Abstract
The production of Escherichia coli K1 serotype capsule was investigated using direct stochastic optical reconstruction microscopy with live bacteria and graphene oxide-coated coverslips, overcoming many morphological artifacts found in other high-resolution imaging techniques. Super-resolution fluorescence images showed that the K1 capsular polysaccharide is not uniformly distributed on the cell surface, as previously thought. These studies demonstrated that on the cell surfaces the K1 capsule at the poles had bimodal thicknesses of 238 ± 41 and 323 ± 62 nm, whereas at the equator, there was a monomodal thickness of 217 ± 29 nm. This bimodal variation was also observed in high-pressure light-scattering chromatography measurements of purified K1 capsular polysaccharide. Particle tracking demonstrated that the formation of the capsule was dominated by the expansion of lyso-phosphatidylglycerol (lyso-PG) rafts that anchor the capsular polysaccharide in the outer membrane, and the expansion of these rafts across the cell surface was driven by new material transported through the capsular biosynthesis channels. The discovery of thicker capsules at the poles of the cell will have implications in mediating interactions between the bacterium and its immediate environment.
Introduction
Escherichia coli infections can cause disease in humans. A major factor in their virulence is often the coatings of the bacteria, which are called capsules. Recent developments in fluorescence microscopy allowed us to make much higher resolution images of capsules on living bacteria than were previously possible, and we studied E. coli that causes urinary tract infections. We developed the technique of super-resolution fluorescence microscopy (STORM) with graphene oxide (GO)-coated coverslips to image the capsules.1 Without the graphene oxide coating, it would be very challenging to image the bacteria using antibody labeling due to the large background of the nonspecifically bound fluorophore. This background fluorescence is conveniently extinguished by the graphene oxide (GO) using resonant energy transfer, without requiring invasive cleaning or blocking techniques. The bacteria are cylindrical with hemisphere caps (super-ellipsoidal) in shape, and the lengths of the molecules in the capsules were also measured using chromatography. Statistical analysis of atomic force microscopy (AFM) measurements with spherical colloidal probes (including control experiments) was then used to explore the forces the capsules on the bacteria exert on their surroundings. A model based on the polymeric nature of the capsules then enabled us to explain the origin of the force regimes measured.
Many clinically important bacteria have polysaccharide capsules attached to their surfaces, which increase their virulence.2,3 The primary physical mechanism behind this virulence is thought to be the creation of a repulsive steric potential that allows the bacterial cells to resist phagocytosis by the immune cells of the host organisms. The exact molecular mechanisms involved in the physical chemistry of these repulsive steric brushes are not well understood, and there are also a number of gaps in our understanding of their biochemistry, such as how the capsular polysaccharides are synthesized and transported through the inner and outer cell membranes.4−7
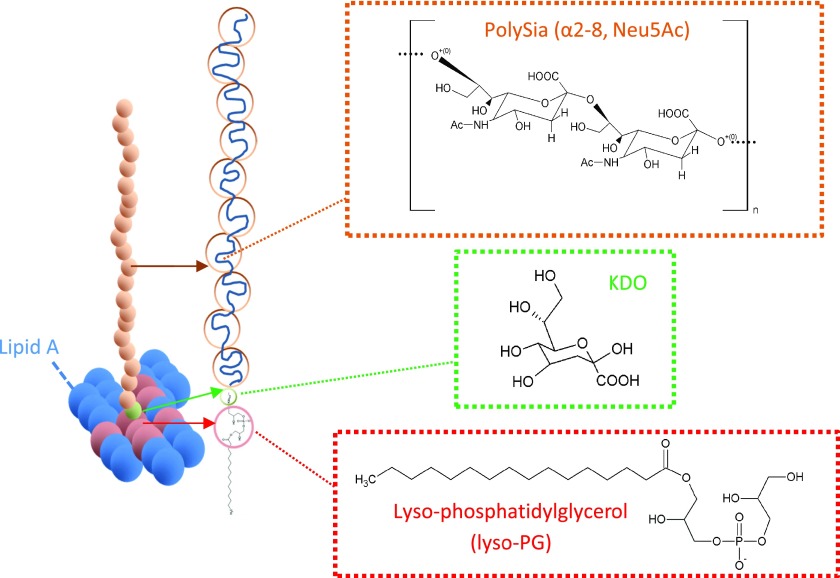
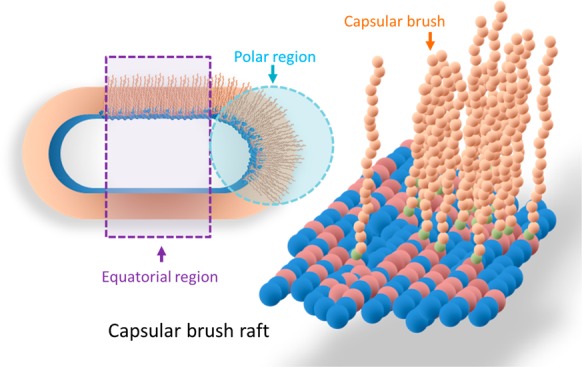
K1 capsular polysaccharide is one of more than 80 K-antigen serotypes8,9 that are found on the surface of pathogenic E. coli and K1 capsules occur on bacteria that cause urinary tract infections.10−12 The K1 capsular polysaccharide chains are made of nine-carbon N-acetylneuraminic monomers, with α-2,8 keto-glycosidic linkages (with a degree of polymerization of less than 200) linked with a β-linked poly-3-deoxy-d-manno-oct-2-ulosonic acid (KDO) linker on lyso-phosphatidylglycerol (lyso-PG)13 (Figure 1).
Figure 1.
Schematic diagram of a single chain of polysialic acid composed of sialic monomers (orange circles) with α-2,8 keto-glycosidic linkages linked with a few β-linked poly-3-deoxy-d-manno-oct-2-ulosonic acid (KDO) linkers (green circle); a monomer structure is shown in the green box, anchored onto lyso-phosphatidylglycerol (lyso-PG), in the red circle; a lyso-PG structure is shown in the red box. The lyso-PG is surrounded with lipid A in the bacterial outer membrane (the O-antigens are omitted for clarity).
Capsular polysialic polysaccharides on bacterial surfaces have been investigated previously in terms of both their biosynthesis and for applications in terms of targets for new antimicrobials and as vaccine candidates.12 However, detailed descriptions of capsular polysaccharide morphology and studies of their dynamics on live bacterial surfaces are relatively limited.
Due to the high virulence of bacteria with capsular polysaccharides, the manipulation of polysaccharide production could have important medical benefits. Most studies have been focused on the biochemical synthesis of capsular polysaccharides and the molecular machines used to control the biosynthesis pathways.14 Recent models have concluded that in the case of the E. coli K1 polysaccharide the capsular polysaccharide chains are made inside bacterial cells before transport across the periplasmic space onto the outer membrane.15 In addition to biosynthesis mechanisms, bacterial capsular morphology has been observed on fixed bacterial surfaces using scanning electron microscopy, transmission electron microscopy,16,17 and atomic force microscopy.18,19 These studies confirmed a physical model of the bacterial capsule as a capsular polyelectrolyte brush anchored onto the underlying phospholipid membrane. Brush is a term adopted from soft condensed matter physics, meaning an array of surface-tethered polymer chains. Polymer brushes are commonly used in chemistry to stabilize colloidal phases of matter against aggregation, for example, in paints, to reduce surface adsorption, such as in antibiofouling coatings, and to reduce frictional forces between surfaces in lubricants.20,21
A recent paradigm in membrane structure is that of lipid rafts, i.e., the lipids that form cellular membranes are not uniformly mixed and have a well-defined heterogeneous structuration.22 The idea of lipid raft formation has been recently applied to bacterial cells.23 In the current study, we explore the formation of K1 capsular lyso-PG rafts that merge together to form the capsular structure during its maturation. We find that their nucleation occurs randomly across the bacterial surface and the rafts do not move significantly from their initial point of nucleation.
Synthetic polymeric brushes have been extensively studied in the literature both theoretically and experimentally.24,25 Models can describe polymer brush morphology and the forces they experience. Extensions have also been made to when the polymers are charged (polyelectrolytes), and in this case, the counterions can also play a major role.26−28 Such models for polyelectrolyte brushes have been adapted to describe capsular polysaccharides and allowed intracellular forces measured in atomic force microscopy (AFM) to be explained. There have been only a small number of quantitative studies of bacterial capsules with AFM, which used Klebsiella pneumoniae.(29,30) Here, we study E. coli capsules for the first time, using a colloidal probe cantilever (that improves the signal-to-noise ratio over standard cantilevers), and demonstrate quantitative agreement with super-resolution fluorescence measurements (previously, the brush thickness was used as an unsubstantiated fit parameter with K. pneumoniae).
Super-resolution fluorescence microscopy techniques are rapidly becoming a standard tool in biological physics laboratories, such as STORM,31,32 PALM,33 SIM,34 and STED.35 These methods can provide subdiffraction-limited images with little or no disruption of biological processes. Samples can be hydrated at room temperature and require minimally invasive sample preparation procedures.36 Recent developments can provide relatively fast data acquisition (fast cameras, bright fluorophores, and optimal software analysis) enabling live cells to be imaged, and these developments have been used in the current research on E. coli. Super-resolution studies on bacteria are still relatively limited, but research has been performed on chromosome organization,37 DNA repair,38 and peptidoglycan architecture.39
Due to the small thickness of K1 polysaccharide capsules (≤200 nm), the resolution of conventional optical microscopy is at its very limit (R = λ/2NA ≈ 200 nm, where λ is the wavelength and NA is the numerical aperture). Thus, to visualize the morphology of bacterial capsules using conventional staining techniques is challenging, and the fine structure of bacterial capsules cannot be resolved due to the diffraction limit. However, conventional fluorescence microscopy does allow the dynamics of diffraction-limited capsular polysaccharide rafts to be tracked, as they move around bacterial surfaces, and we therefore performed it in parallel with other studies. Furthermore, confocal microscopy could be used to provide a √2 improvement on the resolution (∼200/√2 nm), contrast enhancement, and improved three-dimensional sectioning, although finer details in the images were still not resolvable. Therefore, we used super-resolution fluorescence, direct stochastic optical reconstruction microscopy (dSTORM) to image live bacteria.31,40 The stochastic emission of excited fluorophores with specific imaging buffers allowed us to create static super-resolved images (50 nm resolution). We are not aware of any detailed previous studies of live bacterial capsules using super-resolution fluorescence imaging in the literature, and the work builds on our previous study using graphene oxide coatings with dSTORM that allows high-contrast quantitative imaging.41 Transparent GO films spin-coated onto coverslips extinguished nonspecifically bound fluorophores, which otherwise would provide a large fluorescence background and low contrast (low SNR) on images of the capsules.
In the current article, we provide an integrative picture of capsule formation based on a combination of fluorescence microscopy techniques and AFM. Lyso-PG capsular rafts were initiated at a relatively small number of nuclei (∼10) in each bacterium and grew to cover the whole bacteria with limited diffusional motion. The thicknesses of the lyso-PG rafts at the bacterial poles were substantially larger than those at the equator from dSTORM, which was in agreement with chromatography experiments on identical specimens. Quantitative agreement was found between the AFM and dSTORM experiments, implying that microscopy experiments can now be used to predict the mesoscopic forces experienced by the bacteria. The repulsive interparticle potential provided by the capsules will thus be longer ranged on the poles than on the equator of the bacteria by a significant factor (49%), providing an anisotropic modulation of bacterial interactions, e.g., they are less likely to adhere to any other surfaces (including another bacterium) at their poles than on their equators.
Materials and Methods
K1-Encapsulated E. coli Strains and Their Cultivation
E. coli strains: EV36 (capsule-deficient) and EV136 (encapsulated, generously provided by Eric Vimr) were cultured in two different media: Luria-Bertani (LB)42 and M9 (minimal)43 with supplements of 0.4% (v/v) glycerol and 0.1% (w/v) casein to observe bacterial polysaccharide capsule formation. For fully encapsulated bacteria, EV36 were cultured in LB at 37 °C overnight before inoculation in fresh LB media and further incubated at 37 °C until they were imaged. To observe bacterial capsule expansion over time, 1:100 of the bacteria were cultured in LB at 20 °C overnight and inoculated in fresh M9 media at 20 °C. When the OD600 reached 0.3, 20 mL of cells was harvested by centrifugation (3500 rpm for 15 min), the pellet was transferred to a fresh prewarmed M9 media, and then the temperature was upshifted to 37 °C to induce capsule formation.
Immunofluorescent Techniques for Labeling K1 Capsular Polysaccharide
Live cells were collected at well-defined time points, washed three times with phosphate-buffered saline (PBS), and deposited on poly-l-lysine-and-graphene oxide (PLLGO)-coated 8-well chambers. This PLLGO coating improves the signal-to-noise ratio (i.e., the contrast) for fluorescence imaging by a factor of 100.41
The K1 α-2,8 N-acetylneuraminic (polysialic) acid in the polysaccharide capsules can be recognized by mAb735 (monoclonal IgG2A) antibodies.44−46 For live cell imaging using wide-field fluorescence microscopy, cells were incubated with 1% (w/v) BSA in PBS and then with 1:400 dilution from a 4 mg/mL stock in a blocking buffer and then washed before incubation with 1:500 dilution with a secondary antibody (antimouse Fab′2 AF647, Invitrogen #A-21237). In addition, confocal and super-resolution (dSTORM) microscopies were used without the washing step with the immunostaining methods by incubating both primary (1:8000) and secondary (1:10 000) antibodies at a lower concentration with the bacteria due to the benefits of using PLLGO-coated slides, i.e., the PLLGO coating removed the background due to nonspecifically bound fluorophores and provided a 100 factor of improvement in contrast.1 The polylysine coating has a negligible effect on the proliferation of the bacteria over a period of 3 h (the time for a complete capsule to form, Supporting Information (SI 5)).
Optical Fluorescence Microscopes and Imaging Buffers
For wide-field fluorescence microscopy, ready-to-visualize cells were supplemented with Mowiol buffer to stabilize them. Samples were sealed to prevent evaporation. In confocal fluorescence microscopy, cells were deposited on coverslips of 8-well-chamber slides with M9 media (supplemented with 0.4% (v/v) glycerol and 0.1% (w/v) casein) mixed with both primary (1:8000) and secondary (1:10 000) antibodies for the observation of bacterial capsule development. These 8-well-chamber covers were sealed to prevent evaporation when observed at 37 °C.
In addition to M9 media and antibodies (no washing step was used), super-resolution fluorescence microscopy (dSTORM) also required an imaging buffer, which was GLOX:47 10 mM MEA (cystamine), 1/100 dilution from 20 mg/mL catalase, and 1/10 dilution of 150 mg/mL glucose oxidase. This helped prevent photobleaching of the fluorophores. Since the acidity of the GLOX buffer reduces significantly after a few hours, fresh GLOX buffer and M9 media were refilled every 1 h in longer experiments to prevent photobleaching, toxicity, and acidification (SI 4).
Image Acquisition with the Fluorescence Microscopes
For capsular polysaccharide brush formation, a wide-field fluorescence microscope was used: an Axio Imager.M2 Upright Zeiss microscope with a 63× Plan Apochromat (Oil, DIC) objective lens at a low laser power with an appropriate filter set. In confocal microscopy, images were collected using a Leica TCS SP8 AOBs inverted gSTED microscope in a confocal mode with a 100× HC PL APO (Oil; STED WHITE) objective lens; a super-continuum source at 250 mW was used. In the case of dSTORM imaging, a bespoke STORM microscope48 was used with 405 nm (0.6 mW) and 647 nm (77 mW) excitation lasers and a 100× oil immersive with 1.49NA, TIRF objective lens (Olympus UAPON 100× OTIRF). The depth of focus was calibrated using 100 nm polystyrene beads coated with Cy3B, deposited on glass slide-coated poly-l-lysine. The z-stack images were taken at 10 nm/step for 200 steps, using a piezo stage with an acquisition rate of 100 fps, and analyzed with a virtual light sheet plug-in.49
To observe the dynamics of the capsular lyso-PG brushes, lyso-PG rafts were tracked with fluorescence microscopy using an sCMOS, Hamamatsu ORCA-Flash 4.0 V2, camera. Fluorophores were tracked after excitation by 647 nm laser, with a power of less than 10 mW for long-duration tracking.
dSTORM was used to observe a gradual expansion of the capsular polysaccharide rafts during their growth. Up to 10 000 frames were used to construct each dSTORM image, which was followed by times with no illumination to reduce photobleaching. Thus, the expansion of K1 capsular brushes on bacterial membranes could be investigated by taking images at defined times.
High-Pressure Light-Scattering Chromatography (HPLC)
Purified K1 capsular polysaccharide was analyzed using multiangle light scattering (Wyatt). The purified polysaccharide was diluted in 500 μL of PBS, 1 h prior to loading, and then centrifuged at 14k rpm for 10 min. The extraction method of Clarke et al. was used.50 A superdex 200 (separation range of 5000–600 000 Da) was equilibrated in PBS, and the capsular polysaccharide was injected using an NGC (BioRad) HPLC onto the column. The detectors used were a Wyatt Helios 8-angle light-scattering detector, an Optilab rEX refractometer, and a QELS dynamic light-scattering detector. The molar mass of the lyso-PG was acquired using this system.
Bacterial Capsule Analysis and Image-Processing Methods
Several pieces of software were used to construct images and interpret the results. The Thunderstorm plug-in in Fiji was used to localize the blinking fluorophores and reconstruct super-resolution images.51 Origin (OriginLab, MA) was predominantly used for plotting and fitting functions. Capsular brush thicknesses, mobilities, and expansions were analyzed via Matlab (The MathWorks, Inc., MA). A newly written Matlab code was used to find the capsules’ boundaries and the distribution of capsule thicknesses. The polysaccharide rafts were tracked using the TrackMate plug-in,52 and MatLab code was used to construct the mean square displacement (MSD) as a function of lag time. The depth of focus of the microscope was analyzed with the virtual light sheet plug-in49 and found to be 347 ± 22 nm (SI 2).
Nanomechanical Measurements Using AFM with a Spherical Probe
Bacteria were prepared overnight in M9 media and then deposited on a poly-l-lysine-mica-coated glass slide, 1 h before an experiment. Nanomechanical forces were observed using a Bruker Catalyst AFM system with spherical borosilicate colloids, 2.5 μm in diameter (sQUBE, 2017NM310/1), with a cantilever spring constant (k) of 0.3 N/m. The probes together with their cantilevers were thermally calibrated before they were used to characterize the hydrated capsular brush. Bacterial cells were submerged and observed under UHQ water within 4 h. Nanomechanical forces between the capsular brush and the probe were optimized, and multiple measurements were made on each position of the bacterial surface. Raw data were analyzed using the NanoScope Analysis toolbox, Matlab codes, and Origin. The contact positions of the AFM probe with the capsules were found using exponential fits (SI 3).
Results
Morphology of the E. coli K1 Capsule
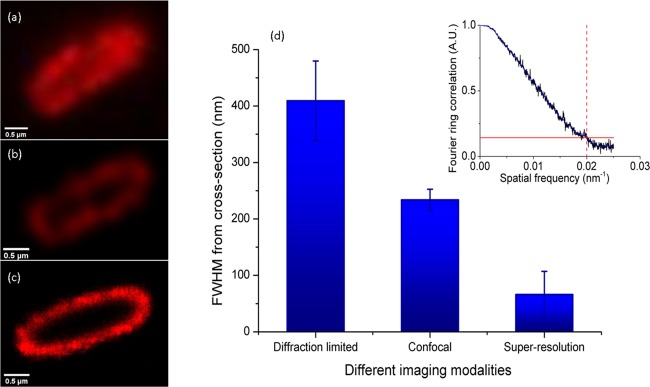
The K1 capsule is constructed via the assembly of K1 polysaccharides (polyneuraminic acid) arranged on the cell surface, which are not chemically cross-linked but may be bridged by multivalent cations. To visualize the presence of a capsule on a bacterial surface, capsules can be dyed and observed using bright-field microscopy,53 stained with ruthenium red and visualized by electron microscopy or immunoelectron microscopy17,54,55 or detected via fluorescence microscopy using specific antibodies.46 Initially, E. coli K1 capsules were observed with different spatial resolutions: a diffraction-limited image at around 400 nm from conventional fluorescence microscopy (Figure 2a), a subdiffraction-limited image at around 200 nm from confocal fluorescence microscopy (Figure 2b), and a super-resolution imaging from dSTORM (Figure 2c) (see also Figure S1 in Suppoting Information). The super-resolution image has a 50 nm spatial resolution (analyzed using the Fourier ring correlation (FRC) method56 and shown as an inset in Figure 3d) and the capsule thickness from the polar ends of the E. coli is seen to be larger than that at the equatorial ends. We only considered the thickness of bacterial capsules that were sectioned through the middle of the cells. When the sectioning plane was not in the middle of the cells, the curvature surface of the bacterial capsule could be observed (Figure S1b), and the measurement of capsule thickness was omitted in these cases. There is also a possibility of an axial projection discrepancy; however, the axial sectioning was calibrated with 100 nm polystyrene beads dyed with Cy3B. Within the 300 nm depth of focus (calibrated using virtual light sheet method49), the variation of bead size due to an axial projection within this sectioning region is negligible, as shown in Figure S3. Thus, the effect on bacterial projections will also be negligible.
Figure 2.
Images of bacterial capsules labeled with anti-K1 antibodies conjugated with AF647 created using (a) diffraction-limited fluorescence microscopy, (b) confocal fluorescence microscopy, and (c) dSTORM super-resolution fluorescence microscopy. (d) shows the spatial resolution of the bacterial capsule images from the different microscopes calculated using the Fourier ring correlation (FRC) technique;56 the inset shows the FRC as a function of spatial frequency from the super-resolution images (the red lines show the threshold value of 1/7). The scale bars on (a)–(c) are 1 μm. The spatial resolution of image (c) from the FRC was 50 nm.
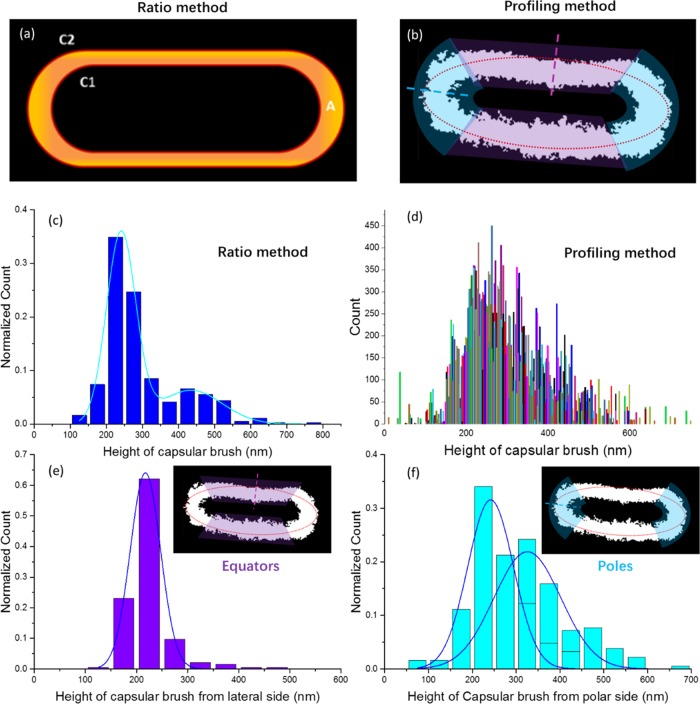
Figure 3.
Histograms of bacterial capsule thicknesses were created using two different methods: (a) ratio between the area (yellow) and the averaged contour lengths (C1 + C2)/2 (red) called the ratio method and (b) normal vector profiles from elliptical fits called the profiling method: the purple and cyan dashed lines correspond to equatorial and polar regions, respectively. (c) Histogram of the thicknesses of the capsular brush from the ratio method shown in (a). A bimodal distribution can be seen. (d) Histogram of the thicknesses of the whole capsular area created using the profiling method (b). The histogram bars are randomly colored to improve visibility. (e) Histogram of the thicknesses from the equatorial regions (purple region, purple arrow indicated, in the inset) and (f) histogram of the thicknesses of the polar regions (cyan region in the inset), both created using the profiling method (b). The polar regions contain a bimodal population of thicknesses that contains longer polysaccharide chains in the brushes than the monomodal population observed at the equator.
Furthermore, we explored the thickness distribution of the polysaccharide brushes around the capsules. Two image-processing methods were used to quantify the thickness (H) of the bacterial capsules from the dSTORM images: (1) a method based on the capsule area called the ratio method, i.e., H = 2A/(C1 + C2) (A is the area of the capsule, and C1 and C2 are the inner and outer ellipsoidal contour lengths) and (2) elliptical fits with line sectioning normal to the ellipse called the profiling method. Although we assumed that the bacterial capsule thickness covered the bacterial surface with a uniform thickness in the first method, the results from the second method showed two qualitatively different domains of bacterial capsular brush height: at the poles (long) and at the equator (short).
Using the assumption of uniform thickness (the ratio method), we calculated the capsule thickness (H) from a ratio between area (A) of the projected capsule image in two dimensions and the average contours from inside and outside the capsule boundaries ((C1 + C2)/2) (Figure 3a). The average thickness of the K1 capsule was 200–300 nm, and the distribution is shown in Figure 3c. In addition, using normal vectors scanning along elliptical fits, in which purple and cyan mark the equatorial and the polar regions, respectively (the profiling method, Figure 3b), a histogram of bacterial capsule thicknesses could be constructed at different positions around the capsule (Figure 3d). The outermost extent of the bacterial capsule at the equatorial regions of the cell is around 200 nm from the cell surface (Figure 3e), whereas at the polar regions, the capsule extends further out with a broader range of 250–400 nm (Figure 3f). The results of Figure 3e,f were calculated from more than 200 K1-encapsulated E. coli cells.
The dSTORM technique provided high-resolution images of the capsular brush to probe their morphology. The uneven surfaces were predominantly due to variations of the brush itself, i.e., the molecular weight of the capsular polysaccharide anchored on lyso-PG. The variations in the images were reproducible, and defects in immunofluorescent labeling are thought to be a secondary effect.
The variation of the thickness of the capsular brush (Figure 3f) was similar to its polydispersity measured by HPLC (Figure 4). Two populations of polysaccharide lengths were measured peaking at (ii) 24.4 ± 1.3 kDa and (i) 47.2 ± 4.3 kDa (magenta curve), where 1 g/mol corresponds to 1.008 Da. These results reveal the potential of using super-resolution technique to quickly identify the polydispersity of polysaccharides on living cells since both HPLC and dSTORM methods demonstrate a similar bimodal distribution.
Figure 4.
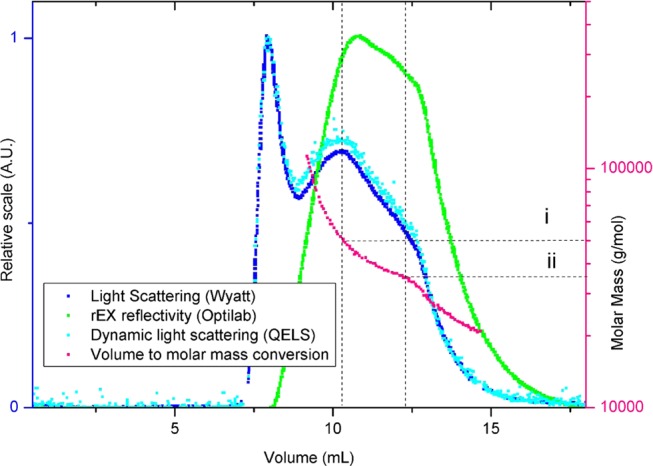
Chromatography results from purified K5 capsular polysaccharide, showing the two main fractions of lipopolysaccharide molecular weights (i and ii indicate molar masses of 47 and 27 kDa, respectively, according to the different injected volumes). These scattering signals were detected using different detectors: a Wyatt Helios 8-angle light-scattering detector (blue), an Optilab rEX refractometer (green), and a QELS dynamic light-scattering detector (cyan). The molar mass (Mw) was interpreted using Zimm plot analysis70,71 (magenta).
Nanomechanical Properties of Capsular Brushes Measured with AFM
A borosilicate spherical probe (diameter 2.5 μm) was attached to the AFM cantilever, which interacted with the bacterial capsular brushes during force measurements. The force curves (Figure 5a) for the capsule-deficient strain EV36 (red) and the encapsulated strain EV136 (blue) show that the encapsulated bacteria had much longer ranged force curves of up to 500 nm compared with 225 nm for the capsule-deficient strain. Additionally, the encapsulated data are replotted in Figure 5b to show different nanomechanical force regimes: (i) the double-layer charge interaction, (ii) the osmotic brush regime, (iii) the Hertzian regime, and (iv) the Hookean regime.19,29 The double-layer interaction occurs at the longest distances (∼400 nm) due to screened (i.e., in a salty environment) electrostatic repulsion between the AFM probe and the charge lyso-PG brush. The osmotic brush regime occurs when the probe contacts the lyso-PG and experiences a steric repulsion. It is concluded that the capsular brush beneath these bacteria is completely collapsed, since it provides consistent thicknesses between AFM and STORM, and was observed in the previous studies.57 In the Hertzian regime, the probe considerably perturbs the structure of the brush and is modeled as a deformable elastic continuum. Finally, in the Hookean regime, the probe indents the cell membrane and is resisted by the cytoplasmic turgor pressure.
Figure 5.
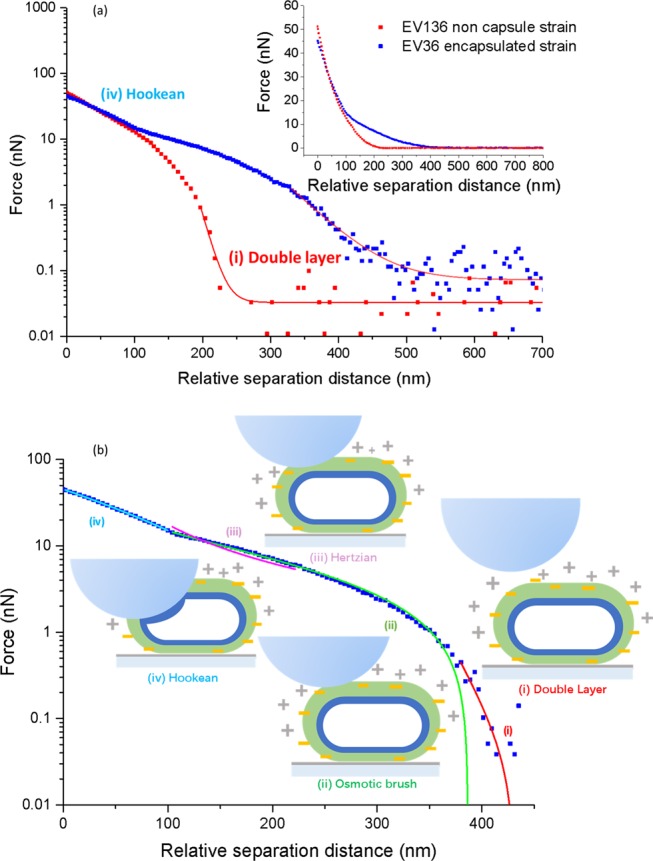
Force as a function of separation distance from AFM measurements, showing the nanomechanical properties of capsular brushes. (a) Comparison between encapsulated and capsular-deficient strains on a log–linear scale (the inset shows a linear–linear scale). (b) Nanomechanical regimes of the interaction between the spherical probe and the encapsulated bacteria. The insets show the schematic diagrams of the position of the probe and the encapsulated bacteria from noncontact (i) to contact modes (ii–iv). Different colors in (b) show the fits to the different regimes: (i) the double-layer charge interaction (red), (ii) the osmotic brush model (green), (iii) the Hertzian regime (magenta), and (iv) the Hookean regime (cyan).
The bacterial capsule thicknesses and their nanomechanical interaction were observed from fully encapsulated bacteria that were cultured at 37 °C. The K1-encapsulated E. coli can switch on and off their capsules in response to temperature changes. Upshifting the temperature from 20 to 37 °C stimulates the transcription of the kps and neu genes, leading to the synthesis and transport of the K1 capsular polysaccharide to the cell surface.8,58 To observe the emergence of the capsules on the bacterial surfaces and their dynamics, both upshifting and constant temperature-controlled experiments were used.
Dynamics of Bacterial Capsular Raft Formation
The linkage of K1 capsular polysaccharides to lyso-PG in the outer membrane means that this array of negatively charged molecules can be regarded as having brush-like properties.26,59 These K1 capsular polysaccharides, covalently anchored to the bacterial cell surface via Lyso-PG molecules, were specifically labeled with anti-K1 mAb735 after upshifting the temperature from 20 to 37 °C for 1 h (Figure 7a) in M9 media. We define lyso-PG capsular brush rafts as aggregated regions of lyso-PG that form discrete regions on the bacteria.22,23 Implicitly with the definition of a membrane raft is the possibility of microphase separation, defining the sizes of the rafts, i.e., they are aggregated because the interaction between lyso-PG chains is more favorable than that with other membrane components.23 Once the capsular brush had been labeled, the dynamics of the early-stage capsular brush rafts were observed in different regions of the bacterial surface and then tracked with subpixel resolution using standard fluorescence microscopy. The tracked rafts are shown in yellow in Figure 7b. In Figure 7c,d, the mean square displacement (MSD) of the rafts with lag time (τ) was analyzed from their tracks at 1 h and 1 h 30 min time points. The motion of the labeled capsular brush was in a subdiffusive regime over the entire duration of the movies based on a power law fit, ⟨r2⟩ = 4Dατα, where r is the displacement, Dα is the fractional diffusion coefficient, and α is the power law exponent. The power law exponents of the capsular brush are shown in Figure 7e from different time points after temperature upshifting. The average is α = 0.33 ± 0.23 at 1 h and α = 0.36 ± 0.20 at 1.5 h. After 180 min (the time for a complete capsule to form), extrapolation of the averaged MSD as a function of lag time implies the capsular raft would have moved around 114 nm. This is a much smaller distance than that required to cover the whole bacterial surface of approximately 2 μm. Thus, the position of the lyso-PG rafts appears to be relatively invariant over the time scale of capsule growth, and the capsular covering is created by raft nucleation at multiple random sites, rather than diffusive transfer from isolated sites.
Figure 7.
dSTORM images of the capsular brush rafts after temperature upshifting from 20 to 37 °C. The nucleation and expansion of the capsular brush rafts are shown in the sequential images (a) and (b). (c) shows the rafts from three sequential times superposed. Red is the initial time point (20 °C), yellow 10 min after temperature upshifting (37 °C), and magenta 20 min after temperature upshifting (37 °C). The scale bars are all 1 μm. (d) Radius of gyration (Rg) of the lyso-PG rafts as a function of time after upshifting the temperature. The expansion process is well described with a logistic function (red line). The error bars are from repeat experiments and are due to variations of capsule emergence on the bacterial cells.
Capsule formation could be imaged from the point at which there was no capsule to the fully encapsulated state. With a typical fluorescence microscope, the quantification of capsule size is overestimated due to the diffraction limit. The gradual growth of the capsular brushes was therefore calculated from super-resolution microscopy (dSTORM). The radius of gyration (Rg) of the antibody-labeled regions on the dSTORM images (Figure 6) provides a robust metric to describe the size of the irregularly shaped lyso-PG rafts. In Figure 6d, the radius of gyration of the capsular brush rafts is plotted with time. A logistic function (red) provided a good fit (eq 7) (shown later). With M9 media as a nutrient source, after the temperature was upshifted to 37 °C, E. coli EV36 became fully encapsulated after 3 h. To minimize the toxicity of dSTORM imaging buffer, we discarded and refilled media mixed imaging buffer after every hour.
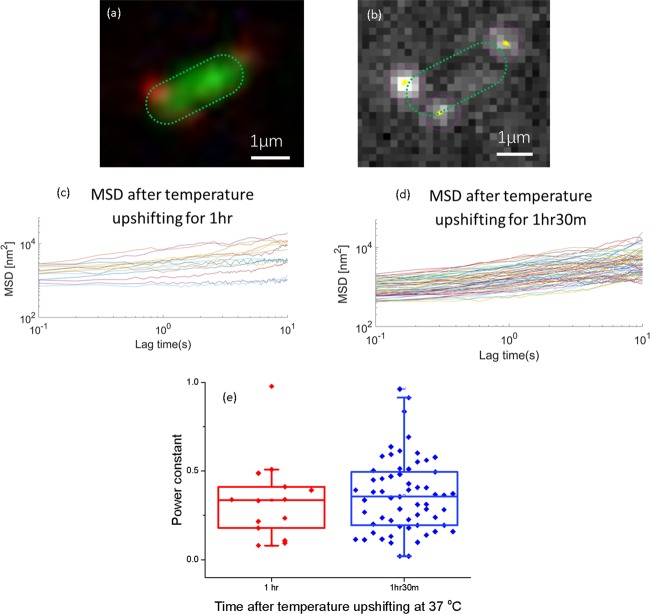
Figure 6.
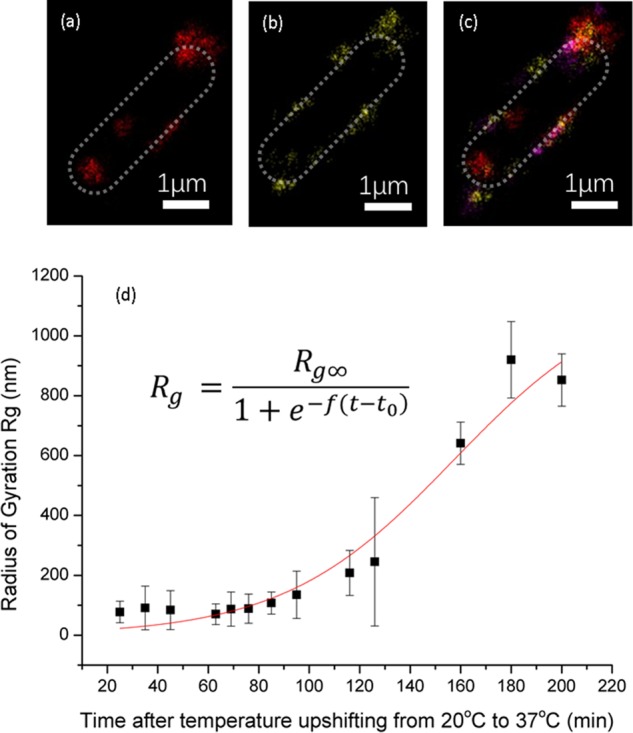
Particle tracking of fluorescently labeled capsular polysaccharide rafts in standard fluorescence microscopy experiments. (a) K1 capsular rafts (labeled with anti-K1 conjugated with antimouse AF647, red) emerge from E. coli cell labeled with GFP (green). (b) Pixelated image used for subpixel tracking. Tracked lyso-PG rafts are shown in yellow. The position of the E. coli outer membrane is shown by the green dotted line. (c) and (d) show the mean square displacement (MSD) as a function of lag time for times of 1 h and 1 h 30 min, respectively; different colors show the results from individual K1 rafts. (e) Subdiffusive power law coefficients, α (from the scaling of the mean square displacement, <r2> ∼ τα, where τ is the lag time), with different times after temperature upshifting. The scale bars are 1 μm on (a) and (b).
Despite the relatively low number of photons emitted and the photobleaching of the fluorophores that labeled the early capsular brush rafts, the rafts could be observed with subdiffraction resolution after localization and image reconstruction processes. In Figure 6a,b, the capsular brush rafts emerged in different locations within a 10 min time difference and Figure 6c is a combined image after 30 min, developing from red (from t0 time point), yellow (at t0 +10 min), and magenta (at t0 +20 min). In addition to this emergence of the lyso-PG rafts, the further expansion of capsular brush rafts could be observed until the capsule completely covered the bacterium (Figure 6d).
Discussion
To date, a few recent studies using the indentation of an AFM cantilever have examined the nanomechanics of K. pneumoniae capsules to quantify their thickness and explored the influence of fimbria on capsular brushes that induce biofilm formation.29,60 These results confirm the morphological structure of these capsules as polyelectrolyte brushes anchored on the lipid membranes. The current super-resolution imaging experiments on live E. coli in a hydrated environment overcome many morphological artifacts from sample preparation and cell damage found in other high-resolution imaging techniques, such as in scanning electron microscopy and transmission electron microscopy experiments.16 By using super-resolution fluorescence images, particle tracking, and AFM with a large colloidal probe, the morphology and dynamics of bacterial capsular brushes could be investigated in more detail than previously possible.
The bacterial capsule morphology found in super-resolution images (Figure 2c) reveals details of how the capsular polysaccharide brush is organized with a resolution beyond the diffraction limit. The variation of brush density along the bacterial surface relates to the localization of the membrane translocation proteins and enzymes (biosynthetic export complexes) for capsular polysaccharide production that are arranged at discrete points on the E. coli membranes. Although mathematical models26−28 for synthetic brushes have examined the variation of polyelectrolyte brush heights attached to nanoparticles as a function of surface curvature, we expect the capsular polysaccharide brush to be independent of curvature due to its high grafting density, large radius of curvature, and relatively short capsular polysaccharide chains (in comparison to the radius of curvature). Thus, the height differences in Figure 3e,f from the equatorial and the polar regions of the bacteria are deduced to be predominantly due to different lengths of K1 capsular polysaccharides and not their curvature or grafting density effects (i.e., the surface density of lyso-PG chains). Specifically, the osmotic brush model indicates that the lyso-PG chains are in fully stretched conformations (see the SI: Figure S1) and the chain size does not depend on membrane curvature or grafting density. Thus, changes in the brush size measured on images are directly due to changes in lyso-PG lengths.
The morphology of polyelectrolyte brushes anchored onto phospholipid membranes can be described by the Pincus polyelectrolyte model for an osmotic brush,26 which uses a balance of the osmotic pressure (fosm) with the loss of entropy during its expansion. This model was extended by Zhulina and Borisov,28 who considered the capsular brushes grafted onto a nanoparticle of small curvature (e.g., a bacterial surface) with a high charge density (e.g., provided by the sialic acid groups), and so the layer of counterions was much smaller compared to the thickness of capsular brush (H). The compressive pressure in the capsular brush tends to stretch the polymer chains and is balanced by the osmotic pressure of the counterions inside the brush, which gives
| 1 |
where N is the degree of polymerization of the chains, T is the temperature, H is the thickness of the polyelectrolyte brush, and α is the charge fraction of the polyelectrolyte chains. Conveniently, the thickness of the capsular polyelectrolyte brush can be calculated independently of the tethering density of the capsular brush in this osmotic brush regime using
| 2 |
where a is the monomer length.
A single chain of K1 α-2,8 polysialic acid that is extended from the point where its lyso-PG is anchored is shown in Figure 1 together with a polymeric blob model representation (the basis of the osmotic brush model). We observed thicknesses from Figure 2a,c,d of 242 ± 18 nm for the average areal approximation, 217 ± 29 nm for the equatorial region, and 238 ± 41 and 323 ± 62 nm for the bimodal polar regions. These regions are highlighted in Figure 8. Thus, the number of monomers per chain could be approximated as 200–300 units. This length of polymer would predict a molecular weight closer to 70 kDa (chromatography gave two fractions at 47 and 27 kDa, Figure 4). This apparent discrepancy is likely a consequence of the arrangement of the capsular polysaccharide on the bacterial surface. Specifically, ionic bridges between the capsular polysaccharide and the lipopolysaccharide molecules also decorate the bacterial cell surface.61 These interactions will cause swelling of the capsule62 and hence the discrepancy in the perceived capsule width, as determined via imaging and the actual molecular weight of the polysaccharide. Previous studies have demonstrated that purified E. coli K5 capsular polysaccharide also has two molecular weight peaks.63 This earlier observation may now in part be explained by the observations in this paper in terms of polar and equatorial distributions of E. coli capsular polysaccharides. This size difference of polysaccharide from the poles to the equators of the cell may reflect differences in the transport of the polysaccharide at these two sites in the cell and different protein complexes involved in this polysaccharide biosynthesis. Several reports on subcellular localization had explained how proteins tend to gather around polar regions,64 including proteins involving in capsular biosynthesis.65−67 Additionally, a model in which polysaccharide export is initiated while biosynthesis is still taking place means that slower transport could result in increased polymer length before the molecule is finally extruded, i.e., different reaction kinetics at the poles and the equators cause the changes in the lyso-PG lengths. Further quantitative comparison to the osmotic brush model is included in the Suppoting Information Figures S1 and S3 and additional information on the biosynthesis is shown in Figure S7.
Figure 8.
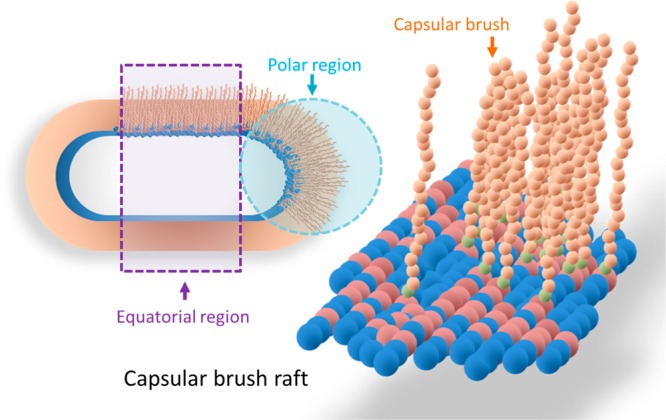
Schematic diagram of the polysaccharide anchored onto phospholipids of the external bacterial membrane forming the K1 capsular lyso-PG raft. A single type of K1 capsule anchored on lyso-PG is shown to form a raft. The capsular brush rafts created at the polar and the equatorial regions (the dashed circle/rectangle shows the different regions of the capsular brush rafts) and then spread out to cover the whole bacterial surface.
In addition to noncontact observations using super-resolution fluorescence imaging of live bacterial cells, AFM with large spherical probes provides insights into the nanomechanical properties of the capsular brushes. The superior signal-to-noise ratio compared with the Strugnall AFM studies29,68 was attributed to the use of a colloidal probe, which averages over a larger region of the capsule, effectively increasing the measured signal. Not only could the data differentiate between capsule-deficient and encapsulated strains of E. coli (in Figure 5a), four force regimes involved in the interaction of the encapsulated bacteria could be determined (Table 1).
Table 1. Force–Distance Regimes Used To Describe AFM Data in Figure 5ba.
| regimes | approximate fitting function | parameters | definition | ||
|---|---|---|---|---|---|
| (I) double layer |
|
λD | Debye screening length (nm) | ||
| (II) osmotic brush |
|
δL0 = exp(a1/b1) | the onset of linear compliance, capsule thickness: H ≈ 2δL0 (nm) | ||
| (III) Hertzian |
|
E | Young’s modulus (Pa) | ||
| (IV) Hookean |
|
kbac | elastic constant of bacteria cells (N/m) |
A0 describes the short-range van der Waals force (negligible when compared with the electrostatic force in this scenario), A1 is the force magnitude for the long-range electrostatic interaction, d is the separation distance, d0 is used to correct the relative separation distance, a1 and b1 are the reduced parameters of the full osmotic brush model for grafted polyelectrolyte brushes29 (where ln(δ0L) = a1/b1), δ is the indented distance after probe contact, c is the correction term of the indented distance from a relative separation distance, R is the effective radius (Rprobe, Rbac/(Rprobe + Rbac)), where Rprobe, Rbac are the radius of the probe and the indented bacteria, respectively, and v is the Poisson ratio of the bacterial cells, e.g., assumed equal to 0.5 as a first approximation for an isotropic elastic solid.60 The main parameters that we investigated were the bacterial capsule thickness, the Debye screening length (λD), the effective Young’s modulus (Eeff), and the elastic constant of the K1 bacterial cells (k).
From the noncontact regime to the contact regime (regime I, the indentation of both the capsular brush and the bacterial membrane, eq 3, Table 1), the Debye screening length (λD) was found to be equal to 39.4 ± 2.1 nm, which is the length scale over which the electrostatics is felt. In regime II from the compression of the brush, the bacterial capsule thickness was approximated using δL0 = exp(a/b), which gave a total thickness (H) of 241 ± 34 nm (Table 1). It is assumed that the lower capsular brush in contact with the poly-l-lysine coating is completely collapsed, as observed in the previous studies.57 The bacterial capsule thickness is in reasonably good agreement with the thickness of the single component equatorial brush measured with dSTORM, i.e., 217 ± 29 nm. In regime III, Young’s modulus was found to be 11.36 ± 0.22 kPa, indicating a soft effective modulus for the compression of the membrane. In regime IV, the elastic constant was k = 4.14 ± 0.06 mN/m, which is related to the cell turgor pressure. Although the spherical probe was large compared to the size of the bacteria, identical force curves were observed after several measurements, and so their reproducibility was good. See the Suppoting Information Figure S3 for AFM images of encapsulated bacteria.
In addition to static morphological observations, the dynamics of bacterial capsular brush formation were investigated after temperature upshifting from 20 to 37 °C. In Figure 7, the mean square displacements show that the K1 capsular brush had low mobility and the rafts of the capsular brush were unable to diffuse appreciably between regions of the bacterial surface over time scales of bacterial capsular raft growth (∼114 nm displacement over 3 h). We conclude that the lyso-PG capsular brushes nucleate from the polysaccharide transport channels and then spread over the bacterial surfaces, as additional material is pushed through the channels, but their position remains closely correlated with that of the original channels.65 These results on the low mobility of the capsule rafts are in good agreement with separate fluorescence correlation spectroscopy experiments of our group performed with a confocal microscope (including GFP-labeled proteins in the synthetic pathway that act as raft nucleation sites) and will be presented in more detail in the future.
Considering the rate of capsular brush expansion (Figure 6), we calculated the increase of the brush sizes using Rg, the radius of gyration of the capsular brush raft. The number of lyso-PG translocation channels (NABC) was relatively low at ∼10 channels (Figure S4) when lyso-PG was produced after temperature upshifting. There was a slow growth of bacterial capsule initially, and the full growth curve could be described by a logistic model,i.e.,
| 7 |
where the time constant (t0) was 157 ± 18 min, Rg∞ is the radius of a fully formed capsule at long times, and f is the steepness parameter. The smallest Rg that was included in this trend was 76.7 ± 11.2 nm, which was set by the resolution of the dSTORM technique. The bacterial capsular brush rafts randomly emerged at positions on the bacterial surfaces, around 30 min after temperature upshifting. However, due to the low emission and photobleaching from the high power of the dSTORM acquisition process, the very early stage rafts were omitted from the analysis. The imaging buffer perturbed the growth of bacterial cells (Figure S4), causing a substantial decrease in proliferation at long time scales (>2 h). To reduce this problem, we replenished cells with M9 media and fresh imaging buffer every hour to prevent the acidification process, reducing bacterial division. Within these limitations, bacterial capsule expansion was investigated, mainly focusing on the later-stage development of large rafts. The bacteria can switch from no capsule to fully covered capsules within 3 h. In addition to the rate of bacterial capsule production, the polysaccharide translocation channels are important for creating a relatively uniform capsule that covers the whole bacterium. We observed a random distribution of the nucleation of capsular biosynthesis on the bacterial surfaces,65,69 which also occurred at the septum when the bacteria divided.67
A number of fundamental questions still exist on the export of the K1 capsular polysaccharides through the two external membranes of Gram-negative bacteria (SI 6). More work is needed to understand the polymer translocation of large capsular polysaccharide molecules from their site of synthesis in the cytoplasm through the external membranes onto the cell surface, which is likely to involve a multiprotein complex.8,9,13,15 Treatments could be found to compromise the steric potential created by the capsular brushes and thus help the immune system to combat an infection more quickly, e.g., the addition of multivalent cations. It would also be useful to employ genetic synthetic biology techniques to modulate the size of the capsular brush to create a quantitative model of capsular physiology.
Conclusions
K1 bacterial capsules labeled with anti-K1 antibodies allowed the characterization of capsular polysaccharide polydispersity, using super-resolution fluorescence microscopy (dSTORM) with graphene oxide-coated coverslips. The distribution of capsule thicknesses varied with localization on different regions of the bacterial surfaces. Capsular polysaccharide brushes were longer at the poles than at the equator of the bacterial cells by a factor of 49%. The capsular brush could be indented by a large spherical probe attached to an AFM to determine the nanomechanical properties of the bacterial capsule, and results were in agreement with dSTORM images based on the osmotic brush model for the steric forces created by a polyelectrolyte brush.
For dynamic investigations, capsular brushes emerged from a few tens of capsular biosynthesis channels to cover whole bacterial cells during their growth. The capsular polysaccharides could be rapidly labeled after transportation to the bacterial surface. Super-resolution fluorescence microscopy provides the noninvasive observation of live bacteria that could visualize dynamic events during lyso-PG raft production when the rafts emerged from their channels on the E. coli surfaces.
Acknowledgments
The authors would like to thank P. March and N. Hodson for guidance with the fluorescence microscopy and the AFM. M. Goldrick, J. Hart, J. Blee, and H. Cox provided laboratory support and general problem-solving tips. The authors would also like to acknowledge E. Vimr for his generous gift of the bacterial strains and H. Claus for the gift of mAb735. The Development and Promotion of Science and Technology Talents Project (Royal Government of Thailand), DPST, is thanked for the support of S.P. The MRC and EPSRC funded this research through a research grant (EP/F062966/1).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.langmuir.8b04122.
Additional wide-field super-resolution images of encapsulated E. coli; demonstration of the calibration of the axial projections in super-resolution images using virtual light sheet software; scaling calculation for the thickness of polyelectrolyte brushes; addition force–distance curves and images from AFM; effect of the oxygen scavenger system with the imaging buffer on bacterial proliferation; effect of poly-l-lysine-coated graphene oxide coverslips on bacterial proliferation; proteins involved in the production of K1 polysaccharide capsules.
The authors declare no competing financial interest.
Supplementary Material
References
- Li R.; Georgiadis P.; Cox H.; Phanphak S.; Roberts I.; Waigh T. A.; Lu J. R. Quenched stochastic optical reconstruction microscopy (qSTORM) with graphene oxide films. Sci. Rep. 2018, 8, 16928 10.1038/s41598-018-35297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.The Physiology and Biochemistry of Prokaryotes, 3rd ed.; OUP, 2011. [Google Scholar]
- Whitt D. D.; Winkler M. E.; Wilson B. A.; Salyers A. A.. Bacterial Pathogenesis: A Molecular Approach.; ASM Press, 2011. [Google Scholar]
- Flores-Mireles A. L.; Walker J. N.; Caparon M.; Hultgren S. J. Urinary tract infections: epidemiology, mechanics of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269. 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonolini M.Imaging and Intervention in Urinary Tract Infections and Urosepsis; Springer, 2018. [Google Scholar]
- Goller C. C.; Seed P. C. Revisiting the E. coli polysaccharide capsule as a virulence factor during urinary tract infection. Virulence 2010, 1, 333–337. 10.4161/viru.1.4.12388. [DOI] [PubMed] [Google Scholar]
- Valle J.; Da Re S.; Henry N.; Fontaine T.; Balestrino D.; Latour-Lambert P.; Ghigo J. M. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 12558. 10.1073/pnas.0605399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I. S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996, 50, 285–315. 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides. Annu. Rev. Biochem. 2006, 75, 39–68. 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Flores-Mireles A. L.; Walker J. N.; Caparon M.; Hultgren S. J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kraker M. E. A.; Davey P. G.; Grundmann H. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: Estimating the burden of antibiotic resistance in europe. PLoS Med. 2011, 8, e1001104 10.1371/journal.pmed.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress B. F.; Englaender J. A.; He W.; Kasper D.; Linhardt R. J.; Koffas M. A. G. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol. Rev. 2014, 38, 660–697. 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis L. M.; Whitfield C. Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 2013, 378, 35–44. 10.1016/j.carres.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Lizak C.; Worrall L. J.; Baumann L.; Pfleiderer M. M.; Volkers G.; Sun T.; Sim L.; Wakarchuk W.; Withers S. G.; Strynadka N. C. J. X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid. Sci. Rep. 2017, 7, 5842 10.1038/s41598-017-05627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R.; Steenbergen S. M. Early molecular-recognition events in the synthesis and export of group 2 capsular polysaccharides. Microbiology 2009, 155, 9–15. 10.1099/mic.0.023564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R.; Kalivoda K. A.; Deszo E. L.; Steenbergen S. M. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132–53. 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev A. V.; McCrossan M. V.; Wren B. W. Demonstration of polysaccharide capsule in Campylobacter jejuni using electron microscopy. Infect. Immun. 2001, 69, 5921–5924. 10.1128/IAI.69.9.5921-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y.; Zhang T. Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl. Microbiol. Biotechnol. 2011, 92, 381–392. 10.1007/s00253-011-3551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularski A.; Wilksch J. J.; Hanssen E.; Strugnell R. A.; Separovic F. Atomic force microscopy of bacteria reveals the mechanobiology of pore forming peptide action. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 1091–1098. 10.1016/j.bbamem.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Israelachvili J. N.Intermolecular and Surface Forces, 3rd ed.; Academic Press, 2011. [Google Scholar]
- Berg J. C.An Introduction to Interfaces and Colloids: the Bridge to Nanoscience; World Scientific, 2010. [Google Scholar]
- Lingwood D.; Simons K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Bramkamp M.; Lopez D. Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 81–100. 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gennes P. G. Conformations of polymers attached to an interface. Macromolecules 1980, 13, 1069–1075. 10.1021/ma60077a009. [DOI] [Google Scholar]
- Rubinstein M. C. R. H.Polymer Physics; OUP, 2003. [Google Scholar]
- Pincus P. Colloid stabilization with grafted polyelectrolytes. Macromolecules 1991, 24, 2912–2919. 10.1021/ma00010a043. [DOI] [Google Scholar]
- Zhulina E. B.; Birshtein T. M.; Borisov O. V. Curved polymer and polyelectrolyte brushes beyond the Daoud-Cotton model. Eur. Phys. J. E 2006, 20, 243–256. 10.1140/epje/i2006-10013-5. [DOI] [PubMed] [Google Scholar]
- Zhulina E. B.; Borisov O. V. Polyelectrolytes grafted to curved surfaces. Macromolecules 1996, 29, 2618–2626. 10.1021/ma9515801. [DOI] [Google Scholar]
- Wang H.; Wilksch J. J.; Lithgow T.; Strugnell R. A.; Gee M. L. Nanomechanics measurements of live bacteria reveal a mechanism for bacterial cell protection: the polysaccharide capsule in Klebsiella is a responsive polymer hydrogel that adapts to osmotic stress. Soft Matter 2013, 9, 7560. 10.1039/c3sm51325d. [DOI] [Google Scholar]
- Mularski A.; Wilksch J. J.; Wang H.; Hossain M. A.; Wade J. D.; Separovic F.; Strugnell R. A.; Gee M. L. Atomic force microscopy reveals the mechanobiology of lytic peptide action on bacteria. Langmuir 2015, 31, 6164–6171. 10.1021/acs.langmuir.5b01011. [DOI] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–795. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M.; Huang B.; Dempsey G. T.; Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 2007, 317, 1749–1753. 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Schermelleh L.; Carlton P. M.; Haase S.; Shao L.; Winoto L.; Kner P.; Burke B.; Cardoso M. C.; Agard D. A.; Gustafsson M. G. L.; et al. Subdiffraction mulicolor imaging of the nuclear periphery with 3D structure illumination microscopy. Science 2008, 320, 1332–1336. 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S. W.; Siebert M.; Mertel S.; Knoche E.; Wegener S.; Wichmann C.; Matkovic T.; Muhammad K.; Depner H.; Mettke C.; Bückers J.; Hell S. W.; Müller M.; Davis G. W.; Schmitz D.; Sigrist S. J. Far-field optical nanoscopy. Science 2007, 316, 1153–1158. 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Chao Y.; Zhang T. Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl. Microbiol. Biotechnol. 2011, 92, 381–392. 10.1007/s00253-011-3551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Li G. W.; Chen C.; Xie X. S.; Zhuang X. Chromosome organization by a nucleoid associated protein in live bacteria. Science 2011, 333, 1445–1450. 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphoff S.; Reyes-Lamothe R.; Garza de Leon F.; Sherratt D. J.; Kapanidis A. N. Single-molecule DNA repair in live bacteria. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8063–8068. 10.1073/pnas.1301804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R.; Mesnage S.; Boneca I. G.; Hobbs J. K.; Foster S. J. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol. Microbiol. 2011, 82, 1096–1109. 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- van de Linde S.; Löschberger A.; Klein T.; Heidbreder M.; Wolter S.; Heilemann M.; Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011, 6, 991–1009. 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- Li R.; Georgiades P.; Cox H.; Phanphak S. R. I.; Waigh T. A.; Lu J. R. Quenched stochastic optical reconstruction microscopy (qSTORM) with graphene oxide. Sci. Rep. 2018, 16928 10.1038/s41598-018-35297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R.; Sambrook J.. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press, 2012. [Google Scholar]
- Vimr E. R.; Troy F. A. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 1985, 164, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosch M.; Görgen I.; Boulnois G. J.; Timmis K. N.; Bitter-Suermann D. NZB mouse system for production of monoclonal antibodies to weak bacterial antigens: isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc. Natl. Acad. Sci. U.S.A. 1985, 82, 1194–1198. 10.1073/pnas.82.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M.; Ikeda A.; Hane M.; Hanashima S.; Kitajima K.; Sato C.; Yamaguchi Y. Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: Insight into the binding preference for Polysialic acid. J. Biol. Chem. 2013, 288, 33784–33796. 10.1074/jbc.M113.496224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. E.; Aal Owaif H. A.; Jia J.; Roberts I. S. Phenotypic heterogeneity in expression of the K1 polysaccharide capsule of uropathogenic Escherichia coli and downregulation of the capsule genes during growth in urine. Infect. Immun. 2015, 83, 2605–2613. 10.1128/IAI.00188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahidiazar L.; Agronskaia A. V.; Broertjes J.; Van Broek B. D.; Jalink K. Optimizing imaging conditions for demanding multi-color super resolution localization microscopy. PLoS One 2016, 11, e0158884 10.1371/journal.pone.0158884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P.; Allan V. J.; Wright G. D.; Woodman P. G.; Udommai P.; Chung M. A.; Waigh T. A. The flexibility and dynamics of the tubules in the endoplasmic reticulum. Sci. Rep. 2017, 7, 16474 10.1038/s41598-017-16570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palayret M.; Armes H.; Basu S.; Watson A. T.; Herbert A.; Lando D.; Etheridge T. J.; Endesfelder U.; Heilemann M.; Laue E.; Carr A. M.; Klenerman D.; Lee S. F. Virtual-‘light sheet’ single-molecule localisation microscopy enables quantitative optical sectioning for super-resolution imaging. PLoS One 2015, 10, e0125438 10.1371/journal.pone.0125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B. R.; Esumeh F.; Roberts I. S. Cloning, expression and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J. Bacteriol. 2000, 182, 3761–3766. 10.1128/JB.182.13.3761-3766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný M.; Křížek P.; Borkovec J.; Švindrych Z.; Hagen G. M. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 2014, 30, 2389–2390. 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J. Y.; Perry N.; Schindelin J.; Hoopes G. M.; Reynolds G. D.; Laplantine E.; Bednarek S. Y.; Shorte S. L.; Eliceiri K. W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Stoderegger K. E.; Herndl G. Visualization of the exopolysaccharide bacterial capsule and its distribution in oceanic environments. Aquat. Microb. Ecol. 2001, 26, 195–199. 10.3354/ame026195. [DOI] [Google Scholar]
- Allen P. M.; Roberts I.; Boulnois G. J.; Saunders J. R.; Hart C. A. Contribution of capsular polysaccharide and surface properties to virulence of Escherichia coli K1. Infect. Immun. 1987, 55, 2662–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov O.; Korenevsky A.; Beveridge T. J.; Dutcher J. R. Use of atomic force microscopy and transmission electron microscopy for correlative studies of bacterial capsules. Appl. Environ. Microbiol. 2008, 74, 5457–5465. 10.1128/AEM.02075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen R. P. J.; Lidke K. A.; Bates M.; Puig D. L.; Grünwald D.; Stallinga S.; Rieger B. Measuring image resolution in optical nanoscopy. Nat. Methods 2013, 10, 557. 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Wilksch J. J.; Strugnell R. A.; Gee M. L. Role of capsular polysaccharides in biofilm formation: an AFM nanomechanics study. ACS Appl. Mater. Interfaces 2015, 7, 13007–13013. 10.1021/acsami.5b03041. [DOI] [PubMed] [Google Scholar]
- Whitfield C.; Roberts I. S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999, 31, 1307–1319. 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- Csajka F. S.; Seidel C. Strongly charged polyelectrolyte brushes: A molecular dynamics study. Macromolecules 2000, 33, 2728–2739. 10.1021/ma990096l. [DOI] [Google Scholar]
- Wang H.; Wilksch J. J.; Strugnell R. A.; Gee M. L. Role of capsular polysaccharides in biofilm formation: an AFM nanomechanics study. ACS Appl. Mater. Interfaces 2015, 7, 13007–13013. 10.1021/acsami.5b03041. [DOI] [PubMed] [Google Scholar]
- Fresno S.; Jimenez N.; Izquierdo L.; Merino S.; Corsaro M. M.; De Castro C.; Parrilli M.; Naldi T.; Regue M.; Tomas J. M. The ionic interaction of Klebsiella pneumoniae K2 capsule and core lipopolysaccharide. Microbiology 2006, 152, 1807–1818. 10.1099/mic.0.28611-0. [DOI] [PubMed] [Google Scholar]
- Bayer M. E. Visualisation of the bacterial polysaccharide capsule. Curr. Top. Microbiol. Immunol. 1990, 150, 129–159. 10.1007/978-3-642-74694-9_7. [DOI] [PubMed] [Google Scholar]
- Manzoni M.; Rollini M.; Piran E.; Parini C. Preliminary characterisation of an E. coli K5 lyase-deficient strain producing the K5 polysaccharide. Biotechnol. Lett. 2004, 26, 351–356. 10.1023/B:BILE.0000015473.89280.ce. [DOI] [PubMed] [Google Scholar]
- Rudner D. Z.; Losick R. Protein subcellular localization in bacteria. Cold Spring Harbor Perspect. Biol. 2010, 2, a000307 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty C.; Thompson J.; Barrett B.; Lord L.; Andersen C.; Roberts I. S. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: The role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol. Microbiol. 2006, 59, 907–922. 10.1111/j.1365-2958.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- Javens J.; Wan Z.; Hardy G. G.; Brun Y. V. Bypassing the need for subcellular localization of a polysaccharide export-anchor complex by overexpressing its protein subunits. Mol. Microbiol. 2013, 89, 350–371. 10.1111/mmi.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques M. X.; Rodrigues T.; Carido M.; Ferreira L.; Filipe S. R. Synthesis of capsular polysaccharide at the division septim of Streptoccoccus pneumoniae is dependent on a bacterial tyrosine kinase. Mol. Microbiol. 2011, 82, 515–534. 10.1111/j.1365-2958.2011.07828.x. [DOI] [PubMed] [Google Scholar]
- Gaboriaud F.; Gee M. L.; Strugnell R.; Duval F. L. Coupled electrostatic, hydrodynamic, and mechanical properties of bacterial interfaces in aqueous media coupled electrostatic, hydrodynamic, and mechanical properties of bacterial interfaces in aqueous media. Langmuir 2008, 24, 10988–10995. 10.1021/la800258n. [DOI] [PubMed] [Google Scholar]
- Nourikyan J.; Kjos M.; Mercy C.; Cluzel C.; Morlot C.; Noirot-Gros M.-F.; Guiral S.; Lavergne J.-P.; Veening J.-W.; Grangeasse C. Autophosphorylation of the bacterial tyrosine-kinase CpsD connects capsule synthesis with the cell cycle in Streptococcus pneumoniae. PLoS Genet. 2015, 11, e1005518 10.1371/journal.pgen.1005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona M. P.; Saiz E. Combination of SEC/MALS experimental procedures and theoretical analysis for studying the solution properties of macromolecules. J. Biochem. Biophys. Methods 2003, 56, 95–116. 10.1016/S0165-022X(03)00075-7. [DOI] [PubMed] [Google Scholar]
- Zimm B. H. The scattering of light and the radial distribution function of high polymer solutions. J. Chem. Phys. 1948, 16, 1093–1099. 10.1063/1.1746738. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.