Abstract
Calibration and reproducibility of quantitative 18F-FDG PET measures are essential for adopting integral 18F-FDG PET/CT biomarkers and response measures in multicenter clinical trials. We implemented a multicenter qualification process using National Institute of Standards and Technology–traceable reference sources for scanners and dose calibrators, and similar patient and imaging protocols. We then assessed SUV in patient test–retest studies. Methods: Five 18F-FDG PET/CT scanners from 4 institutions (2 in a National Cancer Institute–designated Comprehensive Cancer Center, 3 in a community-based network) were qualified for study use. Patients were scanned twice within 15 d, on the same scanner (n = 10); different but same model scanners within an institution (n = 2); or different model scanners at different institutions (n = 11). SUVmax was recorded for lesions, and SUVmean for normal liver uptake. Linear mixed models with random intercept were fitted to evaluate test–retest differences in multiple lesions per patient and to estimate the concordance correlation coefficient. Bland–Altman plots and repeatability coefficients were also produced. Results: In total, 162 lesions (82 bone, 80 soft tissue) were assessed in patients with breast cancer (n = 17) or other cancers (n = 6). Repeat scans within the same institution, using the same scanner or 2 scanners of the same model, had an average difference in SUVmax of 8% (95% confidence interval, 6%–10%). For test–retest on different scanners at different sites, the average difference in lesion SUVmax was 18% (95% confidence interval, 13%–24%). Normal liver uptake (SUVmean) showed an average difference of 5% (95% confidence interval, 3%–10%) for the same scanner model or institution and 6% (95% confidence interval, 3%–11%) for different scanners from different institutions. Protocol adherence was good; the median difference in injection-to-acquisition time was 2 min (range, 0–11 min). Test–retest SUVmax variability was not explained by available information on protocol deviations or patient or lesion characteristics. Conclusion: 18F-FDG PET/CT scanner qualification and calibration can yield highly reproducible test–retest tumor SUV measurements. Our data support use of different qualified scanners of the same model for serial studies. Test–retest differences from different scanner models were greater; more resolution-dependent harmonization of scanner protocols and reconstruction algorithms may be capable of reducing these differences to values closer to same-scanner results.
Keywords: 18F-FDG PET/CT, test–retest, SUV, reproducibility, quantitative imaging
Quantitative 18F-FDG PET/CT can measure molecular changes at multiple tumor sites and has been used to evaluate early response to cancer therapy (1). Biologic variability, such as body weight, glucose levels, and lesion location, is a fundamental source of 18F-FDG SUVmax quantitation error that cannot be controlled. However, other sources of variability related to patient preparation, image acquisition, and scanner calibration can be controlled and minimized. Previous same-scanner test–retest studies have achieved average variability of 10%–12% (2,3). Scanner qualification (4) and standardization of patient preparation and imaging protocols (5,6) may reduce measurement error. Consistency in scanner protocol parameters such as uptake time, image reconstruction, and scanner maintenance may limit machine error to less than 10% (7–9), but inconsistent or nonoptimized protocols can add error ranging from 18% to more than 40% (7,10,11). In addition, deviations from standards are common even under the scrutiny of a test–retest study (12–14). Measurement error and bias in quantitative PET measures will influence sample size and other study characteristics (15–18).
Published patient test–retest studies have used the same scanner (2,3,13,14), or scanners at the same institution from the same manufacturer (19); guidelines for using 18F-FDG PET/CT to assess response to therapy in multicenter trials strongly recommend using the same scanner for serial measurements (6,20). Allowing serial measurements from different PET/CT scanners would remove a barrier to accrual. For example, a second pretreatment scan could be avoided if a diagnostic scan from a community site could be used as the baseline scan for a phase I study (where intensive monitoring requires treatment at an academic site). However, allowing serial scans from different sites would require prospective multicenter validation of 18F-FDG uptake quantification. We have described a rigorous qualification process using National Institute of Standards and Technology–traceable reference sources for scanners and dose calibrators (21). This study assesses differences in SUV in tumors and in normal liver from test–retest studies scanning patients with the same or different scanners or sites uniformly calibrated and following a similar imaging protocol. We hypothesize that this approach will yield acceptable levels of test–retest precision in 18F-FDG uptake measures.
MATERIALS AND METHODS
Multicenter Consortium and PET/CT Scanner Qualification
Five scanners were used from within the University of Washington Medical Center/Seattle Cancer Care Alliance network: 2 GE Healthcare Discovery STE PET/CT scanners (“same model”); and a Philips Gemini TF 64, a Siemens Biograph 6 and a Siemens Biograph 20 mCT at network sites. Scanner characteristics are listed in Table 1. Before patient scans, sites underwent qualification. Scanner and dose calibrator performance were assessed with repeat measurements of National Institute of Standards and Technology–traceable, long-lived reference sources (68Ge, half-life of 271 d) (21,22). In each round of measurements, a cylindric scanner source (phantom) was scanned using a clinical whole-body protocol, and a smaller source was measured in the dose calibrator using 18F-FDG settings. Performance measurements were completed every 3 mo and submitted for assessment of signal bias. Scanners were considered qualified after 3 successive rounds of measurements showed stable bias (< ∼5% variation). Details of 68Ge/68Ga PET dose calibrator and scanner cross-calibration kit and scan results are reported elsewhere (21).
TABLE 1.
Scanning Protocol Characteristics
| Characteristic (PET emission images) | Discovery STE (both) | Gemini TF 64 | Biograph 20 mCT | Biograph 6 |
| N | 17 3D, 18 2D | 6 | 2 | 3 |
| Slice thickness (mm) | 3.27 | 4 | 5 | 5 |
| Pixel size (mm) | 5.47 | 4 | 4.07 | 4.06 |
| Pixel volume (cm3) | 0.098 | 0.064 | 0.083 | 0.083 |
| Reconstruction diameter (mm) | 700 | 576 | 815 | 683 |
| Array size (pixels) | 128 × 128 | 144 × 144 | 200 × 200 | 168 × 168 |
| Bed-position duration (min) | 5, 7 | 4 | 2, 3.5 | 3 |
| Average coverage (total cm/total min) | 2.1, 2.4 | 2.3 | 2.8, 4.9 | 4.0, 4.1 |
| Reconstruction method | OSEM 3D/2D | BLOB-OS-TOF | PSF+TOF 2i21s | PSF, 3i24s |
| Scatter correction method | Model-based | SS-SIMUL | Model-based | Model-based |
3D = 3-dimensional; 2D = 2-dimensional; OSEM = ordered-subset expectation maximization; TOF = time-of-flight; PSF = point-spread function; 2i21s = 2 iterations, 21 subsets; 3i24s = 3 iterations, 24 subsets; SS-SIMUL = single-scatter simulation.
All scans were in inferior-to-superior direction.
In addition to scanner qualification, a nuclear medicine technologist traveled to each site to observe patient preparation protocols. Sites agreed to adhere to clinical protocol guidelines (similar to the eventual Uniform Protocols for Imaging in Clinical Trials [UPICT] protocol (6)) for parameters that might affect SUV bias, such as time between injection and image acquisition, patient fasting requirements, and injected dose (Supplemental Table 1; supplemental materials are available at http://jnm.snmjournals.org).
Patient Eligibility
Patients with pathologically confirmed solid malignancies who were undergoing an 18F-FDG PET/CT scan for tumor staging or restaging were eligible for the study. Patients were required to have either no cancer treatment at the time of imaging or chronic treatment that had not changed for at least 3 mo. Study enrollment and informed consent were required for the second scan, since the first scan was clinically indicated. This study was approved by the institutional review committee for imaging at all network sites, and all patients in the study signed an informed consent form.
18F-FDG Imaging
Two 18F-FDG PET/CT scans were scheduled on 2 separate days within 2 wk. The location of the second scan was dependent on scanner availability and the patient’s willingness to travel to another site. 18F-FDG dose (259–407 MBq recommended) was measured in a dose calibrator. The injection syringe and intravenous catheter were measured for residual activity after removal. The emission scan was started at 1 h ± 10 min after injection.
Image Analysis
A single certified nuclear medicine physician reviewed each image set, recording anatomic location, SUVmax, and slice location of the SUVmax pixel for each lesion. A second nuclear medicine radiologist verified each lesion location. Discrepancies (surgical inflammation; additional lesions to report) were resolved by consensus before data analysis. Cubic regions of interest of 3 × 3 pixels were drawn over the portion of each identified lesion with the most uptake on 3 consecutive slices (for measuring tumor SULpeak). Up to 25 lesions were analyzed, selecting the most 18F-FDG–avid. A spheric region of interest (3-cm diameter) drawn on the liver (right lobe) assessed SUVmean in normal soft tissue (1).
Statistical Analysis
For each region of interest, both difference in uptake (Eq. 1) and percentage uptake difference (Eq. 2) between 18F-FDG scans were calculated. For test–retest at different institutions, difference scores are positive if the community-based network scanner SUV is higher. Difference in log(SUVmax) (Eq. 3) was used to calculate the repeatability coefficient (RC) as previously reported, for the most 18F-FDG–avid lesion at the first scan and for the average SUVmax of up to 7 lesions (13,14), using the 7 most 18F-FDG–avid lesions. The RC was also calculated, accommodating multiple lesions per patient in the analysis using a variance estimate (sum of between-subject and within-subject variance (23)) from a linear mixed-effects regression model of dRC with random intercept.
| Eq. 1 |
| Eq. 2 |
| Eq. 3 |
Three groups were of interest: group A was patients studied on the same scanner, group B was patients studied on different scanners of the same model within the same institution, and group C was patients studied on different scanner models at different institutions. Because only 2 patients were in group B, we anticipated combining groups for statistical comparisons.
This study addresses reproducibility across different scanners, where an average bias of zero is not assumed. Therefore, the primary analysis emphasizes Bland–Altman limits of agreement (centered around average difference) rather than the RC (centered around zero). For testing group differences, |D| was selected as the primary endpoint to facilitate interpretation as absolute percentage difference (11). Linear mixed-effects regression models were fitted to measure associations between test–retest difference (|D|) and scanning group, patient-level, and lesion-level characteristics. A common offset (random intercept) accommodated multiple lesions per patient, and deletion diagnostics checked that primary results were not unduly influenced by data from any individual patient. The dependent variable was log-transformed to satisfy linearity assumptions (Eq. 4).
| Eq. 4 |
Log-transformed absolute percentage difference was also used to evaluate the concordance correlation coefficient, a measure of agreement encompassing both bias and variability (24). When directionality as well as magnitude was part of the relationship between outcome and predictor (as for differences in uptake time), difference in log(SUVmax) (Eq. 3) was the dependent variable. Statistical analyses used SAS/STAT software, version 9.4 (SAS Institute, Inc.) (25).
RESULTS
Twenty-three patients were included (20 female, 3 male) (Table 2), of 26 patients enrolled from 2012 to 2015. Two excluded patients had no 18F-FDG PET/CT–evaluable lesions (1 patient with no lesions, 1 with diffuse uptake only); the third withdrew before the repeat scan because of distress over the clinical scan results. Most patients had breast cancer, but patients with other cancers were also enrolled. The median time between scans was 9 d (range, 1–15 d). Ten patients were studied in the same scanner or site, whereas 13 were studied in different scanners or sites (2 within the same institution, 11 at different institutions with different scanner manufacturers and technologists). One site did not maintain scanner qualification and was disqualified from enrolling additional patients. The same institution injected 3 patients with greater than the protocol-specified maximum of 407 MBq. Another site allowed study entry for a patient with 182 mg/dL blood glucose (175 mg/dL protocol-specified maximum).
TABLE 2.
Patient Characteristics
| Characteristic | Same site/scanner (n = 10) | Same institution, different scanner (n = 2) | Different site/scanner (n = 11) | All patients (n = 23) |
| Age (y) | 53.5 (32–67) | 58 (45–71) | 66 (43–76) | 60 (32–76) |
| Body mass index at scan 1 (kg/m2) | 28.3 (18.3–37.6) | 38.6 (31.4–45.7) | 28.4 (22.5–43.2) | 28.4 (18.3–45.7) |
| Time between scans (d) | 8 (2–15) | 7 (1–13) | 10 (7–14) | 9 (1–15) |
| Lesions* (n) | 5 (1–9) | 17 (9–25) | 5 (1–17) | 5 (1–25) |
| Sex (n) | ||||
| Male | – | – | 3 | 3 (13%) |
| Female | 10 | 2 | 8 | 20 (87%) |
| Diagnosis (n) | ||||
| Breast cancer | 10 | 2 | 6 | 18 (78%) |
| Other† | – | – | 5 | 5 (22%) |
| Ongoing treatment between scans (n) | ||||
| None | 2 | – | 5 | 7 (30%) |
| Bisphosphonates or biologic only | 1 | 1 | 1 | 3 (13%) |
| Endocrine therapy‡ | 4 | – | 3 | 7 (30%) |
| Chemotherapy§ | 3 | 1 | 2 | 6 (26%) |
| PET/CT scanner (n) | ||||
| Discovery STE (both) | 10 | 2 | – | 12 (52%) |
| Ingenuity TF | – | – | 6 | 6 (26%) |
| Biograph 6 | – | – | 3 | 3 (13%) |
| Biograph 20 mCT | – | – | 2 | 2 (9%) |
All identified, but ≤25 lesions/patient used in analysis.
1 each: colorectal, head/neck, stage IV lung, stage III melanoma, neuroendocrine/Merkel cell cancer.
2 also bisphosphonates; 3 also biologic.
4 also biologic; 1 also endocrine. Biologics: erlotinib, trastuzumab, everolimus, pertuzumab, denosumab, ado-trastuzumab emtansine. Cytotoxic agents: capecitabine, cyclophosphamide, doxorubicin.
Continuous data are expressed as median and range.
Scan and lesion characteristics are summarized in Supplemental Table 2 for 162 lesions (82 bone, 80 soft tissue). The average injected dose was approximately 370 MBq (10 mCi). Uptake time ranged from 54 to 70 min and did not differ by more than 11 min between scans. Mean glucose level was 93 mg/dL for the first scan and 94 mg/dL for the second scan (overall range, 78–182 mg/dL). The average weight was 76.2 kg (range, 49–133.2 kg). Two of the 3 greatest differences were from the 2 patients recruited from 1 community site; the weights at that site were lower (by 4.8 and 2.6 kg) than measurements from the academic site.
Lesion SUVmax Difference
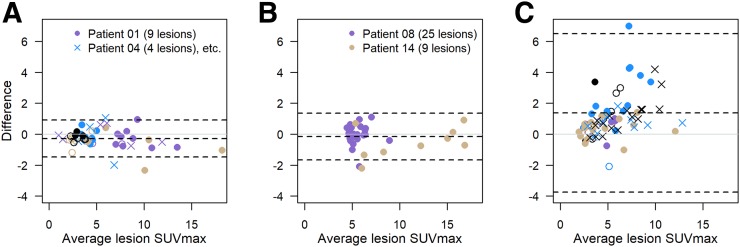
Median percentage uptake difference in SUVmax (Eq. 2) was 11.9% (range, 0.1%–97.0%), and median difference in SUVmax units was 0.6 (range, 0.0–19.1). Figure 1 shows Bland–Altman plots for SUVmax for the 2 scans, separately for 10 patients with repeat scans using the same scanner (panel A), 2 patients imaged with different scanners of the same model (panel B), and 11 patients imaged on 2 different scanners at 2 different sites (academic and network site, panel C). SUVmax for the 162 lesions ranged from 1.0 to 28.8 (average for the repeated scans). Test–retest agreement appears to be better for the same scanner model (panels A and B) than for different models at different sites (panel C).
FIGURE 1.
Bland–Altman plots of difference in SUVmax vs. average SUVmax: 10 patients (51 lesions) with repeat scans using same scanner (A); 2 patients (34 lesions) using different scanners from same academic institution (B); and 11 patients (77 lesions) using different scanners from different sites (C). Within each panel, plotting character/color is same for multiple lesions in single patient. Dashed lines = average difference and 95% limits of agreement. The 2 lesions from melanoma patient (SUVmax, 38.3 and 25.0 on first scan and 19.2 and 16.4 on second scan) are not shown in C but contribute to limits-of-agreement calculations.
Although the median SUVmax was almost 1 unit lower for the same scanner condition (A) than for different scanners (B and C) (Supplemental Table 2), lesions with low 18F-FDG avidity (<3), medium avidity (3–7), and high avidity (>7) were present in both conditions. However, the 2 patients in panel B had no lesions with an SUVmax of less than 4.4. Most (73%) of the absolute differences were less than 1 SUVmax unit. Fourteen of 23 patients (61%) did not have any lesions with an SUVmax difference of 1 unit or more.
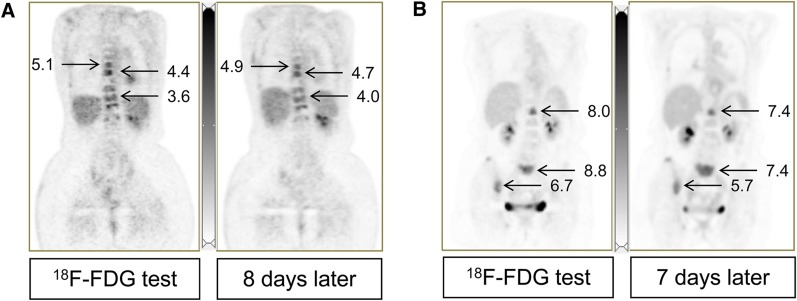
Figure 2 shows image examples: 1 patient with 9 bone lesions studied twice in the same scanner model, and another with 17 mixed bone and soft-tissue lesions studied in a Discovery STE and a Biograph 20 mCT.
FIGURE 2.
(A) Coronal images from 60-y-old woman with stage IV ductal breast carcinoma (blue circles in Fig. 1A, same scanner). SUVmax for 9 evaluable lesions ranged from 3.4 to 5.1 (average, 4.0) for first scan and from 3.1 to 4.9 (average, 4.2) for second scan. Percentage difference was −16% to +16% (average, 3.9%); SUV unit difference was −0.62 to +0.64 (average, 0.15). (B) A 73-y-old woman with stage IV mixed ductal/lobular breast carcinoma (yellow circles in Fig. 1C, different institutions). SUVmax for 17 evaluable lesions was 2.0–12.2 (average, 4.8) for first scan and 1.9–12.0 (average, 4.4) for second scan. Percentage difference was −24% to +25% (average, −7.1%); SUV unit difference was −1.4 to +1.0 (average, −0.39). Normal liver SUVmean was 2.5 (A) and 2.6 (B) in both scans.
Predictors of Test–Retest Differences in Lesion SUVmax
Mixed-effects models are summarized in Table 3. Model 1 shows a fitted linear mixed-effects model for the 3 scanning scenarios (Fig. 1), suggesting that the 2 patients scanned on different scanners of the same model can be combined with the same-model patients for further analysis. This analysis (model 2) finds an average difference in SUVmax of 8% (95% confidence interval, 6%–10%) for test–retest studies on the same scanner model at the same institution, and an average difference of 18% (13%–24%) when the test–retest scans are performed at different qualified sites and on different scanner models. The overall concordance correlation coefficient was 0.91 (95% bootstrap confidence interval, 0.85–0.94), 0.97 for the same site (0.95–0.98), and 0.84 for different sites (0.74–0.90).
TABLE 3.
Linear Mixed-Effects Models (Linear Regression for Liver)
| Model | Fitted % difference in repeat scans | 95% confidence interval |
| Model 1 (SUVmax)* | ||
| A. Same scanner (n = 10) | 8% | 6%–11% |
| B. Same institution, different scanner (n = 2) | 6% | 3%–11% |
| C. Different institution and scanner (n = 11) | 18% | 13%–24% |
| Model 2 (SUVmax)† | ||
| Same scanner or institution | 8% | 6%–10% |
| Different institution and scanner | 18% | 13%–24% |
| Model 3 (liver SUVmean)‡ | ||
| Same scanner or institution | 5% | 3%–10% |
| Different scanner and institution | 6% | 3%–11% |
C > A (P = 0.0015), C > B (P = 0.003), A and B not different on average (P = 0.66) (Tukey–Kramer adjustment for pairwise comparisons).
P < 0.001, Wald test.
P = 0.85, Wald test (n = 23).
Group differences are back-transformed from log(absolute percentage difference + 1); n = 162 tumors in 23 patients.
The model 2 estimates shown in Table 3 were robust to sensitivity analysis, such as removing the melanoma patient’s 2 tumors that had an extremely high SUVmax. They were also similar for SULpeak (Supplemental Fig. 1).
Exploratory subgroup analyses examining patient and scanner factors are summarized in Supplemental Table 3. Controlling for scanning site, bone lesions had test–retest reproducibility at least as good as for soft-tissue lesions. Other patient and scanner factors did not appear to affect the magnitude of test–retest differences.
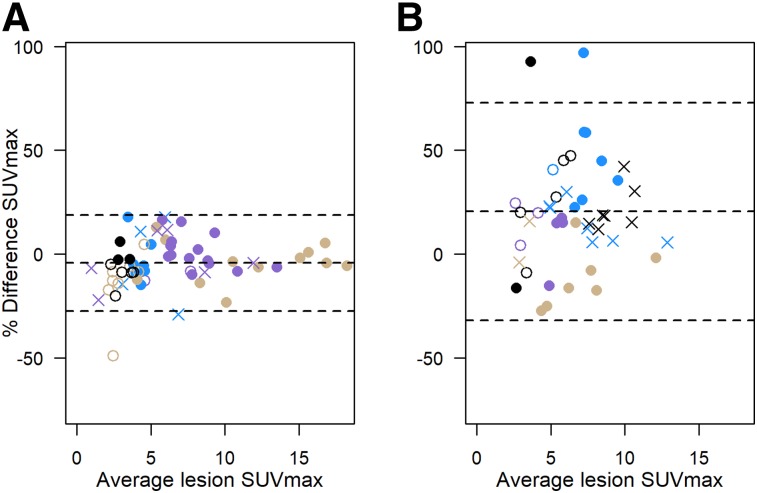
Figure 3 shows Bland–Altman plots for (signed) percentage difference in SUVmax. The magnitude of test–retest variability and differences between same-model and different-model conditions are similar to the results shown in Table 3 and Figure 1: percentage SUVmax differences were generally lower for lesions in patient studies in the same scanner than for those on 2 different scanner models. Estimated 95% RCs and coefficient of variation (from log-transformed SUVmax, as previously published (13,14)) are summarized in Supplemental Table 4 and shown graphically in Supplemental Figure 2, along with the Quantitative Imaging Biomarkers Alliance (QIBA) profile SUVmax 95% limits of same-scanner repeatability (14,20).
FIGURE 3.
Percentage difference in SUVmax vs. average SUVmax: 12 patients (85 lesions) with repeat scans using same scanner or different scanners from same unit (combined data from Figs. 1A and 1B) (A); 11 patients (77 lesions) using different scanners from different sites (B). Plotting character/color identifies multiple lesions in single patient, as for Figure 1. Dashed lines = average percentage difference and 95% limits of agreement.
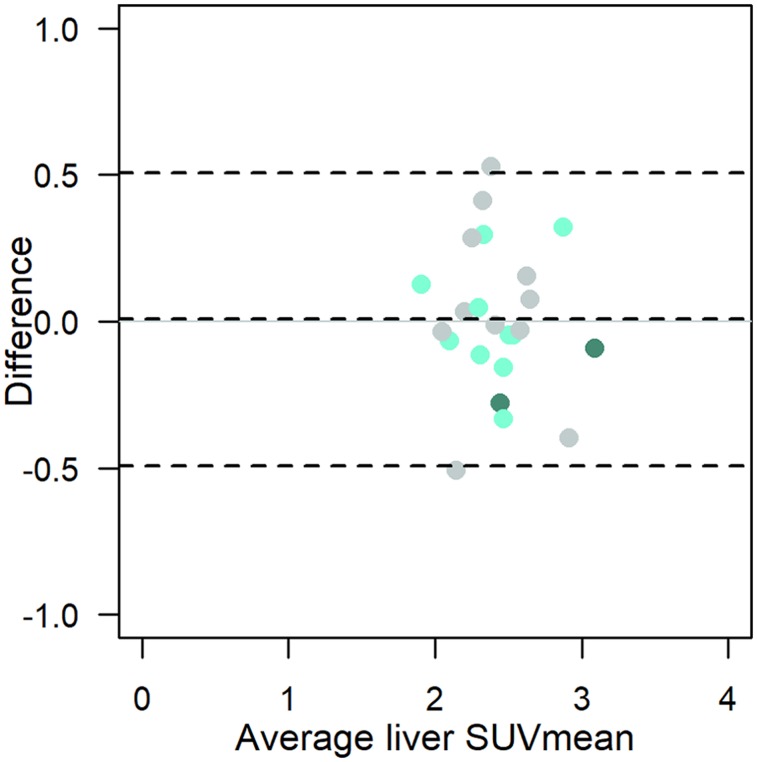
Liver SUVmean
Mean liver uptake (Fig. 4) was consistent, with little between-patient variation around the average SUVmean of 2.4, and differences within 0.5 units for repeat within-patient scans. Linear regression (with log-transformed absolute value of percentage difference, as above for the lesion-level analysis) found the average percentage difference to be similar, 5%–6% for both the same scanner or site and different sites (Table 3). A linear mixed-effects model controlling for site did not support an association between magnitude of percentage difference in liver SUVmean and lesion SUVmax (P = 0.12, with higher liver test–retest differences predicting slightly lower tumor test–retest).
FIGURE 4.
Bland–Altman plot of liver SUVmean (n = 23). Light-green circles = same scanner; dark-green circles = different scanners from same site; gray circles = different scanner models from different sites; dashed lines = average difference and 95% limits of agreement.
DISCUSSION
After qualification including calibration with a common reference object, SUVmax was highly reproducible for 10 breast cancer patients with test–retest studies on the same scanner and for 2 breast cancer patients scanned on different scanners of the same model (with shared service personnel and imaging protocols). The estimated within-subject coefficient of variation of 9% (Supplemental Table 4) was lower than the average of 11% for other same-scanner test–retest studies in oncology patients (Table 3 in Lodge (11)). In contrast, 11 patients with repeat scans on different scanner models showed a within-subject coefficient of variation of 22%, with observation of both bias (each lesion with higher SUVmax on one scan than the other) and variability (different lesions with higher and lower SUVmax between scans for the same patient) (Fig. 1). The 95% RC of (−21%, 26%) for same-model test–retest is within the (−28%, 39%) QIBA 18F-FDG PET/CT profile limits for single-center studies using the same scanner (20), whereas the 95% RC of (−42%, 73%) for different models does not appear to meet the QIBA profile standards (Supplemental Fig. 2). No patient, lesion, or scanning protocol features clearly predicted test–retest variability, in part because of rigorous control of factors such as uptake time.
The SUV in a normal region of liver is a standard method to assess the validity of tumor 18F-FDG uptake estimates (1). Our average liver SUVmean of 2.4 with an average absolute difference of 0.19 (SD, 0.16) was similar to the results of previous studies (26,27). We did not adjust SUVmax for uptake time or for normal liver or blood uptake but would expect results similar to those of a recent study (28), in which lack of variability in uptake times and blood uptake diminished the impact of adjustment algorithms in improving test–retest agreement of tumor uptake measurements.
Uptake in large, uniform regions such as the liver is not affected by resolution effects such as partial-volume errors. Scanner calibration would be expected to minimize test–retest variance even between different makes and models of scanners, as we observed: test–retest variability in normal liver uptake appeared similar in the same and different scanner models (Fig. 4), unlike the greater variability in lesion uptake measured in different scanners (Fig. 1). Most lesions do not have uniform 18F-FDG uptake over a large area, so they are known to have size-dependent resolution effects (29). Variation in size-dependent bias for different types of scanners motivates the ongoing work in harmonization of reconstruction algorithms and other scanner features in multicenter trials (30,31).
Although the true activity is known for the National Institute of Standards and Technology–traceable sources, measured PET image activity for the epoxy calibration phantom may be biased by manufacturer-dependent CT-based attenuation and scatter correction effects. The calibration phantoms could therefore not evaluate absolute scanner calibration; however, their spatial uniformity and temporal stability still permitted precise monitoring of scanner calibration consistency (21). By monitoring every 3 mo, we could evaluate the effects of periodic scanner recalibration and identify any long-term drifts in scanner bias. Low variability in test–retest liver uptake measures, regardless of manufacturer, supports the efficacy of our scanner calibration.
A limitation of this study is that it had a relatively small sample size and that the same site/institution group included only breast cancer patients. In addition, because no patients had both 18F-FDG PET/CT scans outside the academic institution we could not assess same-scanner test–retest agreement at network sites. An exploratory subgroup analysis did not identify lesion or scanning protocol factors with strong effects on test–retest SUVmax agreement (Supplemental Table 3). However, these analyses were not powered to assess lesion location (e.g., propensity for motion artifacts or subcutaneous nodules with compromised attenuation) or lesion type (e.g., high-uptake, inflammatory melanoma lesions). Some scanner characteristics, such as a voxel size greater than 4 mm, did not fall within the eventual UPICT standard (Supplemental Table 1). Finally, we did not control spatial resolution between scanners, nor did we attempt to quantify the effect of variable noise or image reconstruction parameters on SUVmax. A higher average SUVmax for community scanners that mostly used ultra-high-definition reconstruction (Fig. 1C; Table 1) is consistent with studies with multiple reconstructions of the same images (32).
CONCLUSION
This study shows that 18F-FDG PET/CT scanner calibration and qualification, with consistent imaging protocols, can yield highly reproducible SUV measurements; test–retest error for the same scanner or same scanner model (within the same institution) is similar to or lower than estimates in prior test–retest studies. If our findings for different scanners of the same model are confirmed, clinical trials that apply these qualification, calibration, and quality control criteria could increase patient recruitment by allowing serial measurements from similar scanner models at different sites. Additionally, reducing test–retest variation reduces the required number of patients for a given study power (15,18). Before considering use of different scanner models for serial measurements, though, future studies should incorporate modern guidelines such as the UPICT protocol (6) or the QIBA profile (20) and explore harmonization techniques proposed to overcome inherent differences in acquisition and reconstruction methods (31).
DISCLOSURE
This work was supported by NIH grants U01CA148131, U01CA190254, R50CA211270, P30CA015704, P30CA047904 (Biostatistics), R01CA169072, and NCI-SAIC-24XS036-004. No other potential conflict of interest relevant to this article was reported.
Supplementary Material
Acknowledgments
We thank the Seattle Cancer Care Alliance Network, as well as the physicians, technologists, and physicists from the University of Washington Medical Center, the Seattle Cancer Care Alliance, Harborview Medical Center, Tacoma General, and Skagit Valley Medical Center who helped make this study possible. We also thank Nuclear Medicine Technologists Lisa Dunnwald, Amy Quinn, and Patrick Clark for network site visits; Rebecca Christopfel for administrative assistance; and the patient volunteers. We also acknowledge helpful discussions with QIBA and National Cancer Institute Quantitative Imaging Network (QIN) members.
REFERENCES
- 1.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med. 2009;50(suppl 1):122S–150S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber WA, Ziegler SI, Thodtmann R, Hanauske AR, Schwaiger M. Reproducibility of metabolic measurements in malignant tumors using FDG PET. J Nucl Med. 1999;40:1771–1777. [PubMed] [Google Scholar]
- 3.Nahmias C, Wahl LM. Reproducibility of standardized uptake value measurements determined by 18F-FDG PET in malignant tumors. J Nucl Med. 2008;49:1804–1808. [DOI] [PubMed] [Google Scholar]
- 4.Scheuermann JS, Saffer JR, Karp JS, Levering AM, Siegel BA. Qualification of PET scanners for use in multicenter cancer clinical trials: the American College of Radiology Imaging Network experience. J Nucl Med. 2009;50:1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boellaard R, Delgado-Bolton R, Oyen WJ, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015;42:328–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham MM, Wahl RL, Hoffman JM, et al. Summary of the UPICT protocol for 18F-FDG PET/CT imaging in oncology clinical trials. J Nucl Med. 2015;56:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nucl Med. 2009;50(suppl 1):11S–20S. [DOI] [PubMed] [Google Scholar]
- 8.Doot RK, Scheuermann JS, Christian PE, Karp JS, Kinahan PE. Instrumentation factors affecting variance and bias of quantifying tracer uptake with PET/CT. Med Phys. 2010;37:6035–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahey FH, Kinahan PE, Doot RK, Kocak M, Thurston H, Poussaint TY. Variability in PET quantitation within a multicenter consortium. Med Phys. 2010;37:3660–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Langen AJ, Vincent A, Velasquez LM, et al. Repeatability of 18F-FDG uptake measurements in tumors: a metaanalysis. J Nucl Med. 2012;53:701–708. [DOI] [PubMed] [Google Scholar]
- 11.Lodge MA. Repeatability of SUV in oncologic 18F-FDG PET. J Nucl Med. 2017;58:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V, Nath K, Berman CG, et al. Variance of SUVs for FDG-PET/CT is greater in clinical practice than under ideal study settings. Clin Nucl Med. 2013;38:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velasquez LM, Boellaard R, Kollia G, et al. Repeatability of 18F-FDG PET in a multicenter phase I study of patients with advanced gastrointestinal malignancies. J Nucl Med. 2009;50:1646–1654. [DOI] [PubMed] [Google Scholar]
- 14.Weber WA, Gatsonis CA, Mozley PD, et al. Repeatability of 18F-FDG PET/CT in advanced non-small cell lung cancer: prospective assessment in 2 multicenter trials. J Nucl Med. 2015;56:1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurland BF, Doot RK, Linden HM, Mankoff DA, Kinahan PE. Multicenter trials using 18F-fluorodeoxyglucose (FDG) PET to predict chemotherapy response: effects of differential measurement error and bias on power calculations for unselected and enrichment designs. Clin Trials. 2013;10:886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doot RK, Pierce LA, II, Byrd D, Elston B, Allberg KC, Kinahan PE. Biases in multicenter longitudinal PET standardized uptake value measurements. Transl Oncol. 2014;7:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurland BF, Muzi M, Peterson LM, et al. Multicenter clinical trials using 18F-FDG PET to measure early response to oncologic therapy: effects of injection-to-acquisition time variability on required sample size. J Nucl Med. 2016;57:226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doot RK, Kurland BF, Kinahan PE, Mankoff DA. Design considerations for using PET as a response measure in single site and multicenter clinical trials. Acad Radiol. 2012;19:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamibayashi T, Tsuchida T, Demura Y, et al. Reproducibility of semi-quantitative parameters in FDG-PET using two different PET scanners: influence of attenuation correction method and examination interval. Mol Imaging Biol. 2008;10:162–166. [DOI] [PubMed] [Google Scholar]
- 20.QIBA Profile: FDG-PET/CT as an imaging biomarker measuring response to cancer therapy—version 1.13. QIBA website. http://qibawiki.rsna.org/images/1/1f/QIBA_FDG-PET_Profile_v113.pdf. Published November 18, 2016. Accessed December 27, 2018. [DOI] [PMC free article] [PubMed]
- 21.Byrd DW, Doot RK, Allberg KC, et al. Evaluation of cross-calibrated 68Ge/68Ga phantoms for assessing PET/CT measurement bias in oncology imaging for single- and multicenter trials. Tomography. 2016;2:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman BE, Cessna JT. Development of a traceable calibration methodology for solid 68Ge/68Ga sources used as a calibration surrogate for 18F in radionuclide activity calibrators. J Nucl Med. 2010;51:448–453. [DOI] [PubMed] [Google Scholar]
- 23.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–160. [DOI] [PubMed] [Google Scholar]
- 24.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 25.Crawford SB, Kosinski AS, Lin HM, Williamson JM, Barnhart HX. Computer programs for the concordance correlation coefficient. Comput Methods Programs Biomed. 2007;88:62–74. [DOI] [PubMed] [Google Scholar]
- 26.Boktor RR, Walker G, Stacey R, Gledhill S, Pitman AG. Reference range for intrapatient variability in blood-pool and liver SUV for 18F-FDG PET. J Nucl Med. 2013;54:677–682. [DOI] [PubMed] [Google Scholar]
- 27.Viner M, Mercier G, Hao F, Malladi A, Subramaniam RM. Liver SULmean at FDG PET/CT: interreader agreement and impact of placement of volume of interest. Radiology. 2013;267:596–601. [DOI] [PubMed] [Google Scholar]
- 28.Hofheinz F, Apostolova I, Oehme L, Kotzerke J, van den Hoff J. Test-retest variability in lesion SUV and lesion SUR in 18F-FDG PET: an analysis of data from two prospective multicenter trials. J Nucl Med. 2017;58:1770–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber WA. Use of PET for monitoring cancer therapy and for predicting outcome. J Nucl Med. 2005;46:983–995. [PubMed] [Google Scholar]
- 30.Lasnon C, Salomon T, Desmonts C, et al. Generating harmonized SUV within the EANM EARL accreditation program: software approach versus EARL-compliant reconstruction. Ann Nucl Med. 2017;31:125–134. [DOI] [PubMed] [Google Scholar]
- 31.Aide N, Lasnon C, Veit-Haibach P, Sera T, Sattler B, Boellaard R. EANM/EARL harmonization strategies in PET quantification: from daily practice to multicentre oncological studies. Eur J Nucl Med Mol Imaging. 2017;44:17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhnert G, Boellaard R, Sterzer S, et al. Impact of PET/CT image reconstruction methods and liver uptake normalization strategies on quantitative image analysis. Eur J Nucl Med Mol Imaging. 2016;43:249–258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.