthetic target arrays for lineage tracing; gRNA, guide RNA; indels, small insertions or deletions; MEMOIR, memory by engineered mutagenesis with optical in situ readout; mSCRIBE, mammalian SCRIBE; RSM, recombinase state machine; SCRIBE, synthetic cellular recorders integrating biological events; ssDNA, single-stranded DNA; TRACE, temporal recording in arrays by CRISPR expansion.
Signal detection and transformation
Signal and input types
While there are a variety of biological signals present within a cell, the dynamic regulation of gene expression through mRNA transcription is one of the most important and prevalently measured class of cellular signals. The ensemble of transcription levels across all of its genes can represent a simplified ‘state’ of a cell. Beyond transcriptional states, proteins and metabolites, both intracellular and extracellular, represent other classes of cellular signals that can change during cellular growth, development and maintenance in different environments. Both the concentration and identities of these molecules can serve as inputs into a biological recording system. Finally, physiologically important characteristics of the intracellular and extracellular environment such as temperature, pH, oxidative stress, radiation levels, or electrochemical and electromagnetic gradients can also be inputs for sensing and recording. For all of these input types, the presence or absence of the signal (digital state) and its intensity or magnitude (analogue state) are important recordable information, as well as their variation across space and time.
Signal sensing
Cells possess numerous native mechanisms to assess transcriptional states that can be coopted for cellular memory devices. For instance, the transcription level of a gene of interest can be measured by linking its upstream promoter to a recording system to capture transient regulatory changes. Indeed, early bacterial gene expression screens utilized a strategy where native promoters are fused to a recombinase-based reporter that permanently altered a genomic site to identify virulence pathways20. Recording certain combinations of genes and their expression levels can capture even more complex cellular phenotypes of interest such as growth rate or cellular burden21.
The levels of intracellular and extracellular chemical, metabolite, RNA, or protein-based signals can be detected with a growing toolbox of engineered biosensors with high signal specificity. These modular sensors can convert a myriad of signal types such as cancer-associated antigens22, pathogen derived peptides23, xenobiotic metabolites24 and light25. Many sensors, such as transcription factors, two-component systems, and more complex signalling cascades, couple binding of an input ligand to a sensory protein with altered transcription from a specific output promoter, which can then be readily linked to recording systems26,27. Alternatively, RNA-based sensors such as RNA aptamers and riboswitches recognize specific metabolites and alter expression of an output gene by diverse mechanisms (e.g. tuning of translation)28. Beyond chemical and protein ligands, RNA signals such as mRNA levels of endogenous genes or microRNAs can also be sensed via riboregulators which bind target RNA molecules and alter expression of an output29,30.
Signal transformation
Once sensed, a signal of interest must be converted into a format that is capable of specifically activating a recording system. For many systems, this step simply involves expression of the recording machinery to mediate DNA modification. Alternatively, a transformation of the input signal into a different format may be required. For example, a transcriptional signal can be converted to an altered abundance of intracellular DNA by using a copy number inducible plasmid system, which subsequently is recorded into genomic arrays by CRISPR integrases as short spacers31. Signal transformation can be represented as a transfer function of signal input to the resulting recording activity; its detection threshold, dynamic range, and response characteristics (analogue versus digital) must match the desired application. Synthetic biology and genetic engineering techniques can be utilized to rationally alter and optimize this transformation, for instance by tuning expression levels of recording machinery or altering sensor detection thresholds by protein engineering32.
Synthetic gene circuits can be interfaced with biosensors for more complex tuning of signal transformation or to add more sophisticated functionality such as signal integration and computation27,33. For example, signal processing circuits can be linked to biosensors to achieve digital or analogue responses to an input signal34,35. In order to alter signal response dynamics and record rapidly fluctuating signals, positive feedback and memory modules can be utilized17. In more complex eukaryotic signalling cascades, scaffold proteins can be shuffled or linked to redirect pathway outputs and achieve diverse response characteristics and dynamics36. Finally, transcriptional or post-translational synthetic circuits implementing complex logic operations can be rapidly designed to integrate and perform signal processing on multiple environmental signals37,38.
Most transcription-based biosensors inherently suffer from a lower temporal resolution due to slow signal transduction and gene expression processes (>102 seconds). By contrast, enzyme-based post-translational sensors can respond to signals much quicker (<10−2 seconds), which may be necessary to capture transient or fast biological processes39. In order to rapidly capture signals into DNA, the activity of recording modules must be directly linked to a signal of interest, for example through chemically inducible dimerization40 or post-translational modification41. Importantly, DNA polymerization can occur at >500 base pairs per second in vivo, which at least theoretically can match the signal transduction speeds of fast biosensors42.
Writing onto DNA medium
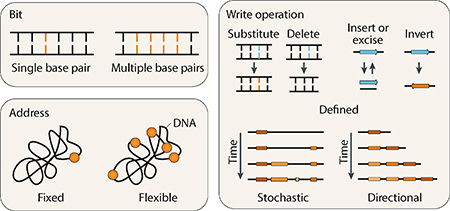
Natural and engineered DNA targeting and modifying enzymes, which include recombinases, polymerases, integrases, nucleases, and multi-functional variants, can be leveraged as writing modules in DNA memory systems. Many new molecular tools to manipulate DNA in cells have emerged, with increased programmability, ease, precision, and accuracy43,44. The biochemical characteristics of a DNA writer and its accessory factors (exogenous or from the host) define the ‘recording syntax’ of the system, including the base pair unit of information storage (bit), the sequence location of DNA writing (address) and type of DNA modification employed (write operation) (Box 2).
Box 2. Recording syntax of DNA writing.
In information recording, understanding the architecture and structure of data storage is crucial to defining the overall functionality and capability of recording. The syntax of DNA-recording systems can be assessed across three primary properties (see the figure):
1). Bit: the base-pair unit constituting information storage
In modern digital memory architectures, information is stored in units of binary bits (0,1). Data storage in DNA can leverage its expanded alphabet of 4 distinct nucleotides (A, C, G or T). A single base-pair constitutes the most simple unit or bit of storage. Alternatively, some systems may designate multiple base-pairs containing a large amount of information (such as a targeted sequence) as the base unit of memory bits. Recording bits may also encode functional biological information such as promoters or specific reporter genes.
2). Address: the specific sequence location where DNA writing occurs
Modifications can occur at a fixed address due to sequence-specific properties of specific molecular machinery. Alternatively, DNA writing can occur at a flexible address, which can be specified to different locations of interest by utilizing sequence-specific directing machinery, such as Cas9 or zinc-finger nuclease (ZFN) approaches. Single or multiple addresses can be targeted within a cell to increase storage capacity or record multiplex signals simultaneously.
3). Write operation: the structure and nature of DNA modification
Defined write operations result in single or multiple base-pair alteration operations such as substitution, deletion, insertion, excision and inversion of a target DNA sequence (e.g. by specific recombinases). Stochastic write operations result in structured destruction or evolution of a target sequence (e.g. by programmable nucleases). Finally, directional write operations, which are unique in that they encompass the addition of sequence to create DNA information, can be utilized to sequentially write new DNA in a directional manner (e.g. polymerases or CRISPR spacer acquisition). The timescales and efficiency of DNA modification are key parameters that define the performance of the recording system.
Fixed-address writers
Fixed-address writers are targeted to specific biological sequences based on biochemical properties of the DNA-modifying enzyme, and work by treating the orientation or presence/absence of specific target DNA sequences as bits or states. Site-specific recombinase systems [G], which are widely used in gene expression and knockout applications45, enable the inversion, excision or integration of specific target DNA sequences depending on the orientation of flanking recognition sites46, thereby enabling manipulation of these DNA bits. For example, 11 pairs of orthogonal recombinase systems were mined from metagenomic databases, allowing the creation of a memory array in which each bit is represented by the presence or absence of specified DNA sequences targeted by each recombinase. This system was capable of storing 1.375 bytes of information in the genome of Escherichia coli47, and was further ported to a commensal gut bacterium, Bacteroides thetaioataomacron, for sensing dietary components in the murine gut48. As the recombination event is irreversible, integrase–excisionase pairs49 or complementary recombinase pairs50 can be utilized to reset the orientation of target addresses. These orthogonal recombinase systems can further be interleaved and layered to achieve more complex functionalities such as counting51, signal amplification and digitization52 or two-input Boolean logic functions53,54.
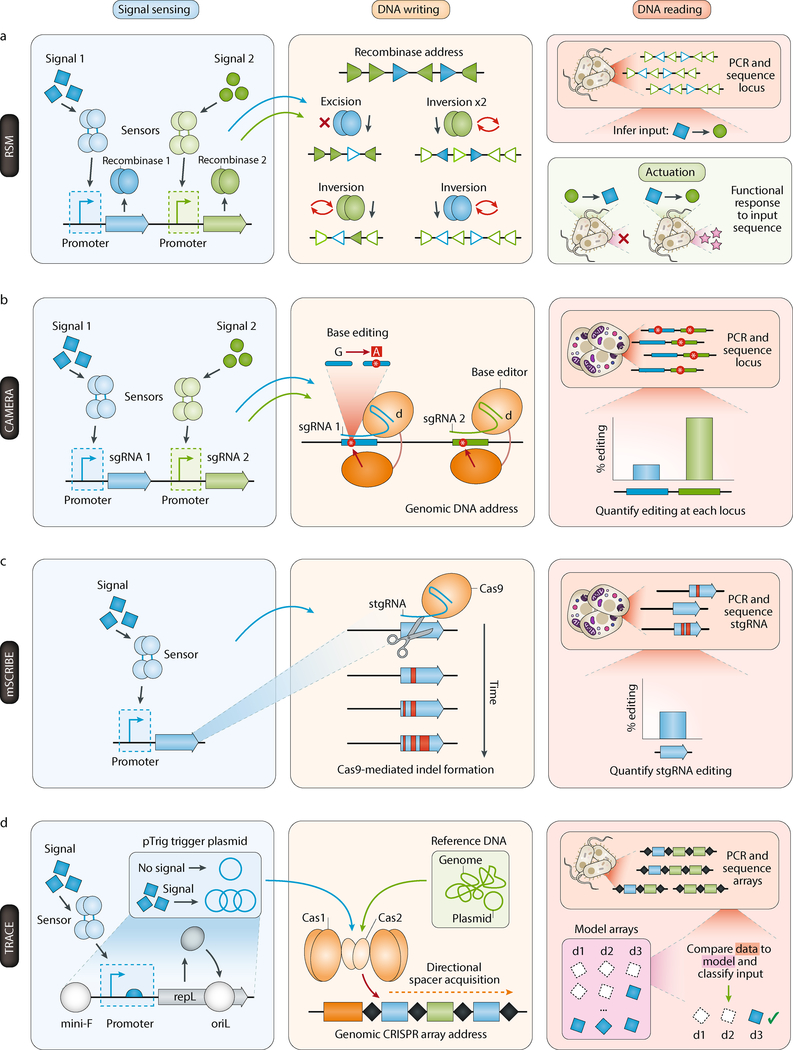
The complex set of possible combinatorial recombinase target arrangements was recently formalized for three orthogonal recombinase systems in the recombinase state machine (RSM [G] ) framework55 (Figure 2a). As the recombination process can be stochastic, layered recombinase systems can be utilized to encode information such as the ordering and duration of inputs within a population through the frequency of different recombination states within the population56. Finally, complex recombinase arrangements and circuits can be implemented in mammalian systems, demonstrating the portability of fixed-address writing approaches57.
Figure 2. Example DNA recording devices.
The functionality of four exemplary DNA-based recorders are illustrated following the recording device architecture. a | Recombinase state machine (RSM) fixed-address writer55. Orthogonal recombinases are expressed in response to a signal, and they mediate excision or inversion events at a designed recombinase address (filled triangles are unrecombined sites, unfilled triangles are recombined sites). Based on the ordering of inputs, different resulting address sequences can be achieved, which are read out by sequencing, or which can mediate functional responses by interleaving genetic parts (promoters, expression cassettes, or terminators) within the recombinase address. b | CRISPR-mediated analog multi-event recording apparatus (CAMERA) flexible writer59. Single-guide RNAs (sgRNAs) are expressed in response to a signal, and direct a base editor (dCas9-BE) to mutagenize specific loci within a genomic address, which can be read out by sequencing. c | Mammalian synthetic cellular recorder integrating biological events (mSCRIBE [G]) stochastic writer79. A self-targeting guide RNA (stgRNA) is expressed in response to an input signal, and directs Cas9-mediated editing and generation of a small insertion or deletion (indel) at the same stgRNA address, resulting in continuous editing and sequence evolution. The resulting stgRNA address can be read out by sequencing. d | Temporal recording in arrays by CRISPR expansion (TRACE) directional writer31. A signal is converted into altered DNA abundance through use of a copy-number inducible trigger plasmid (pTrig). Short spacers can be incorporated into a genomic array address in a directional manner, either from the trigger sequence or at a constant rate from genomic or plasmid reference sequences. Resulting arrays can be sequenced and the order and source of spacers can be compared to a model of CRISPR expansion to classify the signal input sequence over time.
Flexible-address writers
Unlike fixed-address writers, which are targeted to predefined sequence locations, flexible-address writers are capable of writing to arbitrarily specified and programmable target locations, yielding precise single or multiple base pair changes. This specifiable nature of flexible-address writers enables a higher density of data storage and more direct interfacing with host programs and physiology. One implementation is the synthetic cellular recorders integrating biological events (SCRIBE [G] ) system demonstrated in bacteria58. In SCRIBE, a single-stranded DNA (ssDNA) is first generated by a retron [G] in response to a biological signal. Then, ssDNA allelic replacement mediated by a recombinase can occur at a defined DNA address, yielding a low frequency but defined genomic mutation. The degree of editing at the storage address across a recording cell population can be used to determine the intensity of the input signal exposed to the population as well as its duration. In addition, since the address is pre-defined, reporter genes can be targeted to elicit a functional response within cells, such as production of a colorimetric reporter or alteration of antibiotic resistance58.
Another flexible-address writing implementation is CRISPR-mediated analog multi-event recording apparatus (CAMERA [G]), which employs engineered base editors (BE) to generate C•G-to-T•A mutations that encode information bits at designated DNA addresses with single-nucleotide specificity59 (Figure 2b). Base editing [G] is mediated by transcription of both a catalytically dead Cas9 [G] (dCas9) fused to a cytidine deaminase60 and guide RNAs (gRNAs) that target to the DNA memory address. The presence of edited bases and their frequency across the population encode both digital and analogue information (i.e. signal identity and intensity). Since the sequence of the resulting edited memory addresses are reproducibly generated, additional layers of editing can occur in a sequential manner to encode temporal information, which enables more complex recording architectures61. Even more excitingly, recently demonstrated adenine base editors (ABE) that generate A•T-to-G•C mutations62 can work in the opposite mutational direction to cytosine base editors. In future systems, cytidine and adenine base editors could be utilized in combination to enable a powerful capability to rewrite DNA addresses repeatedly.
Stochastic writers
Stochastic writers record biological information by continually altering a target DNA sequence in a semi-random manner. By analyzing the extent and nature of sequence changes, the intensity of a signal can be inferred. For instance, the programmable site-specific nuclease Cas9 [G] 43,63–66 can be used to generate a double stranded break at a target DNA address, which is then repaired by endogenous non-homologous end joining [G] (NHEJ) processes which at a low probability may yield sequence insertions or deletions (indels)67. The resulting indels are diverse, hence information is generated at the modified DNA address.
In one class of such stochastic writers, Cas9 is used to target designed DNA addresses consisting of multiple identical target sites (known as arrays or scratchpads) that are stochastically and irreversibly modified during continuous cellular recording68–70. This approach has been utilized for large-scale recording and lineage reconstruction in entire animals68. Beyond recording cell lineage information, these writers could be extended to record analogue signals such as the amount of gene expression over time, by coupling Cas9 expression to a cellular signal of interest. A variety of other nucleases such as Cpf171, zinc-finger nucleases (ZFNs)72–74 and transcription activator-like effector nucleases (TALENs)75–77 could be used in a similar manner.
A recursive stochastic writing approach can also be used for continuous cellular recording, with the potential advantage that recording is linked to stochastic evolution rather than collapse of a target sequence. In this class, gRNAs that direct a Cas9-based writer to the DNA can be designed to target themselves, i.e. a self-targeting gRNA (stgRNA [G])78 (Figure 2c). Over time, the DNA address will undergo continuous mutagenesis, which encodes the magnitude of a biological signal of interest. Such recording devices have been demonstrated in mammalian cells to record inflammation levels in a xenograft model79.
Directional writers
In contrast to the above approaches where DNA addresses have pre-defined storage capacities and DNA is specifically edited or stochastically altered, directional writers [G] have the ability to create new DNA sequences through addition of nucleotides in a directional manner. As such, these directional writers are well-suited for recording temporally changing biological signals. In general, a temporal data recorder (e.g. audio recorder) functions by transforming time-varying signals into physical spacing on a substrate (e.g. a magnetic tape strip). Similarly, in directional DNA writing, the duration in the time domain is represented by physical distances between recorded data in base pairs.
One such system is a proposed polymerase-based ticker tape, which is an engineered DNA polymerase [G] that writes temporal signals in the form of misincorporated bases as it directionally replicates across a DNA template80. The polymerase error rate can be made sensitive to a signal of interest, such as ion concentrations during recording of neuronal activity, thus allowing for temporal encoding of these signals onto DNA memory substrates81.
Alternatively, CRISPR acquisition systems that catalyze the incorporation of short DNA spacers in a unidirectional manner into expanding CRISPR arrays82,83,84 can be used to record signals. Such systems have been used to record oligonucleotide sequences that are electroporated into a bacterial population85. Because the ordering of incorporated spacers reflects their exposure to the cells, analysis of the resulting arrays across a population of cells allows for reconstruction of exposure ordering. This approach has been further scaled for the recording and storage of a 2.6 kilobyte animated image in the genomes of a bacterial population86. We recently described a system, temporal recording in arrays by CRISPR expansion (TRACE [G]), that utilizes CRISPR spacer acquisition to record biological signals by linking a transcriptional signal of interest within a cell to a copy number inducible plasmid31 (Figure 2d). With this approach, the temporal exposure history over four days could be accurately reconstructed, and temporal recordings could be further multiplexed to record three signals across a population of cells. In a conceptually similar manner to these CRISPR integrase approaches, recombinases can be also used to recursively integrate sequences into a genomic array, with the added benefit of larger and more specific sequences that can be incorporated87.
Reading from stored data on DNA
The appropriate method for extracting the stored DNA information is dependent on the recording syntax, base pair resolution and throughput needed to decode the data. Often, the extracted data may need to be further analyzed, interpreted, or deconvolved using method-specific in silico reconstruction tools and algorithms to yield the final useful information.
DNA-sequencing-based readers
DNA sequencing is the most direct way to extract information from DNA-based recording devices. Sanger sequencing can provide low-throughput but high accuracy sequences of ~800 bp. Nucleotide polymorphism frequencies across a population at specific DNA addresses can also be determined from Sanger chromatograms88. Alternatively, NGS can determine the sequence of DNA addresses at a much larger scale, and progress in this arena14 has enabled analysis of many recent recording devices. Short-read sequencing-by-synthesis (from Illumina) can currently provide the highest throughput and read quality, albeit with a maximum read length of ~600 bp89. For DNA addresses with longer lengths (e.g. large recombinase-targeted loci87,90), long-read sequencing technologies such as single-molecule real-time sequencing (SMRT, from Pacific Biosciences) or nanopore sequencing (from Oxford Nanopore Technologies) are necessary. Although long-read sequencing modalities currently have a relatively lower throughput and lower quality compared to more mature short-read NGS platforms, portable instruments such as the MinION nanopore sequencer offer exciting real-time readout of DNA data storage91.
Molecular and imaging-based readers
For writers with defined addresses, the presence or absence of specific DNA sequences can be directly determined using simple molecular biology tools such as allele-specific PCR92, restriction digestion assays, and fluorescence resonance energy transfer [G] (FRET)-based reporters93. Alternatively, direct imaging-based techniques enable probing of recorded data from individual cells in their native spatial context. For example, in the memory by engineered mutagenesis with optical in situ readout (MEMOIR [G]) stochastic writer, single-molecule RNA fluorescence in situ hybridization (smFISH) of edited CRISPR array addresses enables in situ readout of cellular lineage and endogenous gene expression during cellular differentiation70. In addition, a number of emerging in situ sequencing approaches94,95, as well as bulk imaging advancements such as expansion microscopy96, will support higher resolution spatial readout of a wide range of recording systems.
Data analysis and reconstruction
The scale, complexity and stochastic nature of DNA recording pose new challenges for data analysis and information reconstruction. Quantitative and statistical modelling of the recording performance is essential for mechanistic understanding of the underlying process and failure modes. For instance, in the mammalian SCRIBE stochastic writing system, sequential sequence changes to stgRNAs was analyzed by calculating the transition probability between sequence states79. Analysis of these data enabled quantitative understanding of key properties of Cas9-mediated DNA editing and the recording process, as well as the identification of editing events that led to undesired inactivation of the device.
Modelling essential recording processes can also aid quantitative data reconstruction and information interpretation. In the TRACE directional writing system, a model of CRISPR spacer expansion from either reference or trigger DNA sources was developed and parameterized using control experiments. This model enabled simulation of all possible temporal input states that were then compared to measured data in a classification scheme, which led to accurate predictions of the temporal input signal31. Alternatively, parallel DNA writing systems can be utilized for temporal signal reconstruction. For example, in the MEMOIR stochastic writing system, a model of the recording process suggested that multiple DNA addresses, which are either edited at a constant rate or in response to a signal, can be utilized to reconstruct temporal exposure histories by comparing the resulting writing across these addresses70.
Actuation from recorded data
Beyond simply retrieving recorded information from the DNA, an important feature of in vivo DNA-based recording is the possibility of transforming recorded data directly into biological responses. Various genetic circuits can be embedded within the architecture of DNA memory, allowing for direct functional responses when data are written and matched to a pre-defined pattern. For example, promoters and genes of interest can be interleaved within recombinase circuits, allowing for actuation of responses such as expression of multiple fluorescent reporter genes only after the cells are exposed to a specific series of inputs and the target address achieves a specific configuration55. A recording device can also directly alter the genotype of a cell upon storage of a specific dataset. In the SCRIBE flexible-address writing system, inactivating mutations (i.e. a premature stop codon) in genes of interest were added or removed, resulting in alteration of cellular phenotypes, such as antibiotic resistance, across a cell population58. These cellular actuation strategies enable new classes of programmable genetic circuits that can both chronicle biological conditions and respond to them directly by generating heritable DNA changes and not just transient transcriptional responses.
Assessing performance of recording devices
A DNA recorder’s design architecture and biochemical machineries dictate its performance characteristics (i.e. temporal resolution, capacity, and accuracy of recording) and system capabilities (e.g. host portability, and multiplexing). Critical and quantitative assessment of different recording modalities is needed to identify their strengths and weaknesses, suitability for a given application, and opportunities for further optimization. Here, we outline key performance metrics and assessment criteria to help stratify and evaluate emerging DNA-based recording devices (Table 1).
Quantitative performance metrics
Temporal resolution of recording
Different recording architectures can resolve biological signals at different temporal resolutions, which can be quantified in terms of the frequency of input signal per unit time (i.e. in hertz). Recording is fundamentally limited by the timescales of sensing machinery, signal transformation and the speed and efficiency of DNA writing. For example, a fixed-address writing system, which must sense a metabolite and respond by expressing a recombinase protein that mediates inversion of a target DNA sequence, has a lower temporal resolution than a polymerase-based directional writing system that directly records ion concentrations close to the rate of DNA polymerization. Importantly, the temporal resolution of DNA writers can be optimized with rational engineering approaches. To match temporal tracking with organismic development for example, the genome editing of synthetic target arrays for lineage tracing (GESTALT [G]) stochastic writing system employed various engineered Cas9 array configurations that reduced editing efficiency thus lengthening the timescales of recording68.
Capacity and density of information storage
Storage capacity can be quantified in terms of data size in bits per cell. Most systems (e.g. defined and stochastic writers) contain a fixed data capacity that is limited by the size of the predefined DNA target address. By contrast, directional writers can increase their storage capacity on-the-fly as new sequences are written. Together with the recording syntax, the base pair editing resolution defines the data density or the amount of stored data in bits per base pair. Single base pair editing modalities such as Cas9 base-editor flexible-address writers thus offer a higher information storage density. Information can also be distributed across a population to increase storage capacity; for example in CRISPR integrase directional writers where individual cells on average contain a small amount of information, a population is required to reconstruct the signal data.
Accuracy and stability of data storage
Accurate data recording and stable data retention over time are crucial for long-term information storage. DNA recorders with higher writing efficiency can, in general, yield more accurate signal reconstructions because data are more efficiently transformed and stored in the DNA. A distinct characteristic of biological recording is the reliance on stochastic DNA writing and continuous DNA replication and propagation that occur with high, yet still imperfect, fidelity. The origin and location of DNA storage addresses can also affect long-term stability. Different replication systems and sequences may also have different replication fidelity97, and recording syntaxes utilizing arrays with higher sequence similarities may have increased levels of recombination that result in loss of data98,99. To improve stability, different error-correction strategies can be used, such as redundant data storage across a population and reconstruction of consensus information in CRISPR integrase-based recordings of image information86.
Cross-species portability and cellular burden
A recording system’s enzymatic machinery governs its portability, which is defined as the degree of functionality in diverse hosts. Many DNA writing modules may depend on specific host factors or processes. For example, stochastic writers rely on Cas9-mediated indels generated by NHEJ repair processes that are prevalent in eukaryotes but rare in prokaryotes100,101. The SCRIBE system requires expression of a species-specific recombinase to mediate DNA writing in bacteria, and CRISPR integrase-based writing requires an accessory integration host factor (IHF) for spacer integration in E. coli102. On the other hand, base editing DNA writers directly record data by deaminating DNA bases60, relying on highly conserved cellular replication and repair processes found in both eukaryotes and prokaryotes. Indeed, these base editing systems have been demonstrated in both E. coli and mammalian cells59,61, suggesting high portability of the approach across different hosts.
Recording may also place a burden on native host processes, which can manifest as changes in growth rate, cell physiology or evolutionary stability. Expression of recording machinery may redirect precious cellular resources, whereas the act of DNA writing itself may induce cellular stress responses. In addition, undesired DNA writing, such as Cas9 off-target cleavage103,104 or CRISPR integration at non-target sites105 may introduce lethal genomic mutations that reduce cell fitness. Finally, the DNA address itself could place an additional burden on the cell to harbour and maintain a larger amount of DNA. These effects may be accentuated over long multi-generational timescales, during which a recording device may acquire inactivating mutations that reduce this burden. For example, characterization of a recombinase-based writer revealed host adaptation to reduce expression of the recombinase, thus inactivating the device50. For robust and long-term functionality, the cellular burden of a recording device must minimized.
Multiplexing and scalability of biological recording
Recording devices can be multiplexed, thus enabling simultaneous measurement and comparison of a large number of biological signals. As most recording devices can be modularly linked to transcriptional input signals, various endogenous and engineered transcriptional sensory systems have been linked to recording systems in parallel. If orthogonal recording machinery exists, or if recording can be directed to distinct DNA addresses, multiple channels of recording can be implemented within a single cell47,55. Alternatively, the same recording machinery could be linked to different input sensors in different barcoded cells to allow multiplex data storage across a population, such as in the TRACE system31.
Recording systems may be scaled to store different information modalities or link to complementary biological readouts. Constitutive recording at a basal rate, for example with stochastic writers, enables applications in lineage tracing19. The recorded information can be read out in parallel to other readout modalities. For instance, these same approaches can be readily combined with single-cell RNA sequencing (scRNA-seq) methods. In this example, cell type is inferred from the transcriptome, and lineage information is provided by additionally sequencing the DNA address where recording occurs (or RNA transcript expressed from the address) to compare the molecular identity of a cell with its previous lineage106–108.
Applications of cellular recording
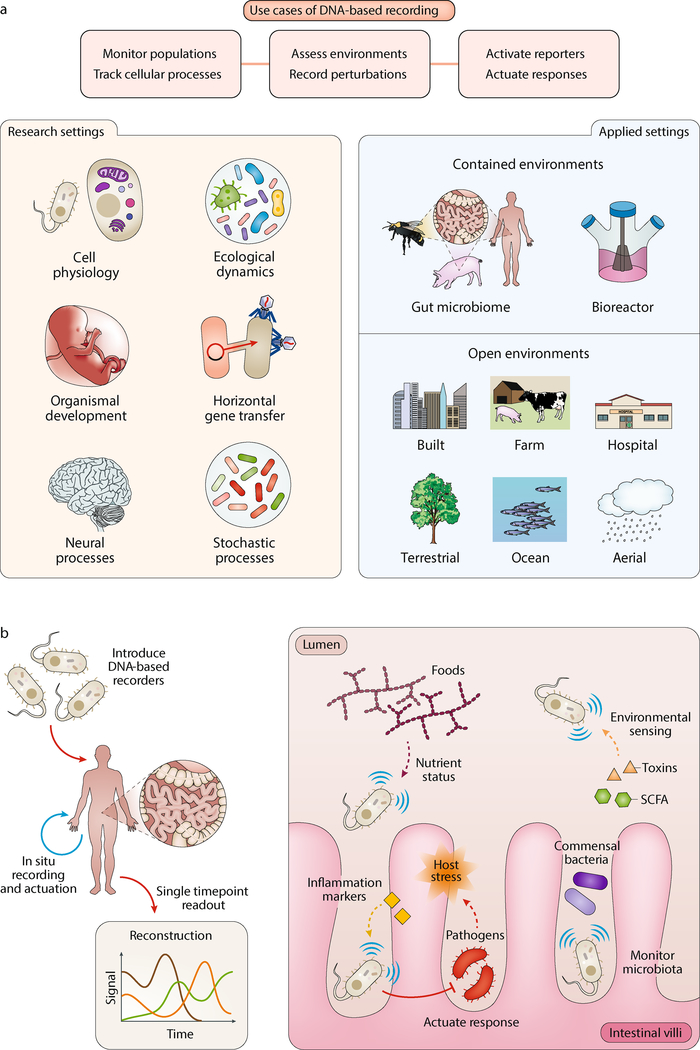
DNA-based cellular memory systems can be deployed in a variety of useful ways in basic research and applied fields (Figure 3). Applications where measurement and tracking of biologically relevant information at locations that are otherwise difficult, if not impossible, to access are particularly well-suited for DNA-based recording systems. To implement these systems in contained environments such as individual bioreactors and host-associated microbiomes, or open settings such as agricultural crops or buildings, different considerations will need to be evaluated and integrated, such as the mode of signal transformation, the spatiotemporal sensitivity and capacity of recording, and the stability of data storage.
Figure 3. Applications of DNA-based biological recorders.
a | Use cases of DNA based recording (top) as well as applications across research and applied settings (bottom). b | Example utility of DNA-based recorders in the gut microbiome. Engineered cellular memory devices could be utilized for noninvasive multiplex temporal recording of important signals such as nutrient status and microbial- and host-derived metabolites. In addition, these recorders could mediate functional actions in response to specific signals or profiles of inputs. SCFA, short-chain fatty acids.
Mapping biological processes
Direct, large-scale, and high-resolution cellular recording enables fundamentally new measurements of biological processes that are normally unobtainable. These new datasets will be crucial for improving our understanding of many complex, interconnected, and spatiotemporally diverse biological systems and ecologies. In the microbial biosphere where communities can exist at very high density (e.g. 1011 cells per gram of fecal matter109), measuring and tracking every cell is infeasible. Using microbial DNA-based recorders, one could probe and chronicle colonization and gene expression in specific microbial populations within and between hosts (e.g. humans, animals or insects) to gain new and greater insights into their ecology and dynamics110. Tracking temporal changes of metabolites such as nutrients in these microbiomes could further reveal facets of microbial physiology and metabolic interactions111. Furthermore, delineating exposures to phages and mobile DNA using CRISPR-based recorders could be a powerful new approach for analyzing horizontal gene transfer processes112 in different environments in real time.
As DNA recorders can be deployed in single cells and analyzed across populations, relative spatial and historical information can be stored in cells of complex tissues and organs during growth, maintenance, and ageing. In developmental biology, DNA-based lineage tracing strategies have already enabled the mapping of organismal development at unprecedented scales and resolutions68. Extending these approaches to record relevant biological signals will yield new insights in population and developmental biology, potentially down to the single-cell level. For example, DNA recording approaches have been applied to measure the relationship of cell state transition processes and lineage in embryonic stem cells70. Extensions of such frameworks to the nervous system of complex animals could enable large-scale biological recording and readout of massively complex signalling networks in neurons to probe complex spatiotemporal processes in the brain113,114. DNA-based recording could also be implemented in emerging cell therapy applications such as chimeric antigen receptor (CAR) T cells to improve actuation in response to complex input signals and track activation history115. Beyond measurements of absolute and relative levels of biological signals, DNA-based recorders could also measure variance of these signals across populations, which often govern key community-wide properties such as stochastic gene expression116,117 and microbial persistence phenotypes118.
Ubiquitous cellular sentinels
A wide range of synthetic biology applications exist for cellular sentinels that utilize DNA-based recording systems. Engineering cells in an ecosystem to passively and continuously monitor intracellular and extracellular states and changes (i.e. a black box recorder) over large areas and long periods of time constitutes a powerful strategy for ubiquitous sensing and reporting. However, a key limitation of such sentinel cells thus far has been the reliance on colorimetric, fluorescence or luminescence reporter molecules, which require continuously operating detectors that are generally not portable and scalable. DNA-based recorders are poised to substantially impact this arena, creating an entirely new class of environmental sentinel applications. Various recording paradigms could be implemented in engineered organisms, including bacteria, invertebrates (e.g. worms), insects (e.g. mosquitos and bees119), plants, and mammals and their host-associated microbiomes, in open and contained settings.
To monitor open environments (e.g. terrestrial, aquatic or aerial), engineered recorders could track the persistence and levels of pathogen-associated quorum signals120,121, toxic heavy metals122,123, and other biotic signatures of interest for various industries to ensure the health of crops, livestock and fisheries. For such open-environment sensing applications, the safety and dissemination of such synthetic recording devices must be rigorously assessed and the proper regulatory frameworks must be developed. DNA-based sentinels that can be applied to different surfaces could be used in biosurveillance and forensic applications to monitor the flow of materials (e.g. goods or contrabands) and controlled substances (e.g. explosives124,125) across the globe. A distinct advantage of cellular fingerprinting and recording strategies over existing inert chemical markers126 is the ability to track transient or fluctuating changes in environmental conditions (e.g. temperature or humidity), which may occur during transportation.
Other settings such as host-associated environments (e.g. humans, livestock and insects) are highly relevant application areas for DNA-based sentinels. DNA-based recording approaches have recently been applied to commensal Bacteroides thetaiotaomicron to record the availability of dietary nutrients such as rhamnose in the gut48. This ability to monitor host function and health status using engineered probiotics in the mammalian gut could enable new healthcare applications to non-invasively detect and record infections127,128 and biomarkers of inflammation levels129,130. Combining these approaches with actuation systems that are directly linked to these memory modules could yield smarter live-cell diagnostic and therapeutic probiotics that are capable of recording and responding to the spatial distribution and dynamics of difficult to measure biomarkers and metabolites131–133.
For contained environments such as microbial and mammalian fermentation reactors or bioremediation systems, engineered cellular sensors and recorders could provide real-time monitoring and diagnosis of cell physiology and metabolism to enhance the productivity of cell factories of different chemicals and drugs, as well as provenance tracking of valuable or sensitive strains. These active monitoring and recording approaches could be applied to a variety of built-up environments such as hospitals, airports and schools to examine the spread of contagious and infectious agents. In the future, DNA-based recording devices could interface with silicon-based electronics to interconvert biologically encoded data with digitally stored information134. Combined with fast and economical read–write DNA technologies, these approaches could enable direct control and information transfer between biological and electronic systems.
Outlook and conclusions
We envision that DNA-based memory systems will constitute a powerful new modality of biological measurement, enabling fundamentally new insights into complex cellular and organismal behaviours and next-generation surveillance applications. However, a number of key technical challenges and knowledge gaps still need to be addressed, spanning the engineering, implementation and analysis of these biological memory devices (Box 3).
Box 3. Key future challenges for DNA-based recording.
Highly multiplexed recording of diverse input signals
New engineered sensors and sensing strategies will be required to detect biological signals of interest that cannot currently be sensed or transformed into recordable information. Approaches to scale the number of channels that can be simultaneously recorded in individual cells and increase storage capacity should be explored, such as specifying the recording of different signals to specific target addresses. Synthetic biology circuits could be applied to integrate multiple cellular signals and report on complex cellular phenotypes.
Recording fast signals
How can cellular signals be quickly and modularly linked to activate recording into DNA on post-translational timescales? Timescales of native cellular processes (e.g. DNA replication) must be taken into account to capture rapidly fluctuating signals. Synthetic circuit modules such as positive feedback loops could be utilized to capture fast or transient inputs.
Quantifying variability
Can recording systems be applied to measure variability of a signal across a population? Statistical modelling and machine learning frameworks could be applied to infer the distribution of a signal magnitude or dynamics across cells and improve the accuracy of data reconstruction.
Stability of recording systems
Any engineered recording system will place a burden on cell fitness. The resource requirements and cellular impact of systems must be minimized to avoid altering cellular functionality and improve long-term stability.
Rapid and low-cost data readout
For practical or field applications of cellular recording, reading and interpreting raw data from DNA must be possible with minimal resources and in a fast manner. Low-cost and deployable nucleic acid assays or ubiquitous sequencing paradigms such as nanopore sequencing could be utilized to enable practical or field applications of cellular recording.
Complex in vivo data processing
New cellular computation paradigms and genetic circuits must be developed to interface with and interpret the results of cellular recording. These new approaches could close the loop between data recording and actuation of cellular responses as a result of complex multi-signal and dynamic input patterns.
Existing systems and recording syntaxes can be systematically improved to increase performance. Directed evolution or mutagenesis can alter the functionality or increase the enzymatic efficiency of DNA writing machinery135,136 or generate systems for parallel recording modalities85. Indeed, efforts have already yielded improved system components such as Cas9 variants with increased specificity and relaxed protospacer adjacent motif (PAM) requirements, which could be utilized to expand recording capabilities137–139. In addition, variants of system components can be metagenomically mined from the vast natural biological diversity for new properties. For example, CRISPR–Cas12a (Cpf1) displays staggered nuclease activity yielding a 4–5bp overhang compared to blunt ends generated by Cas971, which could enable alternative recording syntaxes. The storage capacity of existing systems could be increased by using more recording addresses such as additional genomic CRISPR arrays or Cas9-targeted array sites. Recording new input signal types may be possible with new system components, for example with reverse transcriptase (RT) Cas12 CRISPR integrase [G] variants that directly record RNA as an input signal into genomic CRISPR arrays140.
Entirely new classes of DNA-modifying biochemical modalities with improved performance characteristics almost certainly exist in nature that could be leveraged for recording applications. An ideal DNA recording syntax would consist of biochemical steps to write DNA with single base pair resolution in a structured manner (i.e. directionally) with high efficiency and in a manner that can be robustly modulated. Correspondingly, biological processes and corresponding enzymatic machinery with aspects of these features (i.e. non-templated polymerases141,142, terminal deoxynucleotidyl transferases [G] (TdTs)143,144) should be investigated and leveraged for next-generation recording applications. Other strategies not relying directly on the four natural base pairs can also be investigated; for example, unnatural bases could be used to expand the information density and capacity of recording145.
New measurement modalities drive novel scientific understanding of the fascinating behaviours of the natural world. Biological systems span many length and time scales, posing a challenge to traditional direct measurement paradigms that cannot practically be applied to directly measure and record the trillions of cells within developing organisms or environmental microbiomes. DNA-based recording devices offer an exciting new platform to surmount these challenges with a fundamentally different approach. By leveraging the self-replication and large numbers inherent to biological life, these systems could scale rapidly to record signals of previously immeasurable size and resolution, from mapping signal processing networks in the brain to understanding complex ecological niche utilization strategies in densely populated gut microbiomes. Highly optimized recording architectures, novel DNA-writing approaches, and continued progress in the scale and ease of sequencing DNA will further drive rapid progress in engineering recording systems that are capable of capturing larger amounts of information and highly multiplex signals. We envision that such DNA memory devices will catalyze a new field of basic research and applied endeavours to understand and probe complex populations or entire organisms.
Acknowledgements
We apologize to colleagues whose work could not be cited due to space limitations. H.H.W. acknowledges funding from the NIH (1R01AI132403–01), ONR (N00014–17-1–2353, N00014–15-1–2704), NSF (MCB‐1453219) and Burroughs Welcome Fund PATH (1016691). R.U.S. is supported by a Fannie and John Hertz Foundation Fellowship and a NSF Graduate Research Fellowship (DGE-11–44155).
Glossary
- Site-specific recombinase systems
Systems composed of a recombinase enzyme and flanking target recognition sites around a target sequence. These systems enable inversion, excision or integration of the target sequence based on the orientation of recognition sites
- RSM
(Recombinase state machine). A fixed-address writer encompassing a formalized architecture of genetic programs created from combinations of three orthogonal recombinase systems
- SCRIBE
(Synthetic cellular recorder integrating biological events). A single-stranded DNA (ssDNA)-recombination-based flexible writing approach
- Retron
A bacterial reverse transcriptase system that produces a molecule that is a hybrid of RNA and single-stranded DNA (ssDNA) called multicopy ssDNA (msDNA)
- CAMERA
(CRISPR-mediated analogue multi-event recording apparatus). A base-editing-based flexible writing approach
- Base editing
A Cas9-based genome engineering approach in which a catalytically dead Cas9 (dCas9) with no nuclease activity is linked to a deaminase (dCas9-BE), enabling single base-pair genomic mutation at desired locations
- Catalytically dead Cas9
(dCas9). A modified version of Cas9 that lacks endonuclease activity via engineered point mutations. It can be linked to other effector domains for diverse sequence-specific genome engineering applications
- Cas9
Clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9, a genome engineering nuclease tool enabling cleavage of desired genomic sites specified by a single-guide RNA (sgRNA)
- Non-homologous end joining
(NHEJ). An endogenous pathway enabling repair of double-strand breaks (DSBs)
- stgRNA
(Self-targeting guide RNA). a single-guide RNA (sgRNA) that that is targeted to its own sequence, which enables stochastic sequence evolution over time
- Directional writers
DNA writing relying on directional addition of single or multiple base pairs
- DNA polymerase
A type of enzyme that replicates DNA polymers based on an existing template DNA by serial addition of individual nucleotides
- TRACE
(Temporal recording in arrays by CRISPR expansion). A Cas1/2-based CRISPR spacer acquisition system to record biological signals over time
- Fluorescence resonance energy transfer
(FRET). A biochemical mechanism of energy transfer between two chromophores which can be utilized for sequence-specific DNA detection applications
- MEMOIR
(Memory by engineered mutagenesis with optical in situ readout). A Cas9-nuclease-based stochastic writing approach with spatial readout by single molecule RNA fluorescence in situ hybridization (smFISH)
- GESTALT
(Genome editing of synthetic target arrays for lineage tracing). A Cas9-nuclease-based stochastic writing approach enabling large-scale lineage tracing applications
- Cas12 CRISPR integrase
Conserved machinery in CRISPR immune systems mediating integration of short spacers from intracellular DNA sources into genomic arrays in a directional manner
- Terminal deoxynucleotidyl transferases
(TdTs). DNA polymerases that can add nucleotides to DNA without a template
- mSCRIBE
(Mammalian SCRIBE). A Cas9-nuclease-based stochastic writing approach
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Antebi YE, Nandagopal N & Elowitz MB An operational view of intercellular signaling pathways. Curr Opin Syst Biol 1, 16–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masel J & Siegal ML Robustness: mechanisms and consequences. Trends in Genetics 25, 395–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purvis JE & Lahav G Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church GM, Gao Y & Kosuri S Next-generation digital information storage in DNA. Science 337, 1628 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Goldman N et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erlich Y & Zielinski D DNA Fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Grass RN, Heckel R, Puddu M, Paunescu D & Stark WJ Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. Engl 54, 2552–2555 (2015). [DOI] [PubMed] [Google Scholar]
- 8.van der Woude MW & Baumler AJ Phase and antigenic variation in bacteria. Clinical Microbiology Reviews 17, 581–611 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marraffini LA CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Nemazee D Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol 6, 728–740 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Medhekar B & Miller JF Diversity-generating retroelements. Current Opinion in Microbiology 10, 388–395 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haselkorn R Developmentally regulated gene rearrangements in prokaryotes. Annu. Rev. Genet 26, 113–130 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Nowacki M, Shetty K & Landweber LF RNA-Mediated Epigenetic Programming of Genome Rearrangements. Annu Rev Genomics Hum Genet 12, 367–389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shendure J et al. DNA sequencing at 40: past, present and future. Nature 550, 345–353 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Kosuri S & Church GM Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keung AJ, Joung JK, Khalil AS & Collins JJ Chromatin regulation at the frontier of synthetic biology. Nat. Rev. Genet 16, 159–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrill DR & Silver PA Making cellular memories. Cell 140, 13–18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newby GA et al. A Genetic Tool to Track Protein Aggregates and Control Prion Inheritance. Cell 171, 966–979.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodworth MB, Girskis KM & Walsh CA Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet 18, 230–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camilli A & Mekalanos JJ Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Molecular Microbiology 18, 671–683 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceroni F et al. Burden-driven feedback control of gene expression. Nat. Methods 15, 387–393 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Roybal KT et al. Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell 167, 419–432.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrov N et al. A modular yeast biosensor for low-cost point-of-care pathogen detection. Science Advances 3, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor ND et al. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidl SR, Sheth RU, Wu A & Tabor JJ Refactoring and Optimization of Light-Switchable Escherichia coliTwo-Component Systems. ACS Synth. Biol 3, 820–831 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Stock AM, Robinson VL & Goudreau PN Two-component signal transduction. Annu. Rev. Biochem 69, 183–215 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Lim WA Designing customized cell signalling circuits. Nat Rev Mol Cell Biol 11, 393–403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaacs FJ, Dwyer DJ & Collins JJ RNA synthetic biology. Nature Biotechnology 24, 545–554 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Green AA, Silver PA, Collins JJ & Yin P Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wroblewska L et al. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nature Biotechnology 33, 839–841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheth RU, Yim SS, Wu FL & Wang HH Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461 (2017).By utilizing a copy-number-inducible plasmid, the CRISPR–Cas integrase system is utilized to record and reconstruct temporally changing biological signals.
- 32.Landry BP, Palanki R, Dyulgyarov N, Hartsough LA & Tabor JJ Phosphatase activity tunes two-component system sensor detection threshold. Nature Communications 9, 1433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brophy JAN & Voigt CA Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel R, Rubens JR, Sarpeshkar R & Lu TK Synthetic analog computation in living cells. Nature 497, 619–623 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Rubens JR, Selvaggio G & Lu TK Synthetic mixed-signal computation in living cells. Nature Communications 7, 11658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bashor CJ, Helman NC, Yan S & Lim WA Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319, 1539–1543 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Liu Y et al. Directing cellular information flow via CRISPR signal conductors. Nat. Methods 13, 938–944 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Nielsen AAK et al. Genetic circuit design automation. Science 352, aac7341 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Olson EJ & Tabor JJ Post-translational tools expand the scope of synthetic biology. Current Opinion in Chemical Biology 16, 300–306 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Stanton BZ, Chory EJ & Crabtree GR Chemically induced proximity in biology and medicine. Science 359, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deribe YL, Pawson T & Dikic I Post-translational modifications in signal integration. Nat Struct Mol Biol 17, 666–672 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Pham TM et al. A single-molecule approach to DNA replication in Escherichia coli cells demonstrated that DNA polymerase III is a major determinant of fork speed. Molecular Microbiology 90, 584–596 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Doudna JA & Charpentier E The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096–1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Kim H & Kim J-S A guide to genome engineering with programmable nucleases. Nat. Rev. Genet 15, 321–334 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Wirth D et al. Road to precision: recombinase-based targeting technologies for genome engineering. Current Opinion in Biotechnology 18, 411–419 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Grindley NDF, Whiteson KL & Rice PA Mechanisms of site-specific recombination. Annu. Rev. Biochem 75, 567–605 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Yang L et al. Permanent genetic memory with >1-byte capacity. Nat. Methods 11, 1261–1266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mimee M, Tucker AC, Voigt CA & Lu TK Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Systems 1, 62–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonnet J, Subsoontorn P & Endy D Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. U.S.A 109, 8884–8889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Rodriguez J, Yang L, Gorochowski TE, Gordon DB & Voigt CA Memory and Combinatorial Logic Based on DNA Inversions: Dynamics and Evolutionary Stability. ACS Synth. Biol 4, 1361–1372 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Friedland AE et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Courbet A, Endy D, Renard E, Molina F & Bonnet J Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Science Translational Medicine 7, 289ra83–289ra83 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Bonnet J, Yin P, Ortiz ME, Subsoontorn P & Endy D Amplifying Genetic Logic Gates. Science 340, 599–603 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Siuti P, Yazbek J & Lu TK Synthetic circuits integrating logic and memory in living cells. Nature Biotechnology 31, 448–452 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Roquet N, Soleimany AP, Ferris AC, Aaronson S & Lu TK Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016).Recombinase-based genetic circuits are formalized in a computer science state machine framework, enabling design synthetic circuits that discriminate the ordering of chemical inputs.
- 56.Hsiao V, Hori Y, Rothemund PW & Murray RM A population‐based temporal logic gate for timing and recording chemical events. Molecular Systems Biology 12, 869–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinberg BH et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nature Biotechnology 35, 453–462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farzadfard F & Lu TK Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science (2014). doi: 10.1126/science.1256272A framework for writing at genomic addresses utilizing ssDNA recombination is demonstrated, enabling recording of input signal intensity and duration, and interfacing with host responses in E. coli.
- 59.Tang W & Liu DR Rewritable multi-event analog recording in bacterial and mammalian cells. Science 360, (2018).The authors develop base editing approaches for cellular recording applications in both E. coli and mammalian cells.
- 60.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farzadfard F et al. Single-Nucleotide-Resolution Computing and Memory in Living Cells. biorxiv.org doi: 10.1101/263657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jinek M et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jinek M et al. RNA-programmed genome editing in human cells. Elife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lieber MR The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5 (2008). [DOI] [PubMed] [Google Scholar]
- 68.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).Cas9-nuclease-based stochastic editing of target arrays is utilized to reconstruct the lineage of cells and zebrafish embryos.
- 69.Schmidt ST, Zimmerman SM, Wang J, Kim SK & Quake SR Quantitative Analysis of Synthetic Cell Lineage Tracing Using Nuclease Barcoding. ACS Synth. Biol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017).Cas9-nuclease-based stochastic editing of target arrays is combined with smFISH spatial readouts to reconstruct spatial lineage and could be applied to reconstruct spatiotemporal gene expression.
- 71.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YG, Cha J & Chandrasegaran S Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A 93, 1156–1160 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bibikova M, Beumer K, Trautman JK & Carroll D Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Miller JC et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature Biotechnology 25, 778–785 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Boch J et al. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 326, 1509–1512 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Moscou MJ & Bogdanove AJ A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Christian M et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalhor R, Mali P & Church GM Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2016).The authors couple recursive editing of sgRNA sequences to an in situ sequencing readout for spatial lineage tracing applications
- 79.Perli SD, Cui CH & Lu TK Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 353, aag0511 (2016).The authors demonstrate recursive editing of sgRNA sequences, allowing for recording of signal intensity and duration in mammalian cells.
- 80.Glaser JI et al. Statistical analysis of molecular signal recording. PLoS Comput Biol 9, e1003145 (2013).The authors propose a statistical framework for temporal recording of ion concentration utilizing polymerase directional writing.
- 81.Zamft BM et al. Measuring cation dependent DNA polymerase fidelity landscapes by deep sequencing. PLoS ONE 7, e43876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Jackson SA et al. CRISPR-Cas: Adapting to change. Science 356, eaal5056–11 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Sternberg SH, Richter H, Charpentier E & Qimron U Adaptation in CRISPR-Cas Systems. Molecular Cell 61, 797–808 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Shipman SL, Nivala J, Macklis JD & Church GM Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175 (2016).In this work, the CRISPR–Cas integrase system is utilized to record the temporal ordering of oligonucleotide sequences electroporated into cell populations.
- 86.Shipman SL, Nivala J, Macklis JD & Church GM CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547, 345–349 (2017).CRISPR–Cas-integrase-based oligonucleotide recordings are scaled to store an animated frame in the genomes of living bacteria.
- 87.Shur A, bioRxiv RM 2018. Proof of concept continuous event logging in living cells. biorxiv.org doi: 10.1101/225151 [DOI] [Google Scholar]
- 88.Kluesner M et al. EditR: A novel base editing quantification software using Sanger sequencing. biorxiv.org doi: 10.1101/213496 [DOI] [Google Scholar]
- 89.Bentley DR et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quick J et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gaudet M, Fara A-G, Beritognolo I & Sabatti M Allele-specific PCR in SNP genotyping. Methods Mol. Biol 578, 415–424 (2009). [DOI] [PubMed] [Google Scholar]
- 93.Didenko VV DNA probes using fluorescence resonance energy transfer (FRET): Designs and applications. Biotechniques 31, 1106–+ (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee J-H et al. Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen X, Sun Y-C, Church GM, Lee J-H & Zador AM Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Research 46, e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kunkel TA & Bebenek R DNA replication fidelity. Annu. Rev. Biochem 69, 497–529 (2000). [DOI] [PubMed] [Google Scholar]
- 98.Deveau H et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. Journal of Bacteriology 190, 1390–1400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gudbergsdottir S et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Molecular Microbiology 79, 35–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weller GR et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297, 1686–1689 (2002). [DOI] [PubMed] [Google Scholar]
- 101.Pitcher RS, Wilson TE & Doherty AJ New insights into NHEJ repair processes in prokaryotes. Cell Cycle 4, 675–678 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Nuñez JK, Bai L, Harrington LB, Hinder TL & Doudna JA CRISPR Immunological Memory Requires a Host Factor for Specificity. Molecular Cell 62, 824–833 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Pattanayak V et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature Biotechnology 31, 839–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fu Y et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nivala J, Shipman SL & Church GM Spontaneous CRISPR loci generation in vivo by non-canonical spacer integration. Nature Microbiology 3, 310–318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nature Biotechnology 40, 181–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 108.Spanjaard B et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nature Biotechnology 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sender R, Fuchs S & Milo R Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 14, e1002533–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abel S et al. Sequence tag–based analysis of microbial population dynamics. Nat. Methods 12, 223–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nicholson JK et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 112.Smillie CS et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011). [DOI] [PubMed] [Google Scholar]
- 113.Kording KP Of toasters and molecular ticker tapes. PLoS Comput Biol 7, e1002291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marblestone AH et al. Physical principles for scalable neural recording. Front Comput Neurosci 7, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lim WA & June CH The Principles of Engineering Immune Cells to Treat Cancer. Cell 168, 724–740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eldar A & Elowitz MB Functional roles for noise in genetic circuits. Nature 467, 167–173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Balázsi G, van Oudenaarden A & Collins JJ Cellular decision making and biological noise: from microbes to mammals. Cell 144, 910–925 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fisher RA, Gollan B & Helaine S Persistent bacterial infections and persister cells. Nature Reviews Microbiology 15, 453–464 (2017). [DOI] [PubMed] [Google Scholar]
- 119.Leonard SP et al. Genetic Engineering of Bee Gut Microbiome Bacteria with a Toolkit for Modular Assembly of Broad-Host-Range Plasmids. ACS Synth. Biol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gupta S, Bram EE & Weiss R Genetically Programmable Pathogen Sense and Destroy. ACS Synth. Biol. 2, 715–723 (2013). [DOI] [PubMed] [Google Scholar]
- 121.Hwang IY et al. Reprogramming microbes to be pathogen-seeking killers. ACS Synth. Biol 3, 228–237 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Tauriainen S, Karp M, Chang W & Virta M Luminescent bacterial sensor for cadmium and lead. Biosensors and Bioelectronics 13, 931–938 (1998). [DOI] [PubMed] [Google Scholar]
- 123.Stocker J et al. Development of a Set of Simple Bacterial Biosensors for Quantitative and Rapid Measurements of Arsenite and Arsenate in Potable Water. Environ. Sci. Technol 37, 4743–4750 (2003). [DOI] [PubMed] [Google Scholar]
- 124.Antunes MS et al. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE 6, e16292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Belkin S et al. Remote detection of buried landmines using a bacterial sensor. Nature Biotechnology 35, 308–310 (2017). [DOI] [PubMed] [Google Scholar]
- 126.Gooch J, Daniel B, Abbate V & Frascione N Taggant materials in forensic science: A review. TrAC Trends in Analytical Chemistry 83, 49–54 (2016). [Google Scholar]
- 127.Hwang IY et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nature Communications 8, 15028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Danino T et al. Programmable probiotics for detection of cancer in urine. Science Translational Medicine 7, 289ra84–289ra84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Daeffler KNM et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Molecular Systems Biology 13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Riglar DT et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nature Biotechnology 35, 653–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Landry BP & Tabor JJ Engineering Diagnostic and Therapeutic Gut Bacteria. Microbiol Spectr 5, (2017). [DOI] [PubMed] [Google Scholar]
- 132.Riglar DT & Silver PA Engineering bacteria for diagnostic and therapeutic applications. Nature Reviews Microbiology 16, 214–225 (2018). [DOI] [PubMed] [Google Scholar]
- 133.Din MO et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature Publishing Group 536, 81–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tschirhart T et al. Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nature Communications 8, 14030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ghadessy FJ et al. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nature Biotechnology 22, 755–759 (2004). [DOI] [PubMed] [Google Scholar]
- 136.Heler R et al. Mutations in Cas9 Enhance the Rate of Acquisition of Viral Spacer Sequences during the CRISPR-Cas Immune Response. Molecular Cell 1–9 (2016). doi: 10.1016/j.molcel.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kleinstiver BP et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Silas S et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351, aad4234–aad4234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Clark JM Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Research 16, 9677–9686 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zyrina NV, Antipova VN & Zheleznaya LA Ab initiosynthesis by DNA polymerases. FEMS Microbiology Letters 351, 1–6 (2013). [DOI] [PubMed] [Google Scholar]
- 143.Lee HH et al. Enzymatic DNA synthesis for digital information storage. biorxiv.org doi: 10.1101/348987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Palluk S et al. De novo DNA synthesis using polymerase-nucleotide conjugates. Nature Biotechnology 36, 645–650 (2018). [DOI] [PubMed] [Google Scholar]
- 145.Zhang Y et al. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]