Abstract
Mutations in the KCNA1 gene encoding the voltage-gated potassium (K+) channel Kv1.1 have been linked to rare neurological syndromes, episodic ataxia type 1 (EA1) and myokymia. In 2009, a KCNA1 mutation was identified in a large family with autosomal dominant hypomagnesemia. Despite efforts in establishing a genotype-phenotype correlation for the wide variety of symptoms in EA1, little is known on the serum magnesium (Mg2+) levels in these patients. In the present study, we describe a new de novo KCNA1 mutation in a Polish patient with tetany and hypomagnesemia. Electrophysiological and biochemical analyses were performed to determine the pathogenicity of the mutation. A female patient presented with low serum Mg2+ levels, renal Mg2+ wasting, muscle cramps, and tetanic episodes. Whole exome sequencing identified a p.Leu328Val mutation in KCNA1 encoding the Kv1.1 K+ channel. Electrophysiological examinations demonstrated that the p.Leu328Val mutation caused a dominant-negative loss of function of the encoded Kv1.1 channel. Cell surface biotinylation showed normal plasma membrane expression. Taken together, this is the second report linking KCNA1 with hypomagnesemia, thereby emphasizing the need for further evaluation of the clinical phenotypes observed in patients carrying KCNA1 mutations.
Keywords: Magnesium, Potassium channel, Genetics, Patch clamp, Hypomagnesemia
Introduction
Hypomagnesemia (serum magnesium [Mg2+] <0.7 mmol/L) is a common electrolyte disorder that is caused by insufficient Mg2+ intake, the use of certain types of drugs, and/or genetic defects [1]. Over the last 15 years, studying the hereditary forms of hypomagnesemia has resulted in the identification of genes involved in the regulation of Mg2+ homeostasis [1, 2]. Many of the hypomagnesemia-causing gene mutations result in the defective reabsorption of Mg2+ in the distal convoluted tubule (DCT) of the kidney. As renal Mg2+ reabsorption does not take place beyond the DCT, defects in this segment inevitably result in renal Mg2+ wasting.
In 2009, we identified a c.763A>G mutation in KCNA1, encoding the voltage-gated potassium (K+) channel Kv1.1, in a large family with tetany, muscle cramps, and severe hypomagnesemia [3]. The identified variant results in a p.Asn255Asp missense mutation that completely abolishes the channel activity despite normal plasma membrane localization [3, 4]. Within the kidney, Kv1.1 is exclusively found at the apical membrane of DCT cells [3, 5]. It is hypothesized that Kv1.1 activity sets the driving force for active Mg2+ reabsorption via the Mg2+-permeable channel TRPM6.
Since the description of the first family in 2009, new hypomagnesemia-causing KCNA1 mutations have not been reported. Originally, KCNA1 mutations have been associated with episodic ataxia type 1 (EA1; MIM #160120), which is a rare neurological disorder characterized by motor discoordination, involuntary muscle contraction (myokymia), and seizures. While a recent genotype-phenotype correlation study in a large cohort of EA1 patients demonstrated high inter- and intrafamilial variability of symptoms, the penetrance of the hypomagnesemia phenotype has not been assessed in EA1 [6].
Here, we present a novel KCNA1 mutation in a family with tetany and hypomagnesemia for the first time since the initial report. We determined the pathogenicity of the mutation using electrophysiological and biochemical analyses.
Subjects and Methods
Patients
All patient studies were performed in accordance with the guidelines of the Declaration of Helsinki. The local Ethics Committee approved the study and the patient and her relatives provided written informed consent for genetic analysis. Serum and urine electrolyte measurements were performed by general laboratory screenings.
Microsatellite Analysis for Fatherhood Determination
To ascertain fatherhood, blood genomic DNA samples from the patient and both parents were PCR-amplified using primers surrounding polymorphic microsatellite loci D15S1018 (AFMb332yc5), D9S1784 (AFMa136xa5), D12S1604 (AFMa189yd9), D12S1590 (AFMa119wh5), D17S1529 (AFM022xb6), D8S405 (UT5312), and D9S172 (AFM199xf10). Primer sequences are provided in online supplementary Table S1 (for all online suppl. material, see www.karger.com/doi/10.1159/000488954). Samples were subjected to capillary electrophoresis; data were analyzed with the GeneMapper software version 5 (Applied Biosystems).
Whole Exome Sequencing
DNA from whole blood was extracted according to standard protocols. Exome sequencing was performed from 200 ng of genomic DNA using the standard protocol SureSelectXT Automated Target Enrichment for Illumina paired-end multiplexed sequencing, and the Agilent Bravo automated liquid handling platform. After validation (2200 TapeStation; Agilent Technologies, CA, USA) and quantification (Qubit System; Invitrogen, Waltham, MA, USA), pools of 6 libraries each were generated and subsequently sequenced on an Illumina HiSeq 4000 sequencing instrument using a paired-end 2 × 75 bp protocol.
For data analysis, the VARBANK pipeline version 2.15 (unpublished) and the corresponding filter interface was used. Raw reads were mapped to the human genome reference-build hg19 using the Burrows Wheeler Aligner alignment algorithm with a base quality threshold of 15 for read trimming (parameter: -q 15) [7]. The resulting binary alignment map files were further processed by Picard version 1.64 (http://broadinstitute.github.io/picard/) to mark duplicate reads and GATK version 1.6 to perform local realignment around short insertions and deletions and to recalibrate the base-calling quality scores [8]. A mean coverage of 76× was achieved with 84.6% of target regions covered more than 30×. The data were filtered for high-quality rare (MAF < 0.001) autosomal variants and attention was focused on genes with known involvement in renal Mg2+ handling.
Sanger Sequencing
The presence of the KCNA1 mutation p.Leu328Val was checked by the polymerase chain reaction using primers designed on the second exon of the human KCNA1 gene (NM_000217.2, F: 5′-GGCCATCCTCAGGGTCATCC-3′, R: 5′-AGCATCGGG GAT ACTGGAGAA-3′). Amplified sequences were Sanger sequenced according to standard methods.
DNA Constructs
The human KCNA1 pCIneo IRES GFP plasmid (Kv1.1 wild type) was obtained as previously described [3]. The p.Leu328Val mutation was introduced using the QuickChange site directed mutagenesis following the manufacturer's protocol (Stratagene, La Jolla, CA, USA). The empty pCIneo IRES GFP construct was used as a control (mock) in the experiments.
Cell Culture
HEK293 (human embryonic kidney 293) cells were cultured in Dulbeccos modified Eagles medium (DMEM, Bio Whittaker Europe, Vervier, Belgium) supplemented with 10% (v/v) fetal calf serum (PAA, Linz, Austria), 2 mM L-glutamine, and 10 μL/mL nonessential amino acids at 37°C in a humidity controlled incubator with 5% (v/v) CO2. The cells were transiently transfected with the respective DNA constructs using Lipofectamin 2000 (Invitrogen-Life Technologies, Bleiswijk, The Netherlands). The cells were cultured for 48 h prior to the start of experiments. For cell surface biotinylation, HEK293 cells were seeded on poly-L-lysine-coated 6-well plates.
Electrophysiology
Whole-cell recordings from transfected HEK293 cells were obtained at room temperature (20–25°C) using an EPC10 patch clamp amplifier controlled by Patchmaster software (HEKA Elektronik, Lambrecht, Germany). Currents were digitized at 10 kHz and digitally filtered at 2.9 kHz. Patch pipettes were pulled from thin-walled borosilicate glass (Harvard Apparatus, March-Hugstetten, Germany) with tip resistance of 2–4 MΩ when filled with pipette solution. Cell capacitance and access resistance were monitored using the automatic capacitance compensation of the Patchmaster software. Transfected cells were identified by their green fluorescence when illuminated at 480 nm. The pipette solution contained (mM): 140 KCl, 1 MgCl2, 0.1 CaCl2, 2 EGTA, 10 HEPES, and 1 Na2-ATP (pH adjusted to 7.3 with KOH). The extracellular solution contained (mM): 138 NaCl, 5.4 KCl, 1.2 MgCl2, 1 CaCl2, 10 EGTA, and 10 glucose (pH adjusted to 7.3 with NaOH). Recorded Kv1.1 currents were evoked by 150 ms voltage steps to potentials between −100 and +50 mV from a holding potential of −80 mV. Voltage steps were delivered in 10 mV increments at 10 s intervals. Current-voltage relations were constructed using the sustained amplitude at the end of test pulses. Current densities were obtained by normalizing the current amplitude to the cell membrane capacitance. No leakage subtraction was performed and cells with visible changes in leakage currents during the course of study were excluded from further analysis. The analysis and display of the data were performed using Igor Pro software (WaveMetrics, Lake Oswego, OR, USA).
Cell Surface Biotinylation
At 48 h after transfection, cells were washed twice with PBS-CM (PBS supplemented with 1 mM MgCl2, 0.5 mM CaCl2) following biotinylation of cell surface proteins using sulfo-NHS-LC-LC-biotin (0.5 mg/mL, Pierce, Etten-Leur, The Netherlands) in PBS-CM for 30 min at 4°C. Unreacted biotin was quenched using 0.1 % (w/v) BSA in PBS-CM. Next, cells were washed once with PBS (pH 7.5) and lysed in lysis buffer 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium-glycerophosphate, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 0.27 M sucrose, and the freshly added protease inhibitors pepstatin A (1 μg/mL, MP Biomedicals, Santa Ana, CA, USA), PMSF (1 mM, Sigma Aldrich, St Louis, MO, USA), leupeptin (5 μg/mL, MP Biomedicals), and aprotinin (1 μg/mL, Brunschwig Chemie, Basel, Switzerland). Cell lysates were collected and centrifuged (16,000 g) at 4°C for 15 min. Protein concentration was determined using the Bradford method. Equal amounts of protein were incubated with neutravidin-agarose beads (Pierce) for 2 h at 4°C. The beads were washed 3 times with lysis buffer and Kv1.1 was eluted with Laemmli sample buffer containing 100 mM DTT.
Immunoblotting
Kv1.1 expression was analyzed by immunoblot analysis for the input and the plasma membrane fraction using the Kv1.1 antibody (1: 3,000 Neuromab, Davis, CA, USA) and GFP antibody (1: 5,000, Sigma). Equal protein amounts were subjected to 10% (w/v) SDS-PAGE gel, transferred to PDVF membrane (Millipore), and incubated overnight with Kv1.1 antibody in TBS buffer supplemented with 0.1% Tween-20 (TBS-T) containing 5% (w/v) non-fat dry milk. Next day, the membranes were washed with TBS-T and incubated for 60 min with horseradish peroxidase conjugated secondary goat anti-mouse antibody (1: 10,000, Chemie Brunschwig) and goat anti-rabbit antibody (1: 10,000, Sigma) for 1 h at room temperature. Subsequently, the membranes were washed with TBS-T, and proteins were visualized with the Biorad ChemiDoc XRS using chemiluminescence SuperSignal West reagent (Thermo Fisher Scientific, Breda, The Netherlands).
Statistical Analysis
Data are shown as mean ± SEM. Statistical significance was determined by analysis of variance Dunnett's post hoc analysis using Prism software (GraphPad, San Diego, CA, USA). Differences in means with p < 0.05 were regarded as statistically significant.
Results
Tetany and Hypomagnesemia in a Female Patient
The patient presented at age 4 with mild tetanic episodes, whose frequency and intensity slowly progressed during follow-up. Significant hypomagnesemia (0.52 mmol/L) was first noted during pregnancy at the age of 24 years. Biochemical workup established a renal loss of Mg2+ (fractional excretion 3.9%, despite hypomagnesemia [normal < 1%]). No other biochemical abnormalities were observed, as the glomerular filtration rate, iPTH, plasma levels, and urinary excretion rates for Na+, Cl−, K+, Ca2+ and phosphate were in the normal range. She was treated with intravenous and oral Mg2+ and Ca2+ supplements. At present, at the age of 27 years, she receives oral Mg2+ supplements (390 mg of elemental Mg2+ per day), 1,000 mg of Ca2+ as well as 1000IE of vitamin D3. In addition, she is treated with hydrochlorothiazide and amiloride. However, therapy is only partially effective. Despite a normalization of plasma Mg2+ (0.78 mmol/L) and total Ca2+ levels (2.43 mmol/L), she continues to be affected by ongoing tetanies of mouth, neck, and hands. In addition, she has repeated cramps of the lower extremities and constant muscular weakness. She occasionally receives tizanidine, a muscle relaxant. However, this treatment is insufficient to control persisting neuromuscular symptoms. Her parents, siblings, and daughter were examined for hypomagnesemia and tetanic symptoms (Fig. 1a). However, no abnormalities were found.
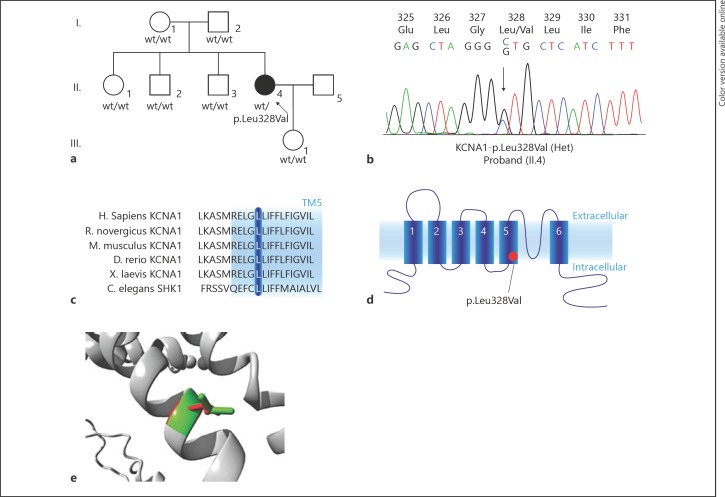
Fig. 1.
KCNA1 mutation in a family with hypomagnesemia. a Pedigree of the family. Filled symbols represent affected individuals, the arrow indicates the proband. b Mutation analysis chromatogram of the proband. The mutation is indicated by an arrow. c Multiple alignment analysis shows conservation of the Leu328 amino acid (blue bar) among species. The blue background indicates a transmembrane region (TM5, based on human sequence). d Schematic overview of a subunit of the Kv1.1 channel. The red dot indicates the location of the mutation. e Localization of the p.Leu328Val mutation in the Kv1.1 protein structure (Uniprot Q09470).
Microsatellite Analysis
Fatherhood was confirmed by analysis of 7 polymorphic microsatellite loci (for details see online suppl. Table 1).
De novo KCNA1-p.Leu328Val Mutation
To identify genetic defects that could explain hypomagnesemia, we performed whole exome sequencing. After sequential filtering (frequency cut off: 0.001) and quality control, homozygous variants in 7 genes, compound-heterozygous variants in 5 genes, as well as heterozygous variants in 223 genes were identified (for details see online suppl. Material). Filtering of the variants including only hypomagnesemia-causing genes resulted in the identification of a heterozygous non-synonymous variant in KCNA1 (OMIM #176260, c.982C>G, p.Leu328Val; Fig. 1b). Especially, TRPM6, FXYD2, SLC12A3, CLDN16, CLDN19, EGF, CNNM2, HNF1B, and KCNJ10 were negative for rare variants. The KCNA1-c.982C>G variant was absent in dbSNP, ExAC, and gnomAD databases and was predicted to be deleterious by SIFT and PROVEAN [9, 10, 11]. Sanger sequencing of the parents did not detect mutations in KCNA1, indicating a de novo origin of the variant. KCNA1-p.Leu328 is highly conserved among species (Fig. 1c). The KCNA1 gene codes for the voltage-gated K+ channel Kv1.1, which is composed of 4 subunits each containing 6 transmembrane segments (S1–S6) and intracellular amino- and carboxy-terminal tails (Fig. 1d). The p.Leu328Val mutation is located at the base of the 5th transmembrane domain (S5), which is part of the channel pore (Fig. 1d). Previously, a homology model was developed (http://www.cmbi.ru.nl/∼hvensela/Kv1.1/, [4]) that was based on the resolved chimaeric Kv1.2-Kv2.1 structure (PDB 2R9R [12]). Figure 1e shows the Kv1.1 homology model and depicts the transmembrane helices surrounding Leu328. This residue is located on the surface of the S5 helix and was not found to be in contact with other domains. Of note, the mutant valine residue is smaller than the original leucine residue (Fig. 1e, green: original Leu328, red: mutant Val328).
KCNA1-p.Leu328Val Results in Non-Functional Channel
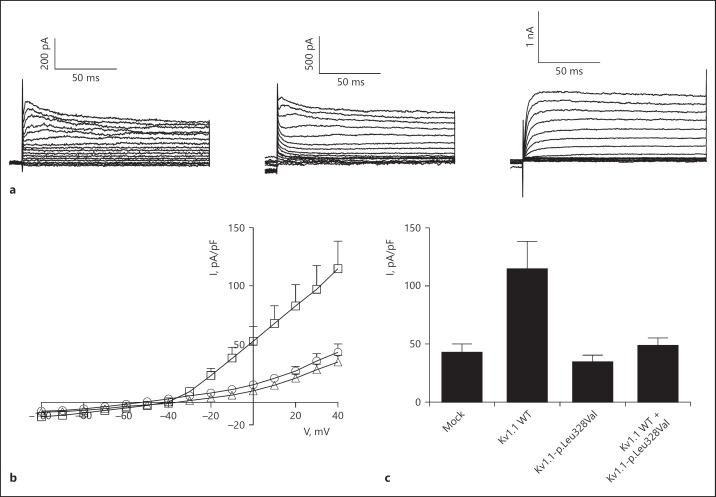
The effect of the p.Leu328Val mutation on the function of the encoded Kv1.1 channel was investigated by whole-cell patch clamp analysis. HEK293 cells were transiently transfected with mock, Kv1.1 wild type or Kv1.1-p. Leu328Val. Cells expressing Kv1.1 wild type channels produced characteristic outwardly rectifying K+ currents, while the expression of Kv1.1-p.Leu328Val resulted in small currents similar to control (mock; Fig. 2a, b). The current amplitude of Kv1.1-p.Leu328Val, extracted at +50 mV, was significantly reduced compared to Kv1.1 wild type (Fig. 2c). Considering the heterozygosity of this de novo mutation, a potential dominant negative effect was investigated by co-transfecting HEK293 cells with equal amounts of Kv1.1 wild type and Kv1.1-p.Leu328Val. The current amplitude of cells co-expressing Kv1.1 wild type and Kv1.1-p.Leu328Val was significantly reduced compared to that of Kv1.1 wild type alone (p < 0.05; Fig. 2c).
Fig. 2.
Electrophysiological analysis of Kv1.1 wild type and Kv1.1-p.Leu328Val channels. a Representative original traces of outward K+ currents of HEK293 cells expressing mock, Kv1.1 wild type, or Kv1.1-p.Leu328Val. The insert on the right top shows the voltage protocol consisting of voltage steps from −100 to +50 mV in 10-mV increments, applied from a holding potential of −80 mV, every 10 s. b The current-voltage (I/V) relationships of mock (circles), Kv1.1 wild type (squares), Kv1.1-p.Leu328Val (triangles). Mean values are shown. c Histogram presenting the averaged current densities at +50 mV of mock (n = 5), Kv1.1 wild type (n = 12), Kv1.1-p.Leu328Val (n = 6), and Kv1.1 wild type + Kv1.1-p.Leu328Val (n = 9). Asterisk indicates p < 0.05, compared to Kv1.1 wild type. Mean ± SEM is shown.
KCNA1-p.Leu328Val is Expressed at the Plasma Membrane
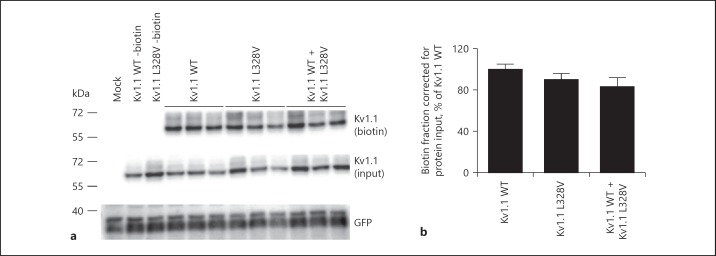
To examine whether the reduced function of the Kv1.1-p.Leu328Val channel is due to trafficking failure toward the plasma membrane, cell surface biotinylation experiments were performed. Figure 3a shows a representative immunoblot of the biotinylation demonstrating an equal amount of Kv1.1 wild type and Kv1.1-p.Leu328Val channels at the plasma membrane (upper panel). Furthermore, cells co-expressing Kv1.1 wild type and Kv1.1-p.Leu328Val showed similar plasma membrane abundance as Kv1.1 wild type or Kv1.1-p.Leu328Val alone (Fig. 3a). Quantification of 3 independent experiments confirmed no significant differences between the conditions (Fig. 3b), suggesting that similar amounts of either homotetrameric (wild type or mutant) channels or heterotetrameric (wild type plus mutant) channels were located at the plasma membrane. Of note, no difference in Kv1.1 expression was observed in the total cell lysates (Fig. 3a, middle panel). green fluorescent protein (GFP) expression was used as transfection control and was equal in all conditions tested (Fig. 3a, lower panel).
Fig. 3.
Expression of wild-type and mutant Kv1.1 channels. a Cell surface biotinylation of HEK293 cells expressing either mock, Kv1.1 wild type, Kv1.1-p.Leu328Val, or co-expressing Kv1.1 wild type and Kv1.1-p.Leu328Val. Kv1.1 expression was analyzed by immunoblotting for plasma membrane fraction and input from the total cell lysates. Representative immunoblot of 4 independent experiments is shown. b Bar graph representing the quantification of the cell surface biotinylation experiments. It depicts the relative plasma membrane expression compared to input, and is shown as percentage of Kv1.1 wild type. Mean ± SEM is shown.
Discussion
Here, we describe the first new KCNA1 mutation that is causative for hypomagnesemia since the identification of the initial family in 2009. A de novo c.982C>G mutation in KCNA1 was identified in a patient with hypomagnesemia and tetany. Electrophysiological and biochemical analyses show that the resulting Kv1.1-p.Leu328Val amino acid change causes loss of function, despite normal plasma membrane expression.
The proband presented with hypomagnesemia, tetany, muscle weakness, and tremor, which is highly comparable to the Brazilian family described previously [3]. In line with this family, the proband showed inappropriately normal urinary Mg2+ excretion despite hypomagnesemia indicating a renal leak [3]. Treatment with Mg2+ supplements was only partially successful, as it restored the serum Mg2+ concentration to the low-normal range but did not completely relieve clinical symptoms. Interestingly, the mutation identified here occurred de novo, as its presence was excluded in all other family members tested including the parents. Furthermore, it was not transmitted to the unaffected daughter of the patient. These findings confirm the causality of KCNA1 mutations for hypomagnesemia and demonstrate that this aspect is not mutation-specific for p.Asn255Asp.
KCNA1 mutations have been widely described as cause for EA1, with thus far 38 different mutations involved [13, 14]. The majority of the mutations result in reduced channel function, weakened surface expression, or changes in the biophysical characteristics that lead to a dominant-negative effect upon association with wild-type subunits [14]. The latter is also observed in our study. The phenotype of EA1 patients is highly heterogeneous and may include myokymia, cataplexy, epilepsy, neuromyotonia, paroxysmal dyspnea, and also skeletal defects [13, 15, 16, 17, 18, 19, 20]. Of note, episodic ataxia is not present in all patients. Despite the identification of the p.Asn255Asp mutation as cause for hypomagnesemia in 2009 [3], hypomagnesemia has not been reported in (other) patients affected by EA1 due to mutations in KCNA1. Therefore, it is still difficult to assess the penetrance of the hypomagnesemia phenotype in patients with KCNA1 mutations.
The novel identified KCNA1-p.Leu328Val mutation has never been described before. However, closely located mutations p.Glu325Asp and p.Leu329Ile have been reported in EA1 patients, in which serum Mg2+ levels are unknown [19, 20, 21]. The Leu328, as well as these latter 2 residues, are located on the cytoplasmic side of the transmembrane helix S5. The KCNA1-p.Glu325Asp, p.Leu328Val, and p.Leu329Ile mutations result only in small changes of glutamic acid to aspartic acid and leucine to valine or isoleucine, respectively, which are amino acids that are similar in size, hydrophobicity, and charge. Based on our homology model, they are likely to interact with the lipids in the plasma membrane and provide stability of the protein complex. Moreover, the amino acid changes might disrupt the S5 helix, which then disturbs the channel pore structure and results in reduced channel function as apparent in our study. Likewise, the KCNA1 p.Glu325Asp was shown to significantly alter channel function characteristics [22]. Interestingly, the other hypomagnesemia-causing p.Asn255Asp mutation was also located at the cytoplasmic interface of the plasma membrane, and demonstrated a similar dominant-negative loss of channel function observed in the present study [3]. Despite recognition of the wide distribution throughout the protein and altered effects on channel properties, there is still no clear association reported between the broad spectrum of clinical phenotypes and specific mutations. Detailed analysis will be necessary to elucidate how functional consequences could explain the disease diversity.
Although expression levels of Kv1.1 in the kidney are low, immunohistochemical stainings have shown that Kv1.1 is exclusively expressed at the apical membrane of the DCT [3, 5]. In the DCT, intracellular Mg2+ levels are comparable to the Mg2+ concentration in the pro-urine and therefore a chemical gradient for Mg2+ reabsorption is suggested to be virtually absent. It has therefore been hypothesized that K+ secretion via Kv1.1 provides an electrical gradient that drives Mg2+ reabsorption via TRPM6 [3, 4]. Indeed, our previous studies showed that Kv1.1 activity determined the membrane potential in HEK293 cells [3]. This is in line with studies in rabbit distal tubule cells showing that the apical K+ conductance sets the membrane potential but has been recently contested by experiments in mouse DCT segments that did not show apical K+ transport [23, 24]. Recent expression studies suggest that expression of Kv1.1 and other K+ channels, like ROMK, depend on the specific conditions including K+ and Mg2+ availability [25, 26]. Future studies should aim at resolving the inconsistencies on the exact nature of the apical K+ channel in the DCT.
In conclusion, this study reports a patient with hypomagnesemia and tetany caused by a de novo KCNA1 mutation, unequivocally showing that KCNA1 mutations are causative for hypomagnesemia.
Ethics Statement
The study was performed in accordance with the guidelines of the Declaration of Helsinki. The local Ethics Committee approved the study. Informed consent was obtained from the patient and her relatives.
Disclosure Statement
The authors declare that there are no conflicts of interest to disclose.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgments
We are indebted to the patient and the family who participated in this research. This work was supported by grants from the Netherlands Organization for Scientific Research (NWO Veni 863.13.010) to Dr. Jenny van der Wijst and (NWO Rubicon 825.14.021, NWO Veni 016.186.012) to Dr. Jeroen H.F. de Baaij, from the Dutch Kidney Foundation (Kolff 14OKG17) to Dr. Jeroen H.F. de Baaij, and the European Union Seventh Framework Program (EURenOmics, FP7/2007-2013 No. 305608).
References
- 1.de Baaij JH, Hoenderop JG, Bindels RJ. Magnesium in man: implications for health and disease. Physiol Rev. 2015;95:1–46. doi: 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 2.de Baaij JH, Hoenderop JG, Bindels RJ. Regulation of magnesium balance: lessons learned from human genetic disease. Clin Kidney J. 2012;5:i15–i24. doi: 10.1093/ndtplus/sfr164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaudemans B, van der Wijst J, Scola RH, Lorenzoni PJ, Heister A, van der Kemp AW, Knoers NV, Hoenderop JG, Bindels RJ. A missense mutation in the Kv1.1 voltage-gated potassium channel-encoding gene KCNA1 is linked to human autosomal dominant hypomagnesemia. J Clin Invest. 2009;119:936–942. doi: 10.1172/JCI36948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Wijst J, Glaudemans B, Venselaar H, Nair AV, Forst AL, Hoenderop JG, Bindels RJ. Functional analysis of the Kv1.1 N255D mutation associated with autosomal dominant hypomagnesemia. J Biol Chem. 2010;285:171–178. doi: 10.1074/jbc.M109.041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhlen M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 6.Graves TD, Cha YH, Hahn AF, Barohn R, Salajegheh MK, Griggs RC, Bundy BN, Jen JC, Baloh RW, Hanna MG;, CINCH Investigators Episodic ataxia type 1: clinical characterization, quality of life and genotype-phenotype correlation. Brain. 2014;137:1009–1018. doi: 10.1093/brain/awu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG;, Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 13.Browne DL, Gancher ST, Nutt JG, Brunt ER, Smith EA, Kramer P, Litt M. Episodic ataxia/myokymia syndrome is associated with point mutations in the human potassium channel gene, KCNA1. Nat Genet. 1994;8:136–140. doi: 10.1038/ng1094-136. [DOI] [PubMed] [Google Scholar]
- 14.D'Adamo MC, Hasan S, Guglielmi L, Servettini I, Cenciarini M, Catacuzzeno L, Franciolini F. New insights into the pathogenesis and therapeutics of episodic ataxia type 1. Front Cell Neurosci. 2015;9:317. doi: 10.3389/fncel.2015.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brownstein CA, Beggs AH, Rodan L, Shi J, Towne MC, Pelletier R, Cao S, Rosenberg PA, Urion DK, Picker J, Tan WH, Agrawal PB. Clinical heterogeneity associated with KCNA1 mutations include cataplexy and nonataxic presentations. Neurogenet. 2016;17:11–16. doi: 10.1007/s10048-015-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eunson LH, Rea R, Zuberi SM, Youroukos S, Panayiotopoulos CP, Liguori R, Avoni P, McWilliam RC, Stephenson JB, Hanna MG, Kullmann DM, Spauschus A. Clinical, genetic, and expression studies of mutations in the potassium channel gene KCNA1 reveal new phenotypic variability. Ann Neurol. 2000;48:647–656. [PubMed] [Google Scholar]
- 17.Kinali M, Jungbluth H, Eunson LH, Sewry CA, Manzur AY, Mercuri E, Hanna MG, Muntoni F. Expanding the phenotype of potassium channelopathy: severe neuromyotonia and skeletal deformities without prominent Episodic Ataxia. Neuromuscul Disord. 2004;14:689–693. doi: 10.1016/j.nmd.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Shook SJ, Mamsa H, Jen JC, Baloh RW, Zhou L. Novel mutation in KCNA1 causes episodic ataxia with paroxysmal dyspnea. Muscle Nerve. 2008;37:399–402. doi: 10.1002/mus.20904. [DOI] [PubMed] [Google Scholar]
- 19.Browne DL, Brunt ER, Griggs RC, Nutt JG, Gancher ST, Smith EA, Litt M. Identification of two new KCNA1 mutations in episodic ataxia/myokymia families. Hum Mol Genet. 1995;4:1671–1672. doi: 10.1093/hmg/4.9.1671. [DOI] [PubMed] [Google Scholar]
- 20.Tristan-Clavijo E, Scholl FG, Macaya A, Iglesias G, Rojas AM, Lucas M, Castellano A, Martinez-Mir A. Dominant-negative mutation p.Arg324Thr in KCNA1 impairs Kv1.1 channel function in episodic ataxia. Movement Disord. 2016;31:1743–1748. doi: 10.1002/mds.26737. [DOI] [PubMed] [Google Scholar]
- 21.Knight MA, Storey E, McKinlay Gardner RJ, Hand P, Forrest SM. Identification of a novel missense mutation L329I in the episodic ataxia type 1 gene KCNA1 - a challenging problem. Hum Mut. 2000;16:374. doi: 10.1002/1098-1004(200010)16:4<374::AID-HUMU15>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Adelman JP, Bond CT, Pessia M, Maylie J. Episodic ataxia results from voltage-dependent potassium channels with altered functions. Neuron. 1995;15:1449–1454. doi: 10.1016/0896-6273(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 23.Yoshitomi K, Shimizu T, Taniguchi J, Imai M. Electrophysiological characterization of rabbit distal convoluted tubule cell. Pflugers Arch. 1989;414:457–463. doi: 10.1007/BF00585057. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Wang L, Thomas S, Wang K, Lin DH, Rinehart J, Wang WH. Src family protein tyrosine kinase regulates the basolateral K channel in the distal convoluted tubule (DCT) by phosphorylation of KCNJ10 protein. J Biol Chem. 2013;288:26135–26146. doi: 10.1074/jbc.M113.478453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San-Cristobal P, Lainez S, Dimke H, de Graaf MJ, Hoenderop JG, Bindels RJ. Ankyrin-3 is a novel binding partner of the voltage-gated potassium channel Kv1.1 implicated in renal magnesium handling. Kidney Int. 2014;85:94–102. doi: 10.1038/ki.2013.280. [DOI] [PubMed] [Google Scholar]
- 26.Wade JB, Fang L, Coleman RA, Liu J, Grimm PR, Wang T, Welling PA. Differential regulation of ROMK (Kir1.1) in distal nephron segments by dietary potassium. Am J Physiol Renal Physiol. 2011;300:F1385–F1393. doi: 10.1152/ajprenal.00592.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data