Abstract
Purpose of Review
The metabolic syndrome is a pathological state in which one of the key components is insulin resistance. A wide spectrum of body compartments is involved in its pathophysiology. Genetic and environmental factors such as diet and physical activity are both related to its etiology. Reversible modulation of gene expression without altering the DNA sequence, known as epigenetic modifications, has been shown to drive this complex metabolic cluster of conditions. Here, we aim to examine some of the recent research of specific epigenetically mediated mechanisms and microbiota-induced epigenetic modifications on the development of adipose tissue and obesity, β-cell dysfunction and diabetes, and hepatocytes and non-alcoholic fatty disease.
Recent Findings
DNA methylation patterns and histone modifications have been identified in this context; the integrated analysis of genome, epigenome, and transcriptome is likely to expand our knowledge of epigenetics in health and disease. Epigenetic modifications induced by diet-related microbiota or metabolites possibly contribute to the insulin-resistant state.
Summary
The identification of epigenetic signatures on diabetes and obesity may give us the possibility of developing new interventions, prevention measures, and follow-up strategies.
Keywords: Epigenetics, Microbiota, T2D, Insulin resistance, Adipose tissue, Metabolic syndrome, Obesity, NAFLD
Introduction
Insulin Resistance, the Metabolic Syndrome, Type 2 Diabetes, and Non-alcoholic Fatty Disease
The underlying pathophysiology of the metabolic syndrome remains to be completely elucidated. This syndrome has been called by at least eight different names like metabolic trisyndrome, syndrome X, deadly quartet, or insulin resistance syndrome and has included different features like gout, hyperlipidemia, impaired glucose tolerance, type 2 diabetes (T2D), hypertension, and central adiposity [1]. The Indian surgeon Susruta was one of the first known physicians who in 600 BC linked obesity and diabetes and who used to prescribe exercise to minimize its consequences [2]. Hippocrates, who lived 460–370 BC, wrote that “corpulence is not only a disease itself but the harbinger of others” [3].
A current widely accepted definition of metabolic syndrome proposed by a consensus from the International Diabetes Federation and the American Heart Association/National Heart, Lung, and Blood Institute is a constellation of three or more from five risk factors: abdominal obesity, high triglycerides, low level of high-density lipoprotein cholesterol, high blood pressure, and elevated fasting blood glucose [4].
Insulin resistance (IR) is believed to be a common factor that is strongly correlated to most components of the metabolic syndrome [5]. It is a state in which the target tissues like skeletal muscle, adipose tissue, and the liver respond inadequately to insulin [6].
Points of Attention Reading Epigenetic Studies
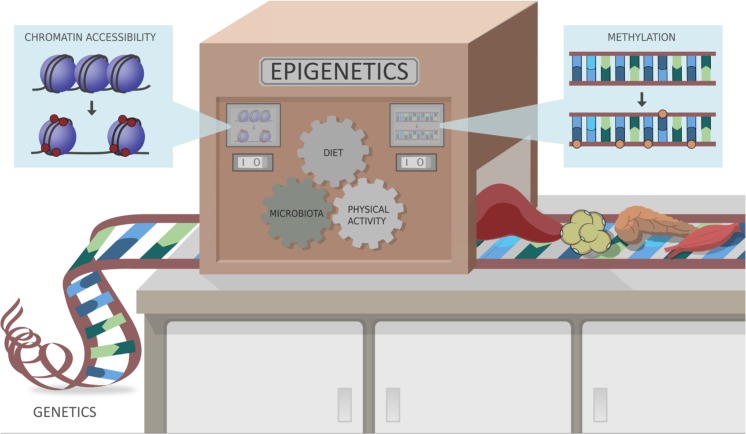
The field of epigenetics studies the contribution of endogenous and exogenous factors (diet, gut microbiota, environment, and medication) on human phenotype changes that do not involve mutation in the DNA sequence, but rather affect transcription (e.g., via acetylation or methylation of histone proteins, or methylation of DNA itself). In this review, we will therefore try to offer a glimpse of the current knowledge of epigenetic factors and their role on the insulin-resistant metabolic state. We will also review gut microbiota and their derived metabolites and their effect on epigenetic markers (Fig. 1).
Fig. 1.
Epigenetic is the collective of heritable changes in phenotype that occur independent of the primary DNA sequence, altering the gene expression. Epigenetic modifications can alter chromatin accessibility, through for example acetylation and methylation of DNA and histones. These modifications can be induced by different agents such as diet, microbiota, metabolites, and physical activity
As mentioned, the field of epigenetics studies the modulation of gene expression and function that occurs without altering the DNA sequence. Epigenetic modifications can be caused by alterations in DNA methylation, modifications to histone proteins, remodeling of chromatin, and RNA-based mechanisms, such as non-coding RNAs [7]. There is some controversy in considering all of these as epigenetic marks, as some believe that a property needed to define a true epigenetic mark is to be able to carry information through cell division, while some argue that some histone modifications and high-order chromatin structure frequently lack this property and therefore their mention as epigenetic elements should be qualified [8]. Modifications of histone tails are, however, considered as critical mechanisms in the activation and repression of gene transcription [9]. The epigenome is the epigenetic information in a cell, comprising DNA methylation, post-translational modifications of histones, and higher-order chromatin structure [10].
In this regard, an important question is how epigenetic information is stored and inherited [11], also known as “epigenetic memory.” D’Urso and Brickner refer to “epigenetic memory” as the propagation of a change in gene expression that can possibly happen through different mechanisms: cellular memory, transcriptional memory, and transgenerational memory [12].
When analyzing epigenetic research, one should not forget that there is cross-talk between these modifications. This combination will probably be determinant of the overall modulation of transcriptional outcome, by activating or repressing gene expression [13]. For example, recently it has been demonstrated that the trimethylation of lysine 36 in histone H3 (by convention, H3K36me3) stimulates acetylation of lysine 16 in histone 4 (H4K16ac) [14]. Some histone modifications are also associated with DNA methylation at CpG islands, CpG-rich regions that are present in the promoters of many genes [9]. A simultaneous analysis of the genome, epigenome, and transcriptome (the set of genes expressed in a given state) [15] enriches the chances of elucidating how an epigenetic marker might influence the expression of a specific gene. When we consider DNA methylation, it is crucial to note that its effect on gene expression varies according to different genomic contexts, such as transcriptional start sites with or without CpG islands, in gene bodies, at regulatory elements, and at repeat sequences, leading to either up- or downregulation of gene expression [16].
Another point of attention while reading epigenetic studies is the question of the use of surrogate tissue, a situation in which the health or condition of an inaccessible “target” tissue (e.g., liver or β-cells) is determined by analyzing an accessible or “surrogate” tissue such as peripheral blood leukocytes [17]. We will highlight this in this paper when we discuss some recent studies that have analyzed genome-wide or specific gene DNA methylation and histone modification in relation to bacterial metabolite-induced epigenetic marks.
Obesity and Epigenetic Modulation
With a rising prevalence especially in Western world countries, obesity results from an interplay between genetic susceptibility, diet, epigenetics, metagenomics, and the environment [18]. To investigate whether blood can be used as a surrogate tissue for adipose tissue, one recent population-based epigenome-wide study collected data, from adipose tissue from the upper outer quadrant of the buttock and blood, in 143 healthy subjects, to try to extend knowledge regarding the similarity between adipose tissue and blood. In general, subcutaneous adipose tissue samples were hypomethylated compared with blood samples. Genes were divided into two groups: concordant genes (with smaller gene methylation variation between the two groups) and discordant (with a higher variation). Gene ontology analysis was performed to elucidate their role in obesity and showed that whereas concordant genes (identified as mainly responsible for maintaining basic cellular functions) had constant gene expression pattern across tissues, discordant genes (critical for tissue-specific biological functions) had distinct epigenetic and transcription patterns [19]. In another study, DNA methylation from peripheral blood leukocytes and adipocytes derived from the upper outer quadrant of the buttock was performed in 106 middle-aged men and women. Although there was no significant association between blood leukocyte DNA methylation and adiposity, adipose tissue DNA methylation profiles were associated with measures of adiposity, including centrally located fat, body fat distribution, and body mass index, reinforcing the concept that tissue-specific DNA methylation patterns influence adipose tissue regulation in human obesity [20].
In this regard, mammals possess two types of adipose tissue: white adipose tissue (WAT) and brown adipose tissue (BAT) [21]. WAT functions like energy stores while BAT is a major site of thermogenesis, thus regulating body temperature and energy expenditure. Beige (brite) adipocytes are the brown adipocytes appearing in WAT [22]. WAT and BAT are contained in the subcutaneous and visceral compartments and are capable of reciprocal reversible transdifferentiation according to physiological needs: the need for thermogenesis induces browning and positive energy balance induces whitening [23]. A study in mice who were exposed to cold has confirmed the notion that rodent inguinal WAT is the adipose depot most prone to browning. An increased transient level of zinc finger (Zic1 mRNA) expression was found during the early browning process. Although overall DNA methylation did not appear to be related to this gene expression, a repressive H3K9me histone mark was found as a possible epigenetic feature involved in the early stages of this white-to-brown differentiation (Table 1) [21]. It is thought that DNA methylation in human subcutaneous adipose tissue and omental visceral adipose tissue from non-obese vs. obese individuals has depot-specific differences. Keller et al. have indeed identified those differences in new candidate genes and in previously known obesity-related genes. Some of the differentially methylated genes were HAND2, HOXC6, PPARG, SORBS2, CD36, and CLDN1. Specifically, the PPARG promoter region was differently methylated in omental visceral adipose tissue with decreased gene expression when compared with subcutaneous adipose tissue. PPARG is a key regulator of adipogenesis and adipocyte differentiation (Table 1) [24].
Table 1.
Examples of epigenetic modifications and microbiome-related modifications in obesity, type 2 diabetes, and NAFLD
| Obesity | Type 2 diabetes | NAFLD | |
|---|---|---|---|
| Epigenetics |
- H3K9me in WAT - Differently methylated obesity-related genes: HAND2, HOXC6, PPARG, SORBS2, CD36, CLDN1 - Differently methylated PPARG promoter region in omental visceral compared with subcutaneous adipose tissue |
- Differently methylated genes: TCF7L2, THADA, KCNQ1, FTO, IRS1 - H3K27me3 modification of myocytes - Metformin indirectly induced hypermethylation of tumor-promoting pathway genes and inhibits cell proliferation |
- Hypermethylation PNPLA3 - Differently methylated MT-DN6 gene |
| Microbiome | Butyrate inhibition of HDAC3 and HDAC1 reduced methylation in obesity-associated genes FTO and MC4R upon probiotic supplementation | Butyrate inhibition of HDAC differently methylated palmitate-treated human islets in genes TCF7L2, GLIS3, HNF1B, and SLC30A8 | Bacterial dysbiosis leading to increased permeability, endotoxemia, and increased pro-inflammatory cytokines |
H3K9me, methylation of lysine 9 on histone 3; WAT, white adipose tissue; HDAC, histone deacetylase; HDAC3, histone deacetylase 3; HDAC1, histone deacetylase 1; MT-ND6, mitochondrially encoded NADH dehydrogenase 6; H3K27me3, trimethylation of lysine 27 on histone 3; PNPLA3, patatin-like phospholipase domain containing 3
Gut Microbiota–Derived Metabolites, Epigenetics, and Obesity
The gut microbiota refers to the microbes that collectively inhabit the intestinal tract of an animal organism, and the microbiome is the collection of all genomes from microbes in an ecosystem [25]. Sender et al. have elegantly estimated the total number of bacteria in the colon, in a 70-kg “reference male,” to be approximately 3.8 × 1013 [26]. The human gut microbiota is mainly composed of four phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria [27]. However, besides bacteria, feces also contain viruses, fungi, and human intestinal epithelial cells, and it is thought that all of these components drive overall microbiome composition [28]. In this regard, metabolites derived from gut microbiota or consumed foods may influence epigenetic mechanisms contributing to human disease including obesity and IR [29]. Butyrate, for example, is a short-chain fatty acid (SCFA) that is the primary fuel source for colon enterocytes, thus influencing gut homeostasis. This SCFA is derived from microbiota and originates from the fermentation of dietary fiber, with Roseburia, Eubacterium hallii, and Faecalibacterium prauznitzii among the most common bacterial strains producing this SCFA [30]. Butyrate also inhibits histone deacetylase 3 (HDAC3), an enzyme that modulates histone acetylation in intestinal epithelial cells thereby affecting metabolic control. As expected, wild-type C57BL6 mice fed with a high-fat diet gained weight and became obese, while HDAC3 knockout mice did not develop obesity despite being on the same high-fat diet, and had less liver fat and smaller adipocytes. In addition, butyrate supplementation inhibits histone deacetylase 1 (HDAC1), leading to downregulation of the inflammatory pathway in rodents with colitis (Table 1). Inhibition of HDAC1 downregulates the IL-6/STAT3/IL-17 pathway and promotes Foxp3 expression leading to an altered Th17/Treg ratio, causing an anti-inflammatory effect [31]. As diet-induced obese mice had significant weight reduction after oral SCFA butyrate administration [32], taken together, these data underscore the potential role of epigenetic modifications in developing an obese phenotype. The question however remains whether beneficial effects on metabolism are also seen in human obesity.
Recently, we have reported a comparative study of oral butyrate supplementation in lean individuals and in subjects with the metabolic syndrome. Intriguingly, oral butyrate treatment improved metabolism in lean individuals, yet we observed no effect in participants with the metabolic syndrome. We had to use the maximum allowed dose of 4 g/day, a limit based on previous literature [33]. This possible “sub-therapeutic dose” not adjusted by weight might explain why a positive effect on insulin sensitivity was only seen in lean subjects. However, it may also be possible that intestinally produced SCFAs are handled differently in the obese insulin-resistant state, to regulate glucose and lipid metabolism [34]. In support of this hypothesis, we have previously shown the beneficial effect on glucose metabolism of fecal microbiota transplantation from lean donors to subjects with the metabolic syndrome [35]. As these changes were associated with alterations in fecal SCFA levels, it is possible that epigenetic mechanisms underlie these beneficial effects, a hypothesis currently under investigation.
Another example of the microbiome and epigenetic interactions was shown by Vähämiko et al., who showed that DNA methylation was reduced in the promoters of the obesity-associated genes FTO and MC4R in women who had probiotic supplementation with Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 and their children during pregnancy (Table 1) [36]. In this regard, early-life (intra-uterine) exposure to these gut microbiota and their diet-derived metabolites might influence the development of the infant’s microbiome post-partum and the long-term regulation of metabolism [37]. Future research focusing on the epigenome of pregnant women and their children and its relationship with the composition of amniotic fluid is needed.
Role of Microbiota-Derived Metabolite Epigenetic Regulation in NAFLD-NASH
Regarding IR and the liver, an association has been observed between IR and non-alcoholic fatty liver disease (NAFLD). NAFLD refers to a wide spectrum of liver damage, ranging from simple steatosis to steatohepatitis, advanced fibrosis, and cirrhosis [38]. NAFLD (including non-alcoholic fatty liver “NAFL” and subsequent non-alcoholic steatohepatitis “NASH”) is regarded as a common complication of IR and T2D in obese subjects [39]. Whether IR precedes or is a consequence of NAFLD is however still a matter of debate [40].
Genetic variation has also been associated with the presence of NAFLD; for example, variation in the patatin-like phospholipase domain containing 3 (PNPLA3) gene is linked to differences in hepatic fat content and susceptibility to NAFLD [41]. A variant of the TM6SF2 gene has also been linked to increased liver fat [42]. Other gene variants have been linked to NAFLD like LYPLAL1, GCKR, APOB, MTTP, LPIN1, DOS2, UCP2, ENPP1, IRS1, IL28B, MERTK, and irisin [7]. Another recent finding was a reduced risk from progression from steatosis to steatohepatitis in patients with a loss-of-function variant in the HSD17B13 gene [43]. Interestingly, DNA methylation seems to regulate PNPLA3 gene expression. For example, the regulatory region of PNPLA3 was hypermethylated in human liver biopsy samples, with PNPLA3 mRNA levels being lower in patients with advanced NAFLD compared with those with mild NAFLD, demonstrating a relationship with the severity of the disease (Table 1) [44].
Moreover, genome-wide DNA methylation analysis study using peripheral blood leukocytes has identified six differentially methylated CpG sites in patients with NAFLD (simple steatosis and NASH) compared with healthy controls. Serum liver enzymes and plasma cholesterol levels were directly correlated with the level of DNA methylation as well as the presence of simple steatosis or NASH [45].
As liver mitochondrial dysfunction seems to play an important role in disease progression, mitochondrial DNA methylation was evaluated in liver biopsies of 23 subjects with simple steatosis, 22 subjects with biopsy-proven NASH, and 18 subjects with near-normal liver histology. An association with histological severity was found in hepatic DNA methylation and the transcriptional activity of the mitochondrially encoded NADH dehydrogenase 6 (MT-ND6), a key protein involved in mitochondrial function (Table 1) [46]. Subsequently, another study showed that genes encoding liver proteins were also differently methylated in patients with advanced NAFLD (fibrosis stage 3–4) compared with those with mild (fibrosis stages 0–2) NAFLD [47]. Moreover, De Mello et al. have shown that differential liver DNA methylation was not only associated with NASH compared with normal liver and simple steatosis but also particularly correlated with fasting plasma insulin, suggesting a relationship with the expression of genes involved in hepatic insulin signaling [48].
Different mechanisms have been studied regarding the potential etiological role of the microbiome and of DNA methylation in NAFLD. Bacterial dysbiosis contributes to lowering the expression of tight junction proteins in the intestinal epithelium, leading to increased permeability, bacterial translocation, and endotoxemia. These endotoxins cause an increase in pro-inflammatory cytokines, IR, and hepatic lipid accumulation (Table 1) [49]. In support of the potential association between altered gut microbiota composition and liver inflammation in NAFLD, Faecalibacterium prauznitzii was inversely correlated with CD45+ and CD163+ in NAFLD, whereas Prevotella was negatively correlated with CD20+ [49].
Epigenetics, Microbiota, and T2DM
Since the first genome-wide association study (GWAS) in T2D in 2007, over 400 genome-wide significant loci have been identified. Most (but not all) are also associated with impaired β-cell function. However, these genetic variants explain only ~ 20% of T2D heritability, in part possibly due to the inclusion of participants of mostly European descent in these studies. Future research on ethnic-specific variants, copy number variants such as small deletions or insertions, and rare functional variants of strong effects may increase our understanding of T2D heritability using large-scale biobanks in diverse populations [6, 50, 51].
Indeed, the association between DNA methylation and T2D has been reported repeatedly over the last years. For example, Dayeh et al. performed a genome-wide DNA methylation analysis of human pancreatic islets harvested from donors with and without T2D. The authors selected differentially methylated genes for functional analysis of insulin and glucagon in vitro using clonal β and α cells in which those genes were silenced or overexpressed. They identified 853 unique genes with differential DNA methylation, which included 17 genes previously identified in GWAS as affecting the risk of T2D, such as TCF7L2, THADA, KCNQ1, FTO, and IRS1 (Table 1). This reinforces the idea that genetic and epigenetic mechanisms may interact to affect pancreatic β-cell function, development of IR, and T2D [52]. Furthermore, in 2017, Volkov et al. expanded their first study, performing their analysis with whole-genome bisulfite sequencing, a method that offers a more complete picture of the human islet methylome in T2D. Interestingly, the authors also integrated their findings of differentially methylated regions with previously published maps of histone modifications and enhancer regions in human islets and found that demethylation regions within the genome were associated with specific histone markers [53••]. This reinforces the importance of cross-talk between genome, transcriptome, and epigenome in islets. Varshney et al. found genetic variants that are linked to disruption of regulatory factor X binding, a family of transcription factors involved in glucose metabolism [54]. Because skeletal muscle is a key regulator of insulin sensitivity [55], it has also been examined in epigenetic research. The notion of a muscular epigenetic memory has been proposed, defined as “the capacity of skeletal muscle to respond differently to environmental stimuli in an adaptive or maladaptive manner if the stimuli have been previously encountered” [56]. Trying to identify intrinsic myocyte properties in T2D a gene-set analysis of histone modifications was performed in muscle biopsies taken from 24 subjects, divided into four groups (normal glucose tolerant or insulin resistant/T2D, both in obese and non-obese). In all groups, a significant histone modification was found in H3K27me3 (trimethylation of lysine 27 on histone 3), with a subsequent downregulation of genes involved in muscle function and upregulation of genes involved in inflammation in T2D (Table 1) [57].
With regard to specific food-derived compounds that could drive these epigenetic changes, the trimethylamine N-oxide (TMAO) pathway is involved in the development of IR by affecting certain epigenetic mechanisms. Trimethylamine is derived from dietary choline (meat, milk, grain, egg and their derived products, composite dishes, and fish) and l-carnitine (red meat and milk) by certain microbiota species and further oxidized to TMAO in the liver. The POUNDS Lost Trial included 510 overweight and obese individuals exposed to a low-calorie diet for two years and found that a decrease in choline and l-carnitine levels was significantly related to weight loss, indicating it might serve as a predictive marker of response to weight-loss treatment [58, 59,60]. The same study documented a significant improvement in IR associated with decreases in choline and l-carnitine [61]. Moreover, non-esterified fatty acids are known to induce IR and thus impair β-cell function [62]. To better understand the functional impact of free fatty acids on β-cell functionality, approximately 1000 islets from 13 human donors were cultured for 48 h with either palmitate or control media, and genome-wide mRNA expression and DNA methylation were assessed. Differences in expressed genes and global DNA methylation levels were found in palmitate-treated human islets, including in T2D-associated genes such as TCF7L2, GLIS3, HNF1B, and SLC30A8 (Table 1) [63]. Finally, we previously mentioned that SCFAs including butyrate could also induce HDAC changes and thus serve as a potential epigenetic biomarker. Consistent with this notion, oral administration of gamma-aminobutyric acid (GABA)–enriched rice bran to obese rats showed an attenuation of metabolic syndrome induced by high-fat diet, which was driven by alterations in fecal SCFA levels with increased levels in butyrate and propionate and decreased levels of acetate. Also, an increase in butyrate-producing bacteria (particularly those that use acetate as a substrate, Anaerostipes and Anaerostipes spp.) was seen, which was subsequently associated with an increased release of GLP-1 [64]. Since plasma GABA levels were increased upon lean donor fecal microbiota transplantation, we postulate that specific dietary compounds can regulate metabolism via GABA derived from specific intestinal bacterial strains [65].
More specifically related to T2D, a recent study using peripheral blood mononuclear cells showed that HDAC3 activity and HDAC3 mRNA levels were positively correlated with IR [66]. Although the direct effect of other diabetes medications on the epigenome is not known, a recent intriguing publication by Zhong et al. revealed that the biguanide T2D drug metformin might affect DNA methylation. Metformin indirectly increases DNA methylation of specific tumor-promoting pathway genes and inhibits cell proliferation (Table 1) [67••].
Dietary Effects of Epigenetic Modulation
Nutrition exerts its effect on epigenetic markers from early life. Good evidence regarding the lifelong impact of such epigenetic programming comes from the Dutch Hunger Winter Families Study. Compared with their non-famine-exposed siblings, subjects who had periconceptional exposure to famine were characterized by lower methylation of the insulin growth factor 2 (IGF2) gene 60 years later [68]. Moreover, the offspring of mothers who had been exposed to famine in utero had increased neonatal adiposity [69]. The underlying pathophysiological mechanisms might be driven by saturated fatty acid overload, which is known to induce distinct epigenetic changes in human adipose tissue. Indeed, DNA methylation can predict weight increase in response to overfeeding in humans [70]. Further evidence of a role for bacterial species in this adiposity phenotype comes from a short-term 12-week low-fat/low-calorie diet intervention in 75 patients with NASH who were treated either with probiotics or placebo. The authors showed that probiotic treatment had a significant decrease in BMI and liver stiffness in patients with NASH, with a concomitant change in the composition of gut microbiota [71].
Conclusions
A crucial characteristic of epigenetic modification is that it is reversible and modifiable, and thus a possible target for therapeutic intervention for IR and T2D [72] either via dietary, microbiota, or pharmacological therapy-based interventions. For example, changes in microbiota or microbiota-derived metabolite production can be a way to achieve some of the desired specific epigenetic modifications and to reverse deleterious insulin-resistant obesity. Moreover, several other dietary small compounds have been shown to target proteins or genetic regulatory regions, altering epigenetic programming of organs involved in metabolism. However, a myriad of challenges must be overcome to perform high-quality research in epigenetics. Multiple factors have to be taken into consideration, such as differences between animals and humans, target tissues and cells, heterogeneity of cell composition, sample size of studies, age, sex, medication use, epigenome coverage, and interplay between the abovementioned epigenetic mechanism [11, 73]. To investigate the causality of epigenetic markers in obesity and IR/T2D, both interventional trials and long-term prospective studies on integromics (combining microbiome, epigenome, transcriptome, and plasma metabolomics) should be done in humans.
Funding Information
Max Nieuwdorp is supported by a ZONMW-VIDI grant 2013 [016.146.327] and a Dutch Heart Foundation CVON IN CONTROL Young Talent Grant 2013. The study reported here was additionally supported by Le Ducq consortium grant 17CVD01 and a Novo Nordisk Foundation GUT-MMM grant 2016.
Compliance with Ethical Standards
Conflict of Interest
Daniela Stols-Gonçalves, Luca Schiliró Tristão, and Peter Henneman declare that they have no conflict of interest. Max Nieuwdorp is on the Scientific Advisory Board of Caelus Pharmaceuticals, The Netherlands.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Pediatric Type 2 and Monogenic Diabetes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Daniela Stols-Gonçalves, Email: d.stolsgoncalves@amc.uva.nl.
Luca Schiliró Tristão, Email: lucatristao@gmail.com.
Peter Henneman, Email: p.henneman@amc.uva.nl.
Max Nieuwdorp, Email: m.nieuwdorp@amc.uva.nl.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Sarafidis PA, Nilsson PM. The metabolic syndrome: a glance at its history. J Hypertens. 2006;24(4):621–626. doi: 10.1097/01.hjh.0000217840.26971.b6. [DOI] [PubMed] [Google Scholar]
- 2.Tipton CM. Susruta of India, an unrecognized contributor to the history of exercise physiology. J Appl Physiol. 2008;104(6):1553–1556. doi: 10.1152/japplphysiol.00925.2007. [DOI] [PubMed] [Google Scholar]
- 3.Haslam DW, James WPT. Lecture 4 obesity. Lancet. 2005;366(9492):1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 4.Alberti KGMM, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; National heart, lung, and blood institute; American heart association; World heart federation International. Circulation. 2009;120(16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 5.Eckel RH, Alberti KGMM, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet (London, England) 2010;375(9710):181–183. doi: 10.1016/S0140-6736(09)61794-3. [DOI] [PubMed] [Google Scholar]
- 6.Kwak SH, Park KS. Review article pathophysiology of type 2 diabetes in Koreans. Endocrinol Metab. 2018;33:9–16. doi: 10.3803/EnM.2018.33.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol European Assoc Study Liver. 2018;68(2):268–279. doi: 10.1016/j.jhep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Spadafora R, Stonestrom AJ. The key role of epigenetics in human disease. N Engl J Med. 2018;379(4):400–401. doi: 10.1056/NEJMc1805989. [DOI] [PubMed] [Google Scholar]
- 9.Riancho J, del Real A, Riancho JA. How to interpret epigenetic association studies: a guide for clinicians. Bonekey Rep. 2016;5(May):797. doi: 10.1038/bonekey.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med. 2018;378(14):1323–1334. doi: 10.1056/NEJMra1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean C. What holds epigenetic memory? Nat Rev Mol Cell Biol Nat Publ Group. 2017;18(3):140. doi: 10.1038/nrm.2017.15. [DOI] [PubMed] [Google Scholar]
- 12.D’Urso A, Brickner JH. Mechanisms of epigenetic memory maintenance. Trends Genet. 2014;30(6):230–236. doi: 10.1016/j.tig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tollefsbol Trygve O. Handbook of Epigenetics. 2017. An Overview of Epigenetics; pp. 1–6. [Google Scholar]
- 14.Li L, Wang Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem. 2017;292(28):11951–11959. doi: 10.1074/jbc.M117.788224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velculescu VE, Velculescu VE, Zhang L, Zhang L, Zhou W, Zhou W, et al. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/S0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet Nat Publ Group. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 17.Rockett JC, Burczynski ME, Fornace AJ, Herrmann PC, Krawetz SA, Dix DJ. Surrogate tissue analysis: monitoring toxicant exposure and health status of inaccessible tissues through the analysis of accessible tissues and cells. Toxicol Appl Pharmacol. 2004;194(2):189–199. doi: 10.1016/j.taap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Pigeyre M, Yazdi FT, Kaur Y, Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin Sci. 2016;130(12):943–986. doi: 10.1042/CS20160136. [DOI] [PubMed] [Google Scholar]
- 19.Huang YT, Chu S, Loucks EB, Lin CL, Eaton CB, Buka SL, Kelsey KT. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigen Taylor Francis. 2016;11(3):227–236. doi: 10.1080/15592294.2016.1146853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agha GE, Houseman A, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol. 2015;44(4):1277–1287. doi: 10.1093/ije/dyu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perugini J, Bordoni L, Venema W, Acciarini S, Cinti S, Gabbianelli R, et al. Zic1 mRNA is transiently upregulated in subcutaneous fat of acutely cold-exposed mice. J Cell Physiol. 2018;234(3):2031–2036. doi: 10.1002/jcp.27301. [DOI] [PubMed] [Google Scholar]
- 22.Giralt M, Villarroya F. White, brown, beige/brite: different adipose cells for different functions? Endocrinology. 2013;154(9):2992–3000. doi: 10.1210/en.2013-1403. [DOI] [PubMed] [Google Scholar]
- 23.Cinti S. Adipose organ development and remodeling. Compr Physiol. 2018;8(4):1357–1431. doi: 10.1002/cphy.c170042. [DOI] [PubMed] [Google Scholar]
- 24.Keller M, Hopp L, Liu X, Wohland T, Rohde K, Cancello R, Klös M, Bacos K, Kern M, Eichelmann F, Dietrich A, Schön MR, Gärtner D, Lohmann T, Dreßler M, Stumvoll M, Kovacs P, DiBlasio AM, Ling C, Binder H, Blüher M, Böttcher Y. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol Metab. 2017;6(1):86–100. doi: 10.1016/j.molmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 26.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and Bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar H, Lund R, Laiho A, Lundelin K, Ley RE, Isolauri E, et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. MBio. 2014;5(6):1–10. doi: 10.1128/mBio.02113-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bojanova DP, Bordenstein SR. Fecal transplants: what is being transferred? PLoS Biol. 2016;14(7):1–12. doi: 10.1371/journal.pbio.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16(5):559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Hartstra AV, Bouter KEC, Bäckhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. 2015;38(1):159–165. doi: 10.2337/dc14-0769. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, Chen X, Tang D, Xu L, Yin Y, Pan Y, Zhou Q, Zhou Y, Yu C. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm Bowel Dis. 2018;24(9):1926–1940. doi: 10.1093/ibd/izy182. [DOI] [PubMed] [Google Scholar]
- 32.Whitt J, Woo V, Lee P, Moncivaiz J, Haberman Y, Denson L, et al. Disruption of epithelial HDAC3 in intestine prevents diet-induced obesity in mice. Gastroenterology. 2018;155(2):501–513. doi: 10.1053/j.gastro.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Sabatino A, Morera R, Ciccocioppo R, Cazzola P, Gotti S, Tinozzi FP, et al. Oral butyrate for mildly to moderately active Crohn’s disease. Aliment Pharmacol Ther. 2005;22(9):789–794. doi: 10.1111/j.1365-2036.2005.02639.x. [DOI] [PubMed] [Google Scholar]
- 34.Bouter K EC, Bakker G J, Levin E., Hartstra A V, Kootte R S, Udayappan S D, Katiraei S., Bahler L., Gilijamse P. W., Tremaroli V., Stahlman M., Holleman F., van Riel N. A. W., Verberne H J, Romijn J A, Dallinga-Thie G M, Serlie M J, Ackermans M T, Kemper E M, van Dijk Willems K., Backhed F., Groen A K, Nieuwdorp M. Differential metabolic effects of oral butyrate treatment in lean versus metabolic syndrome subjects. Clinical and Translational Gastroenterology. 2018;9(5):e155. doi: 10.1038/s41424-018-0025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916.e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 36.Vähämiko S, Laiho A, Lund R, Isolauri E, Salminen S, Laitinen K. The impact of probiotic supplementation during pregnancy on DNA methylation of obesity-related genes in mothers and their children. Eur J Nutr. 2018;0(0):1–11. doi: 10.1007/s00394-017-1601-1. [DOI] [PubMed] [Google Scholar]
- 37.Wise A, Robertson B, Choudhury B, Rautava S, Isolauri E, Salminen S, et al. Infants are exposed to human milk oligosaccharides already in utero. Front Pediatr. 2018;6(4):457–467. doi: 10.3389/fped.2018.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346(16):1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 39.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gastaldelli A. Insulin resistance and reduced metabolic flexibility: cause or consequence of NAFLD? Clin Sci. 2017;131(22):2701–2704. doi: 10.1042/CS20170987. [DOI] [PubMed] [Google Scholar]
- 41.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet Nat Publ Group. 2014;46(4):352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, Liu Y, Kozlitina J, Stender S, Wood GC, Stepanchick AN, Still MD, McCarthy S, O’Dushlaine C, Packer JS, Balasubramanian S, Gosalia N, Esopi D, Kim SY, Mukherjee S, Lopez AE, Fuller ED, Penn J, Chu X, Luo JZ, Mirshahi UL, Carey DJ, Still CD, Feldman MD, Small A, Damrauer SM, Rader DJ, Zambrowicz B, Olson W, Murphy AJ, Borecki IB, Shuldiner AR, Reid JG, Overton JD, Yancopoulos GD, Hobbs HH, Cohen JC, Gottesman O, Teslovich TM, Baras A, Mirshahi T, Gromada J, Dewey FE. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378(12):1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamoto T, Kitamoto A, Ogawa Y, Honda Y, Imajo K, Saito S, et al. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol Eur Assoc Study Liver. 2015;63(2):494–502. doi: 10.1016/j.jhep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 45.Wu J, Zhang R, Shen F, Yang R, Zhou D, Cao H, Chen G, Pan Q, Fan J. Altered DNA methylation sites in peripheral blood leukocytes from patients with simple steatosis and nonalcoholic steatohepatitis (NASH) Med Sci Monit. 2018;24:6946–6967. doi: 10.12659/MSM.909747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pirola CJ, Fernández Gianotti T, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 47.Hotta K, Kitamoto T, Kitamoto A, Ogawa Y, Honda Y, Kessoku T, Yoneda M, Imajo K, Tomeno W, Saito S, Nakajima A. Identification of the genomic region under epigenetic regulation during non-alcoholic fatty liver disease progression. Hepatol Res. 2018;48(3):E320–E334. doi: 10.1111/hepr.12992. [DOI] [PubMed] [Google Scholar]
- 48.de Mello VD, Matte A, Perfilyev A, Männistö V, Rönn T, Nilsson E, Käkelä P, Ling C, Pihlajamäki J. Human liver epigenetic alterations in non-alcoholic steatohepatitis are related to insulin action. Epigenet Taylor Francis. 2017;12(4):287–295. doi: 10.1080/15592294.2017.1294305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwenger KJP, Bolzon CM, Li C, Allard JP. Non-alcoholic fatty liver disease and obesity: the role of the gut bacteria. Eur J Nutr. Springer Berlin Heidelberg; 2018;0(0):0. [DOI] [PubMed]
- 50.Langenberg C, Lotta LA. Genomic insights into the causes of type 2 diabetes. Lancet Elsevier Ltd. 2018;391(10138):2463–2474. doi: 10.1016/S0140-6736(18)31132-2. [DOI] [PubMed] [Google Scholar]
- 51.Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med Springer US. 2018;24(10):1526–1531. doi: 10.1038/s41591-018-0160-1. [DOI] [PubMed] [Google Scholar]
- 52.Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volkov P, Bacos K, Ofori JK, Esguerra JLS, Eliasson L, Rönn T, et al. Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis. Diabetes. 2017;66:1074–1085. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 54.Varshney A, Scott LJ, Welch RP, Erdos MR, Chines PS, Narisu N, et al. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc Natl Acad Sci U S A. 2017;114(9):2301–2306. doi: 10.1073/pnas.1621192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Defronzo RA, Gunnarsson R, Hospital H. Effects of insulin on peripheral and splanchnic glucose metabolism. J Clin Invest. 1985;76(July):149–155. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharples AP, Stewart CE, Seaborne RA. Does skeletal muscle have an ‘epi’-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell. 2016;15(4):603–616. doi: 10.1111/acel.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Väremo L, Henriksen TI, Scheele C, Broholm C, Pedersen M, Uhlén M, Pedersen BK, Nielsen J. Type 2 diabetes and obesity induce similar transcriptional reprogramming in human myocytes. Genome Med. 2017;9:47. doi: 10.1186/s13073-017-0432-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribas GS, Vargas CR, Wajner M. L-carnitine supplementation as a potential antioxidant therapy for inherited neurometabolic disorders. Gene. 2014;533(2):469–476. doi: 10.1016/j.gene.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 59.Vennemann FBC, Ioannidou S, Valsta LM, Dumas C, Ocké MC, Mensink GBM, Lindtner O, Virtanen SM, Tlustos C, D’Addezio L, Mattison I, Dubuisson C, Siksna I, Héraud F. Dietary intake and food sources of choline in European populations. Br J Nutr. 2015;114(12):2046–2055. doi: 10.1017/S0007114515003700. [DOI] [PubMed] [Google Scholar]
- 60.Heianza Y, Sun D, Smith SR, Bray GA, Sacks FM, Qi L. Changes in gut microbiota- related metabolites and longterm successful weight loss in response to weight-loss diets: the POUNDS lost trial. Diabetes Care. 2018;41(3):413–419. doi: 10.2337/dc17-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heianza Y, Sun D, Li X, Didonato JA, Bray GA, Sacks FM, et al. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: the POUNDS lost trial. Gut. 2018;3:1–8. doi: 10.1136/gutjnl-2018-316155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 63.Hall E, Volkov P, Dayeh T, Bacos K, Rönn T, Nitert MD, et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Medicine. 2014;12:103. doi: 10.1186/1741-7015-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Si X, Shang W, Zhou Z, Shui G, Lam SM, Blanchard C, Strappe P. Gamma-aminobutyric acid enriched rice bran diet attenuates insulin resistance and balances energy expenditure via modification of gut microbiota and short-chain fatty acids. J Agric Food Chem. 2018;66(4):881–890. doi: 10.1021/acs.jafc.7b04994. [DOI] [PubMed] [Google Scholar]
- 65.Kootte RS, Levin E, Salojärvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611–619. doi: 10.1016/j.cmet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 66.Sathishkumar C, Prabu P, Balakumar M, Lenin R, Prabhu D, Anjana RM, et al. Augmentation of histone deacetylase 3 (HDAC3) epigenetic signature at the interface of proinflammation and insulin resistance in patients with type 2 diabetes. Clin Epigenetics. 2016;8:125. doi: 10.1186/s13148-016-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong T, Men Y, Lu L, Geng T, Zhou J, Mitsuhashi A, et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene. 2017;36:2345–2354. doi: 10.1038/onc.2016.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Painter R, Osmond C, Gluckman P, Hanson M, Phillips D, Roseboom T. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG An Int J Obstet Gynaecol. 2008;115(10):1243–1249. doi: 10.1111/j.1471-0528.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- 70.Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, Nilsson E, Risérus U, Ling C. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105(4):991–1000. doi: 10.3945/ajcn.116.143164. [DOI] [PubMed] [Google Scholar]
- 71.Manzhalii E, Virchenko O, Falalyeyeva T, Beregova T, Stremmel W. Treatment efficacy of a probiotic preparation for non-alcoholic steatohepatitis: a pilot trial. J Dig Dis. 2017;18(12):698–703. doi: 10.1111/1751-2980.12561. [DOI] [PubMed] [Google Scholar]
- 72.Kwak SH, Park KS. Recent progress in genetic and epigenetic research on type 2 diabetes. Exp Mol Med Nat Publ Group. 2016;48(3):e220–e228. doi: 10.1038/emm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heijmans BT, Mill J. Commentary: the seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41(1):74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]