Epilepsy characterized by highly heterogeneous treatments is one of the most common neurological disorders in the world 1, 2. A number of antiepileptic drugs (AEDs) were developed to treat this disease; however, the drug response was remarkably variable among individuals. Therefore, predicting each patient's response to AEDs is important for the personalized treatment, which will improve the therapeutic efficiency. Data mining (DM) is the process of discovering knowledge embedded in large data set 3. It is capable of simultaneously modeling multiple factors; thus, some DM approaches were developed to establish prediction models. We proposed that this method can also be used to establish AEDs response prediction model. Here, we firstly genotyped 31 SNPs in a total of 699 patients with epilepsy. Then, nine DM approaches were employed to establish prediction model for AEDs response with these SNPs and three clinical factors. Finally, the performance of these models was validated in an independent population.
Our protocol was approved by the Ethics Committee of Xiangya School of Medicine, Central South University. All individuals provided a written informed consent in compliance with the code of ethics of the World Medical Association (Declaration of Helsinki) before this study was initiated. We applied this study for clinical admission in the Chinese Clinical Trial Register (Registration Number: ChiCTR‐TCH‐0000813). The clinical characteristics of all subjects are summarized in Table S1. The gene and SNP selection was mainly based on our previous two published studies 4, 5. A total of 31 SNPs in 10 genes were included (Table S2). Nine DM approaches were employed in this study to establish prediction model using WEKA software as described previously, including Bayesian net (BN), logistic regression (LR), artificial neural network (ANN), k‐nearest neighbor (k‐NN), support vector machine (SVM), decision tree (DT), random forest (RF), adaptive boosting (AB), and bagging (BAG) 6, 7. The outputs consisted of response versus nonresponse, which were all binary variables. For all methods, 10‐fold cross‐validation was employed to evaluate the model prediction accuracy. The data set were randomly and alternately divided into ten groups: nine groups were assigned as training sets used to estimate the classification accuracy, and one group was assigned as evaluation set used to test the prediction accuracy of established models. The process was repeated ten times to make sure each group was assigned once as an evaluation set. The overall model prediction accuracy was the averaged value across all ten trials. The P value was two‐sided, and P<0.05 was considered statistically significant. All statistical analyses were performed using PLINK and SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) 8.
In our samples, carbamazepine was one of the most commonly used drugs and we firstly established response prediction models for this drug. These models were established using nine DM approaches with 31 SNPs and three clinical factors. The results are indicated in Figure 1A; except BN and KNN models, the sensitivity was higher than specificity in all other models. The DT model achieved the highest sensitivity of 0.94; however, it also had the lowest specificity of 0.23. The specificity is generally low in carbamazepine models. The highest came from ANN and KNN models, and they both achieved 0.68. The AB model had the best overall performance, and it reached a sensitivity of 0.82, a specificity of 0.59, a prediction accuracy of 0.82 for responders, a prediction accuracy of 0.59 for nonresponders, and an overall accuracy of 0.71 (Table S3). Its quality was also assessed by the ROC curve, with the AUC of 0.79 (Figure 1B). This model was validated in an independent cohort of 100 patients, and compared with derivation cohort; both the sensitivity (0.95) and specificity (0.60) were higher in the validation population (Table 1). This result showed that this model was successfully validated in another cohort.
Figure 1.
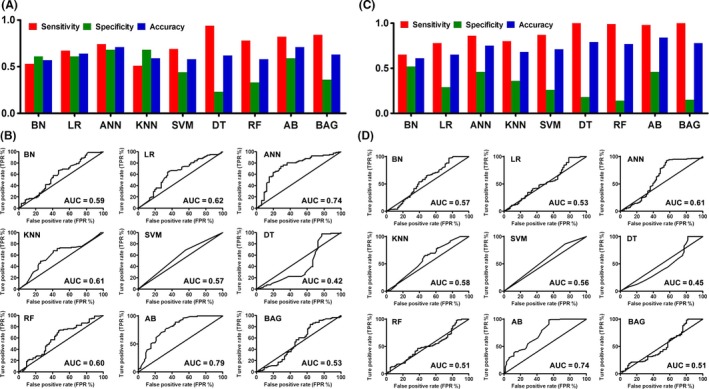
The performance and ROC curves of prediction models established by different DM algorithms for carbamazepine and valproic acid. (A) and (C): The sensitivity (red bar), specificity (green bar), and overall accuracy (blue bar) of models established for carbamazepine (A) and valproic acid (C) by nine DM algorithms (BN, LR, ANN, KNN, SVM, DT, RF, AB, and BAG) were indicated. (B) and (D): The nine established models' performance of carbamazepine (B) and valproic acid (D) was evaluated by ROC curve, which was plotted by true‐positive rate (TPR) against false‐positive rate (FPR). Area under curve (AUC) was also shown for each curve. BN, Bayesian net; LR, logistic regression; ANN, artificial neural network; k‐NN, k‐ nearest neighbor; SVM, support vector machine; DT, decision tree; RF, random forest; AB, adaptive boosting; BAG, bagging.
Table 1.
Performance of models established by AB algorithms in derivation and validation cohorts
| TP | TN | FP | FN | SE | SP | PRE | OA | AUC | MAE | |
|---|---|---|---|---|---|---|---|---|---|---|
| Carbamazepine | ||||||||||
| Derivation cohort | 66 | 39 | 27 | 15 | 0.82 | 0.59 | 0.71 | 0.71 | 0.79 | 0.35 |
| Validation cohort | 52 | 27 | 18 | 3 | 0.95 | 0.60 | 0.81 | 0.79 | 0.83 | 0.32 |
| Valproic acid | ||||||||||
| Derivation cohort | 182 | 30 | 36 | 4 | 0.98 | 0.46 | 0.84 | 0.84 | 0.74 | 0.28 |
| Validation cohort | 135 | 25 | 40 | 0 | 1.00 | 0.39 | 0.85 | 0.80 | 0.75 | 0.30 |
TP, true positives; TN, true negatives; FP, false positives; FN, false negatives; SE, sensitivity; SP, specificity; PRE, precision; OA, overall accuracy; AUC, area under curve; MAE, mean absolute difference.
Valproic acid is another mostly used drug in this study, and we thus next established the prediction model for this drug. The used methods and involved factors were the same as above‐mentioned models. All results are summarized in Figure 1C, and similar to carbamazepine prediction models, the sensitivity is higher than specificity for all algorithms. Also, DT, RF, AB, and BAG models achieved the highest sensitivity, which were 1.00, 0.99, 0.98, and 1.00, respectively (Table S3). However, except AB model, their specificity is very low. Both AB and ANN models achieved the specificity of 0.46; however, the highest (0.52) came from BN model. Overall, AB model had the best performance, and it achieved a sensitivity of 0.98, a specificity of 0.46, a prediction accuracy of 0.98 for responders, a prediction accuracy of 0.46 for nonresponders, and an overall accuracy of 0.84. The AUC of ROC curve is 0.74 (Figure 1D). We next tested this model's performance in an independent cohort of 200 patients. As indicated in Table 1, the sensitivity is the same as that of derivation cohort, but specificity decreased. The validation study showed that this model's sensitivity is quite reliable; however, the specificity still needs to be improved.
In summary, we genotyped 31 SNPs of 10 genes in a total of 699 patients with epilepsy. Then, we established the response prediction models using nine DM approaches with these SNPs and three clinical factors for carbamazepine and valproic acid. The established models were further validated in an independent population. The results indicated that for both drugs, the prediction model established using adaptive boosting algorithm had the best performance. The carbamazepine response prediction model achieved a sensitivity of 0.82, a specificity of 0.59, an overall accuracy of 0.71, and ROC curve AUC of 0.79. The valproic acid response prediction model achieved a sensitivity of 0.98, a specificity of 0.46, an overall accuracy of 0.84, and ROC curve AUC of 0.74. However, it is noteworthy that our models still need to be validated in other large sample size populations before they can be used in clinical practice.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Table S1: Clinical characteristics of enrolled epilepsy patients.
Table S2: Characteristics and annotation of genotyped SNPs.
Table S3: Performance of models established by different DM algorithms.
Acknowledgments
This work was supported by National High‐tech R&D Program of China 863 Program Grant (2012AA02A517) and National Natural Science Foundation of China Grants (81373490, 81573508, 81573463).
References
- 1. Wright J, Pickard N, Whitfield A, Hakin N. A population‐based study of the prevalence, clinical characteristics and effect of ethnicity in epilepsy. Seizure 2000;9:309–313. [DOI] [PubMed] [Google Scholar]
- 2. Glauser T, Ben‐Menachem E, Bourgeois B, et al. ILAE treatment guidelines: Evidence‐based analysis of antiepileptic drug efficacy and effectiveness as initial monotherapy for epileptic seizures and syndromes. Epilepsia 2006;47:1094–1120. [DOI] [PubMed] [Google Scholar]
- 3. Han J, Kamber M, Pei J. Data mining: Concepts and techniques. Waltham, MA: Morgan Kaufmann, 2012. [Google Scholar]
- 4. Qu J, Zhou BT, Yin JY, et al. ABCC2 polymorphisms and haplotype are associated with drug resistance in Chinese epileptic patients. CNS Neurosci Ther 2012;18:647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qu J, Zhang Y, Yang ZQ, et al. Gene‐wide tagging study of the association between KCNT1 polymorphisms and the susceptibility and efficacy of genetic generalized epilepsy in Chinese population. CNS Neurosci Ther 2014;20:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: An update. SIGKDD Explor 2009;11:1–18. [Google Scholar]
- 7. Yin JY, Li X, Li XP, et al. Prediction models for platinum‐based chemotherapy response and toxicity in advanced NSCLC patients. Cancer Lett 2016;377:65–73. [DOI] [PubMed] [Google Scholar]
- 8. Purcell S, Neale B, Todd‐Brown K, et al. PLINK: A tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Clinical characteristics of enrolled epilepsy patients.
Table S2: Characteristics and annotation of genotyped SNPs.
Table S3: Performance of models established by different DM algorithms.


