Abstract
Enhancers are regulatory DNA elements that can activate their genomic targets over a large distance. The mechanism of enhancer action over large distance is unknown. Activation of the glnAp2 promoter by NtrC-dependent enhancer in Escherichia coli was analyzed by using a purified system supporting multiple-round transcription in vitro. The data suggest that DNA supercoiling is an essential requirement for enhancer action over a large distance (2,500 bp) but not over a short distance (110 bp). DNA supercoiling facilitates functional enhancer–promoter communication over a large distance, probably by bringing the enhancer and promoter into close proximity.
Keywords: enhancers|Escherichia coli|transcription in vitro|NtrC activator|glnAp2 gene
Transcriptional enhancers are relatively short (30–200 bp) DNA sequences usually composed of several binding sites for activator protein(s). The landmark of enhancers is their ability to communicate with promoters and activate genes over a large distance (up to 60 kb; see ref. 1 for a review). Prokaryotic and eukaryotic enhancers share several key properties such as ability to activate transcription over a large distance, tight coupling of DNA melting with ATP hydrolysis and high stability of the initiation complexes (see refs. 2 and 3 for recent reviews). In contrast to eukaryotic enhancers, prokaryotic enhancers can activate transcription over a large distance in vitro. This ability allows detailed analysis of the mechanism of enhancer action impossible in the case of eukaryotic enhancers.
The mechanism of enhancer action over a large distance is unknown. It is most likely that proteins bound at the enhancer and promoter directly interact such that intervening DNA forms a loop (see refs. 4 and 5 for reviews). The main problem for communication over a large distance is low concentration of the DNA regions in the vicinity of each other (see ref. 4 for a review). Measurements of local concentration of linear DNA ends in the vicinity of each other suggest that it is relatively high (10−7 M) only when the distance between DNA ends is 100–900 bp (6). When distance between the DNA ends is increased to 3 kb, their local concentration is decreased by an order of magnitude (6). Thus, it is remarkable that DNA regions positioned far away from each other (such as an enhancer and a promoter) can communicate efficiently.
Several models have been proposed to explain the mechanism of enhancer action over a large distance. One class of models suggests that initial communication of an enhancer with a promoter leads to formation of a stable DNA–protein complex in the vicinity of the promoter. This stable complex may facilitate subsequent rounds of transcription serving as a “memory” of initial enhancer–promoter interaction (see ref. 1 for a review). Alternatively, the average distance between promoter and enhancer could be considerably decreased if the intervening DNA is supercoiled (sc) or bent (see ref. 4 for a review). It has also been shown that sequence-dependent superhelical DNA inserts can facilitate enhancer action (7).
The mechanism of action of the NtrC-dependent, σ54-dependent transcriptional enhancer has been intensely studied by using the glnAp2 promoter of Escherichia coli as a model (see refs. 8 and 9 for reviews). The enhancer for this promoter, consisting of two high-affinity NtrC-binding sites, is centered at position −110 bp relative to the site of transcript initiation, but can strongly activate transcription when positioned up to at least 15 kb away in vivo (10) and up to at least 0.9 kb in vitro (11). It can also activate transcription in trans (12). NtrC is an activator that binds to the enhancer, and, when phosphorylated by NtrB protein kinase, forms higher order homo-oligomers and is capable of activating the transcription of the glnAp2 gene (13–16). Phosphorylation of NtrC also activates its ATPase activity, which is required to stimulate conversion from the closed initiation complex (RPc) to the open initiation complex (RPo) (13–16). Active enhancer-bound NtrC∼P interacts with the RPc at the promoter (17–19). During enhancer–promoter interaction, intervening DNA is looped out (20, 21). Interaction of NtrC∼P with the σ54 subunit of the holoenzyme probably drives the transition from the closed into the open complex (22–24).
In this work, a multiple-round in vitro transcription assay was used to investigate the role of DNA structure during enhancer action. It was found that when the enhancer is separated from the promoter by a large distance (2.5 kb), enhancer–promoter communication constitutes the rate-limiting step for initiation of transcription on relaxed plasmid DNA. DNA supercoiling greatly facilitates enhancer–promoter communication, probably by bringing enhancer and promoter into close proximity. In contrast, when the enhancer–promoter spacing is shorter (110 bp), the enhancer works equally well on relaxed and supercoiled DNA. Thus, DNA supercoiling is the critical factor for enhancer action over a large distance.
Materials and Methods
Purified Proteins.
All proteins and protein complexes were purified according to protocols previously described for core RNA polymerase (25), σ54 (25), NtrC (26), and NtrB (27). Isolated proteins have been analyzed in SDS/10% PAGE, and their purity was over 95% as estimated by Coomassie blue and silver staining. Purified calf thymus topoisomerase I (topo I) was kindly provided by H.-Y. Wu (Wayne State University).
DNA Templates.
For detailed description of all plasmid templates used in the experiments, see Fig. 1. sc DNA templates were purified by using Qiagen plasmid purification kit (Chatsworth, CA). Plasmids pTH8, pLR100, and pAN6 were described (11). The plasmid pLY10, which has T7 terminator from pTH8 and the 2.5-kb enhancer–promoter spacer from pLR73 (10), was constructed as follows. The primers 5′-ATTAGGTACCTGGAGGAAACACCTGATGGC and 5′-GAGCAGATCTTACTGAGAGTGCACCATATGCGG were used in PCR reaction with the pTH8 plasmid as a template. The 547-bp product was digested to obtain 537-bp KpnI–BglII DNA fragment. The primers 5′-CGCATTAAAGCTTGCCGAACACC and 5′-GTCTGGTACCAGCACCATGTCGGACTCG were used in PCR with the pLR73 plasmid as a template. The 1,026-bp product was then digested to obtain a 1,013-bp HindIII–KpnI DNA fragment. The plasmid pLR73 was digested with HindIII and BglII to obtain a 6,099-bp vector. The three DNA fragments (537, 1,013, and 6,099 bp) were ligated to obtain the pLY10 plasmid.
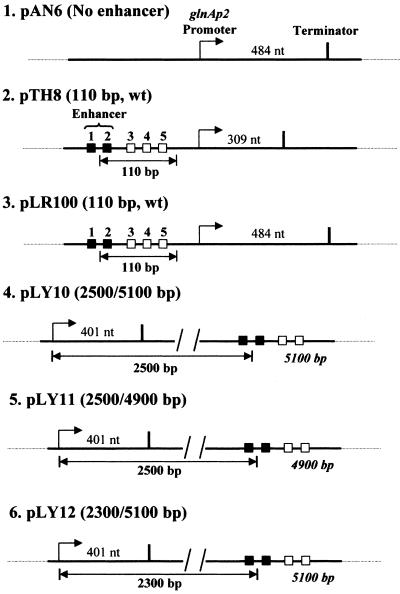
Figure 1.
Plasmid templates for analysis of the mechanism of glnAp2 promoter activation by NtrC-dependent enhancer. Strong and weak NtrC-binding sites are indicated by closed and open boxes, respectively. Under our experimental conditions, only the strong sites are occupied and contribute to the enhancer activity (11). The pTH8 and pLR100 plasmids (3.6 and 3.3 kb in size, respectively) have 110-bp wild type (wt) enhancer–promoter spacing. The pLY10, pLY11, and pLY12 plasmids (7.6, 7.4, and 7.4 kb in size, respectively) have 2.5 kb, 2.5 kb, and 2.3 kb enhancer–promoter distances (the enhancer–promoter distances on the other side of the enhancer are indicated in italics). pAN6 has no strong NtrC-binding sites. The transcripts were terminated at the T7 terminator positioned at different distances downstream of the promoter. The lengths of the transcripts are indicated.
The plasmid pLY11 was constructed as follows: pLY10 was digested with NsiI to excise the 221-bp DNA fragment, then the larger fragment was gel purified and self-ligated to obtain the pLY11 plasmid. The plasmid pLY12 was constructed as follows. The primers 5′-GAAGATCTTCGCAAACCCGACCACCAACTCTTATAAGC and 5′-TCTAATGCCTGAGGCAAAGTTGTG were used in PCR with the pLY10 plasmid as a template. The 1,307-bp product was then digested to obtain 1,295-bp BglII–Bsu36I DNA fragment. The plasmid pLY10 was digested with BglII and Bsu36I to obtain 6,126-bp vector. The DNA fragments 1,295 and 6,126 bp were ligated to obtain the pLY12 plasmid.
In Vitro Transcription.
In vitro transcription was optimized for maximal utilization of promoter using sc pTH8 plasmid as a template. The buffer for the transcription assay contained 50 mM Tris⋅OAc (pH 8.0), 100 mM KOAc, 8 mM Mg(OAc)2, 27 mM NH4OAc, 0.7% PEG 8000, and 0.2 mM DTT. The transcription reactions were conducted in 50-μl aliquots at 2.8 nM DNA. The final (saturating) concentrations of proteins were 500 nM core RNA polymerase, 1,000 nM σ54, 120 nM NtrC, and 400 nM NtrB. The reaction mix was incubated for 15 min at 37°C to form the RPc. When indicated, topo I was added to the reaction to 0.1 units/μl final concentration and incubated at 37°C for 30 min to completely relax sc DNA from linking number difference σ = −0.07 to σ = 0. ATP was added to the reaction to 0.5 mM (single-round) or 2 mM (multiple-round) final concentration, and the reaction was incubated at 37°C for variable time to form RPo. Then NTPs (final concentration 80 μM), 2.5 μCi of [α-32P]UTP (1 Ci = 37 GBq), RNase inhibitor (final concentration 0.2 units/μl) and heparin (final concentration 80 μg per ml, single-round transcription only) were added to the reaction to start transcription. The mixture was incubated at 37°C for various times, and 50 μl of stop solution (200 μg/ml sheared DNA, 40 mM EDTA) was added to terminate the reaction. End-labeled 227-bp DNA fragments (1–2 μl) were added to the mixture as a loading control. The samples were extracted with 100 μl of phenol/chloroform (1:1), precipitated with ethanol, washed with 70% ethanol, and dissolved in 100% formamide. The samples were separated in denaturing 8% urea-containing PAGE, dried, and analyzed by using a PhosphorImager (Molecular Dynamics).
Results
DNA Supercoiling Is Required Only for Enhancer Action Over a Large Distance.
To analyze the mechanism of enhancer action over a large distance, plasmids with different enhancer–promoter spacing or entirely lacking the enhancer (Fig. 1) were used in the experiments described below. It has been shown that transcription from the glnAp2 promoter in our experimental system in vitro requires the presence of the core polymerase, promoter DNA, NtrC, NtrB, and σ54, and occurs in an ATP- and enhancer-dependent manner (data not shown). The reconstituted system supports multiple (up to four) rounds of transcription and enhancer action over at least 2.5 kb (data not shown).
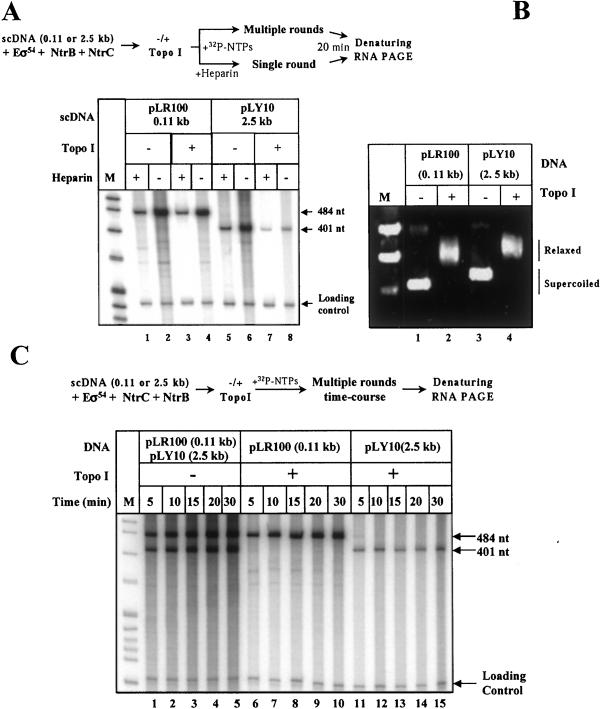
The possibility that DNA supercoiling plays a role during enhancer action was investigated first. The efficiencies of transcription of sc and relaxed templates having enhancer–promoter spacing of 110 bp or 2,500 bp were compared in a single- and multiple-round transcription assays (Fig. 2A). DNA supercoiling had relatively small effect on transcription of the construct having short (0.11 kb) enhancer–promoter spacing (≈2- to 4-fold stimulation in different experiments; Fig. 2A, lane 2 vs. lane 4), in agreement with published data (11). In contrast, the increase of enhancer–promoter spacing from 0.11 to 2.5 kb resulted in a significant (up to 15-fold in the multiple-round assay) decrease of transcription efficiency on relaxed DNA (Fig. 2A, lane 4 vs. lane 8), but had almost no effect on the transcription of sc DNA (Fig. 2A, lane 2 vs. lane 6). Relaxation of both templates (having 0.11- and 2.5-kb enhancer–promoter spacing) by topo I was complete (Fig. 2B). Thus, the observed difference in transcription efficiency was apparently caused by distance between the promoter and enhancer. The observation that DNA relaxation has a small effect on transcription of the template with 0.11-kb spacing also eliminates the possibility of nonspecific inhibition of transcription by incubation in the presence topo I.
Figure 2.
DNA supercoiling is required for efficient enhancer-dependent activation of the glnAp2 promoter over large (2.5 kb) but not short (0.11 kb) distance. (A) DNA supercoiling is required for activation of transcription over 2.5-kb distance. Analysis of labeled transcripts in denaturing PAGE. The experimental approach is outlined at the top. sc plasmid templates having 0.11-kb or 2.5-kb enhancer–promoter spacing were incubated in the presence or in the absence of topo I (+/− topo I); incubation with topo I converts DNA into the relaxed state. Then transcription was conducted under single- (Heparin +) or multiple-round (Heparin−) conditions; all nucleotides were added simultaneously. M, end-labeled pBR322-MspI digest, which was used in all experiments that included analysis of labeled RNA or DNA. The loading control (167/227-bp end-labeled DNA fragment) was added to the reaction mixtures immediately after terminating the reaction. (B) DNA is completely relaxed after incubation with the topo I. Analysis of aliquots of the plasmid templates from the experiment shown in A after incubation with or without topo I and separation in an agarose gel (staining with ethidium bromide). (C) Transcription on relaxed DNA templates occurs at much lower rate only when the enhancer is far from the promoter. Analysis of labeled transcripts in denaturing PAGE. The experimental strategy for comparison of the rates of multiple-round transcription of relaxed (Topo I +) and supercoiled (Topo I −) plasmids with 0.11- or 2.5-kb enhancer–promoter spacing is outlined at the top. Transcription was started by adding complete mixture of nucleotides.
The difference in the efficiency of transcription between relaxed and sc 2.5-kb templates was less pronounced in the single-round assay (≈6-fold) as compared with the multiple-round assay (≈15-fold; Fig. 2A, lanes 5 and 7 vs. 6 and 8), suggesting that the difference was accumulated during multiple-round transcription. This suggestion was confirmed in the experiments comparing time courses of the multiple-round transcription (Fig. 2C). The time courses of transcription of sc and relaxed templates with 0.11-kb spacing and sc template with 2.5-kb spacing were very similar (Fig. 2C, lanes 1–5 and 6–10). In contrast, the rate of accumulation of transcripts from the relaxed template with 2.5-kb spacing was much lower (compare lanes 1–10 and 11–15). One possibility consistent with these observations is that DNA supercoiling facilitates enhancer–promoter communication.
DNA Supercoiling Greatly Facilitates Enhancer–Promoter Communication Over a Large Distance.
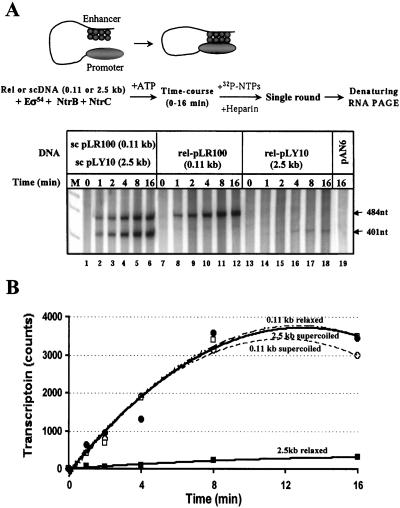
To directly compare the rates of functional enhancer–promoter communication over 0.11 and 2.5 kb, all proteins present in the transcription reaction were preincubated with the templates in the absence of ATP. In this case, both RNA polymerase and NtrC bind to the promoter and the enhancer, respectively, but cannot functionally communicate with each other. Thus, in this case the polymerase cannot form the open complex (17–19). Functional enhancer–promoter communication was initiated by adding ATP to the reaction mixtures, and the rate of transcript initiation was measured in a single-round transcription assay on both relaxed and sc templates (Fig. 3 A and B). sc templates were incubated in the same reaction mixture, but the relaxed templates had to be analyzed in different tubes because relaxed (but not sc) templates strongly competed, which complicated interpretation of the experiment (data not shown). The rates of communication on both sc templates and on relaxed plasmid having short 0.11-kb spacing were nearly identical and high (Fig. 3 A, lanes 1–12, and B). In contrast, the rate on the relaxed plasmid with 2.5-kb spacing was dramatically (≈50-fold) lower (Fig. 3 A, lanes 13–18, and B). Transcription depended on the presence of the enhancer because no transcript was detected from the pAN6 plasmid, which does not contain the enhancer (Fig. 3A, lane 19). In combination with the data on the efficiencies of multiple-round transcription (Fig. 2C), the data suggest that the functional enhancer–promoter interaction is the rate-limiting step in transcription initiation on the relaxed template with large (2.5 kb) promoter–enhancer spacing, and that DNA supercoiling helps to overcome this rate-limiting step. This conclusion was further supported by analysis of time course of escape of the RNA polymerase from the glnAp2 promoter on the relaxed plasmid with 2.5-kb enhancer–promoter spacing after preformation of the open complexes. The experiment suggested that escape is not the rate-limiting step during transcription initiation (data not shown).
Figure 3.
DNA supercoiling strongly facilitates enhancer–promoter communication over a large distance. (A) Time courses of enhancer–promoter communication on relaxed and sc plasmids with 0.11- or 2.5-kb enhancer–promoter spacing. Analysis of labeled transcripts in denaturing PAGE. The experimental strategy for comparison of the rates of enhancer–promoter communication on relaxed (Topo I +) and sc (Topo I −) plasmids with 0.11- or 2.5-kb enhancer–promoter spacing is outlined at the top. Sc plasmids were incubated in the presence or in the absence of topo I to obtain relaxed or supercoiled products, respectively. Then the templates were incubated in the presence of all components of the reaction except ATP to allow binding of the polymerase and NtrC. ATP was added to the reaction to allow functional enhancer–promoter communication, and the rate of promoter opening was measured by using a single-round transcription assay. The control pAN6 plasmid does not contain the enhancer. (B) The rate of enhancer–promoter communication is much lower when enhancer–promoter spacing is large (2.5 kb) and DNA is relaxed. Quantitative analysis of the data shown in A. The intensities of the bands containing 484-nt transcripts were analyzed by using a PhosphorImager. Transcription is saturated after three or four rounds, probably because ATP pool is depleted by NtrC.
It has been shown that NtrC can stimulate binding of purified σ54 subunit to “fork junction” (partially single-stranded) promoter DNA in the absence of ATP (29), implying that NtrC could possibly communicate with the closed complex even without ATP. However preincubation of all components (core polymerase, promoter DNA, NtrC, NtrB, and σ54) for different times without ATP did not change the rate of transcription initiation on either relaxed or sc DNA with 2.5-kb spacing (data not shown). This observation suggests that when double-stranded DNA template and the holoenzyme are used for transcription initiation, enhancer–promoter communication completely depends on the presence of ATP, at least on the relaxed template.
DNA Supercoiling Does Not Induce Precise Alignment of the Enhancer and Promoter.
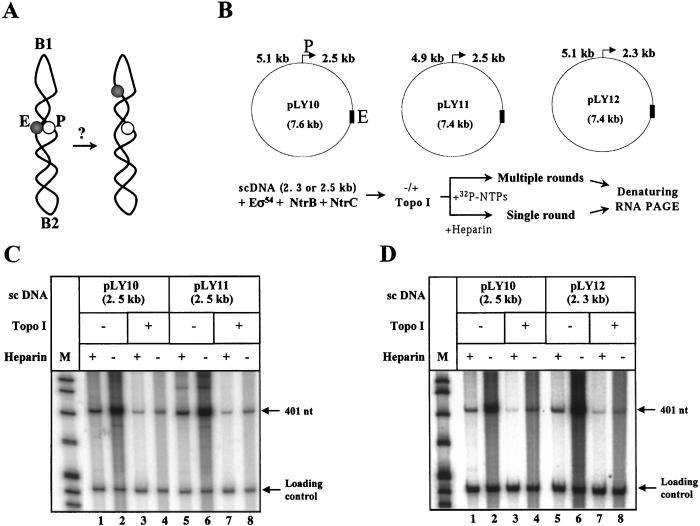
How can DNA supercoiling facilitate functional enhancer–promoter communication over a large distance? High efficiency of transcription on sc DNA could be explained by serendipitous precise alignment of the enhancer and promoter very close to each other in the relatively static sc DNA structure (see Fig. 4A). Thus, the presence of an intrinsically bent DNA region can strongly and uniquely orient sc DNA (30). If an intrinsic DNA bend is positioned between the enhancer and the promoter, it could serendipitously position them close to each other. To have maximal stimulatory effect, the bent DNA region should be positioned precisely in the middle of the DNA region separating the enhancer and promoter on either side of the enhancer to allow close enhancer–promoter alignment.
Figure 4.
Efficient activation of transcription over 2.5 kb is not a result of serendipitous enhancer–promoter juxtaposition. (A) Schematic of the experiment. Enhancer (E) and promoter (P) could be serendipitously positioned next to each other in sc DNA topologically organized by sequence-specific DNA bend(s) (B1 or B2; ref. 30). In this case, one of two asymmetrical deletions of ≈200-bp DNA on either side of the enhancer would be expected to spatially separate enhancer from promoter and decrease activity of the glnAp2 promoter on the sc template. (B) Setup of the experiment. Asymmetric ≈200-bp deletions were introduced into pLY10 on either side of the enhancer to obtain pLY11 and pLY12 plasmids. Design of the experiment is described in Fig. 2A legend and shown at the bottom. (C) Analysis of efficiency of transcription of sc and relaxed pLY11 template. (D) Analysis of efficiency of transcription of sc and relaxed pLY12 template. C and D are labeled as in Fig. 2A. pLY10 was used as a control template in the same experiment.
To investigate this possibility, ≈200-bp asymmetric deletions were introduced on either side of the enhancer (in the 2.5-kb or 5.1-kb DNA separating enhancer and promoter on the pLY10 template; Fig. 4B). If the precise alignment model is correct, one of these deletions would disrupt putative serendipitous enhancer–promoter alignment, increase enhancer–promoter spacing, and result in considerable decrease of transcriptional activity (Fig. 4A). The pLY10 and the pLY11 and pLY12 templates, which have the asymmetric deletions on either side of the enhancer (Fig. 4B), were transcribed with very similar efficiencies in both single- and multiple-round transcription assays (Fig. 4 C and D). These data eliminate the possibility of serendipitous precise alignment of enhancer and promoter and indicate that the activation of the rate of enhancer–promoter communication by DNA supercoiling does not depend on the exact distance between enhancer and promoter.
Discussion
We used the experimental system supporting multiple-round enhancer-dependent transcription from the glnAp2 promoter in vitro and observed that the physiological level of DNA supercoiling was required for enhancer action over a large distance (2.3 or 2.5 kb) but DNA supercoiling was dispensable when enhancer was close to the promoter (0.11 kb). DNA supercoiling greatly facilitates enhancer–promoter communication, the rate-limiting step during transcription initiation on relaxed DNA with large enhancer–promoter spacing. Finally, the effect of DNA supercoiling cannot be explained by serendipitous precise alignment of the enhancer and promoter on sc DNA. Thus, DNA supercoiling is essential for efficient enhancer action over a large distance.
The effect of DNA supercoiling on the efficiency of transcription depends on enhancer–promoter distance. When the promoter was positioned only a short distance (110 bp) from the enhancer, the rate of transcription was high and only moderately (≈2-fold) stimulated by DNA supercoiling. This observation suggests that the local concentrations of the promoter and the enhancer in the vicinity of each other were quite high and could not be further increased by DNA supercoiling. This is consistent with other studies, where measurements of local concentration of linear DNA ends in the vicinity of each other reveal a high concentration (10−7 M) when the distance between DNA ends is 100–900 bp (6). Also, Metropolis–Monte Carlo stimulations suggest that probability of juxtaposition of two DNA sites positioned 100–900 bp from each other is not considerably increased by DNA supercoiling (31).
In contrast, when the promoter and enhancer were separated by a larger distance (2.3 or 2.5 kb), DNA supercoiling increased the rate of functional enhancer–promoter communication (≈50-fold) bringing transcription up to the level characteristic for the constructs with the short spacing. As a result, transcription was strongly activated, essentially irrespective of the distance separating enhancer and promoter (0.11, 2.3, or 2.5 kb), when the template DNA was supercoiled. The observation that the rate of transcription on sc DNA remained very high even when the enhancer–promoter spacing was 2.5 kb suggests that the enhancer probably can activate transcription over much longer distances in vitro. Indeed, NtrC-dependent enhancers can work when separated from the promoter by ≈15 kb in vivo (10).
There are several possible explanations for the effect of DNA supercoiling on the rate of enhancer–promoter communication. Local concentration of DNA sites separated by 2.5 kb is quite low (≈10−8 M; ref. 6) and could be dramatically increased by DNA supercoiling. Metropolis–Monte Carlo simulations of equilibrium DNA conformation suggested that DNA supercoiling increases the probability of juxtaposition of two sites spaced by 3 kb by about two orders of magnitude as compared with relaxed DNA (31). This correlates well with our data suggesting that the rate of communication over 2.5 kb is stimulated by DNA supercoiling ≈50-fold. The good agreement between the theoretical studies and our experimental data suggests that DNA supercoiling probably increases the probability of juxtaposition of the enhancer and promoter. Alternatively, the activator or the holoenzyme could have higher affinity for sc DNA. Thus it has been shown that sequence-dependent superhelical inserts (that do not contain specific NtrC-binding sites) have approximately the same ability to bind NtrC as high affinity specific binding sites for NtrC (7). However, transcription on supercoiled template lacking NtrC binding sites is undetectable (data not shown).
In our previous studies, no σ54 subunit was detected in the elongation complex purified by gel-filtration (data not shown, see also refs. 18 and 32). Moreover, DNase I and KMnO4 footprinting studies did not reveal the presence of any footprint at the glnAp2 promoter after escape of the polymerase from the promoter (data not shown). Our data have also suggested that there is no functional “memory” facilitating enhancer action over a distance during multiple-round transcription (data not shown, see Introduction). Our present data on the role of DNA supercoiling in enhancer action in combination with our previous data on the lack of the “memory” indicate that DNA supercoiling is the primary factor mediating enhancer action over a distance.
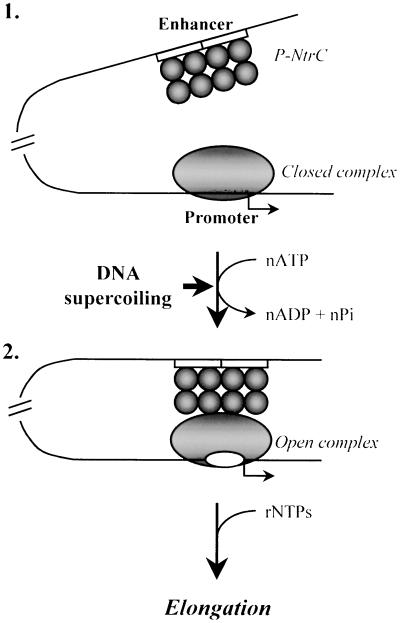
The above data suggest the following mechanism of action of the NtrC-dependent enhancer (Fig. 5). Before activation, nonphosphorylated NtrC is bound at the enhancer and the holoenzyme forms the closed complex at the glnAp2 promoter both in vitro (11, 18, 19) and in vivo (17). Transcription activation starts when NtrC is phosphorylated by NtrB protein (see ref. 33 and references therein; intermediate 1). NtrC∼P interacts with the holoenzyme bound at the promoter such that the intervening DNA forms a loop (intermediate 2; refs. 20 and 21). Enhancer–promoter communication is strongly facilitated by DNA supercoiling when enhancer–promoter distance is 2.3–2.5 kb. Once established, enhancer–promoter interaction greatly stimulates the RPc → RPo transition, the rate-limiting step in the absence of the enhancer (11, 18, 23, 28). In the presence of NTPs, the elongation complex leaves the promoter and the σ54 subunit is displaced into solution, leaving no structural or functional “memory” at the promoter (refs. 18 and 32, and data not shown).
Figure 5.
Mechanism of activation of the glnAp2 promoter by the NtrC-dependent enhancer. (1) Before transcription of NtrC-dependent genes is induced, the Eσ54 holoenzyme forms a closed complex (RPc) at the promoter (localized at the −24 to −12 DNA region) but cannot initiate transcription. NtrC is bound to the enhancer (two 17-bp NtrC-binding sites are indicated by open boxes) but cannot communicate with the promoter. (2) After induction and phosphorylation by NtrB, NtrC forms higher-order homo-oligomers (not shown) and interacts with the holoenzyme, causing looping of the intervening DNA- and ATP-dependent formation of the open complex at the promoter. Enhancer–promoter communication over a distance is greatly facilitated by negative DNA supercoiling. After formation of the open complex is completed, enhancer–promoter interaction is broken and the DNA loop is opened (ref. 21; data not shown). As the RNA polymerase leaves the promoter, the σ54 subunit dissociates into solution (data not shown).
Several features of the prokaryotic Eσ54–NtrC∼P system are remarkably similar to the enhancer-dependent activation of transcription in eukaryotes (see Introduction). At the same time, the lack of experimental systems supporting action of eukaryotic enhancers over a large distance makes direct analysis of distance-dependent aspects of eukaryotic enhancer action in vitro impossible (see ref. 1 for a recent review). Thus, understanding of the mechanism of action of prokaryotic enhancers could be important for better understanding of the mechanism of eukaryotic enhancer action.
Acknowledgments
We thank Dr. H.-Y. Wu for providing purified topo I, Dr. L. Reitzer for providing pLR73, and Drs. D. Clark, M. Kashlev, L. Lutter, R. Needleman, and K. Mizuuchi for valuable discussion and comments on the manuscript. The work was supported in part by the National Institutes of Health Grant GM58650 (to V.M.S.).
Abbreviations
- sc
supercoiled
- topo I
topoisomerase I
- RPc
closed initiation complex
- RPo
open initiation complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Blackwood E M, Kadonaga J T. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 2.Gralla J D. Curr Opin Genet Dev. 1996;6:526–530. doi: 10.1016/s0959-437x(96)80079-7. [DOI] [PubMed] [Google Scholar]
- 3.Buck M, Gallegos M T, Studholme D J, Guo Y, Gralla J D. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rippe K, von Hippel P H, Langowski J. Trends Biochem Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 6.Shore D, Langowski J, Baldwin R L. Proc Natl Acad Sci USA. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahms G, Brahms S, Magasanik B. J Mol Biol. 1995;246:35–42. doi: 10.1016/s0022-2836(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 8.Rombel I, North A, Hwang I, Wyman C, Kustu S. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 9.Studholme D J, Buck M. FEMS Microbiol Lett. 2000;186:1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 10.Reitzer L J, Magasanik B. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 11.Ninfa A J, Reitzer L J, Magasanik B. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 12.Wedel A, Weiss D S, Popham D, Droge P, Kustu S. Science. 1990;248:486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 13.Porter S C, North A K, Wedel A B, Kustu S. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 14.Wedel A, Kustu S. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 15.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 16.Hwang I, Thorgeirsson T, Lee J, Kustu S, Shin Y K. Proc Natl Acad Sci USA. 1999;96:4880–4885. doi: 10.1073/pnas.96.9.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasse-Dwight S, Gralla J D. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popham D L, Szeto D, Keener J, Kustu S. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 19.Buck M, Cannon W. Mol Microbiol. 1992;6:1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- 20.Su W, Porter S, Kustu S, Echols H. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rippe K, Guthold M, von Hippel P H, Bustamante C. J Mol Biol. 1997;270:125–138. doi: 10.1006/jmbi.1997.1079. [DOI] [PubMed] [Google Scholar]
- 22.Wang J T, Syed A, Hsieh M, Gralla J D. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, Wang L, Gralla J D. EMBO J. 1999;18:3736–3745. doi: 10.1093/emboj/18.13.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannon W V, Gallegos M T, Buck M. Nat Struct Biol. 2000;7:594–601. doi: 10.1038/76830. [DOI] [PubMed] [Google Scholar]
- 25.Hunt T P, Magasanik B. Proc Natl Acad Sci USA. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reitzer L J, Magasanik B. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninfa A J, Ueno-Nishio S, Hunt T P, Robustell B, Magasanik B. J Bacteriol. 1986;168:1002–1004. doi: 10.1128/jb.168.2.1002-1004.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 29.Guo Y, Lew C M, Gralla J D. Genes Dev. 2000;14:2242–2255. doi: 10.1101/gad.794800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laundon C H, Griffith J D. Cell. 1988;52:545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- 31.Vologodskii A, Cozzarelli N R. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ninfa A J, Brodsky E, Magasanik B. DNA–Protein Interactions in Transcription. New York: Liss; 1989. pp. 43–52. [Google Scholar]
- 33.Jiang P, Peliska J A, Ninfa A J. Biochemistry. 1998;37:12795–12801. doi: 10.1021/bi9802420. [DOI] [PubMed] [Google Scholar]