P. falciparum malaria can cause multiple disease complications that differ by patient age. Previous studies have attempted to address the roles of parasite adhesion and biomass in disease severity; however, these studies have been limited to single geographical sites, and there is limited understanding of how parasite adhesion and biomass interact to influence disease manifestations. In this meta-analysis, we compared parasite disease determinants in African children and Indian adults. This study demonstrates that parasite biomass and specific subsets of var genes are independently associated with detrimental outcomes in both childhood and adult malaria. We also explored how parasite var adhesion types and biomass play different roles in the development of specific severe malaria pathologies, including childhood cerebral malaria and multiorgan complications in adults. This work represents the largest study to date of the role of both var adhesion types and biomass in severe malaria.
KEYWORDS: cerebral malaria, machine learning, malaria, PfEMP1, Plasmodium falciparum, severe malaria, var gene
ABSTRACT
The clinical presentation of severe Plasmodium falciparum malaria differs between children and adults, but the mechanistic basis for this remains unclear. Contributing factors to disease severity include total parasite biomass and the diverse cytoadhesive properties mediated by the polymorphic var gene parasite ligand family displayed on infected erythrocytes. To explore these factors, we performed a multicohort analysis of the contribution of var expression and parasite biomass to severe malaria in two previously published pediatric cohorts in Tanzania and Malawi and an adult cohort in India. Machine learning analysis revealed independent and complementary roles for var adhesion types and parasite biomass in adult and pediatric severe malaria and showed that similar var profiles, including upregulation of group A and DC8 var, predict severe malaria in adults and children. Among adults, patients with multiorgan complications presented infections with significantly higher parasite biomass without significant differences in var adhesion types. Conversely, pediatric patients with specific complications showed distinct var signatures. Cerebral malaria patients showed broadly increased expression of var genes, in particular group A and DC8 var, while children with severe malaria anemia were classified based on high transcription of DC8 var only. This study represents the first large multisite meta-analysis of var expression, and it demonstrates the presence of common var profiles in severe malaria patients of different ages across distant geographical sites, as well as syndrome-specific disease signatures. The complex associations between parasite biomass, var adhesion type, and clinical presentation revealed here represent the most comprehensive picture so far of the relationship between cytoadhesion, parasite load, and clinical syndrome.
INTRODUCTION
Despite increased malaria control and elimination efforts, severe malaria (SM) from Plasmodium falciparum remains responsible for more than 400,000 deaths every year (1). In areas with high P. falciparum transmission, such as sub-Saharan Africa, SM primarily occurs in pediatric patients, as older children acquire protective immunity to the pathogenic effects of infection. In areas of low transmission, such as Asia and the Americas, SM occurs in both children and adults. This distinction is important, as malaria disease presentation varies between children and adults (2). Whereas cerebral malaria (CM) and metabolic acidosis are common to children and adults, severe malarial anemia is more common in children, and acute kidney injury, jaundice, and acute respiratory distress syndrome are most commonly seen in patients greater than 10 years old (2). The parasite and host factors that contribute to variability in malaria disease presentation remain poorly understood.
A key virulence determinant of P. falciparum is sequestration of infected erythrocytes (IEs) within the microvasculature (3). Extensive microvascular obstruction from sequestered IEs is believed to contribute to metabolic acidosis (4), while high levels of parasite sequestration in brain (5–7) and the placental intervillous space (8) are associated with organ-specific complications. Cytoadhesion of P. falciparum-IEs is mediated by the large and diverse P. falciparum erythrocyte membrane protein 1 (PfEMP1) family, encoded by var genes. PfEMP1 proteins are expressed in a clonally variant fashion on the surface of IEs (9–11). Each parasite genome contains ∼60 var genes that are classified into group A, B, or C based on chromosomal localization and the gene upstream region (12). Molecular insight into PfEMP1 function has been gained by sequence classification of the extracellular Duffy binding-like (DBLα/β/γ/δ/ε/ζ) and cysteine-rich interdomain region (CIDRα/β/γ/δ) adhesion domains (13, 14). CIDRα1 domains bind endothelial protein C receptor (EPCR) (15, 16), CIDRα2-6 domains bind to CD36 (17–19), and CIDRβ/γ/δ domains bind neither receptor (reviewed in reference 20). Additionally, PfEMP1 can simultaneously bind to other coreceptors, as some DBLβ1/3 (group A) and DBLβ5 (group B and C) domains mediate binding to ICAM-1 (21–25). PfEMP1 also includes sets of domains typically found tandemly arrayed in the same protein, termed domain cassettes (DCs). Several DCs encode conserved cytoadhesion traits, including binding to EPCR (DC8 and DC13) (16, 21, 23, 26, 27), ICAM-1 (DC4) (28), and PECAM-1 (DC5) (29). Antigenic switching of PfEMP1 proteins modifies P. falciparum-IE specificity for receptors on endothelial cells (cytoadhesion) or erythrocytes (termed rosetting [30]).
A challenge of studying the epidemiological associations of the var gene family and malaria disease is the immense diversity of var genes in the parasite population (31, 32). The development of primer sets targeting different var domain subtypes represented a breakthrough in malaria pathogenesis research (13, 33). This approach has shown that transcription of var genes encoding predicted EPCR binding activity is elevated in both pediatric (23, 33–36) and adult (37) SM patients. Moreover, deep sequencing of var amplicons has indicated that SM infections comprise a mixed population of parasites expressing different var genes (34, 38). Although these findings implicate specific PfEMP1 subsets in disease severity, these studies were insufficiently powered to assess whether expression of distinct PfEMP1 adhesion types are linked to different organ complications in children and adults. High parasite burden is also thought to play an important role in disease severity. Plasma levels of P. falciparum histidine-rich protein 2 (PfHRP2), a parasite protein released upon merozoite egress, is used as a blood surrogate marker for the combined amount of both circulating ring-stage and sequestered mature-stage P. falciparum-IEs. PfHRP2 levels are associated with severe malaria in both children and adults (39, 40), predict disease progression (41), and are increased in patients with specific complications, including cerebral malaria (42), and metabolic acidosis (43).
This study extends previous work (i) by applying an integrated multicohort analysis to better understand how both parasite burden and binding phenotype are associated with severe malaria in both adults and children and (ii) by examining how parasite burden and parasites expressing different var gene subsets influence disease severity and the spectrum of disease across study sites. To accomplish this, we performed a machine learning meta-analysis of combined var transcript profiles from two previously described African children cohorts (33, 34), along with an expanded Indian adult cohort based on our previously published work (37). This work represents the largest and broadest analysis of these parasite factors in severe malaria to date, thus providing unprecedented power to explore and compare the parasite factors that lead to severe disease.
RESULTS
Characteristics of the study cohorts.
The var gene transcript profiles (var profiles) for this work were obtained from two previously published studies of pediatric malaria in Tanzania (TZ) (33) and Malawi (BLZ) (34) and one adult malaria cohort in Goa, India (GMC). The cohorts included 90 (TZ), 68 (BLZ), and 55 (GMC) SM patients and 32 (TZ), 40 (BLZ), and 37 (GMC) uncomplicated malaria (UM) patients. The GMC cohort expanded our previously published (37) cohort with newly obtained samples, comprising both severe and uncomplicated malaria.
Previous work has shown that malaria disease presentation is influenced by both transmission intensity and patient age (2, 44). The recruited individuals in the three sites had different age profiles (see Table S1 in the supplemental material). Tanzanian UM and SM pediatric patients (median age, 2 years; range, 1 to 3 years) were generally younger than Malawian patients (median age, 4 to 4.5 years; range, 2 to 7 years) (Table S1). Goan adult UM and SM patients had median ages of 25 and 27 years, respectively (Table S1). UM and SM patients in all three cohorts did not show significant differences in circulating parasite density by blood smear. However, UM patients had significantly reduced parasite biomass compared to SM patients, as reflected by plasma PfHRP2 levels (Table S1). PfHRP2 levels were not measured in the TZ cohort. Patient mortality rates for SM were similar at all sites, ranging from 10% in TZ and BLZ patients to 13% in GMC patients (Table S1).
Demographic and clinical characteristics of the study cohorts. Download Table S1, PDF file, 0.1 MB (79.5KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
var transcript profiles and cross-site correlations.
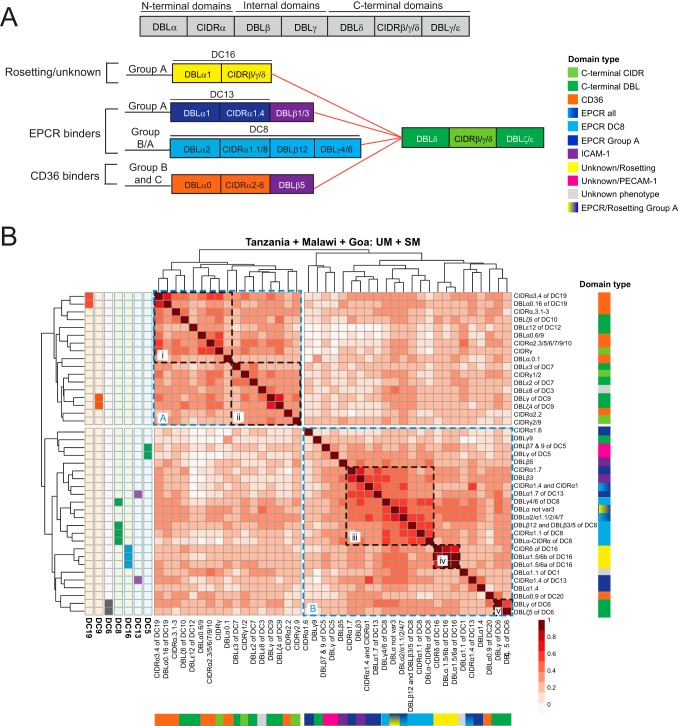
To compare the var adhesion types associated with malaria disease severity at the three sites, var transcript profiling was performed using a set of 41 PCR primer pairs designed to quantify transcripts of defined var domain subtypes (33), many of which have predicted binding phenotypes (33–35, 37) (Fig. 1A). The three cohorts analyzed in this meta-analysis were chosen as they employed the same primer panel enabling comparative analysis across sites. Table S2 in the supplemental material shows the primers used for these analyses, along with a color code based on predicted binding phenotypes of the targeted domain(s) used in all figures throughout this study. To examine how the primer sets performed across different parasite populations in Africa and India, correlations between transcript expression of individual var domain subtypes across every patient were calculated in all three sites and used to hierarchically cluster domains (Fig. 1B, left). Notably, the transcript expression of var domain subtypes grouped according to PfEMP1 domain architecture and var adhesion type. Tandemly arrayed domains (domain cassettes [DCs]) remain closely correlated at all three sites (i.e., DC5, DC6, DC9, DC16, and DC19), all but DC5 retain their close association across every site, and DC5 primers remain the most closely associated in Tanzania (Fig. 1B). For DC8, three of the four tandemly associated domains grouped together, and DC13 is the sole domain cassette whose two characteristic domains are not closely correlated with each other (Fig. 1B), probably reflecting the weaker association of these domain types (DBLγ4/6 [DC8] and DBLα1.7 [DC13]) with their domain cassettes (13). Clustering of DCs also holds true in each individual site, with the exception of DC5 (see Fig. S1 in the supplemental material). This analysis suggests that these protein architectural arrangements are maintained across diverse parasite populations in Africa and India, despite extensive gene recombination in the var gene family (45). This finding also suggests that tandem domain relationships may be under strong biological selection. Additionally, var transcripts clustered according to predicted var adhesion types (Fig. 1B). Cluster A was dominated by var genes encoding predicted CD36 binding domains (preferentially found in subcluster i) and C-terminal domains (preferentially found in subcluster ii). In contrast, cluster B contained var genes encoding predicted EPCR binding domains (subcluster iii) and var genes associated with unknown/rosetting function (subcluster iv). Exceptions in cluster B were subtypes relating to DBLα0.9 of DC20 and DC6 (subcluster v). Although DC6 domains can be found in all three var groups (13), its expression was more closely correlated to the DC8 and group A var genes across the three cohorts. This analysis suggests that transcription of specific var adhesion types is more closely correlated and that DCs are conserved across parasite populations of distant geographical sites.
FIG 1.
Correlation of var domain transcript profiles across the three study sites. (A) Schematic of PfEMP1 domain architecture illustrating the relationship between known binding phenotypes and different var groups. CIDRβ/γ/δ domains (yellow) have unknown binding properties, CIDRα1 domains (light and dark blue) bind EPCR, CIDRα2-6 domains (orange) bind CD36, and DBLβ1/3/5 domains (purple) bind ICAM-1. (B) Transcriptional profiling of var genes was performed with 41 different primer sets targeting different var domain subtypes. Shown is a heat map of correlations (Spearman’s rho) of transcript levels of different var domain subtypes across all samples (SM and UM) from all three sites. Domain transcript levels were hierarchically clustered, and known tandem domain arrangements (e.g., DC19) are indicated to the left of the heat map. The right and bottom legends use the color scheme described in Table S2 to indicate EPCR/CD36/ICAM-1/Rosetting/PECAM-1/unknown/C-terminal domains. Clusters A and B and subclusters i to v are highlighted in dashed boxes.
Clustergrams of var domain subtypes related by expression correlation for each study site, similar to Fig. 1. Heat maps indicate var domain subtypes known to be part of defined domain cassettes. Download FIG S1, EPS file, 2.3 MB (2.1MB, eps) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Broad phenotypic classification of domains targeted by individual var primers and associated color scheme. Download Table S2, PDF file, 0.1 MB (52.4KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cross-site correlations in var transcript profiles.
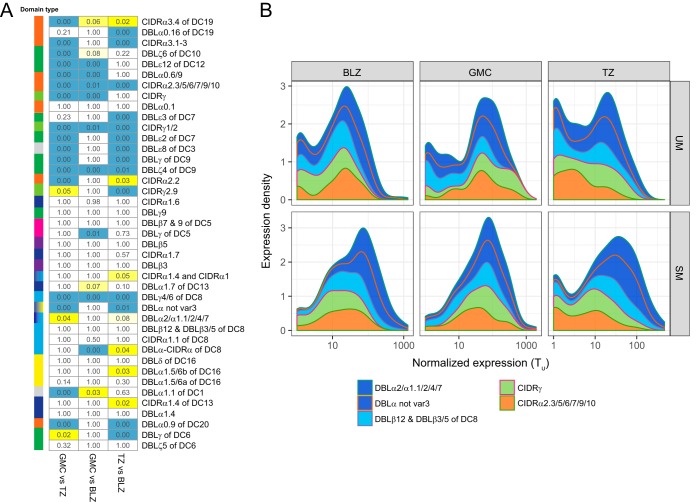
To explore var transcript profiles across the three study sites and to assess the comparability of var profiles derived from each site, we compared var domain subtype expression distributions from each cohort. Overall, var genes encoding EPCR binding (both DC8 and group A var) were the highest expressed transcripts across the three sites (transcription units [Tu] value = 10 to 1,000) (see Fig. S2 in the supplemental material). Differences in expression were tested by using the Kolmogorov-Smirnov test (Fig. 2A). While some domain subtypes presented similar patterns at all sites (P = 1.00), other domain subtypes showed site-specific differences in expression distributions. These differences are not solely explained by differences between patient age and ethnicity. For example, while adult GMC and pediatric BLZ patients shared 26 similarly distributed domain subtypes (P = 1.00), only 14 subtypes met this threshold when comparing pediatric TZ versus BLZ patients. These results were unexpected, as pediatric and adult severe malaria patients present different organ complications.
FIG 2.
Correlations in var transcript levels across the three study sites. (A) Kolmogorov-Smirnov P values comparing var domain subtype distributions between each pair of sites. P values under 0.01 are highlighted in blue, and those under 0.05 are highlighted in yellow. Subtype color annotations and order are identical to those in Fig. 1B. (B) Transcription levels of the top five transcribed var domain subtype transcripts (median Tu, >10) are stratified by site and severity (SM/UM).
Transcript levels of the var domain subtypes targeted by the 41 primer sets vary according by study site and malaria status. Expression density plots for each var domain subtype were stratified by site. BLZ, Malawi; GMC, Goa; TZ, Tanzania. The var domain subtypes are shown in the same order as in Fig. 1B. Download FIG S2, EPS file, 8.1 MB (7.8MB, eps) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
While EPCR-binding domains presented similar expression profiles across sites (P = 1.00), the main site-specific expression differences were observed for CD36-binding and C-terminal domains (Fig. 2A). Figure 2B shows the expression density distributions detected by primer pairs, stratified by study site and severity. We looked for differences in the transcript levels of the most abundantly transcribed var subtypes (median Tu, >10) in the combined severe versus uncomplicated malaria cases. Five primer pairs were identified that passed this threshold: primer pair DBLα2/α1.1/2/4/7 targets all EPCR binders (DC8 and group A), primer pair DBLα not var3 targets group A PfEMP1 (EPCR, rosetting, PECAM-1, unknown), primer pairs DBLβ12 and DBLβ3/5 mostly target DC8 (EPCR), and primer pairs CIDRγ and CIDRα2.3/5/6/7/9/10 target CD36-binding var genes. Notably, the rank order of the top 5 transcribed domains was the same in both SM and UM patients across the three sites, but the var transcript levels tended to be higher in SM patients (Fig. 2B). Two of the three EPCR-binding domains showed significantly higher expression in SM for all sites (DBLα not var3, BLZ, P = 8.8 × 10−6; GMC, P = 0.02; TZ, P = 2.5 × 10−8; DBLβ12 and DBLβ3/5 of DC8 BLZ, P = 1.6 × 10−4; GMC, P = 0.0004; TZ, P = 0.002), while neither of the CD36 high expressers showed any significant difference. In all three sites, UM patients showed a bimodal distribution of the DC8 and group A var transcripts, including two peaks representing low-expression and high-expression individuals (Fig. 2B). However, the low expressers’ peak is absent in SM cases from BLZ and GMC and highly diminished in TZ (Fig. 2B). Thus, even though individual domain subtype expression varies in a site-specific manner, marked increases in abundances of some PfEMP1 domains are a feature of SM across all study sites.
Common var profiles predict severe malaria in both adults and children.
In order to more precisely evaluate the contribution of var transcript profiles to malaria severity, a machine learning approach was applied. Random forest (RF) models were trained on var profiles to classify samples as severe or uncomplicated malaria. This was performed for all sites individually and for Tanzania and Malawi together to derive a model representing all childhood cases.
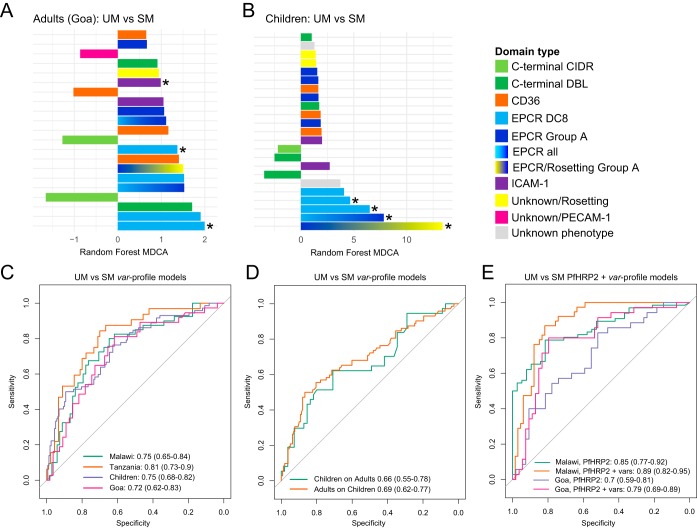
The importance of each adhesion domain to the adult and child RF models is shown in Fig. 3A and B. Importance was measured as the mean decrease in classifier accuracy (MDCA) when the primer is excluded from the model while resampling during model training. As previously reported, in both adults (37) and children (23, 33–36), SM is associated with increased transcription of domains predicted to bind EPCR. Despite comprising only 10% of the var repertoire (13), more than 50% of the top 10 predictive features in both models belong to PfEMP1-EPCR binders (Fig. 3A and B). Both the group A and DC8 subsets of EPCR binding var genes were predictive of pediatric SM (group A: DBLα not var3; DC8: DBLβ12 and DBLβ3/5, DBLα-CIDRα, and CIDRα1.1; and DC8-group A: DBLα2/α1.1/2/4/7). Although we previously reported that only DC8-EPCR binders were predictive of adult SM (37), both DC8 domains (DBLγ4/6, DBLβ12 and DBLβ3/5, DBLα-CIDRα, CIDRα1.1, and DBLα2/α1.1/2/4/7 [see Table S3 in the supplemental material]) and group A-EPCR binders (DBLα not var3 and DBLα2/α1.1/2/4/7 [Table S3]) contributed to the adult SM model in the expanded data set. Additionally, var genes containing the DC19 subset of CD36 binding domains (children, CIDRα3.4; adults, DBLα0.16 [Table S3]) ranked among the top 10 predictive features in both the adult and children SM models. As well, high expression of var genes containing the C-terminal DC6 (DBLγ [Table S3]), which ranked as the top var feature in our previously published adult SM model (37), still ranked as the third most predictive feature in the adult SM model. The presence of several DCs in the top ranks of our models is remarkable. This is because RF analysis may penalize DC domains, as the algorithm recognizes them as a proxy of each other, decreasing the MDCA score. The importance of DC8 (EPCR), DC6 (C-terminal), and DC19 (CD36) in both the adult and children SM models suggest that these domains play an important role in severe disease across age groups.
FIG 3.
Parasite load and var adhesion types independently predict severe malaria in adults and children. (A and B) Bar plots showing var domain subtype model importance (measured as MDCA) for child and adult UM versus SM models. Each bar represents a single domain subtype targeted by one of the 41 primer sets used to generate the RF model, colored by predicted binding phenotype or position in the protein (see also Table S2 and Table S3). Positive MDCA indicates higher expression of a specific domain subtype in SM and vice versa. Bars with asterisks indicate that these domain subtypes showed significant differences in expression (false-discovery rate [FDR] of ≤0.2) using the mProbes algorithm. (C) ROC curves showing out-of-bag predictions of RF models classifying samples as SM or UM. One model was trained per site, along with a total childhood model combining Malawi and Tanzania. Performance is shown as area under the ROC curve, with 95% confidence intervals in parentheses. (D) ROC curves showing blind predictive performance of the child model predicting adult severe malaria, and the adult model predicting child severe malaria. (E) ROC curves showing predictive performance of serum PfHRP2 levels alone to classify severe malaria and PfHRP2 combined with the var profile RF models.
MDCA and mProbes FWER for each var domain subtype in adult and childhood UM versus SM models. Download Table S3, PDF file, 0.1 MB (62.2KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Receiver operating characteristic (ROC) curves and the accompanying 95% confidence intervals (CIs) representing the unbiased out-of-bag (OOB) predictions made during model training are shown in Fig. 3C. All var profile models significantly discriminate UM from SM. Notably, the combined childhood SM model (area under the ROC curve [AUC], 0.75 [95% CI, 0.68 to 0.82]) performs very similarly to the individual Tanzanian and Malawian models (TZ AUC, 0.81 [95% CI, 0.73 to 0.9]; BLZ AUC, 0.75 [95% CI, 0.65 to 0.84]) and the adult Goan model (AUC of 0.72 [95% CI, 0.62 to 0.83]). Taken together, predictive performance was highly consistent across all sites.
To further explore pathogenic mechanisms in children and adults, we made blind predictions, using the adult model to classify the child samples and the childhood model to classify the adult samples. Importantly, both models showed significant ability to cross-classify (Fig. 3D) for both children on adults (AUC, 0.66 [95% CI, 0.55 to 0.78]; P = 0.002) and adults on children (AUC, 0.69 [95% CI, 0.62 to 0.78]; P = 5.3 × 10−7). In other words, adult SM could be predicted by var profiling of pediatric samples and vice versa. This demonstrates that, despite presenting highly distinct severe disease manifestations, similar var subsets are predictive of SM in both adults and children.
Parasite biomass and var profiles are independent and complementary contributors to severe disease in both adults and children.
A recurrent question in the malaria pathogenesis field is the relative contribution of parasite biomass and parasite cytoadhesion to malaria severity. PfHRP2 measurements were available for both the Malawi and Goa cohorts, but not Tanzania. PfHRP2 levels alone significantly discriminated severe from uncomplicated malaria in both Malawi (AUC, 0.85 [95% CI, 0.79 to 0.92]; P = 4 × 10−9) and Goa (AUC, 0.7 [95% CI, 0.59 to 0.81]; P = 0.0018) (Fig. 3E). To evaluate the improvement resulting from the combination of PfHRP2 and the var gene models, logistic regression models of the form severity = PfHRP2 + var signature score were fit. Receiver operating characteristic (ROC) curves for these models are shown in Fig. 3E, and the combined severity predictions significantly improved on both PfHRP2 alone and var profiles alone for both Malawi (χ2 test, P = 0.0005) and Goa (χ2 test, P = 0.006). Statistical analyses indicate substantial improvement in all prediction metrics in both sites when var profile models are combined with PfHRP2 measurements (Malawi, var profile alone odds ratio [OR], 6.6, HRP2 alone OR, 15.9, and var profile plus HRP2 OR, 28.3; Goa, var profile alone OR, 5.77, HRP2 alone OR, 3.75, and var profile plus HRP2 OR, 15) (see Table S4 in the supplemental material). This demonstrates that var profiles and parasite load are independent and complementary biomarkers of malaria severity in two independent study cohorts.
Summary statistics for combining var profile models with PfHRP2 measurements for Malawi and Goa samples. Download Table S4, PDF file, 0.1 MB (42.8KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Parasite load is the primary factor underlying adult SM cases with multiple clinical complications.
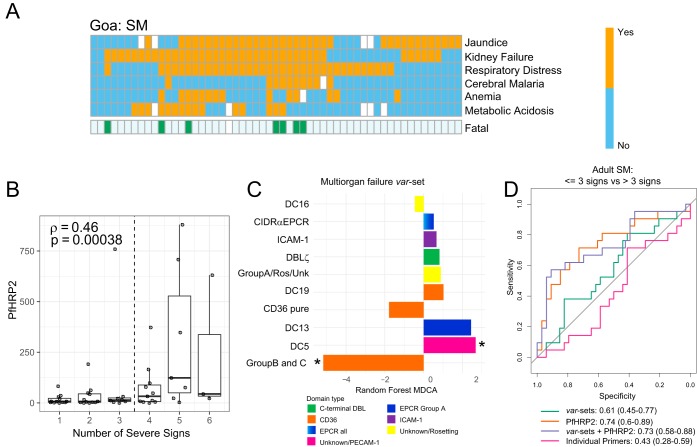
Adult SM is frequently associated with multiorgan complications that increase fatality risk (2). Within our Goan cohort, acute respiratory distress syndrome, acute kidney injury, and jaundice were the most frequent complications and were frequently found together (Fig. 4A). Notably, the number of severe malaria criteria is significantly correlated with PfHRP2 levels (Spearman’s ρ = 0.46; P = 0.00038 [Fig. 4B]), presenting a pronounced increase in individuals with more than three severity criteria. Fatal SM cases were also associated with higher burdens of severe criteria, with six of seven fatal cases in this cohort presenting with more than three severity criteria (Fig. 4A).
FIG 4.
Plasma PfHRP2 levels are associated with the number of severe malaria criteria in adults. (A) Heat map showing prevalence of clinical signs of adult SM in Goa. Fatal SM cases are indicated below. A white box indicates missing information. (B) Box and dot plot of plasma PfHRP2 levels in adult SM, stratified by number of severity criteria. The median is indicated by a horizontal line. (C) Bar plot showing importance of primer sets in the severe criterion count var set model. Negative MDCAs indicate sets with lower expression in patients with >3 severity criteria. (D) ROC curves showing the predictive power of individual var domain subtypes, PfHRP2 levels, var domain subtypes grouped by binding phenotype (var sets), and var sets combined with PfHRP2 levels to classify adult SM patients as having up to three clinical signs versus those with over three clinical severe signs.
An RF model was trained to classify adult severe malaria cases into those with up to three severity criteria and those with over three severity criteria. Notably, the model based only on var profile data showed no discriminatory power (AUC, 0.43 [95% CI, 0.28 to 0.59]), while PfHRP2 levels alone successfully discriminated patients with over three severity criteria (AUC, 0.74 [95% CI, 0.58 to 0.88]) (Fig. 4). As the redundant nature of the primer repertoire used in this study could artificially decrease the importance of some var domains contributing to SM by acting as proxies for each other during modeling, we also trained a model based on primers grouped into related var predicted adhesion types (Fig. 5D; see Table S5 in the supplemental material). Tu values of var sets were summed to construct a summarized matrix of discrete, nonredundant binding phenotype sets. Patients with multiorgan complications presented decreased expression of var genes with a predicted CD36 binding phenotype and increased expression of a subset of group A var genes (DC13 EPCR binders and DC5-containing genes, which bind PECAM-1 in some instances). Conversely, the DC8 EPCR-binding subset did not predict multiorgan complications, as their expression was elevated among all adult SM patients (Fig. 4C). While the var set model showed improved predictive power (AUC, 0.61 [95% CI, 0.45 to 0.77]), it did not reach the threshold for statistical significance. Moreover, there was no improvement over PfHRP2 alone (Fig. 4D). Taken together, this analysis suggests that P. falciparum growth and expansion are the primary parasite factors that influenced presentation of multiple severity signs in adults.
FIG 5.
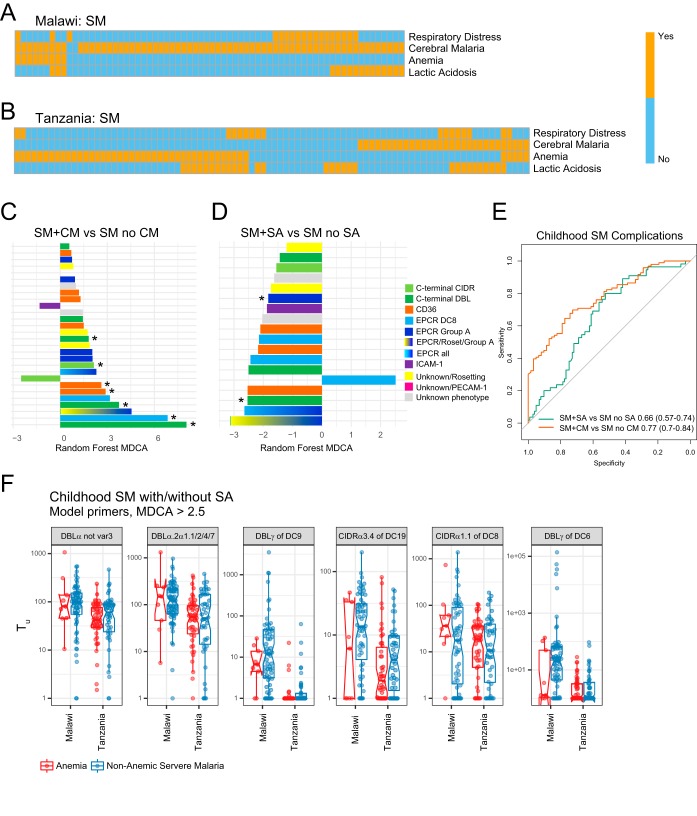
var transcript profiles distinguish childhood coma (CM) and anemia (SA). (A and B) Heat maps showing the prevalence of clinical signs of childhood SM in Malawi and Tanzania, respectively. Patients without a WHO criteria listed on the heat map presented hyperparasitemia. (C and D) Bar plots showing var domain subtype importance (MDCA, negative bars indicate lower primer expression in SM plus CM or SM plus SA patients) for the SM plus CM versus SM with no CM and the SM plus SA versus SM with no SA models. (E) ROC curves of RF models discriminating CM and SA from other SM patients in the combined Malawi and Tanzania sets. (F) Box plots of site-specific expression for the most important (MDCA, >2.5) var domain subtypes in the anemia model.
Classification of targeted domains into summed var sets corresponding to specific domain cassettes, binding phenotypes, and domain positions. Download Table S5, PDF file, 0.1 MB (53KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Specific var profiles are associated with childhood severe malaria and anemia.
To investigate var profile associations with childhood disease complications, we analyzed SM samples from the combined Tanzanian and Malawi data sets. The main SM complications in children are coma (cerebral malaria [CM]), severe anemia (SA), lactic acidosis, and respiratory distress, which can appear alone or in combination (Fig. 5A and B). While the Tanzanian cohort was selected for an even distribution of complications, the Malawian cohort is biased toward CM patients, as it was the main focus of the original study (34). Fitting an RF model to the combined cohorts accurately classified CM from non-CM among SM cases (AUC, 0.77 [95% CI, 0.7 to 0.84] [Fig. 5C to E]). Consistent with previous studies (33, 34, 38), childhood CM is associated with higher levels of transcripts encoding group A and DC8 EPCR binders (group A: DBLα not var3; DC8: DBLγ4/6, DBLβ12 and DBLβ3/5, and CIDRα1.1; and DC8-group A: DBLα2/α1.1/2/4/7) (Fig. 5C; see Table S6 in the supplemental material). We also observed higher levels of transcripts encoding C-terminal domains (DBLγ of DC6 and DC9), as well as the DC19 subset of CD36-binding domains (Fig. 5C; Table S6).
MDCA and mProbes FWER for each var domain subtype in childhood coma (cerebral malaria [CM]) and anemia (SA) models. Download Table S6, PDF file, 0.1 MB (58.1KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
This modeling approach also significantly discriminated SA from non-SA among SM patients (AUC, 0.66 [95% CI, 0.57 to 0.74] [Fig. 5D and E]), with SA being associated with lower expression of most adhesion domains present in the CM model, including group A EPCR binders, the DC6 and DC9 C-terminal domains, and the DC19 subset of CD36 binders (Fig. 5D; Table S6). To further investigate if the performance of the SA model was simply being driven by lower var transcription levels in parasites associated with SA cases or study site-specific biases, we compared transcript levels of the most informative var domain subtypes (MDCA, >2.5) from the SA model (Fig. 5F). This analysis indicated that while most var domains were expressed at lower levels in SA, the DC8 domain, CIDRα1.1 (Table S6), had higher median transcript levels in SA than CM cases in both Malawi and Tanzania. However, CIDRα1.1 DC8 transcripts were also elevated in many CM cases, suggesting that DC8-expressing EPCR-binding parasites are linked to both CM and SA.
DISCUSSION
The var gene family plays a central role in parasite immune evasion and pathogenesis and has extensively diversified under immune pressure. The motivation of this study was to evaluate if the same var subsets were linked to severe malaria in Africa and India and to explore how var adhesion types interact with parasite biomass to influence malaria disease presentation in children and adults. This work represents the most comprehensive analysis to date of parasite factors leading to SM. This has been accomplished by assembling a multisite adult and pediatric data set of var profiles, comprising hundreds of patient samples. The var profiles studied in this meta-analysis were tested with a panel of domain-specific primers that provide broad coverage of the var repertoire (estimated 87% of var genes) (33). While these primers have less overlapping domain coverage of the CD36-binding PfEMP1 subset, which comprise 90% of the P. falciparum var genomic repertoire, independent next-generation sequencing of var amplicons of the pediatric cohorts analyzed in this study (34, 38) validated the use of the var domain-specific primers to determine patient var profiles.
Our central observation is that both parasite biomass and var profiles are important, independent, and complementary in the development of adult and pediatric SM. Furthermore, despite var transcript profiles showing extensive within-host and between-site differences, the var profile signatures associated with severe malaria in African children are highly predictive of severe malaria in Indian adults and vice versa. Most previous var transcript profiling has focused on African children, and less investigation has been conducted in lower-transmission settings where disease affects both children and adults. Despite differences in disease presentation between children and adults, severe malaria in both Indian adults and African children is clearly linked to increased transcription of variants predicted to bind EPCR, including DC8 and group A var genes. This demonstrates the power of this meta-analytical approach to reveal a universal signature of severe malaria and validates the results of previous studies implicating the DC8 and group A var subsets in severe malaria (33–38).
In contrast to the clear associations of specific var transcript profiles with severe malaria, the relationship of specific binding phenotypes and malaria disease syndromes is more complex. Understanding this relationship is complicated by the complex presentation of severe malaria in children and adults, as many patients had more than one severity criterion, especially in our adult Indian cohort. The most definitive link to var adhesion types was observed for childhood malaria, in which children with coma and severe anemia had distinctive var signatures. While predicted EPCR-binding group A var transcripts were lowest in UM cases, intermediate in SA, and highest in CM, DC8 var transcript levels tended to be higher in SA than CM cases, indicating there may be subtle differences in the transcript level or composition of DC8 variants between the two syndromes. However, the incidence of anemia in this study is highly skewed: it is common in our Tanzanian cohort and uncommon in the Malawian cohort. Along with this, the most anemic individuals also were not cerebral malaria patients. This raises the possibility of site-specific and CM expression patterns biasing the signature. Nevertheless, the specific association of DC8 var expression with severe anemia, and of DC8 and group A var with CM, is intriguing. DC8 PfEMP1 binds EPCR and most likely a range of other not yet well understood human receptors (46), while some group A var genes encode dual adhesive properties for EPCR and ICAM-1 (21, 23), as well as other yet undefined human receptors. This is suggestive of a mechanistic hypothesis in which malaria coma is potentially driven by both ICAM-1 and EPCR binding parasites, in contrast with anemia. Further study is needed to disentangle these effects.
In addition to var disease signatures, our study confirms the strong role of parasite biomass in severe disease (39–43). RF models reveal that both factors are independent and complementary biomarkers of severity. The combination of both features increases the sensitivity, specificity, and odds ratio of severe malaria prediction in both pediatric and adult severe malaria. An expansion of the circulating and sequestering parasite population might directly contribute to disease severity by directly increasing endothelial cell activation (47–49) and microvascular obstruction (4, 50).
Whereas EPCR-binding var subsets were linked to severe malaria in adults, progression to multisymptomatic disease (greater than three severity criteria) appeared to be primarily driven by parasite load. However, we did see a possible reduction in CD36 binders in adults with multiple severe malaria complications. This may suggest that an overall increase in EPCR-binding parasites contributes to multiorgan complications in adult SM. Future work is needed to address whether specific var adhesion types are linked to the distinct organ complications of adult SM.
A limitation of this study is that the meta-analysis focuses on P. falciparum transcription differences in severe malaria and lacks experimental assessment of parasite binding. Nevertheless, the cytoadhesion predictions for CD36, EPCR, and ICAM-1 are supported by multiple in vitro studies of both laboratory-adapted P. falciparum lines (21–23, 26–28) and cerebral malaria isolates (16, 51, 52). The consistency in the importance of group A and DC8 var transcripts as a predictor of disease severity in patients of multiple ages and ethnicities, presenting multiple disease syndromes, suggests an important role of EPCR in severe disease. This likely reveals a pathogenic pathway, as IE binding to EPCR is proposed to prevent its vascular homeostatic and protective function (53).
In summary, machine learning meta-analysis has revealed common var signatures of severe malaria in African children and Indian adults and further suggests important interactions between parasite biomass and var adhesion type in disease presentation.
MATERIALS AND METHODS
Ethical approval.
Informed consent was obtained from all study participants. The Indian study was approved by the ethics boards at Goa Medical College and Hospital, the University of Washington, and the Western Institutional Review Board, used on behalf of the Center for Infectious Disease Research/Seattle Children’s Research Institute, as well as by the Government of India Health Ministry Screening Committee. The Malawian study (34) was approved by the institutional review boards at the University of Malawi College of Medicine, Michigan State University, and the Albert Einstein College of Medicine. The Tanzania study (33) was approved by the Tanzania Medical Research Coordinating Committee, and parents or guardians of children provided consent.
Composition of human cohorts and patient recruitment.
Newly recruited patients were obtained from the Goa study site, using the previously published sample collection protocol (37). Additional subjects were recruited between August 2012 and January 2017 from the hospital admissions or outpatient wards at the Goa Medical College and Hospital. P. falciparum infections were identified using Giemsa-stained thick and thin smears for parasitemia determination and species identification. Plasmodium vivax infections were excluded from the study. After informed consent was obtained, blood samples from P. falciparum-infected patients were collected in acid citrate dextrose vacutainers and separated into plasma and red blood cell fractions in RNAlater, before freezing at −80°C. Adult SM was defined as hospitalization with any of the following signs: coma (Glasgow coma score, <10), severe anemia (Hb, <7 g/dl), jaundice (bilirubin, >3 mg/dl), kidney failure (serum creatinine of >3 mg/dl or blood urea nitrogen of >17 mmol/liter), shock (systolic blood pressure of <80 mm Hg plus cold extremities), metabolic acidosis (peripheral venous bicarbonate, <155 mmol/liter), respiratory distress (>20 breaths per min or partial pressure of oxygen in alveoli [PaO2] of <75 mm Hg), or hypoglycemia (blood glucose, <40 mg/dl). Unadmitted patients were classified as UM. For the two African children sites, SM was defined as hospitalization with any of the following presentations: coma (Blantyre coma score, ≤2), severe anemia (Hb, <5 g/dl), and lactic acidosis (plasma lactate, >5 mmol/liter) as published previously (33, 34). Pediatric patients with respiratory distress were classified if any of the following variables was present: deep breathing, grunting, nasal flaring, chest indrawing, and chest retractions (for BLZ) or Kussmaul breathing and being X-ray positive (for TZ).
qRT-PCR measurement of var transcription profiles.
All participants had quantitative reverse transcription-PCR (qRT-PCR)-based var transcript profiling performed as previously described (33). Briefly, TRIzol reagent was used to extract RNA from parasitized red blood cells, and cDNA was synthesized. The var primer threshold cycle (CT) values were quantified by qRT-PCR using a panel of degenerate primers designed to identify known var domain subclasses. Levels of var expression were normalized relative to the housekeeping gene primers adenylosuccinate lyase and seryl-tRNA synthetase (GMC) and aldolase and seryl-tRNA synthetase (TZ and BLZ): ΔCT var primer = CT var primer − mean CT housekeeping primers. Low-abundance transcripts with a ΔCT of >5 were set to 5. Samples where housekeeping primers showed a mean CT of ≥ 25 (TZ) or >30 (BLZ and GMC) were excluded from further analysis. For all subsequent analyses, var expression was represented as transcript units [Tu = 2(5 − ΔCT)], where a Tu value of 1 represents a 0 level of expression.
PfHRP2 plasma quantification.
PfHRP2 quantification was performed as previously described (34, 37). Briefly, BLZ samples were analyzed by a commercial kit (Cellabs, Brookvale, Australia) and for the Goan samples (GMC), a double-site sandwich enzyme-linked immunosorbent assay (ELISA) was performed. The standard curve for Goan samples was established using purified PfHRP2 protein (kindly donated by David Sullivan, Johns Hopkins Bloomberg School of Public Health).
Statistical and machine learning analysis.
All statistical and machine learning analysis was carried out using the R language for statistical computing (54). All primer expression and distribution plots were created using the R ggplot2 library (55).
(i) Clustering and distribution analysis. Primer-primer correlation coefficients were calculated as Spearman’s rho (ρ), and hierarchical clustering was performed using the R hclust function. Significant differences in primer distribution between sites were evaluated using the Kolmogorov-Smirnov test, using the R ks.test function, and P values were adjusted for multiple testing using the Holm approach.
(ii) Machine learning modeling and significance testing. Machine learning models were used to assess the ability of var transcript profiles to classify subjects as SM or UM or to distinguish specific SM severity criteria. RF models were trained to distinguish SM from UM in all three sites using the R randomForest package (56). Missing primer Tu values were set to be equal to 1 (indicating primer expression below the limit of detection). To avoid bias due to overfitting, unbiased model predictive performance was evaluated based on the consensus RF out-of-bag (OOB) predictions generated during model training. Receiver operating characteristic (ROC) curves were constructed based on the OOB predictions. Strength of predictive performance was measured as area under the ROC curve (AUC). This takes the form of a number between 0 and 1, where 0.5 represents random performance and 1 represents perfect classification. The R pROC (57) package was used to estimate 95% confidence intervals (CIs) around the AUC. Models were considered significant when the lower 95% CI was over 0.5 (i.e., the entire 95% confidence interval of the AUC was in the “better than random” area of the ROC curve). P values for ROC curves were calculated by fitting two logistic regression models—one of the form class ∼ signature_score and the other of the form class ∼1—and comparing them with the R anova.glm(test=“Chisq”) function. Sensitivities, specificities, and odds ratios were calculated from confusion matrices of binary predictions made at the optimal point on the ROC curve, defined as the point that minimized the Euclidean distance to a sensitivity and specificity of 1.
To estimate the contribution of individual var subtypes to model performance, the mean decrease in classifier accuracy (MDCA) was used. This statistic is calculated during the resampling training procedure of the RF model and is the percentage of decrease in accuracy in random data subsamples that contain the var subtype versus those that do not. Significance of model var domain subtypes was calculated using the mProbes algorithm (58), which compares model subtypes to randomly shuffled subtypes to calculate a family-wise error rate (FWER).
Creation of summarized var primer sets.
Sets of var domain subtypes (var sets) that combine related information, such as a common binding phenotype (e.g., EPCR or CD36), gene group (e.g., group A, B, or C), domain organization (C-terminal DBL or C-terminal CIDR), or part of a tandemly arrayed domain combination (e.g., DC8/DC13) were developed (Table S5) and applied to summarize the var transcript levels. The summed var transcript abundance (var set) was calculated by summing the reported Tu values for each var domain subtype per sample and subtracting 1 for each additional primer beyond the first. This was done to ensure that the set’s summarized Tu value was equal to 1 if no expression was observed for any var domain subtypes in a particular sample. Machine learning and importance estimates were created for the summarized features, as described above.
ACKNOWLEDGMENTS
This study was made possible by NIH grant U19AI089688 (J.D.S., J.D.A., and P.R.), R01AI034969 (T.E.T.), NIH National Center for Advancing Translational Science (NCATS) Einstein-Montefiore CTSA TL1TR001072 (A.K.), National Center for Dynamic Interactome Research P41 GM109824 (F.D. and J.D.A.), American Heart Association Grant 17POST33670672 (M.B.), the Burroughs Wellcome Fund (A.K.), and Program Grant APP1092789 and Project Grant APP1061993 from the National Health and Medical Research Council of Australia (S.J.R.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
F.D. and M.B. designed the experiments, F.D., M.B., and P.B. performed the experiments, A.K. C.W.W., M.V., L.C., W.L.M., S.J.R., T.E.T., K.B.S., T.L., E.G., K.K., J.L., and P.R. provided published data sets used in the meta-analysis, F.D., M.B., J.D.A., and J.D.S. analyzed the data, and F.D., M.B., J.D.A. and J.D.S. wrote the paper. All coauthors read and approved the manuscript.
Footnotes
Citation Duffy F, Bernabeu M, Babar PH, Kessler A, Wang CW, Vaz M, Chery L, Mandala WL, Rogerson SJ, Taylor TE, Seydel KB, Lavstsen T, Gomes E, Kim K, Lusingu J, Rathod PK, Aitchison JD, Smith JD. 2019. Meta-analysis of Plasmodium falciparum var signatures contributing to severe malaria in African children and Indian adults. mBio 10:e00217-19. https://doi.org/10.1128/mBio.00217-19.
REFERENCES
- 1.World Health Organization. 2018. World malaria report 2018. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Dondorp AM, Lee SJ, Faiz MA, Mishra S, Price R, Tjitra E, Than M, Htut Y, Mohanty S, Yunus EB, Rahman R, Nosten F, Anstey NM, Day NP, White NJ. 2008. The relationship between age and the manifestations of and mortality associated with severe malaria. Clin Infect Dis 47:151–157. doi: 10.1086/589287. [DOI] [PubMed] [Google Scholar]
- 3.Miller LH, Baruch DI, Marsh K, Doumbo OK. 2002. The pathogenic basis of malaria. Nature 415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 4.Dondorp AM, Ince C, Charunwatthana P, Hanson J, van Kuijen A, Faiz MA, Rahman MR, Hasan M, Bin YE, Ghose A, Ruangveerayut R, Limmathurotsakul D, Mathura K, White NJ, Day NP. 2008. Direct in vivo assessment of microcirculatory dysfunction in severe falciparum malaria. J Infect Dis 197:79–84. doi: 10.1086/523762. [DOI] [PubMed] [Google Scholar]
- 5.Marchiafava E, Bignami A. 1895. Two monographs on malaria and the parasites of malarial fever. Edinb Med J 40:911–915. [Google Scholar]
- 6.Taylor TE, Fu WJ, Carr RA, Whitten RO, Mueller JS, Fosiko NG, Lewallen S, Liomba NG, Molyneux ME, Mueller JG. 2004. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med 10:143–145. doi: 10.1038/nm986. [DOI] [PubMed] [Google Scholar]
- 7.MacPherson GG, Warrell MJ, White NJ, Looareesuwan S, Warrell DA. 1985. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am J Pathol 119:385–401. [PMC free article] [PubMed] [Google Scholar]
- 8.Brabin BJ, Romagosa C, Abdelgalil S, Menendez C, Verhoeff FH, McGready R, Fletcher KA, Owens S, D'Alessandro U, Nosten F, Fischer PR, Ordi J. 2004. The sick placenta—the role of malaria. Placenta 25:359–378. doi: 10.1016/j.placenta.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 10.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101–110. doi: 10.1016/0092-8674(95)90056-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 12.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. 2003. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J 2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. 2010. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes—divide and conquer. PLoS Comput Biol 6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. 2000. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol 110:293–310. doi: 10.1016/S0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 15.Lau CK, Turner L, Jespersen JS, Lowe ED, Petersen B, Wang CW, Petersen JE, Lusingu J, Theander TG, Lavstsen T, Higgins MK. 2015. Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host Microbe 17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG. 2013. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, Howard RJ. 1997. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood 90:3766–3775. [PubMed] [Google Scholar]
- 18.Hsieh FL, Turner L, Bolla JR, Robinson CV, Lavstsen T, Higgins MK. 2016. The structural basis for CD36 binding by the malaria parasite. Nat Commun 7:12837. doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson BA, Welch TL, Smith JD. 2003. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol Microbiol 47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- 20.Smith JD, Rowe JA, Higgins MK, Lavstsen T. 2013. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol 15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avril M, Bernabeu M, Benjamin M, Brazier AJ, Smith JD. 2016. Interaction between endothelial protein C receptor and intercellular adhesion molecule 1 to mediate binding of Plasmodium falciparum-infected erythrocytes to endothelial cells. mBio 7:e00615-16. doi: 10.1128/mBio.00615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janes JH, Wang CP, Levin-Edens E, Vigan-Womas I, Guillotte M, Melcher M, Mercereau-Puijalon O, Smith JD. 2011. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog 7:e1002032. doi: 10.1371/journal.ppat.1002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennartz F, Adams Y, Bengtsson A, Olsen RW, Turner L, Ndam NT, Ecklu-Mensah G, Moussiliou A, Ofori MF, Gamain B, Lusingu JP, Petersen JE, Wang CW, Nunes-Silva S, Jespersen JS, Lau CK, Theander TG, Lavstsen T, Hviid L, Higgins MK, Jensen AT. 2017. Structure-guided identification of a family of dual receptor-binding PfEMP1 that is associated with cerebral malaria. Cell Host Microbe 21:403–414. doi: 10.1016/j.chom.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick CJ, Craig A, Roberts D, Newbold CI, Berendt AR. 1997. Intercellular adhesion molecule-1 and CD36 synergize to mediate adherence of Plasmodium falciparum-infected erythrocytes to cultured human microvascular endothelial cells. J Clin Invest 100:2521–2529. doi: 10.1172/JCI119794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith JD, Craig AG, Kriek N, Hudson-Taylor D, Kyes S, Fagan T, Fagen T, Pinches R, Baruch DI, Newbold CI, Miller LH. 2000. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc Natl Acad Sci U S A 97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampath S, Brazier AJ, Avril M, Bernabeu M, Vigdorovich V, Mascarenhas A, Gomes E, Sather DN, Esmon CT, Smith JD. 2015. Plasmodium falciparum adhesion domains linked to severe malaria differ in blockade of endothelial protein C receptor. Cell Microbiol 17:1868–1882. doi: 10.1111/cmi.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillrie MR, Avril M, Brazier AJ, Davis SP, Stins MF, Smith JD, Ho M. 2015. Diverse functional outcomes of Plasmodium falciparum ligation of EPCR: potential implications for malarial pathogenesis. Cell Microbiol 17:1883–1899. doi: 10.1111/cmi.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengtsson A, Joergensen L, Rask TS, Olsen RW, Andersen MA, Turner L, Theander TG, Hviid L, Higgins MK, Craig A, Brown A, Jensen AT. 2013. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J Immunol 190:240–249. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger SS, Turner L, Wang CW, Petersen JE, Kraft M, Lusingu JP, Mmbando B, Marquard AM, Bengtsson DB, Hviid L, Nielsen MA, Theander TG, Lavstsen T. 2013. Plasmodium falciparum expressing domain cassette 5 type PfEMP1 (DC5-PfEMP1) bind PECAM1. PLoS One 8:e69117. doi: 10.1371/journal.pone.0069117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. 1990. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet 336:1457–1460. doi: 10.1016/0140-6736(90)93174-N. [DOI] [PubMed] [Google Scholar]
- 31.Barry AE, Leliwa-Sytek A, Tavul L, Imrie H, Migot-Nabias F, Brown SM, McVean GA, Day KP. 2007. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog 3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruybal-Pesantez S, Tiedje KE, Tonkin-Hill G, Rask TS, Kamya MR, Greenhouse B, Dorsey G, Duffy MF, Day KP. 2017. Population genomics of virulence genes of Plasmodium falciparum in clinical isolates from Uganda. Sci Rep 7:11810. doi: 10.1038/s41598-017-11814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, Seguin-Orlando A, Willerslev E, Gilbert MT, Lusingu J, Theander TG. 2012. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci U S A 109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler A, Dankwa S, Bernabeu M, Harawa V, Danziger SA, Duffy F, Kampondeni SD, Potchen MJ, Dambrauskas N, Vigdorovich V, Oliver BG, Hochman SE, Mowrey WB, MacCormick IJC, Mandala WL, Rogerson SJ, Sather DN, Aitchison JD, Taylor TE, Seydel KB, Smith JD, Kim K. 2017. Linking EPCR-binding PfEMP1 to brain swelling in pediatric cerebral malaria. Cell Host Microbe 22:601–614.e5. doi: 10.1016/j.chom.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mkumbaye SI, Wang CW, Lyimo E, Jespersen JS, Manjurano A, Mosha J, Kavishe RA, Mwakalinga SB, Minja DT, Lusingu JP, Theander TG, Lavstsen T. 2017. The severity of Plasmodium falciparum infection is associated with transcript levels of var genes encoding endothelial protein C receptor-binding P. falciparum erythrocyte membrane protein 1. Infect Immun 85:e00841-16. doi: 10.1128/IAI.00841-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shabani E, Hanisch B, Opoka RO, Lavstsen T, John CC. 2017. Plasmodium falciparum EPCR-binding PfEMP1 expression increases with malaria disease severity and is elevated in retinopathy negative cerebral malaria. BMC Med 15:183. doi: 10.1186/s12916-017-0945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernabeu M, Danziger SA, Avril M, Vaz M, Babar PH, Brazier AJ, Herricks T, Maki JN, Pereira L, Mascarenhas A, Gomes E, Chery L, Aitchison JD, Rathod PK, Smith JD. 2016. Severe adult malaria is associated with specific PfEMP1 adhesion types and high parasite biomass. Proc Natl Acad Sci U S A 113:E3270–E3279. doi: 10.1073/pnas.1524294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jespersen JS, Wang CW, Mkumbaye SI, Minja DT, Petersen B, Turner L, Petersen JE, Lusingu JP, Theander TG, Lavstsen T. 2016. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRalpha1 domains. EMBO Mol Med 8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dondorp AM, Desakorn V, Pongtavornpinyo W, Sahassananda D, Silamut K, Chotivanich K, Newton PN, Pitisuttithum P, Smithyman AM, White NJ, Day NP. 2005. Estimation of the total parasite biomass in acute falciparum malaria from plasma PfHRP2. PLoS Med 2:e204. doi: 10.1371/journal.pmed.0020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendriksen IC, Mwanga-Amumpaire J, von Seidlein L, Mtove G, White LJ, Olaosebikan R, Lee SJ, Tshefu AK, Woodrow C, Amos B, Karema C, Saiwaew S, Maitland K, Gomes E, Pan-Ngum W, Gesase S, Silamut K, Reyburn H, Joseph S, Chotivanich K, Fanello CI, Day NP, White NJ, Dondorp AM. 2012. Diagnosing severe falciparum malaria in parasitaemic African children: a prospective evaluation of plasma PfHRP2 measurement. PLoS Med 9:e1001297. doi: 10.1371/journal.pmed.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox LL, Taylor TE, Pensulo P, Liomba A, Mpakiza A, Varela A, Glover SJ, Reeves MJ, Seydel KB. 2013. Histidine-rich protein 2 plasma levels predict progression to cerebral malaria in Malawian children with Plasmodium falciparum infection. J Infect Dis 208:500–503. doi: 10.1093/infdis/jit176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seydel KB, Fox LL, Glover SJ, Reeves MJ, Pensulo P, Muiruri A, Mpakiza A, Molyneux ME, Taylor TE. 2012. Plasma concentrations of parasite histidine-rich protein 2 distinguish between retinopathy-positive and retinopathy-negative cerebral malaria in Malawian children. J Infect Dis 206:309–318. doi: 10.1093/infdis/jis371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson J, Lam SW, Mahanta KC, Pattnaik R, Alam S, Mohanty S, Hasan MU, Hossain A, Charunwatthana P, Chotivanich K, Maude RJ, Kingston H, Day NP, Mishra S, White NJ, Dondorp AM. 2012. Relative contributions of macrovascular and microvascular dysfunction to disease severity in falciparum malaria. J Infect Dis 206:571–579. doi: 10.1093/infdis/jis400. [DOI] [PubMed] [Google Scholar]
- 44.Snow RW, Marsh K. 1998. New insights into the epidemiology of malaria relevant for disease control. Br Med Bull 54:293–309. doi: 10.1093/oxfordjournals.bmb.a011689. [DOI] [PubMed] [Google Scholar]
- 45.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 46.Magallon-Tejada A, Machevo S, Cistero P, Lavstsen T, Aide P, Rubio M, Jimenez A, Turner L, Valmaseda A, Gupta H, De Las Salas B, Mandomando I, Wang CW, Petersen JE, Munoz J, Gascon J, Macete E, Alonso PL, Chitnis CE, Bassat Q, Mayor A. 2016. Cytoadhesion to gC1qR through Plasmodium falciparum erythrocyte membrane protein 1 in severe malaria. PLoS Pathog 12:e1006011. doi: 10.1371/journal.ppat.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi AK, Sha W, Shulaev V, Stins MF, Sullivan DJ. Jr.. 2009. Plasmodium falciparum-infected erythrocytes induce NF-kappaB regulated inflammatory pathways in human cerebral endothelium. Blood 114:4243–4252. doi: 10.1182/blood-2009-06-226415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillrie MR, Lee K, Gowda DC, Davis SP, Monestier M, Cui L, Hien TT, Day NP, Ho M. 2012. Plasmodium falciparum histones induce endothelial proinflammatory response and barrier dysfunction. Am J Pathol 180:1028–1039. doi: 10.1016/j.ajpath.2011.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pal P, Daniels BP, Oskman A, Diamond MS, Klein RS, Goldberg DE. 2016. Plasmodium falciparum histidine-rich protein II compromises brain endothelial barriers and may promote cerebral malaria pathogenesis. mBio 7:e00617-16. doi: 10.1128/mBio.00617-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanson J, Lee SJ, Hossain MA, Anstey NM, Charunwatthana P, Maude RJ, Kingston HW, Mishra SK, Mohanty S, Plewes K, Piera K, Hassan MU, Ghose A, Faiz MA, White NJ, Day NP, Dondorp AM. 2015. Microvascular obstruction and endothelial activation are independently associated with the clinical manifestations of severe falciparum malaria in adults: an observational study. BMC Med 13:122. doi: 10.1186/s12916-015-0365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storm J, Jespersen JS, Seydel KB, Szestak T, Mbewe M, Chisala NV, Phula P, Wang CW, Taylor TE, Moxon CA, Lavstsen T, Craig AG. 2019. Cerebral malaria is associated with differential cytoadherence to brain endothelial cells. EMBO Mol Med 11:e9164. doi: 10.15252/emmm.201809164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuikue Ndam N, Moussiliou A, Lavstsen T, Kamaliddin C, Jensen ATR, Mama A, Tahar R, Wang CW, Jespersen JS, Alao JM, Gamain B, Theander TG, Deloron P. 2017. Parasites causing cerebral falciparum malaria bind multiple endothelial receptors and express EPCR and ICAM-1-binding PfEMP1. J Infect Dis 215:1918–1925. doi: 10.1093/infdis/jix230. [DOI] [PubMed] [Google Scholar]
- 53.Bernabeu M, Smith JD. 2017. EPCR and malaria severity: the center of a perfect storm. Trends Parasitol 33:295–308. doi: 10.1016/j.pt.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 55.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer Verlag, New York, NY. [Google Scholar]
- 56.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 57.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huynh-Thu VA, Saeys Y, Wehenkel L, Geurts P. 2012. Statistical interpretation of machine learning-based feature importance scores for biomarker discovery. Bioinformatics 28:1766–1774. doi: 10.1093/bioinformatics/bts238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic and clinical characteristics of the study cohorts. Download Table S1, PDF file, 0.1 MB (79.5KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clustergrams of var domain subtypes related by expression correlation for each study site, similar to Fig. 1. Heat maps indicate var domain subtypes known to be part of defined domain cassettes. Download FIG S1, EPS file, 2.3 MB (2.1MB, eps) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Broad phenotypic classification of domains targeted by individual var primers and associated color scheme. Download Table S2, PDF file, 0.1 MB (52.4KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcript levels of the var domain subtypes targeted by the 41 primer sets vary according by study site and malaria status. Expression density plots for each var domain subtype were stratified by site. BLZ, Malawi; GMC, Goa; TZ, Tanzania. The var domain subtypes are shown in the same order as in Fig. 1B. Download FIG S2, EPS file, 8.1 MB (7.8MB, eps) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MDCA and mProbes FWER for each var domain subtype in adult and childhood UM versus SM models. Download Table S3, PDF file, 0.1 MB (62.2KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary statistics for combining var profile models with PfHRP2 measurements for Malawi and Goa samples. Download Table S4, PDF file, 0.1 MB (42.8KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Classification of targeted domains into summed var sets corresponding to specific domain cassettes, binding phenotypes, and domain positions. Download Table S5, PDF file, 0.1 MB (53KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MDCA and mProbes FWER for each var domain subtype in childhood coma (cerebral malaria [CM]) and anemia (SA) models. Download Table S6, PDF file, 0.1 MB (58.1KB, pdf) .
Copyright © 2019 Duffy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.