Abstract
Lower urinary tract symptoms (LUTS) are highly prevalent among the elderly and negatively impact quality-of-life. Since caffeinated beverages are enjoyed worldwide and the relationship between LUTS and caffeine is still not fully understood, it would be of particular interest to examine the underlying mechanisms that drive caffeine’s influence on LUTS development and progression. The aim of this study was to characterize the effects of caffeine on hTert immortalized normal bladder epithelial cells by investigating whether exposure to caffeine can cause potential changes in the bladder proteome and/or biological pathways. Labeled LC-MS/MS proteomic analysis found 57 proteins as being differentially expressed in caffeine-treated bladder epithelial cells, compared to controls; this included 32 upregulated and 25 downregulated proteins. Further functional gene enrichment analysis revealed that caffeine affected major biological pathways, including those for “muscle contraction” and “chromatin assembly”. These findings provide new scientific insights that may be useful in future studies investigating the role of caffeine in bladder dysfunctions.
Keywords: Lower urinary tract symptoms, caffeine, global proteome, bladder epithelial cells, biological network
INTRODUCTION
It is estimated that by the end of 2018, 2.3 billion individuals will be affected by at least one lower urinary tract symptom (LUTS)[1]. These symptoms include urinary storage problems, such as urgency, frequency, nocturia, or voiding problems. LUTS also weighs heavily on overall quality of life; patients report significantly higher mental health issues, lower work productivity, and diminished general health status[2]. The current standard treatment for LUTS involves α-blockers, 5α-reductase inhibitors, and antimuscarinics[3]. However, these are mainly palliative and require consistent maintenance. Consequently, there is a substantial economic burden associated with LUTS[4].
Previous reports have demonstrated that diet and stress play important factors in the development and progression of LUTS[5]. In particular, caffeine, a naturally occurring compound, has been reported to be a potential dietary risk factor in developing LUTS[6]. Caffeine is ubiquitously found in many plants, including cocoa beans, tea leaves, and coffee beans. It is a stimulatory drug that is widely used to prevent sleepiness and can be found in over-the-counter medications, such as some pain remedies. It has also been observed that caffeine may aggravate or worsen urinary symptoms in patients who already have some form of LUTS[7]. In recent years, caffeinated drinks have become a staple of the average diet; more than 85% of adults in the U.S. regularly consume caffeine[8]. A longitudinal study of caffeine intake in young healthy volunteers found that subjects who regularly drank coffee had significant increases in urinary urgency and frequency[9]. Additionally, a separate study observed that greater coffee intake raised the odds of LUTS progression in men and women more than carbonated or citrus beverages[10]. The effects of caffeine have also been studied in the context of bladder cancer (BC); however, rather than finding a negative risk, there have been reported benefits. A case-control study of BC patients in Italy found no causal relationship between caffeine and BC[11]. A separate study found that caffeine may actually benefit BC patients by making cells more susceptible to chemotherapy and apoptosis[12]. Mechanistically, this has been reported to be mediated through caffeine’s effects on the tumor suppressor protein, p53[13].
Despite the potential link between caffeine and LUTS, research into causative mechanisms and functions is lacking. One prior study suggested that caffeine may be facilitating bladder instability and frequent urination by enhancing the activation of neuronal micturition centers through increased expression of transcription factor c-Fos and nerve growth factor[14]. While informative, this study focused primarily on the bladder muscles with little attention on the bladder epithelium, which is more anatomically exposed to urine and its biological/chemical contents. A separate study using a mouse model found that oral caffeine administration resulted in detrusor overactivity and increased bladder sensory signaling[15]. Further studies found similar effects on the detrusor muscle in humans[16]. However, the direct effects of caffeine, its delivery into cells, and its mechanisms remain unestablished.
Our present study sought to examine the cellular effects of caffeine on the bladder epithelium without any pathological conditions. Quantitative and global proteomics analysis found that caffeine can perturb the whole proteome, possibly through the regulation of chromatin assembly in normal bladder epithelial cells.
MATERIALS AND METHODS
Cell culture and cell proliferation assay.
Immortalized normal human bladder epithelial cells, TRT-HU1, were maintained as described previously[17]. The TRT-HU1 cell line was constructed and extensively characterized in previously published papers. The passage number of each cell line was below 10, and mycoplasma contamination was tested for monthly via PCR analysis. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), containing 10% fetal bovine serum (FBS, Invitrogen), 1% penicillin/streptomycin, and 1% L-glutamine (Sigma-Aldrich Corp., St. Louis, MO, USA) in a 37 °C humidified incubator with 5% CO2. To test cell growth during exposure to caffeine, TRT-HU1 cells were seeded onto 6-well plates with a density of 5 × 104 cells/well. Cells were then incubated with standard growth medium with varying doses of caffeine at 0, 0.005 mM, 0.05 mM, or 0.1 mM (C6035, Sigma-Aldrich Corp.) or vehicle for 24, 48, or 72 hrs. Cell proliferation was measured by manually counting cells using a hemocytometer. The averages of each count were used as the total density of the well after each time point. For crystal violet staining, the culture medium was removed, the cells were fixed with 4% paraformaldehyde at room temperature for 5 min and then stained with 0.05% crystal violet for 15 min. The cells were then washed with tap water, after which the water was removed and the cells were dried out on filter paper, and the cell plates were scanned and quantified as described previously[18]. All experiments were run in triplicate for each cell line, and the data are representative of three independent trials.
Antibodies and reagents.
Antibodies against various proteins were obtained from the following sources: ACTG2 (ab189385, Abcam), ACTA2 (ab5694, Abcam), MYH2 (ab124937, Abcam), MYH7B (ab172967, Abcam), HISTH2B (ab52599, Abcam), HIST1H2BM (SAB1301739, Sigma), and β-actin (A1978, Sigma). Commercially available horseradish peroxidase (HRP)-conjugated secondary antibodies (7074, 7076) were obtained from Cell Signaling Technology. All other chemical reagents were procured from Sigma Chemical Corp.
Quantitative proteomics.
Tandem mass tagging (TMT)-based quantitative proteomics was performed as described[19]. Briefly, cellular protein was extracted from caffeine-treated and control cells using 4% SDS-containing buffer. The protein concentration was measured using the Pierce 660nm Assay Kit. From each sample, 60 μg of protein was digested with trypsin using filter-aided sample preparation (FASP) and labeled with TMT6plex reagents in parallel. After TMT labeling, the peptides were merged, desalted with C18 spin columns (Thermo Scientific), and fractionated via high-pH reversed phase liquid chromatography (RPLC) using an Ultimate 3000 XRS System (Thermo Scientific). For high-pH RPLC, about 50 μg TMT-labeled peptides were loaded onto a 100-mm Hypersil GOLD C18 column (2.1 mm inner diameter, 3 μm particle size, 175 Å pore size) (Thermo Scientific), flushed for 3 min with solvent A (10 mM ammonium formate, pH 10), and then separated with a 7-min linear gradient of 0–40% solvent B (10 mM ammonium formate, 95% acetonitrile, pH 10.0). A total of 24 fractions were collected, concentrated into 12 fractions, and dried down in a SpeedVac (Thermo Scientific). Peptides in each fraction were redissolved with 0.2% formic acid and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an EASY-nLC 1000 connected to an LTQ Orbitrap Elite Mass Spectrometer (Thermo Scientific). Briefly, peptides were loaded onto a 2-cm trap column (PepMap 100 C18, 75 μm inner diameter, 3 μm particles, 100 Å pore size) and separated by a 50-cm EASY-Spray column (PepMap RSLC C18, 75 μm inner diameter, 2 μm particles, 100 Å pore size) heated to 55°C. For low-pH RPLC separation, the mobile phase consisted of 0.1% formic acid in water (phase A) or acetonitrile (phase B). The LC gradient was 4–24% B over 200 min, 24–50% B over 20 min, and 50–100% B over 5 min at a flow rate of 150 nL/min, followed by 100% B over 15 min at a flow rate of 300 nL/min.
Mass spectra were acquired in a data-dependent manner, selecting up to the 15 most abundant precursor ions for higher-energy collisional dissociation. The mass resolution for precursor and fragment ions was set to 120,000 and 30,000, respectively. The isolation width was set as 1.5 and the normalized collision energy was set as 40. Database searching and protein quantification was performed by Proteome Discoverer (v2.1), using the SEQUEST algorithm. The acquired raw data were searched against the human Uniprot Protein Sequence Database (released on 01/22/2016, containing 20,985 protein sequences). Searching parameters were set as follows: trypsin, up to two missed cleavage; precursor ion tolerance of 10 ppm, fragment ion tolerance of 0.02 Da; carbamidomethylation of cysteins and TMT6plex modification of lysines and peptide N-term as fixed modifications; acetylation of protein N-term, oxidation of methionine and deamidation of asparagines and glutamines as variable modifications. A standard false discovery rate of 1% was applied to filter peptide-spectrum matches (PSMs), peptide identifications, and protein identifications.
For protein quantification, peptides with >30% precursor ion interference were excluded to minimize erroneous quantification caused by precursor ion interference. PSM level information was extracted using the Proteome Discoverer[20]. After quantifying each PSM intensity, the peptide intensities were summarized, as described by Niu et al. [21]. In brief, this was done in 4 steps: 1) the normalized log2 intensity of the PSMs matched to each peptide by the substrate mean PSM from each reporter intensity was centered, 2) outliers were detected using Dixon’s Q-test and generalized electrostatic discharge (ESD) test, 3) the mean intensity without outliers was taken, and 4) the grand-mean intensities of the three highest abundant PSMs were added before being mean-centered. Bipartite graphs of peptides and protein groups were generated according to the information of the aligned peptides. Among the proteins in the protein group, we defined the representative protein that had the largest number of peptides or unique peptide[22]. When there were more than proteins with the same number of peptides in the same protein group, we selected the protein that had the higher sequence coverage. We then computed the relative intensity of the protein group using a linear-programming formulation, as previously described[23].
Identification of differentially expressed proteins (DEPs).
To identify the DEPs, we selected proteins that have more than two non-redundant peptides. For the selected proteins, we then performed an one sample t-test using log2 fold-changes to compute the significance of t value. For this statistical test, an empirical distribution of the null hypothesis, a protein is not differentially expressed, was estimated by following steps: 1) 100,000 random permutations were applied to the samples, 2) t-values were computed using the log2 fold-changes of the randomly permutated samples, and 3) the Gaussian kernel density estimation method was applied to t-values. The FDRs of each protein from the one sample t-tests were then calculated using Storey’s method[24]. The DEPs were identified as those with a FDR < 0.05 and absolute log2 fold change > 0.58 (1.5 fold). Functional enrichment analysis of DEPs was performed using DAVID software (Ver. 6.8)[25]. Significantly enriched cellular processes were selected for if they had a p-value < 0.05.
Western blot analysis.
Cells were seeded onto 10 cm plates and exposed to 0.05 mM concentrations of caffeine for 72 hrs. Cells were lysed with RIPA buffer (20 mM Tris, 150 mM NaCl, 1% Nonidet, P-40, 0.1 mM EDTA) (Pierce, ThermoFisher) supplemented with a phosphatase inhibitor cocktail (ThermoFisher). The protein concentration of each sample was measured with the Bradford Protein Kit Assay, according to the manufacturer’s instructions (Pierce, ThermoFisher). Equal amounts of protein extract were separated by SDS-PAGE and transferred onto a PVDF membrane. The membranes were then blocked with 5% bovine serum albumin (BSA) or 5% nonfat milk in Tris-buffered saline with 0.1% Tween 20 (TBST [2.42 g/L Tris–HCl, 8 g/L NaCl, and 1 mL/L Tween 20 (pH 7.6)]) and incubated overnight at 4 °C with specific primary antibodies in TBST. The membranes were then incubated with secondary antibodies conjugated with horseradish peroxidase, as described previously. β-actin was used as an internal control. All western blot experiments were run in at least triplicates for each antibody.
Seahorse Respirometry Assay.
TRT-HU1 cells were seeded onto a 24-well Seahorse culture plate at a density of 50,000 cells per well 24 hrs before the Seahorse assay. Media was then changed to XF Base Medium (pH 7.4) supplemented with 10 mM glucose, 1mM sodium pyruvate, and 1mM sodium glutamine. Cells were equilibrated for 1 hr in a non-CO2 incubator at 37 °C before the assay began. Cells were treated with caffeine at 0.05 mM, or 0.5 mM for experiments. Chemical reagents (Sigma) were used at final concentrations, as follows: 1 μM oligomycin—an ATP synthase inhibitor, 1 μM (FCCP) carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone—an uncoupling agent, and a mixture of 0.5 μM antimycin A—a cytochrome C reductase inhibitor and 0.5 μM rotenone—a complex I inhibitor. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) was monitored during the duration of the assay run. OCR and ECAR were monitored before and after addition of caffeine. Results were normalized to protein concentrations determined by BCA assay (Thermo Scientific).
Statistical analysis.
Student’s T-test was performed to confirm differential expression of the proteins between two groups. Variables with normal distribution were presented as mean ± SD. All reported p-values are two-tailed, with p-values < 0.05 indicating statistical significance.
RESULTS
Quantitative proteomic analysis of normal bladder epithelial cells
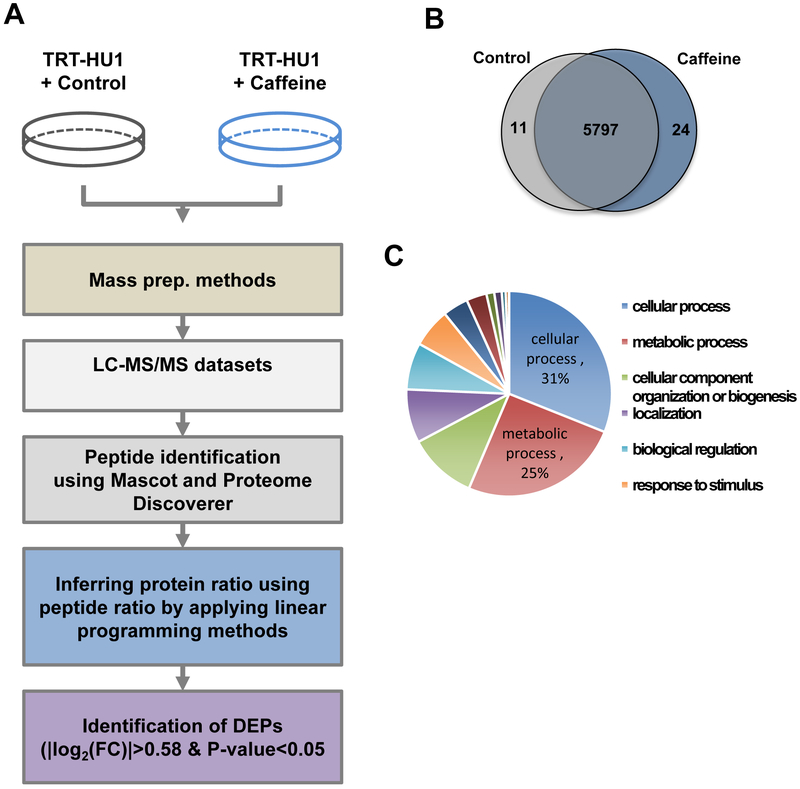
Due to the lack of knowledge regarding the effects of caffeine on biological and proteomic perturbations in the normal bladder epithelium, this study aimed to examine the whole proteome alterations caused by caffeine consumption. To gain insight on the underlying mechanism of caffeine on the bladder epithelium, we treated normal bladder epithelial cells with caffeine and performed TMT-based quantitative proteomic analysis, as outlined in Figure 1A. Based on previous literature, we opted to use caffeine concentrations within normal physiological consumption[12, 26]. Whole-cell lysates in biological duplicates were digested with trypsin. Using LC-MS/MS and followed by bioinformatic analyses, we identified 44,597 peptides corresponding 5,832 proteins with more than 2 peptides in more than two sets of pooled lysates from at least three biological samples per condition. The caffeine-treated group had 5,821 identified proteins with high expression, while the control group had 5,808. (Figure 1B). We then performed functional categorization of the proteins to check whether our quantitative proteomic analysis was biased by any cellular compartments or biological process-related proteins using Panther software[27]. All the identified proteins can be categorized into 14 cellular processes, including cellular process, metabolic process, cellular component organization of biogenesis, localization, biological regulation, response to stimulus, developmental process, multicellular organismal process, biological adhesion, immune system process, locomotion, reproduction, growth, and cell killing, indicating that this proteome reveals major biological functions of bladder epithelial cells in normal physiology (Figure 1C). In addition to this, we examined the cellular localization of the detected proteins, which showed the significant enrichment of 8 cellular compartments including cell part, organelle, macromolecular complex, membrane, extracellular region, cell junction, synapse, and extracellular matrix. (Supplementary Figure 1). Collectively, our quantitative proteomic analysis identified a comprehensive list of proteins in all cellular components and illustrated the cellular functions of highly expressed proteins in normal bladder epithelial cells.
Figure 1. Unbiased proteomics analysis identified proteins in TRT-HU1 cells.
(A) Experimental scheme describing unbiased global proteomics profiling and bioinformatics analysis. (B) Venn diagram depicts number of unique proteins detected in caffeine treated and non-treated normal bladder epithelial cells. (C) Pie chart displays functional categories of all the identified proteins.
Whole proteome in bladder epithelial cells perturbed by caffeine treatment
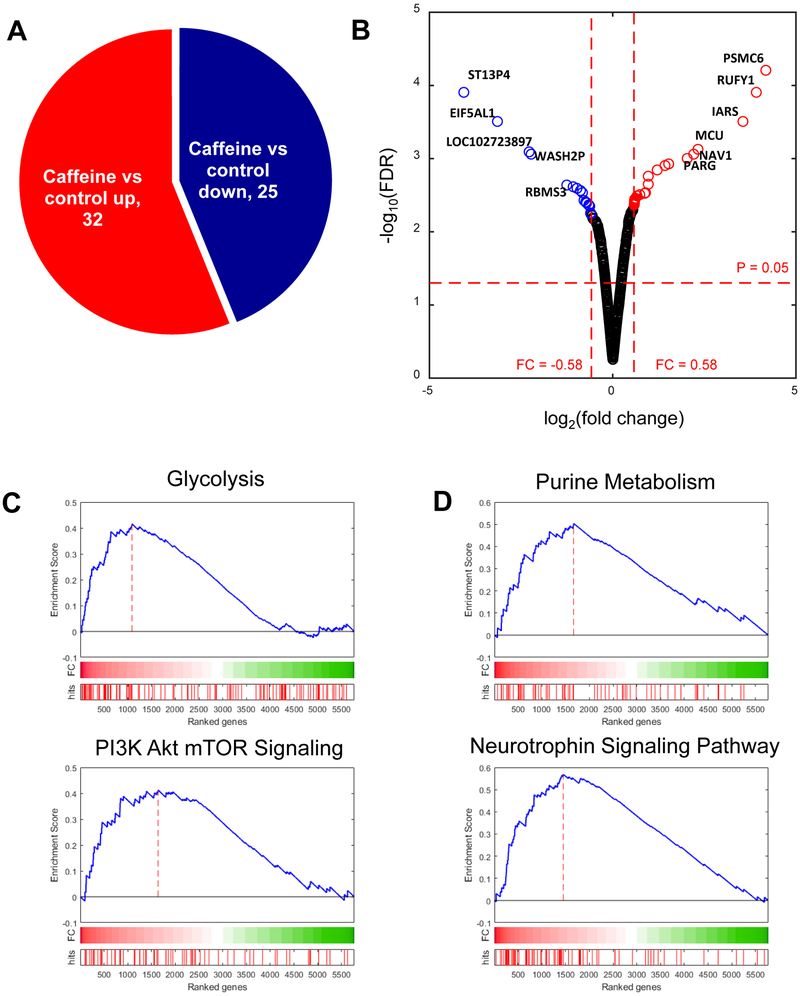
We determined which proteins were differentially expressed after caffeine treatment. DEPs were selected for if they had an absolute log2 fold change greater than 0.58 and p-value less than 0.05. In total, we identified 32 upregulated and 25 downregulated proteins between the control vs. caffeine groups (Table 1, 2, and Figure 2A). As shown in the volcano plot, some DEPs, including PSMC6 (proteasome 26S subunit ATPase 6), RUFY1 (RUN and FYVE domain containing 1), IARS (isoleucyl-tRNA synthetase), MCU (mitochondrial calcium uniporter), NAV1 (neuron navigator 1), RARG (retinoic acid receptor, gamma), and etc, were significantly increased with caffeine treatment, while ST13P4 (ST13, Hsp70 interacting protein pseudogene 4), EIF5AL1 (eukaryotic translation initiation factor 5A-like 1), WASH2P (WAS protein family homolog 2 pseudogene), RBMS3 (RNA binding motif single stranded interacting protein 3), and etc decreased (Figure 1C).
Table 1.
List of upregulated proteins in bladder epithelial cells perturbed by caffeine treatment.
| Symbol | Full name | Fold change (log2) |
P-value |
|---|---|---|---|
| S100A8 | S100 calcium binding protein A8 | 4.17759 | 0.000112 |
| ST13 | ST13, Hsp70 interacting protein | 3.918705 | 0.000224 |
| SERPINB3 | serpin family B member 3 | 3.553147 | 0.000559 |
| SERPINB4 | serpin family B member 4 | 3.553147 | 0.000559 |
| WASH3P | WAS protein family homolog 3 pseudogene | 2.327444 | 0.001342 |
| POTEKP | POTE ankyrin domain family member K, pseudogene | 2.210099 | 0.001565 |
| RBMS1 | RNA binding motif single stranded interacting protein 1 | 2.032904 | 0.001789 |
| KPRP | keratinocyte proline rich protein | 1.526693 | 0.002124 |
| S100A7 | S100 calcium binding protein A7 | 1.4274 | 0.002236 |
| TGM1 | transglutaminase 1 | 1.214751 | 0.002572 |
| MYH7B | myosin heavy chain 7B | 0.970592 | 0.003131 |
| MYH3 | myosin heavy chain 3 | 0.970592 | 0.003131 |
| MYH1 | myosin heavy chain 1 | 0.970592 | 0.003131 |
| MYH7 | myosin heavy chain 7 | 0.970592 | 0.003131 |
| MYH8 | myosin heavy chain 8 | 0.970592 | 0.003131 |
| MYH2 | myosin heavy chain 2 | 0.970592 | 0.003131 |
| MYH13 | myosin heavy chain 13 | 0.970592 | 0.003131 |
| MYH4 | myosin heavy chain 4 | 0.970592 | 0.003131 |
| MYH7B | myosin heavy chain 7B | 0.970537 | 0.004025 |
| MYH3 | myosin heavy chain 3 | 0.970537 | 0.004025 |
| MYH1 | myosin heavy chain 1 | 0.970537 | 0.004025 |
| MYH7 | myosin heavy chain 7 | 0.970537 | 0.004025 |
| MYH8 | myosin heavy chain 8 | 0.970537 | 0.004025 |
| MYH2 | myosin heavy chain 2 | 0.970537 | 0.004025 |
| MYH13 | myosin heavy chain 13 | 0.970537 | 0.004025 |
| MYH4 | myosin heavy chain 4 | 0.970537 | 0.004025 |
| H2BFS | H2B histone family member S | 0.895712 | 0.005255 |
| S100A9 | S100 calcium binding protein A9 | 0.871394 | 0.005367 |
| UVRAG | UV radiation resistance associated | 0.738812 | 0.00559 |
| SULT1A4 | sulfotransferase family 1A member 4 | 0.665067 | 0.005814 |
| SULT1A1 | sulfotransferase family 1A member 1 | 0.665067 | 0.005814 |
| UCHL1 | ubiquitin C-terminal hydrolase L1 | 0.622981 | 0.006261 |
| MAGI3 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | 0.615023 | 0.006373 |
| KRT74 | keratin 74 | 0.598941 | 0.006485 |
| KRT73 | keratin 73 | 0.598941 | 0.006485 |
| ACTA2 | actin, alpha 2, smooth muscle, aorta | 0.598824 | 0.006708 |
| LRRC8E | leucine rich repeat containing 8 VRAC subunit E | 0.594574 | 0.00682 |
| KRT74 | keratin 74 | 0.594473 | 0.006932 |
| KRT73 | keratin 73 | 0.594473 | 0.006932 |
| KRT74 | keratin 74 | 0.594472 | 0.007156 |
| KRT73 | keratin 73 | 0.594472 | 0.007156 |
| FLCN | folliculin | 0.592834 | 0.007379 |
| KRT74 | keratin 74 | 0.589943 | 0.007491 |
| KRT73 | keratin 73 | 0.589943 | 0.007491 |
| CPNE6 | copine 6 | 0.587267 | 0.007715 |
| CPNE4 | copine 4 | 0.587267 | 0.007715 |
Table 2.
List of downregulated proteins in bladder epithelial cells perturbed by caffeine treatment.
| Symbol | Full name | Fold change (log2) |
P-value |
|---|---|---|---|
| ST13P4 | ST13, Hsp70 interacting protein pseudogene 4 | −4.06184 | 0.000224 |
| EIF5AL1 | eukaryotic translation initiation factor 5A-like 1 | −3.14456 | 0.000559 |
| LOC102723897 | - | −2.27955 | 0.001453 |
| WASH2P | WAS protein family homolog 2 pseudogene | −2.27955 | 0.001453 |
| RBMS3 | RNA binding motif single stranded interacting protein 3 | −2.22487 | 0.001565 |
| POTEI | POTE ankyrin domain family member I | −1.25276 | 0.004137 |
| POTEJ | POTE ankyrin domain family member J | −1.25276 | 0.004137 |
| HIST1H2BM | histone cluster 1 H2B family member m | −1.07852 | 0.00436 |
| DES | desmin | −0.97497 | 0.004584 |
| TENM1 | teneurin transmembrane protein 1 | −0.87939 | 0.004919 |
| ACTG2 | actin, gamma 2, smooth muscle, enteric | −0.82627 | 0.005255 |
| HIST1H2BD | histone cluster 1 H2B family member d | −0.78302 | 0.006708 |
| HIST1H2BC | histone cluster 1 H2B family member c | −0.78302 | 0.006708 |
| HIST2H2BF | histone cluster 2 H2B family member f | −0.78302 | 0.006708 |
| HIST1H2BH | histone cluster 1 H2B family member h | −0.78302 | 0.006708 |
| HIST1H2BN | histone cluster 1 H2B family member n | −0.78302 | 0.006708 |
| RPL29 | ribosomal protein L29 | −0.73583 | 0.007044 |
| RNF175 | ring finger protein 175 | −0.71108 | 0.007267 |
| TFAP2B | transcription factor AP-2 beta | −0.65122 | 0.007715 |
| DLG2 | discs large MAGUK scaffold protein 2 | −0.64864 | 0.007826 |
| RAB3A | RAB3A, member RAS oncogene family | −0.62645 | 0.00805 |
| ZDHHC13 | zinc finger DHHC-type containing 13 | −0.60304 | 0.009951 |
| EPHA6 | EPH receptor A6 | −0.60218 | 0.010063 |
| PRC1 | protein regulator of cytokinesis 1 | −0.59503 | 0.010174 |
| MRPL34 | mitochondrial ribosomal protein L34 | −0.5944 | 0.010286 |
Figure 2. Differentially expressed proteins (DEPs) perturbed by caffeine treatment.
(A) Pie chart showing the DEPs in presence of caffeine. (B) A volcano plot showing the up- or downregulated DEPs due to caffeine in bladder epithelial cells. Up- or down-regulated DEPs are marked as red or blue dot. (C) Enriched biological processes and cellular compartment of the DEPs. (D) Bar plot depicts enrichment of KEGG pathways by DEPs.
Next, to understand the function of the perturbed proteins, we performed gene set enrichment analysis (GSEA) using the hallmark gene sets from the Molecular Signature Database (MSigDB)[28]. As a result, we found that “glycolysis” and “PI3K/AKT/MTOR” signaling gene sets were significantly enriched by differential expression due to caffeine treatment (Figure 2C). We also checked the enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the same context and found that the “neurotrophin signaling pathway” and “purine metabolism” were identified to be significantly enriched in the caffeine-treated condition (Figure 2D). This result suggests that caffeine may stimulate bladder epithelial by altering the activation of the PI3K/AKT/MTOR pathway, neurotrophin signaling, and purine metabolism.
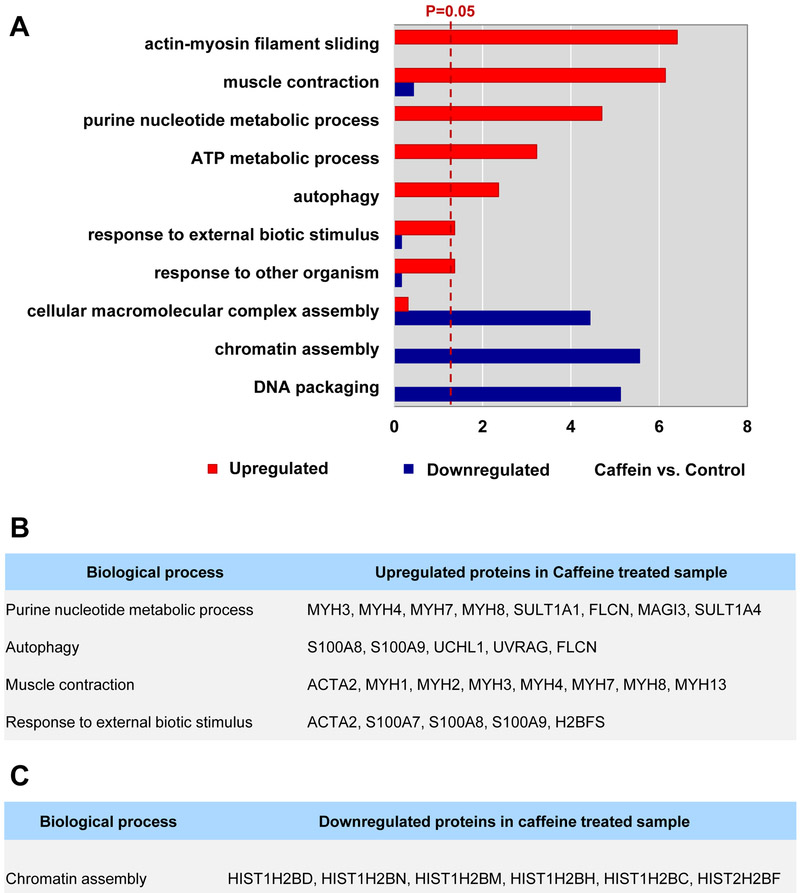
For this, we conducted separate functional enrichment analyses on each of the 32 up- and 25 downregulated proteins using DAVID[25]. The results suggest that upregulated DEPs were significantly enriched for “actin-myosin filament sliding”, “muscle contraction”, “purine nucleotide metabolic process”, “ATP metabolic process”, “autophagy”, “response to external biotic stimulus”, and “response to other organism”. Downregulated DEPs were significantly enriched for “chromatin assembly”, “DNA packaging”, and “cellular macromolecular complex assembly” (Figure 3A). We extracted a list of the most significant upregulated DEPs belonging to “purine nucleotide metabolic process”, “autophagy”, “muscle contraction”, “response to external biotic stimulus”, and “chromatin assembly” downregulated DEPs “chromatin assembly”, respectively (Figure 3B and 3C). While “purine nucleotide metabolic process”, “autophagy”, “muscle contraction”, “response to external biotic stimulus” were enriched for by the upregulated DEPs, “chromatin assembly” was enriched for by downregulated DEPs.
Figure 3. Differential enrichment of cellular processes by up- and downregulated proteins perturbed by caffeine.
(A) Bar plot showing enriched cellular processes in up- or downregulated DEPs. (B) List of proteins involved in the enriched cellular processed by upregulated DEPs. Proteins reported with cancer are in bold. (C) List of proteins involved in the enriched cellular processed by downregulated DEPs.
Biological effects of caffeine treatment on immortalized normal bladder epithelial cells
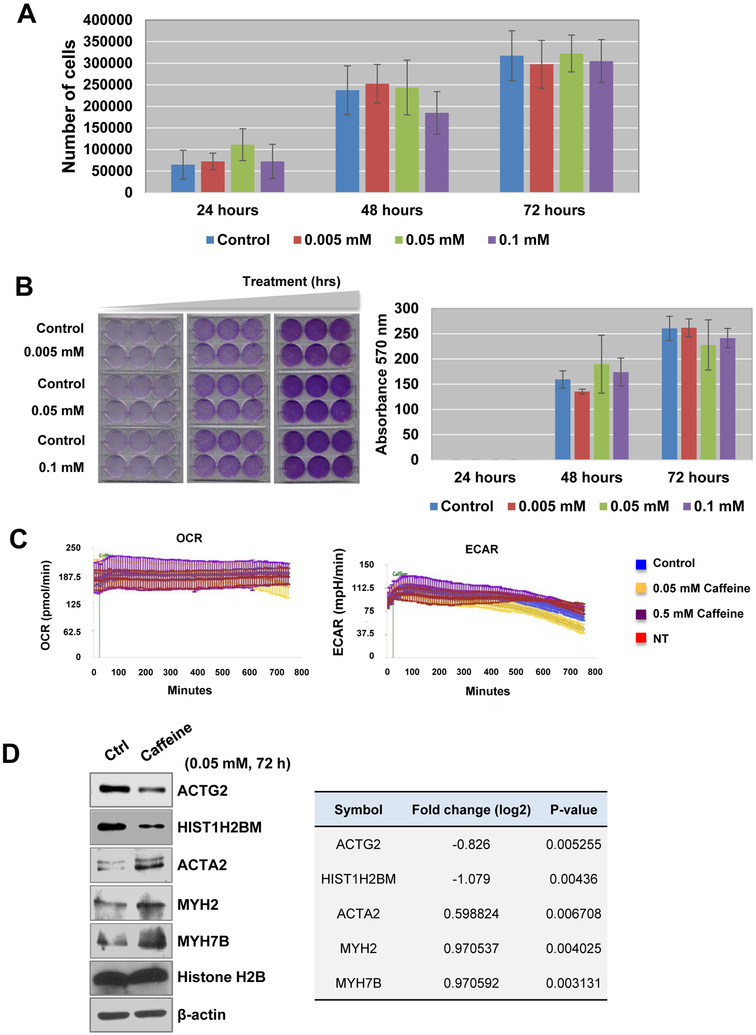
To evaluate the direct effects of caffeine on bladder epithelial cells in vitro, we used TRT-HU1 cells in our study as described in Methods section. After incubating the TRT-HU1 cells in varying concentrations of caffeine (0.005, 0.05, and 0.1 mM) for 24, 48, or 72 hrs, we sought to determine if caffeine affects or controls cell proliferation and metabolism. We found that the cell proliferation rate of caffeine-treated TRT-HU1 cells was not significantly altered, compared to controls (Figure 4A and 4B).
Figure 4. Cell proliferation was suppressed in response to caffeine treatment.
(A-B) TRT-HU1, hTert-immortalized normal bladder epithelial cells, were treated with 0.005, 0.05, and 0.1 mM caffeine for 24, 48, or 72 hrs. (A) Cell counting and (B) crystal violet proliferation assay were conducted as described in Methods. *P<0.05 (two-sided Student’s t-test) compared with the control group. Representative images of TRT-HU1 cells treated with caffeine (lower panels). (C) Seahorse data showed that caffeine treatment did not alter metabolism of normal bladder epithelial cells. Top, oxygen consumption rate (OCR) chart *p < 0.05. Bottom, extracellular acidification rate (ECAR) chart ** p < 0.05. (D) Quantification results from western blot analysis to measure the expression levels of ACTG2, HIST1H2BM, ACTA2, MYH2, MYH7B and histone H2B proteins in the presence or absence of caffeine. β-actin was used loading control. The differentially expressed protein levels obtained from proteomics analysis were shown in Table (right).
To examine the effects of caffeine on cell metabolism, we performed Seahorse Respirometry Analysis. Caffeine treatment over a 12-hr period did not alter the oxygen consumption rate (OCR, mitochondrial respiration) or extracellular acidification rate (ECAR, glycolysis) in TRT-HU1 cells (Figure 4C). Briefly, basal OCR and ECAR measurements (first three time points) were monitored before caffeine (0.05 mM) or vehicle (methanol) was administered. OCR and ECAR continued to be monitored up to 12 hrs post treatment. Non-treated cells (media administration only) were included to control for any effects of the cells being in the analyzer for an extended period (Figure 4C). We could not detect any changes in both the OCR and ECAR, even after a 12-hr treatment with 0.05 mM caffeine. Additional western blot analysis demonstrated that expression of ACTG2 and HIST1H2BM were reduced in the 0.05 mM caffeine-treated cells, while expression of ACTA2, MYH2 and MYH7B were upregulated. The expression of histone H2B and ACTB remained unchanged (Figure 4D).
DISCUSSION
Approximately 60% of adults in the U. S. consume coffee or some other caffeinated beverage. Caffeine, a methylxanthine derived from coffee, is known to affect biological and physiological responses. It is known that low to moderate levels of caffeine intake can benefit liver function, diabetes, neurological diseases, such as Alzheimer’s and Parkinson’s, and certain types of cancer. Caffeine is also known to increase urine calcium excretion via translocation of annexin A1 from the apical surface of cells into the cytoplasm. Therefore, caffeine intake is considered to be associated with lower risk of kidney stones[29]. A previous prospective study on the effects of caffeine in coffee suggested that coffee reduction can be a strategy in preventing urinary symptoms, such as frequency, urgency, and bladder pain syndrome (BPS)[9].
The aim of this study was to evaluate if caffeine consumption is associated with changes to protein expression in the normal bladder using semi-quantitative proteomic analysis. Although there are some reports about anti-lipid accumulation via gene expression suppression of proliferator activated receptor (PPAR) ɣ and CCAAT/enhancer binding protein (C/EBP) α in 3T3-L1 adipocytes [30], or apoptosis induction in various cell lines, the direct effects of caffeine on the bladder remains elusive. Although the amount of caffeine in a cup of coffee varies, it is typically 300–600 mg. Using caffeine levels equivalent to 1–2 cups of coffee in humans, we performed unbiased proteomic analysis using LC-MS/MS on bladder epithelial cells treated with caffeine and identified 32 upregulated and 25 downregulated DEPs (log2 fold-change>0.58; p-value <0.05; Figure 1B, Table 1 and 2). These DEPs were enriched for functions such as actin-myosin filament sliding, muscle contraction, and chromatin assembly proteins (Figure 3A). Alterations to the cytoskeleton and the proteins that interact with it have been linked to a wide range of diseases[31]. Caffeine has similar actions with AMP-activated protein kinase (AMPK), an enzyme whose roles include contraction during energy deprivation in skeletal muscles[32]. Among the proteins involved in the enriched cellular functions, ACTG2, also known as alpha smooth muscle actin, is associated with multiple functions in cell motility, structure, integrity, and intercellular signaling[33]. However, caffeine did not change any of the muscle associated stabilizing proteins which regulate the number, or the length, of microtubules[34]. This suggests that increased expression of ACTG2 by caffeine in the bladder may enhance the contractility of bladder and may also be associated with urinary symptoms. Our experimental data suggested that these proteins, such as ACTG2 and HIST1H2BM, become more redundant in bladder epithelial cells upon exposure to caffeine. Although the function of HIST1H2BM as a regulator of caffeine and its effects has not been studied well, it is originally known to be associated with epigenetic regulation in response to DNA methylation[35].
In this study, we also found that protein expression level of membrane-associated guanylate kinase inverted 3 (MAGI3) was upregulated in presence of caffeine in normal bladder cells. The observation in mass spectrometry analysis was further validated using western blot experiments. MAGI3 localizes to epithelial cell tight junctions, and has been originally identified as a scaffolding protein and tumor suppressor in glioma and breast cancer et al. The previous investigations on the role and molecular mechanism revealed that MAGI3 plays a role in phospholipid signaling pathways and suppress cancer cell proliferation via upregulation of PTEN, a well-known tumor suppressor[36]. MAGI3 has been found to interact with PTEN, via a PDZ Domain of MAGI3, and together contribute to regulation of the kinase activity of AKT/PKB and tumor cell survival. Interestingly, MAGI3 has been reported to be involved in the reduced cell proliferation, arrests of the cell cycle, and inhibition of the migration of glioma cells. Overexpression of MAGI3 resulted in a suppression of β-catenin’s transcriptional activity and inhibition the expression of target genes such as Cyclin D1 in glioma cells[36]. More interestingly, mysregulated or modified MAGI3 can show the different roles in cell proliferation or cancer. For example, when MAGI3 is found fused with AKT3, GAGI3-AKT3 contributes to a constitutive phosphorylation and activation of AKT kinase and its downstream targets, such as GSK3b, in breast cancer[37]. Premature cleavage and polyadenylation in MAGI3, MAGI3pPA, was identified to result in an oncogenic protein in breast cancer[38]. Given that the overexpressed MAGI3 has a functional link to the suppressed oncogenic characteristics, our data support the hypothesis that caffeine might benefit to prevention of cancer development, which consistent with previous reports showing that coffee consumption inversely associated with a decreased risk of cancer including prostate cancer[39].
There are several limitations in this study. We are aware that a single proteomic study conducted on one cell line and MS platform is not enough for conclusive statements on whether in vitro protein perturbation due to caffeine can represent the same perturbation in vivo. Urinary caffeine levels can be quantified by ultra-high performance liquid chromatography tandem mass spectrometry, and those levels are positively associated with consumption frequency of coffee (AUC = 0.849, 95% CI [0.808;0.891])[40]. Also, we conducted two biological replicate under each condition. In general, a small number of replicates show low statistical power. However, we identified DEPs using empirical null distribution estimated by randomly permuted samples, which should reflect distribution of detected proteins. Furthermore, expression of some DEPs was confirmed using western blot analysis (Figure 4C). Despite these limitations, our experimental findings provide evidence for the potential mechanisms through which caffeine intake is associated with LUTS, suggesting that the restriction of caffeine may benefit muscle contraction and functional gene expression through impaired epigenetic regulation.
Supplementary Material
Supplementary Figure 1. Cellular compartments of identified DEPs and sub-classes, (A) Pie chart displays cellular compartments of all the identified proteins. (B-E) Subclasses of the top 4 gene ontology cellular compartments enriched by the detected proteins. Pie chart showing enriched subclass of cell part, organelle, macromolecular complex, and membrane.
Statement of Study Significance.
Caffeine, a methylxantine that is derived from coffee intake, has been reported to be a potential dietary risk factor in developing lower urinary tract symptoms (LUTS). Despite this known potential link, research into causative mechanisms and functions is lacking. Our unbiased proteomics study revealed that caffeine alters the global proteome in human immortalized normal bladder epithelial cells and enriches biological pathways related to muscle contraction and chromatin assembly. Our study is clinically significant because it provides the potential mechanisms through which caffeine can provoke LUTS and suggests that the restriction of caffeine intake may be benefit muscle contraction and functional gene expression.
ACKNOWLEDGEMENTS
The authors acknowledge support from National Institutes of Health grants (1U01DK103260, 1R01DK100974, U24 DK097154, NIH NCATS UCLA CTSI UL1TR000124), Department of Defense grants (W81XWH-15–1-0415), Centers for Disease Controls and Prevention (1U01DP006079), IMAGINE NO IC Research Grant, the Steven Spielberg Discovery Fund in Prostate Cancer Research Career Development Award, and the U.S.-Egypt Science and Technology Joint Fund (to J.K.). J.K. is former recipient of Interstitial Cystitis Association Pilot Grant, a Fishbein Family IC Research Grant, New York Academy of Medicine, and Boston Children’s Hospital Faculty Development. The funders had no role in the experimental design, data collection, analysis, preparation of the manuscript, or decision to publish. In addition, this article is derived from the Subject Data funded in whole or part by National Academies of Sciences, Engineering, and Medicine (NAS) and The United States Agency for International Development (USAID). Any opinions, findings, conclusions, or recommendations expressed in this article are those of the authors alone, and do not necessarily reflect the views of USAID or NAS.
Footnotes
Conflicts of Interests
The authors declare that there are no conflicts of interest regarding the publication of this paper.
REFERENCES
- [1].Irwin DE, Kopp ZS, Agatep B, Milsom I, Abrams P, BJU international 2011, 108, 1132. [DOI] [PubMed] [Google Scholar]
- [2].Coyne KS, Wein AJ, Tubaro A, Sexton CC, Thompson CL, Kopp ZS, Aiyer LP, BJU international 2009, 103 Suppl 3, 4. [DOI] [PubMed] [Google Scholar]
- [3].Silva J, Silva CM, Cruz F, Current opinion in urology 2014, 24, 21. [DOI] [PubMed] [Google Scholar]
- [4].Coyne KS, Wein A, Nicholson S, Kvasz M, Chen CI, Milsom I, Journal of managed care pharmacy : JMCP 2014, 20, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Robinson D, Giarenis I, Cardozo L, Maturitas 2014, 79, 8. [DOI] [PubMed] [Google Scholar]
- [6].He Q, Yang Y, Xia M, Zhang N, Wu S, Xiao Y, Li G, Zhan S, Liu L, Xiao H, Zhao J, Zhonghua yi xue za zhi 2014, 94, 428. [PubMed] [Google Scholar]
- [7].Wells MJ, Jamieson K, Markham TC, Green SM, Fader MJ, Journal of wound, ostomy, and continence nursing : official publication of The Wound, Ostomy and Continence Nurses Society 2014, 41, 371. [DOI] [PubMed] [Google Scholar]
- [8].Anderson BL, Juliano LM, Schulkin J, Journal of women’s health 2009, 18, 1457. [DOI] [PubMed] [Google Scholar]
- [9].Staack A, Distelberg B, Schlaifer A, Sabate J, Neurourology and urodynamics 2017, 36, 432. [DOI] [PubMed] [Google Scholar]
- [10].Maserejian NN, Wager CG, Giovannucci EL, Curto TM, McVary KT, McKinlay JB, American journal of epidemiology 2013, 177, 1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Turati F, Bosetti C, Polesel J, Zucchetto A, Serraino D, Montella M, Libra M, Galfano A, La Vecchia C, Tavani A, Urology 2015, 86, 1179. [DOI] [PubMed] [Google Scholar]
- [12].Bode AM, Dong Z, Cancer letters 2007, 247, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].He Z, Ma WY, Hashimoto T, Bode AM, Yang CS, Dong Z, Cancer Res 2003, 63, 4396. [PubMed] [Google Scholar]
- [14].Cho YS, Ko IG, Kim SE, Hwan L, Shin MS, Kim CJ, Kim SH, Jin JJ, Chung JY, Kim KH, Molecular medicine reports 2014, 10, 2931. [DOI] [PubMed] [Google Scholar]
- [15].Kershen R, Mann-Gow T, Yared J, Stromberg I, Zvara P, The Journal of urology 2012, 188, 1986. [DOI] [PubMed] [Google Scholar]
- [16].Hockey JS, Wu C, Fry CH, BJU international 2000, 86, 531. [DOI] [PubMed] [Google Scholar]
- [17].Kim J, Ji M, DiDonato JA, Rackley RR, Kuang M, Sadhukhan PC, Mauney JR, Keay SK, Freeman MR, Liou LS, Adam RM, In vitro cellular & developmental biology. Animal 2011, 47, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Choi DY, You S, Jung JH, Lee JC, Rho JK, Lee KY, Freeman MR, Kim KP, Kim J, Proteomics 2014, 14, 1845. [DOI] [PubMed] [Google Scholar]
- [19].Qu Y, Zhou B, Yang W, Han B, Yu-Rice Y, Gao B, Johnson J, Svendsen CN, Freeman MR, Giuliano AE, Sareen D, Cui X, Scientific reports 2016, 6, 32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rinas A, Espino JA, Jones LM, Analytical and bioanalytical chemistry 2016, 408, 3021. [DOI] [PubMed] [Google Scholar]
- [21].Niu M, Cho JH, Kodali K, Pagala V, High AA, Wang H, Wu Z, Li Y, Bi W, Zhang H, Wang X, Zou W, Peng J, Analytical chemistry 2017, 89, 2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang B, Chambers MC, Tabb DL, Journal of proteome research 2007, 6, 3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee H, Chae S, Park J, Bae J, Go EB, Kim SJ, Kim H, Hwang D, Lee SW, Lee SY, Molecular & cellular proteomics : MCP 2016, 15, 3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Storey JD, Tibshirani R, Proc Natl Acad Sci U S A 2003, 100, 9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang da W, Sherman BT, Lempicki RA, Nature protocols 2009, 4, 44. [DOI] [PubMed] [Google Scholar]
- [26].Hashimoto T, He Z, Ma WY, Schmid PC, Bode AM, Yang CS, Dong Z, Cancer Res 2004, 64, 3344. [DOI] [PubMed] [Google Scholar]
- [27].Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD, Nucleic acids research 2017, 45, D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, Cell systems 2015, 1, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peerapen P, Thongboonkerd V, Scientific reports 2016, 6, 38536; [DOI] [PMC free article] [PubMed] [Google Scholar]; Ferraro PM, Taylor EN, Gambaro G, Curhan GC, The American journal of clinical nutrition 2014, 100, 1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mitani T, Nagano T, Harada K, Yamashita Y, Ashida H, Journal of nutritional science and vitaminology 2017, 63, 331. [DOI] [PubMed] [Google Scholar]
- [31].Cicchillitti L, Penci R, Di Michele M, Filippetti F, Rotilio D, Donati MB, Scambia G, Ferlini C, Molecular cancer therapeutics 2008, 7, 2070. [DOI] [PubMed] [Google Scholar]
- [32].Egawa T, Hamada T, Kameda N, Karaike K, Ma X, Masuda S, Iwanaka N, Hayashi T, Metabolism: clinical and experimental 2009, 58, 1609. [DOI] [PubMed] [Google Scholar]
- [33].Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C, Molecular biology of the cell 2001, 12, 2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Franklin JL, Mirzaei M, Wearne TA, Homewood J, Goodchild AK, Haynes PA, Cornish JL, Journal of proteome research 2016, 15, 1455. [DOI] [PubMed] [Google Scholar]
- [35].Ek WE, Tobi EW, Ahsan M, Lampa E, Ponzi E, Kyrtopoulos SA, Georgiadis P, Lumey LH, Heijmans BT, Botsivali M, Bergdahl IA, Karlsson T, Rask-Andersen M, Palli D, Ingelsson E, Hedman AK, Nilsson LM, Vineis P, Lind L, Flanagan JM, Johansson A, C. Epigenome-Wide Association Study, Human molecular genetics 2017, 26, 3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ma Q, Zhang Y, Meng R, Xie KM, Xiong Y, Lin S, He ZL, Tao T, Yang Y, Zhao JZ, He JQ, Biomedical and environmental sciences : BES 2015, 28, 502. [DOI] [PubMed] [Google Scholar]
- [37].Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortes ML, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Pina V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio RC, Parkin M, Pho NH, Quintanar-Jurado V, Ramos AH, Rebollar-Vega R, Rodriguez-Cuevas S, Romero-Cordoba SL, Schumacher SE, Stransky N, Thompson KM, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi DC, Richardson AL, Jimenez-Sanchez G, Lander ES, Gabriel SB, Garraway LA, Golub TR, Melendez-Zajgla J, Toker A, Getz G, Hidalgo-Miranda A, Meyerson M, Nature 2012, 486, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ni TK, Kuperwasser C, eLife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pounis G, Tabolacci C, Costanzo S, Cordella M, Bonaccio M, Rago L, D’Arcangelo D, Filippo Di Castelnuovo A, de Gaetano G, Donati MB, Iacoviello L, Facchiano F, i. Moli-sani study, International journal of cancer 2017, 141, 72; [DOI] [PubMed] [Google Scholar]; Sado J, Kitamura T, Kitamura Y, Sobue T, Nishino Y, Tanaka H, Nakayama T, Tsuji I, Ito H, Suzuki T, Katanoda K, Tominaga S, G. Three-Prefecture Cohort Study, Cancer science 2017, 108, 2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Petrovic D, Estoppey Younes S, Pruijm M, Ponte B, Ackermann D, Ehret G, Ansermot N, Mohaupt M, Paccaud F, Vogt B, Pechere-Bertschi A, Martin PY, Burnier M, Eap CB, Bochud M, Guessous I, Nutrition & metabolism 2016, 13, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cellular compartments of identified DEPs and sub-classes, (A) Pie chart displays cellular compartments of all the identified proteins. (B-E) Subclasses of the top 4 gene ontology cellular compartments enriched by the detected proteins. Pie chart showing enriched subclass of cell part, organelle, macromolecular complex, and membrane.