Abstract
External guide sequences (EGSs) are oligonucleotides that consist of a sequence complementary to a target mRNA and recruit intracellular RNase P for specific degradation of the target RNA. In this study, DNA-based EGS molecules were chemically synthesized to target the mRNA coding for the protease of human cytomegalovirus (HCMV). The EGS molecules efficiently directed human RNase P to cleave the target mRNA sequence in vitro. When EGSs were exogenously administered into HCMV-infected human foreskin fibroblasts, a reduction of about 80–90% in the expression level of the protease and a reduction of about 300-fold in HCMV growth were observed in the cells that were treated with a functional EGS, but not in cells that were not treated with the EGS or with a “disabled” EGS carrying nucleotide mutations that precluded RNase P recognition. Moreover, packaging of the viral DNA genome into the capsid was blocked in the cells treated with the functional EGS. These results indicate that HCMV protease is essential for viral DNA encapsidation. Moreover, our study provides direct evidence that exogenous administration of a DNA-based EGS can be used as a therapeutic approach for inhibiting gene expression and replication of a human virus.
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that typically causes asymptomatic infections in healthy individuals but may lead to serious complications in newborns and immunodeficient individuals (1). For example, this virus accounts for one of the most common opportunistic infections in AIDS patients (i.e., CMV retinitis). Moreover, HCMV infection is the leading viral cause of birth defects in newborns and a major cause of morbidity and mortality in bone marrow and solid organ transplant recipients (1). Developing effective antiviral compounds and approaches is central in controlling HCMV infections and preventing HCMV-associated complications.
Antisense molecules are promising gene-targeting agents for specific regulation of gene expression (2, 3). Conventional antisense oligonucleotides have been used as anti-HCMV agents to inhibit the expression of HCMV-essential genes and abolish viral replication (4–6). External guide sequences (EGSs) are antisense oligonucleotides that can be used in conjunction with ribonuclease P (RNase P) or the catalytic RNA subunit of RNase P from Escherichia coli (M1 RNA) to diminish gene expression (7, 8). RNase P is one of the most abundant and active enzymes in cells and is responsible for 5′ termini maturation of tRNAs (9, 10). This enzyme catalyzes a hydrolysis reaction to remove the leader sequence of tRNA precursors by recognizing the common structure shared among all tRNAs (Fig. 1). The EGS-based technology is unique in inducing endogenous RNase P to cleave a target mRNA when the EGS hybridizes to the mRNA to form a structure resembling a tRNA substrate (Fig. 1B), and this approach is highly specific and does not generate nonspecific “irrelevant cleavage” that is observed in RNase H-mediated cleavage induced by conventional antisense phosphothioate molecules (7, 8, 11). Thus, EGSs represent a new class of agents that may lead to highly effective and specific inhibition of gene expression.
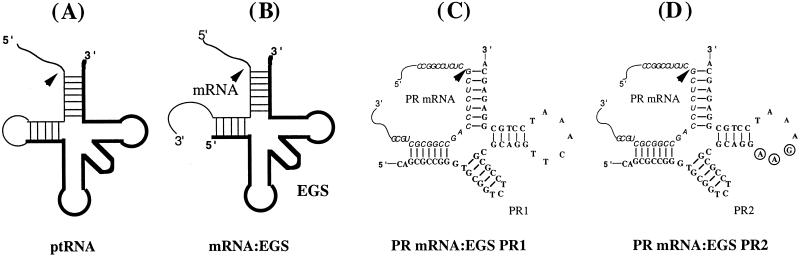
Figure 1.
Schematic representation of substrates for RNase P. (A) A natural substrate (ptRNA). (B) A hybridized complex of a target RNA (e.g., mRNA) and an EGS that resembles the structure of a tRNA. (C and D) Complexes between PR mRNA sequence and EGS PR1 and PR2, respectively. The sequence of these EGSs equivalent to the tRNA sequence was derived from tRNASer and resembles the T-stem and loop, and variable region of the tRNA molecule.
RNA-based EGSs have been expressed endogenously as transgenes in both bacteria and mammalian cells (7, 12), and were effective in inhibiting the gene expression of herpes simplex and influenza virus and in abolishing the replication of influenza virus in human cells (13, 14). In vitro studies have also shown that DNA-based EGSs, as well as EGS molecules with modified nucleotides, can direct M1 RNA or human RNase P to cleave a mRNA sequence, although their targeting efficiencies are lower than those of unmodified RNA-based EGSs (8, 11, 15, 16). However, little is known about whether DNA-based EGSs are functionally active in cultured cells. Equally unclear is whether DNA-based EGSs can be exogenously administered into human cells for inhibiting gene expression and growth of human viruses.
In the study reported here, we provide direct evidence that exogenous administration of chemically synthesized DNA-based EGSs is highly effective in treating HCMV-infected cells and abolishing HCMV replication. The target is the mRNA encoding the HCMV protease (PR). The protease is highly conserved among all human herpesviruses, including herpes simplex virus 1 (HSV-1), Epstein–Barr virus (EBV), and Kaposi sarcoma-associated herpesvirus (KSHV) (1, 17, 18). This enzyme provides a stoichiometric function as a virion structural component and its proteolytic activity is required for processing of the capsid assembly protein (19, 20). It is considered an ideal target for anti-herpes therapy because inhibiting PR expression would shut down both the stoichiometric and enzymatic functions simultaneously and, therefore, may achieve a greater antiviral effect. However, only the HSV-1 protease, UL26, has been demonstrated to be essential for viral capsid assembly, as shown in studies of viral mutants with mutations at the UL26 sequence (21, 22). The functions of HCMV PR, as well as those of EBV and KSHV, in viral lytic replication are currently unknown, because of the difficulty of constructing mutants with deletions of viral essential genes and generating complementing cell lines expressing the essential functions. Therefore, we chose to use the EGS-based technology to investigate the function of HCMV PR. Our results provide direct evidence that HCMV protease is essential for viral DNA encapsidation. Moreover, we show that the DNA-based EGS molecules are highly effective in inhibiting HCMV gene expression and growth in human cell culture and, furthermore, demonstrate the feasibility of using DNA-based EGSs for the studies and treatment of infections caused by human viruses including HCMV.
Materials and Methods
Construction of EGSs and in Vitro Cleavage and Binding Assays.
The DNA template coding for substrate pr39 was generated by annealing oligonucleotides OliT7 (5′-TAATACGACTCACTATAG-3′) and sAP1 (5′-CGGGATCCGCAGCGCCGGCTGGAGAGCGAGAGGCCGGCCTATAGTGAGTCGTAT-TA-3′). EGS PR1 (5′-CAGCGCCGGGTGCGGTCTCCGCGCGCAGGTTCAAATCCTGCGGAGAGCA-3′), PR2 (5′-CAGCGCCGGGTGCGGTCTCCGCGCGCAGGAAGAAATCCTGCGGAGAGCA-3′), and TK1 (5′-TACGTCGGTGCGGTCTCCGCGCGCAGGTTCAAATCCTGCCGCAGACACCA-3′) were chemically synthesized directly by using a DNA synthesizer. Some of these oligonucleotides also contain a 5′ FITC label (Glen Research, Sterling, VA). RNase P was prepared from HeLa cellular extracts as described (7). The EGSs (10 nM) and 32P-labeled pr39 (10 nM) were incubated with human RNase P (2 units). The cleavage reactions were carried out at 37°C for 45 min in buffer A (50 mM Tris, pH 7.4/100 mM NH4Cl/10 mM MgCl2). Cleavage products were separated in denaturing gels and quantitated with a STORM840 phosphorimager (Molecular Dynamics). The procedures to measure the equilibrium dissociation constants (Kd) of the EGS–pr39 complexes were carried out as described (23, 24). In brief, various concentrations of EGS (0.05–50 nM) were preincubated in buffer B (50 mM Tris, pH 7.5/100 mM NH4Cl/100 mM CaCl2/3% glycerol/0.1% xylene cyanol/0.1% bromophenol blue) at 37°C for 10 min before mixing with an equal volume of 0.1–0.5 nM of RNA substrate preheated under identical conditions. The samples were incubated for 15 min to allow binding, then separated on a 5% polyacrylamide gel and was quantitated with a STORM840 phosphorimager.
EGS Internalization and Treatment of Human Cells.
Lipofectamine 2000 reagent (GIBCO) was diluted in 100 μl of Opti-MEM medium with the EGS to give a final concentration of 10 μg/ml lipid-100 nM EGS. The transfection experiments were carried out by using 100 nM EGS. The EGS–lipid mixtures were prepared according to the manufacturer's recommendation (GIBCO) and incubated with cells for 7 h, and then removed. Under these settings, we consistently achieve an optimal transfection efficiency of about 90%. At 7 h posttransfection, cells were fixed with 4% paraformaldehyde, stained with Texas red phalloidin or propidium iodide (Molecular Probes) following the manufacturer's recommendations, and visualized by using a Nikon PCM2000 confocal microscope.
Viruses, Cells, and Antibodies.
HCMV (strain AD169) was propagated in human foreskin fibroblasts in DMEM supplemented with 10% FBS (GIBCO/BRL) as described (25). The polyclonal antibodies against HCMV PR and capsid assembly protein (AP) were kindly provided by Annette Meyer of Parke-Davis Pharmaceutical Research Institute (Pfizer, Ann Arbor, MI). The monoclonal antibodies c1202 and c1203, which react with HCMV proteins UL44 and IE1/IE2, respectively, were purchased from Goodwin Institute for Cancer Research (Plantation, FL). The monoclonal antibodies against gH and human actin were purchased from BioDesign (Kennebunk, Maine) and Sigma, respectively.
Viral Infections and Assays of Viral Gene Expression and Growth in EGS-Treated Cells.
Cells (106) that were first treated with liposome complexes in the absence and presence of EGSs were either mock-infected or infected with HCMV in an inoculum of 1.5 ml DMEM supplemented with 1% FCS. At 12–72 h postinfection, total cellular RNAs and proteins were isolated from cells as described (25). For Northern analyses, the RNA fractions were separated in 0.8–2.5% agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, hybridized with the 32P-radiolabeled DNA probes that contained the HCMV DNA sequences, and analyzed with a STORM840 phosphorimager (25). For Western analyses, the denatured, solubilized polypeptides were separated on 7.5% or 9% (vol/vol) SDS-polyacrylamide gels cross-linked with N,N"-methylenebisacylamide, transferred electrically to nitrocellulose membranes, and reacted to the antibodies against human β-actin, HCMV PR, AP, IE1, UL44, and gH. The membranes were subsequently stained with a chemiluminescent substrate with the aid of a Western chemiluminescent substrate kit (Amersham Pharmacia) and quantitated with a STORM840 phosphorimager (25). To determine the level of the inhibition of viral growth, 5 × 105 human foreskin fibroblasts were first incubated with liposome complexes in the absence and presence of EGSs, and then mock-infected or infected with HCMV AD169 at a multiplicity of infection (moi) of 5. The cells and medium were harvested at 1, 2, 3, 4, 5, 6, and 7 days postinfection and viral stocks were prepared by adding an equal volume of 10% skim milk, followed by sonication. The titers of the viral stocks were determined by infecting 2 × 105 foreskin fibroblasts in 6-well plates and counting the number of plaques 7–10 days postinfection (25). The values obtained were the average from duplicate experiments.
Assaying the Level of Viral Genome Replication and Packaging, and Electron Microscopic Studies.
HCMV-infected cells that were either treated with EGS–lipid complexes or treated with lipid complexes alone were harvested at 72–96 h postinfection. Total and encapsidated (DNase I-treated) DNAs were isolated essentially as described (21) and used as the PCR DNA templates. Viral DNA was detected by PCR amplification of the viral IE1 sequence by using primers CMV3 (5′-CCAAGCGGCCTCTGATAACCAAGCC-3′) and CMV4 (5′-CAGCACCATCCTCCTCTTCCTCTGG-3′) (26). The 5′ and 3′ primers used to amplify the actin sequence (internal control) were Actin5 (5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′) and Actin3 (5′-CTAGAAGCATTGCGGTGGCAGATGGAGGG-3′), respectively. PCR reactions were carried out in the presence of [α-32P]dCTP. The radiolabeled HCMV DNA (481 bp) and actin sequence (610 bp) were separated on 4% nondenaturing polyacrylamide gels and analyzed with a STORM840 phosphorimager (26). A standard (dilution) curve was generated by amplifying different dilutions of the template DNA. The plot of radioactivity for both HCMV and β-actin vs. dilutions of DNA did not reach a plateau for the saturation curve under the conditions described above, indicating that quantitation of viral DNA could be accomplished. The fact that the ratio of viral DNA to β-actin remained constant with respect to each DNA dilution in the standard curve indicated the adequate accuracy and reproducibility of the assay (26). The PCR results were derived from three independent experiments.
Infected cells examined by electron microscope were centrifuged into a small (0.3 ml) pellet, fixed with 2.5% glutaraldehyde into 0.1 M phosphate buffer (pH 7.4) for 24 h at 4°C, and postfixed with 2% OsO4 for 1 h at 24°C. Pellets were then washed, embedded in Epon 812, and cut into sections (70 nm thick) with a Richert Ultracut E ultramicrotome. Sections were stained with a saturated solution of uranyl acetate in methanol (20 min at 60°C) and then with 0.25% lead citrate (2 min at 24°C), and finally examined with a JEOL 100CX transmission electron microscope operated at 100 keV (21).
Results
Because most mRNA species inside cells are usually associated with proteins and are present in a highly organized and folded conformation, it is critical to choose a target region that is accessible to binding of EGSs to achieve efficient targeting. In vivo mapping with dimethyl sulfate (DMS) has been used to determine the accessibility of mRNA and structure of RNAs in cells (27, 28). Using this method, we mapped the region of PR mRNA and chose a position, 395 nucleotides downstream from the translation initiation site, as the cleavage site for human RNase P. This site appears to be one of the regions most accessible to DMS modification and, presumably, would also be accessible to EGS binding. Moreover, its flanking sequence exhibits several features that need to be present to interact with an EGS and RNase P to achieve efficient cleavage. These features include that the nucleotides 3′ and 5′ adjacent to the site of cleavage are a guanosine and a pyrimidine, respectively (7, 24).
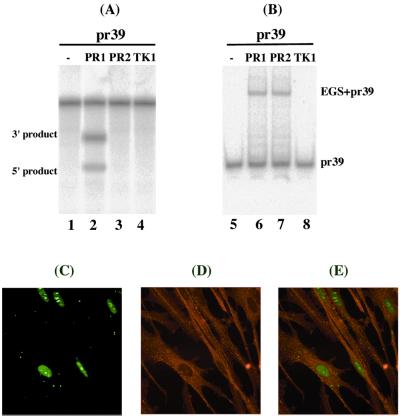
Two EGSs were constructed. PR1, which resembles a part of the tRNASer structure, contains a T-loop and stem, and a variable region but not the anticodon region (Fig. 1C). The anticodon domain has been shown to be dispensable for EGS activity (24). PR2 (Fig. 1D) was derived from PR1 by introducing base-substitution mutations in three positions of the T-loop. The nucleotides in these three positions are highly conserved among tRNA molecules and are important for the recognition of tRNA molecules by RNase P (9, 10, 29). EGSs were chemically synthesized in vitro by using a DNA synthesizer and subsequently incubated with substrate pr39 and human RNase P. Apparent cleavage of pr39 by RNase P was observed in the presence of PR1 and yielded cleavage products of 12 and 27 nucleotides (Fig. 2, lane 2). However, cleavage was barely detected in the presence of PR2 (Fig. 2, lanes 3). To determine whether the differential cleavage efficiencies observed with PR1 and PR2 were possibly due to their different binding affinities to the PR mRNA sequence, the binding between the EGSs and pr39 was assayed in the absence of human RNase P. Similar amounts of EGS–pr39 complexes were observed when the same amount of EGSs was used (Fig. 2B, lanes 5–8). Further detailed assays under different concentrations of the EGSs and pr39 indicated that the binding affinity of PR2 to pr39, measured as the dissociation constant (Kd), is similar to that of PR1 (data not shown). Meanwhile, cleavage products were barely detected in the presence of PR2 even under high concentrations of RNase P and a prolonged incubation period. These observations indicate that the T-loop mutations do not significantly affect the binding affinity between PR2 and PR mRNA sequence but disrupt the recognition of EGS–PR mRNA complex by RNase P. Thus, PR2 may be used as a control for the antisense effect in our experiments in cultured cells (see below).
Figure 2.
(A) Cleavage of 32P-labeled substrate pr39 by human RNase P in the presence of different EGSs. No RNase P was added to the reaction mixture in lane 1. Cleavage reactions were carried out in the presence of PR1 (lane 2), PR2 (lanes 3), or TK1 (lane 4). (B) Binding of PR mRNA substrate by EGSs. Substrate pr39 (0.1 nM) was either incubated alone (lane 5) or in the presence of 0.1 nM of PR1 (lane 6), PR2 (lane 7), or TK1 (lane 8) to allow binding and then loaded on a 5% polyacrylamide gel. (C–E) Internalization of EGSs in human cells. 5′-fluorescein-labeled PR1 (20 nM) was complexed with Lipofectamine 2000 (10 μg/ml) and transfected into human foreskin fibroblasts. The cells were fixed at 7 h postinfection. The images of FITC-conjugated EGSs (C) and cellular actin filaments stained with Texas red phalloidin (D) were used to generate the composite image (E).
The chemically synthesized EGS molecules were complexed with Lipofectamine 2000 liposomes (GIBCO) and delivered into human foreskin fibroblasts. Treatment of cells with the EGS–liposome complexes by using our transfection protocol consistently yielded a transfection efficiency of about 90%. An additional EGS, TK1, which was derived from PR1 and targeted the HSV-1 TK mRNA (13), was also constructed. This EGS was used to determine whether an EGS with an incorrect targeting sequence can direct human RNase P to cleave PR mRNA in tissue culture. No cleavage of pr39 by RNase P was observed in vitro in the presence of TK1 (Fig. 2, lane 4). To determine whether the EGSs were efficiently internalized into the fibroblasts, EGS molecules conjugated with a fluorescein (FITC) label were included in the transfection mixture and were visualized by using confocal microscopy (Fig. 2 C–E). The majority of the EGS localized in regions distinct from those stained with Texas red phalloidin (primarily staining actin filaments in the cytoplasm; Fig. 2 C–E) and colocalized with those stained with propidium iodide (primarily staining genomic DNA in the nuclei; data not shown). Thus, the internalized EGSs appear to localize primarily in the nuclei.
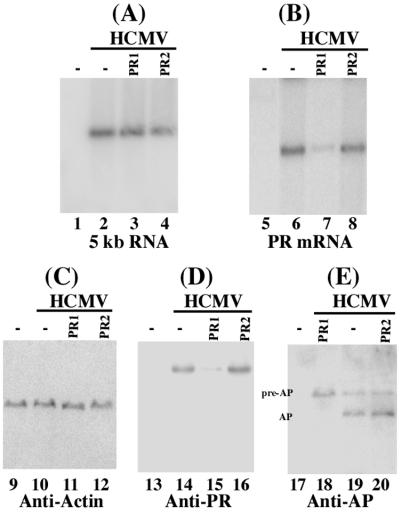
To investigate whether the internalized EGSs inhibit viral PR expression, the cells were treated with EGSs and then infected with HCMV at a moi of 0.1–5, and the levels of viral PR mRNA were determined by Northern analyses (Fig. 3 A and B). The level of the 5-kb viral immediate-early transcript (5 kb RNA) was used as an internal control for the quantitation of expression of PR mRNA (25, 30). Cells that were treated with PR1 exhibited a reduction of 80–90% ± 6–7% in the level of PR mRNA expression, whereas the PR2-treated cells only exhibited a reduction of less than 10% (Table 1). No reduction in the expression level of PR mRNA was observed in cells that were treated with liposome complexes in the absence of EGSs or in the presence of TK1 (data not shown). These results suggest that the significant reduction of PR mRNA expression in the PR1-treated cells was due to targeted cleavage by RNase P. The low level of inhibition observed in the PR2-treated cells was presumably due to the antisense effects of the EGS. The expression level of PR protein was also determined by Western analyses, using expression of cellular β-actin as the internal control (Fig. 3 C and D). A reduction of 81–89% ± 5–7% in the expression level of PR protein was observed in cells treated with PR1, whereas a reduction of less than 10% was found in the PR2-treated cells (Table 1).
Figure 3.
Expression of HCMV mRNAs (A and B) and proteins (C–E) in EGS-treated cells. Human foreskin fibroblasts (1 × 106) were first treated with liposome complexes, then either mock-infected (lanes 1, 5, 9, 13, and 17) or infected with HCMV (moi = 0.5; lanes 2–4, 6–8, 10–12, 14–16, and 18–20), and finally harvested at either 24 (A and B) or 72 h (C–E) postinfection. RNA and protein samples were isolated from cells treated with liposome complexes in the absence of EGSs (lanes 1 and 2, 5 and 6, 9 and 10, 13 and 14, 17, and 19), or in the presence of PR1 (lanes 3, 7, 11, 15, and 18) and PR2 (lanes 4, 8, 12, 16, and 20). In Northern analyses (A and B), equal amounts of each RNA sample (30 μg) were separated on agarose gels, transferred to a nitrocellulose membrane, and hybridized to a 32P-radiolabeled probe that contained the cDNA sequence of the HCMV 5-kb transcript (lanes 1–4) and PR mRNA (lanes 5–8). In Western analyses (C–E), protein samples were separated in two identical SDS-polyacrylamide gels and transferred electrically to two identical membranes. One membrane was allowed to react with a monoclonal antibody (Anti-Actin) against human actin (C), whereas the others were stained with the anti-PR and anti-AP antibodies (D and E).
Table 1.
Inhibition of HCMV gene expression by EGSs
| Viral genes | moi = 0.5
|
moi = 5
|
||||
|---|---|---|---|---|---|---|
| HFFs | PR1 | PR2 | HFFs | PR1 | PR2 | |
| PR mRNA | 0% | 90% ± 6% | 7% | 0% | 80% ± 7% | 8% |
| IE1 mRNA | 0% | 0% | 0% | 0% | 0% | 0% |
| US2 mRNA | 0% | 0% | 0% | 0% | 0% | 1% |
| PR protein | 0% | 89% ± 7% | 5% | 0% | 81% ± 5% | 6% |
| UL44 protein | 0% | 1% | 1% | 0% | 0% | 0% |
| Glycoprotein H | 0% | 2% | 1% | 0% | 0% | 0% |
Levels of inhibition of the mRNA and protein expression of different viral genes in human foreskin fibroblasts that were treated with liposome complexes in the presence of PR1 and PR2, as compared to the levels of inhibition in cells that were treated with the liposome complexes alone in the absence of EGSs [human foreskin fibroblasts (HFFs)]. The values shown are the means from triplicate experiments. The values of standard deviation that were less than 5% are not shown.
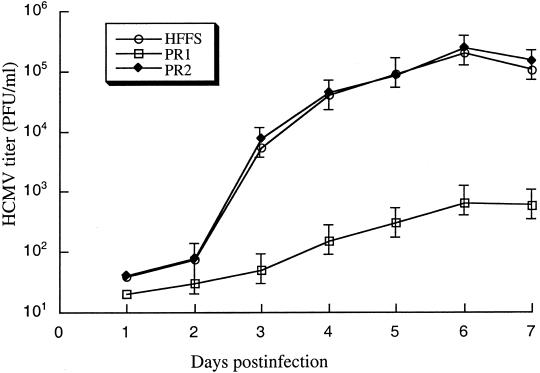
We next investigated whether the inhibition of PR expression abolishes viral growth. Cells were treated with liposome complexes in the absence and presence of EGSs and then infected by HCMV at a moi of 5. Virus stocks were prepared from the infected cultures at 1-day intervals through 7 days postinfection. The count of plaque-forming units (pfu) was determined by measurement of the viral titer on cells. After 4 days postinfection, a reduction of about 300-fold in viral yield was observed in the PR1-treated cells, whereas no significant reduction was found in those that were either treated with PR2 or TK1 (Fig. 4). To further determine whether EGSs can function as therapeutic agents for the treatment of HCMV infection, cells were first infected with HCMV at a moi of 1. At 6 h postinfection, these cells were then treated with liposome complexes in the absence and presence of EGSs every 48 h. After 4 days postinfection, a reduction of about 500-fold in viral yield was observed in the PR1-treated cells. In contrast, no significant reduction was found in those that were either treated with PR2 or TK1 (W.D., J.Z., and F.L., unpublished results). Thus, PR1 is effective in treating HCMV infection and blocking viral growth.
Figure 4.
Growth of HCMV in human foreskin fibroblasts (HFFs) that were treated with liposome complexes in the absence of EGSs, or in the presence of PR1 or PR2. Cells were first incubated with liposome complexes and then infected with HCMV at a moi of 5. Virus stocks were prepared from the infected cells at 1-day intervals through 7 days postinfection and viral titers were determined. These values are the means from duplicate experiments. The standard deviation is indicated by the error bars.
Although the HSV-1 protease has been shown to be required for viral DNA encapsidation (21), it has not been reported whether the HCMV protease is also essential for viral capsid maturation. Meanwhile, it is possible that the observed reduction of viral growth in the PR1-treated cells may be due to other effects of the EGS on viral lytic replication and may not necessarily be due to specific RNase P-mediated cleavage of PR mRNA. To study the function of protease and to determine the antiviral mechanism of the EGS-directed cleavage, we have carried out four sets of experiments. First, we examined the expression of other viral genes in the PR1-treated cells. Inhibition of PR expression is not expected to affect the expression of other viral genes, including immediate-early (α), early (β), and late (γ) genes (1), which are not regulated by the protease as suggested in the studies of HSV-1 protease mutants (21, 22). Northern analyses were carried out to determine the expression levels of the IE1 (an α transcript) and US2 mRNA (a β transcript). Moreover, Western analyses were performed to determine the expression level of viral protein UL44, a viral late (βγ) protein, and gH, a viral late (γ) protein. No significant difference in the expression level of these genes was observed in cells that were treated with liposome complexes in the absence of EGSs or in the presence of TK1, PR1, or PR2 (Table 1). These results suggest that PR1 specifically inhibits the expression of PR and does not affect overall viral gene expression.
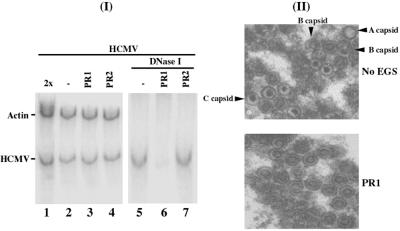
We next examined the level of proteolytic cleavage of the capsid AP, which is catalyzed by the protease (19). Although the expression of the assembly protein was not affected, the cleavage products of the assembly protein were barely detected in the PR1-treated cells (Fig. 3E, lane 18). Thus, reduction of PR expression abolishes the assembly protein processing. In the third set of experiments, we investigated whether viral genomic replication, as well as viral DNA encapsidation, is affected in the PR1-treated cells. Total DNA was isolated from HCMV-infected cell lysates that were either treated with DNase I or not. The encapsidated viral DNAs will be resistant to DNase I digestion, whereas those that are not packaged in the capsid will be susceptible to degradation. The level of intracellular viral DNA was determined by PCR detection of HCMV IE1 sequence, using the level of β-actin DNA as the internal control. When assaying the DNA samples from cell lysates that were not treated with DNase I, we found no significant difference in the level of total intracellular (both encapsidated and uncapsidated) viral DNA in the EGS-treated cells (Fig. 5, lanes 2–4). When the DNase I-treated samples were assayed, however, the “encapsidated” DNA was hardly detected in the PR1-treated cells (Fig. 5, lanes 5–7). These observations suggest that EGS-mediated inhibition of PR expression does not affect the replication of viral DNA but blocks the step of packaging the viral genome into the capsids.
Figure 5.
(I) Intracellular level of viral total and encapsidated DNA, as measured by a semiquantitative PCR assay amplifying HCMV IE1 sequence by using human β-actin as internal control. Total DNA (lanes 1–4) or DNase I-treated DNA samples (lanes 5–7) were isolated from cells that were treated with liposome complexes in the absence of EGSs (lanes 1, 2, and 5) or in the presence of PR1 (lanes 3 and 6) and PR2 (lanes 4 and 7) and were infected with HCMV (moi = 0.5). Twice the amount of the DNA templates was used in the PCR shown in lane 1 as that in lane 2. The amplification by PCR was within the linear range. The radiolabeled PCR products were separated in 4% nondenaturing polyacrylamide gels and quantitated with a STORM840 phosphorimager. (II) Electron micrographs of thin sections of HCMV-infected cells that were treated with liposome complexes in the absence (Upper, No EGS) and presence of PR1 (Lower, PR1). (Magnification, ×40,000.)
In the fourth set of experiments, we examined thin sections of infected cells by electron microscopy. The cells were first treated with EGSs and infected with HCMV, then harvested and fixed with 2.5% glutaraldehyde at 72–96 h postinfection and prepared for electron microscopy. The nuclei of cells that were treated with liposome complexes in the absence of an EGS or in the presence of PR2 or TK1 were found to contain the three expected capsid types: A, B, and C (Fig. 5II Upper; data not shown). A large number of capsids were also found in the nuclei of the PR1-treated cells. However, almost all of these capsids were of the electron-translucent “B capsid-like” (pro-capsid) type (Fig. 5II Lower). The dense-cored capsids (mature, DNA-containing capsid or C capsid) were hardly observed, indicating that viral DNA was not efficiently packaged and capsid maturation was blocked.
Discussion
The EGS-based technology represents an attractive approach for gene inactivation because it utilizes endogenous RNase P to generate highly efficient and specific cleavage of the target RNA. In particular, RNase P is one of the most abundant and active enzymes found in nature because it is responsible for processing of all tRNA molecules, which accounts for 2% of all RNA species within a single cell (9, 10). Moreover, RNase P-mediated cleavage directed by EGSs is highly specific and does not generate “irrelevant cleavage,” which is usually observed with RNase H-mediated cleavage induced by conventional DNA-based oligonucleotides (7, 8, 11). Our study uses exogenous administration of DNA-based EGSs for antiviral applications and demonstrates that RNase P-mediated targeting directed by DNA-based EGS is highly effective in inhibiting HCMV gene expression and growth in cultured cells. We show that these EGS molecules directed human RNase P to cleave the PR mRNA sequence efficiently in vitro. Moreover, we also show that these EGSs were readily delivered in human cell culture. A reduction of about 80–90% in the PR expression was achieved with a functional EGS, PR1, whereas a reduction of less than 10% was observed in cells that were treated with PR2 or TK1. PR2 bound to PR mRNA substrate pr39 in vitro as well as PR1 but contained nucleotide mutations that disrupted RNase P recognition. These results suggest that the overall observed inhibition with PR1 was primarily due to targeted cleavage by RNase P as opposed to the antisense effect or other nonspecific effects of the EGSs.
Our results also suggest that the RNase P-mediated targeting directed by DNA-based EGS is highly specific. The EGSs did not exhibit significant cytotoxicity as cells treated with EGSs are indistinguishable from the cells treated with lipid complexes alone in the absence of the EGSs, in terms of cell growth and viability for 10 days (data not shown). Moreover, the antiviral effect of the EGS treatment (inhibition of viral growth) appears to be due to the reduction of the PR expression. Only the expression of PR mRNA and protein was found to be reduced in cells treated with PR1. We found no significant change in the expression of IE1, US2, UL44, and gH (Table 1). The observed level of inhibition of PR expression correlates with the extent of the reduction of viral DNA encapsidation and growth, and the assembly protein processing. Thus, the EGS is highly specific in inhibiting the expression of its target mRNA.
Delivery of the EGS into the nuclear compartment is essential to the success of the EGS technology because RNase P is exclusively localized in nuclei (10). Our data provide direct evidence that exogenous DNA-based EGSs can be delivered predominantly into the nuclei. The efficient delivery and proper localization of the EGS may be mediated by cellular tRNA-binding proteins, which may interact with the tRNA-like domains of the EGS and target the EGS to the nuclear compartment. Further exploitation of these interactions will facilitate the development of EGSs as novel gene-targeting agents for both basic research and clinical therapeutic applications.
HCMV is a member of the human herpesvirus family, which includes seven other different viruses such as HSV and Epstein–Barr virus (1, 17, 18). The protease is highly conserved among all herpesviruses (19, 20) and is considered an ideal target for anti-herpes therapy because inhibiting PR expression would shut down both the stoichiometric and enzymatic functions of the protease. To develop EGSs as a conventional drug that can be used as an exogenous agent for intracellular delivery in antiviral therapy, EGSs containing modified oligonucleotides are required to increase their stability in vivo (11, 15). It has been recently reported that chemically synthesized RNA-based EGSs with modified nucleotides can be administered exogenously into human cells and inhibit cellular gene expression (11). Our study shows that exogenous administration of DNA-based EGSs is highly effective in inhibiting gene expression and growth of a human virus. Further understanding of how the functional groups in the nucleotides of an EGS interact with human RNase P and the mRNA substrate will lead to the construction of highly active and stable EGSs with either different bases or modifications at these nucleotide positions. Moreover, engineering different designs of EGSs (24, 31) for increasing their targeting activity, as well as developing new means for improving their delivery, are needed to increase the efficacy of the EGSs in vivo. These studies will further facilitate the development of the EGS-based technology for gene-targeting applications in both basic research and clinical therapy, including the studies and treatment of HCMV infections.
Acknowledgments
We thank Annette Meyer for anti-PR and anti-AP antibodies; Hua Zhu for plasmid constructs; Z. Hong Zhou for assistance of electron microscopy studies; and Arash Nassi and Ada Tam for technical assistance. W.D. and P.T. are partially supported by Block Grant Predoctoral Fellowships (University of California, Berkeley). F.L. is a Pew Scholar in Biomedical Sciences and a Scholar of Leukemia and Lymphoma Society. This research was supported by the UC-Berkeley Chancellor's Special Initiative Award and the National Institutes of Health.
Abbreviations
- EGS
external guide sequence
- PR
protease
- HCMV
human cytomegalovirus
- moi
multiplicity of infection
- HSV
herpes simplex virus
- AP
assembly protein
References
- 1.Mocarski E S, Courcelle C T. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 2001. pp. 2629–2674. [Google Scholar]
- 2.Rossi J J. Chem Biol. 1999;6:R33–R37. doi: 10.1016/S1074-5521(99)80001-5. [DOI] [PubMed] [Google Scholar]
- 3.Stein C A, Cheng Y C. Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 4.Mulamba G B, Hu A, Azad R F, Anderson K P, Coen D M. Antimicrob Agents Chemother. 1998;42:971–973. doi: 10.1128/aac.42.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azad R F, Driver V B, Tanaka K, Crooke R M, Anderson K P. Antimicrob Agents Chemother. 1993;37:1945–1954. doi: 10.1128/aac.37.9.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marvick C. J Am Med Assoc. 1998;280:871. [Google Scholar]
- 7.Yuan Y, Hwang E, Altman S. Proc Natl Acad Sci USA. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forster A C, Altman S. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 9.Frank D N, Pace N R. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 10.Altman S, Kirsebom L A. In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 351–380. [Google Scholar]
- 11.Ma M, Benimetskaya L, Lebedeva I, Dignam J, Takle G, Stein C A. Nat Biotechnol. 2000;18:58–61. doi: 10.1038/71924. [DOI] [PubMed] [Google Scholar]
- 12.Guerrier-Takada C, Li Y, Altman S. Proc Natl Acad Sci USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawa D, Wang J, Yuan Y, Liu F. RNA. 1998;4:1397–1406. doi: 10.1017/s1355838298980918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plehn-Dujowich D, Altman S. Proc Natl Acad Sci USA. 1998;95:7327–7332. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma M Y, Jacob-Samuel B, Dignam J C, Pace U, Goldberg A R, George S T. Antisense Nucleic Acid Drug Dev. 1998;8:415–426. doi: 10.1089/oli.1.1998.8.415. [DOI] [PubMed] [Google Scholar]
- 16.Perreault J P, Altman S. J Mol Biol. 1992;226:399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- 17.Roizman B, Knipe D M. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 2001. pp. 2399–2640. [Google Scholar]
- 18.Kieff E, Rickinson A B. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 2001. pp. 2511–2574. [Google Scholar]
- 19.Welch A R, Woods A S, McNally L M, Cotter R J, Gibson W. Proc Natl Acad Sci USA. 1991;88:10792–10796. doi: 10.1073/pnas.88.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Roizman B. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann P J, III, Deckman I, Colonno R J. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preston V G, Rixon F J, McDougall I M, McGregor M, al Kobaisi M F. Virology. 1992;186:87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- 23.Pyle A M, McSwiggen J A, Cech T R. Proc Natl Acad Sci USA. 1990;87:8187–8191. doi: 10.1073/pnas.87.21.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y, Altman S. Science. 1994;263:1269–1273. doi: 10.1126/science.8122108. [DOI] [PubMed] [Google Scholar]
- 25.Trang P, Lee M, Nepomuceno E, Kim J, Zhu H, Liu F. Proc Natl Acad Sci USA. 2000;97:5812–5817. doi: 10.1073/pnas.100101797. . (First Published May 16, 2000; 10.1073/pnas.100101797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Jiang H, Liu F. RNA. 2000;6:571–583. doi: 10.1017/s1355838200992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Altman S. Genes Dev. 1995;9:471–480. doi: 10.1101/gad.9.4.471. [DOI] [PubMed] [Google Scholar]
- 28.Zaug A J, Cech T R. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- 29.Sprinzl M, Dank N, Nock S, Schon A. Nucleic Acids Res. 1991;19,Suppl.:2127–2171. doi: 10.1093/nar/19.suppl.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Cong J P, Shenk T. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner M, Rosa E, Nordstrom J L, Goldberg A R, George S T. RNA. 1998;4:847–855. doi: 10.1017/s1355838298980323. [DOI] [PMC free article] [PubMed] [Google Scholar]