Abstract
Recent experiments have provided new quantitative measurements of the rippling phenomenon in fields of developing myxobacteria cells. These measurements have enabled us to develop a mathematical model for the ripple phenomenon on the basis of the biochemistry of the C-signaling system, whereby individuals signal by direct cell contact. The model quantitatively reproduces all of the experimental observations and illustrates how intracellular dynamics, contact-mediated intercellular communication, and cell motility can coordinate to produce collective behavior. This pattern of waves is qualitatively different from that observed in other social organisms, especially Dictyostelium discoideum, which depend on diffusible morphogens.
Myxobacteria are common components of soil, but their life cycle is far from common. Although they are prokaryotes, their life, in some respects, is similar to that of multicellular organisms (1, 2). Under starvation conditions, a population of myxobacterial cells aggregates by streaming into a number of central foci, eventually forming at the focus a multicellular fruiting body. During this aggregation phase, the cells may pass through a period where the surface is swept by a complex pattern of waves, called the “ripple phase.” These waves are composed of bacteria moving in concert in such a way that colliding waves appear to pass through one another (3). This is quite unlike the seemingly similar phenomenon observed in Dictyostelium discoideum and in chemical waves where colliding wave fronts annihilate one another (4, 5). Here we present a quantitative model for the ripple phase in Myxococcus xanthus that reproduces most of the observed phenomena. A distinguishing feature of this model is that it depends only on intercellular communication by direct cell contact, without any diffusible morphogen signaling.
We shall base our model on the following consequences of experimental observations on Myxobacteria.
(i) Contact Signaling.
Myxobacteria signal via the C-signaling system, which operates only when two cells contact one another nearly end to end (3, 6, 7). The ripple patterns can be altered significantly, or even abolished, by manipulation of external C-signal protein concentration or dilution of wild-type cells by mutants that can receive, but not send, C-signal (3). Therefore, we shall base the model on signaling that depends entirely on direct cell contacts, with no diffusible signaling molecule.
(ii) Reversal Cycle.
Experiments on individual prerippling bacteria under various conditions show that they glide back and forth, reversing their direction spontaneously about every 5–10 min with a variance much smaller than the mean (see table 1 of ref. 8 and table 2 of ref. 9). Thus the times between reversals are not exponentially distributed, i.e., not Markovian. We interpret this to mean that the internal biochemical circuit controlling reversals contains a delay or cycle time for completion.
(iii) Density Dependence.
Measurements show that reversal frequencies depend on the amount of C-signal protein (10–12). Thus cells in a population where C-signaling is transmitted by cell collisions will alter their reversal frequencies in a density-dependent fashion. We will show that this density dependence is nonlinear, indicating a cooperative aspect of C-signaling.
(iv) Refractory Period.
The D. discoideum signaling system passes through a “refractory period” after a response to an external signal during which it is insensitive to subsequent signals. This property is essential to its ability to propagate waves. However, there is currently no direct evidence that, after reversal of direction, the C-signaling system passes through a similar refractory period, although there are indications in the reversal histogram for prerippling cells measured by Welch and Kaiser that show very few reversals at short times (see figure 4 of ref. 10). We shall demonstrate that a refractory period is necessary for the production of ripple waves and discuss possible experiments required to estimate its length.
We incorporate these properties into a mathematical model as follows.
A Model for the Ripple Phase in Myxobacteria
The motion of a single bacterium in the (x, y) plane can be described by the stochastic equation of motion:
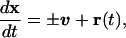 |
1 |
where ±v = (±vx, ±vy) are the instantaneous velocities in the x and y directions, (±) indicates that individual bacteria glide along their long axes and change their direction by simple reversals rather than turning. r(t) = (rx(t), ry(t)) are random terms modeling the variance in individual speeds.
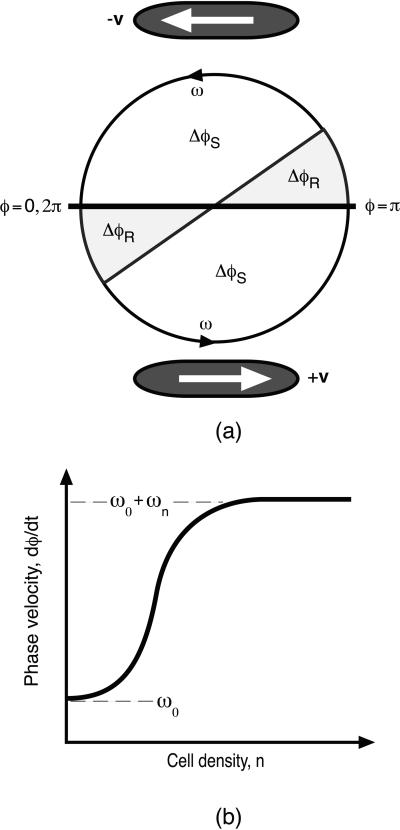
To describe the cyclic internal biochemical state, we define a periodic “phase” variable, 0 ≤ φ ≤ 2π, which locates the state of the C-signal-controlled reversal system in its cycle. We can picture this cycle as shown in Fig. 1, where we have plotted the phase on a circle: 0 ≤ φ < π corresponds to right-moving cells, and π ≤ φ < 2π corresponds to left-moving cells. A cell's state can be pictured as a point moving counterclockwise around the circle at a mean speed ω (see Fig. 1a). Each cell instantly reverses its direction as it crosses φ = 0, π. For an individual cell, φ(t) advances at a rate given by
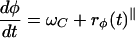 |
2 |
Here ωC is the phase velocity, which is affected by C-signaling because of collisions with other bacteria. As we discuss below, the C-signaling intensity, and thus the phase velocity, depends on the local cell density. The phase velocity, dφ/dt, is stochastic, so that the period is distributed about a mean τ = 2π/ω; we model this by adding a random term, rφ(t), to the phase velocity in Eq. 2.
Figure 1.
(a) The internal phase is plotted as motion on a circle with mean velocity ω. Cells move to the right (+v) when 0 ≤ φ < π and to the left (−v) when π ≤ φ < 2π. On switching directions, a cell is refractory to signaling for a period, ΔφR, after it enters the sensitive phase, ΔφS. (b) The density dependence of the phase velocity ω(n).
After a reversal, a cell does not respond to C-signal, although it can still deliver C-signal, which is denoted by the sectors denoted ΔφR in Fig. 1a. A cell leaving a refractory sector enters a sensitive sector, ΔφS, where it is sensitive to C-signal until φ = π, whereupon it reverses and enters a refractory sector.
When bacteria aggregate in the preripple phase of their life cycle, their behavior changes in response to cell density. Cell populations that are in the early stages of fruiting body development increase their probability of reversing their direction in response to C-signaling. Therefore, the average velocity at which an individual traverses its reversal cycle will depend on the local density of its neighbors (3, 6).** We describe the motion of a population of bacteria by using the density function n(t, x, y, θ, φ), giving the number of bacteria at time t, position (x, y), moving in the direction θ = tan−1 (vy/vx), with phase φ. The density function n(t, x, y, θ, φ) obeys the general conservation law (i.e. the Fokker–Planck equation for Eqs. 1 and 2 of the form
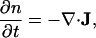 |
where J comprises the spatial, angular, and phase flux of individuals in (x, y, θ, φ) space.
We can simplify considerably this description by taking advantage of the observation that individual cells participating in ripple-phase waves are aligned in the direction of the wave propagation (3, 10). Therefore, we can assume that the cells are aligned parallel to the x axis and so glide only in ±x direction, although they may drift randomly in both x and y directions. This eliminates the angular variable, θ, so that the density equation for n(x, y, φ) is:
 |
3 |
where ω± refers to right- (+) or left- (−) moving cells [i.e., ±1 = sign(π − φ)]. Dx is the effective diffusion coefficient caused by the variance in the gliding speed, Dy is the diffusive flux of cells in the y direction, and Dφ is the effective diffusion coefficient in the biochemical phase space. C-signaling takes place only between cells that meet head-on moving in the opposite direction, so we will need to compute the local spatial density of right- and left-moving cells by integrating the density, n(t, x, φ), over φ:
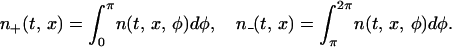 |
4 |
To complete the model, we must specify the dependence of cell reversals on the C-signaling intensity via the local population density; that is, an expression for ωC in Eq. 2. For cells in their sensitive phase, the frequency of reversals increases monotonically with the density of opposite moving cells, so that ω∓ must be a function of n± computed from Eq. 4: ω∓(n±). This density dependence must ultimately saturate; thus, a convenient mathematical representation is given by the Hill function shown in Fig. 1b:‡‡
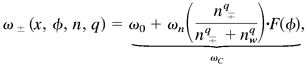 |
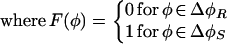 |
5 |
Here ω0 is the phase velocity of a single cell in the absence of other cells, and ωn is the increase in the phase velocity for a cell in its sensitive period, ΔφS, when the local cell density is n. Thus the second term in Eq. 5 gives gliding cells a bigger probability of reversing in regions where there are many opposite-moving cells. At small densities, ω increases as the qth power of density; therefore, this exponent characterizes the cooperativity of the signal processing or transduction; q = 1 corresponds to the case where reversals of a nonrefractory cells increase in proportion to collisions. It turns out that only values of q ≥ 3 can produce rippling, so that the ripple phase is a collective phenomenon. Note that Eq. 5 says that the phase velocity depends only on the local cell density, i.e., we have not introduced any diffusible morphogen analogous to “quorum sensing” in bacteria with diffusible signals (15, 16).
Results
The mean field models described by Eqs. 3 and 5 are solved numerically to produce the characteristic waves observed in the ripple phase. Using the density n to calculate the phase velocity, we can then solve the stochastic models Eqs. 1 and 2 to illustrate the path of individual cells. With parameters estimated directly from the experiments of Welch and Kaiser (10), we performed parameter scans to determine the range that supported rippling. These are explained in the supporting information on the PNAS web site, www.pnas.org. The analysis presented supports the numerical parameter ranges. Thus rippling is not a robust property of bacterial aggregation and so may provide insight into the properties of the C-signaling system. The numerical simulation procedure based on refs. 17–19 is also described in the supporting information. Movies illustrating the various phenomena can be downloaded from the PNAS web site with the supporting information.
Unidirectional Wave Propagation.
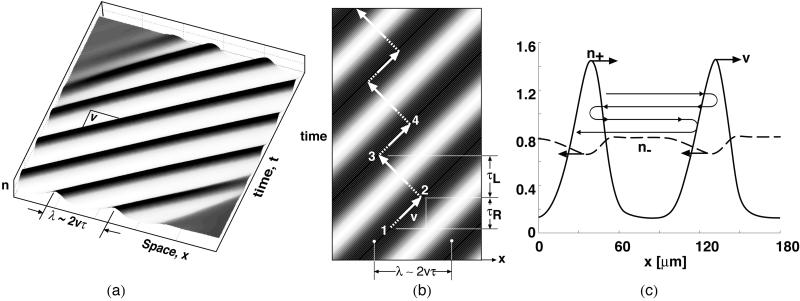
The principles underlying the ripple phase can best be illustrated by examining one-dimensional unidirectional wave propagation obtained by setting in Eq. 3 all properties constant in the y direction and fixed concentration boundary conditions in the x direction. The waves are best appreciated via the movies; however, a static representation can be illustrated in the space–time plots shown in Fig. 2 (See Movie 1, which is published as supporting information on the PNAS web site). Fig. 2a shows that the solution to Eq. 3 generates a unidirectional wave train. The wave amplitude is such that the density of bacteria in a ripple crest is up to 10 times that in the trough, depending on the ratio of convective and diffusive fluxes in Eq. 3. In the supporting information, we show that the wavelength, λ, is approximately
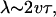 |
6 |
where v is the mean gliding speed and τ the mean period between reversals in a trough (where C-signaling intensity is weakest). The reversal time in the trough ≈4.2 min, as estimated from the right peak of the histogram in Welch and Kaiser (10) The wave speed is ∼11 μm/min, about the same as individual velocity. The resulting wavelength is ∼90 μm, in agreement with table 1 in ref. 10.
Figure 2.
(a) A space–time plot showing a train of right-moving waves (crests are black, troughs white). See Movies 1–3, which are published as supporting information on the PNAS web site. (b) Using Eqs. 1 and 2, one can follow a material point equivalent to a single bacterium as it participates in a wave train moving to the right. The trajectory of a bacterium moves to the right with the wave crest 1→2, then reverses and travels to the left through the trough to the top of the next wave crest, 2→3, then reverses again and travels rightward with the crest 3→4. The dashed portion indicates the refractory period. Because a cell encounters fewer collisions (C-signals) in troughs, the time between reversals is longer when the cell is moving in the troughs against the direction of the wave train. Therefore, there is a slow drift to the left, against the direction of wave propagation. (c) Distributions of right-moving (solid) and left-moving cells (dashed) in a unidirectional wave train moving to the right with velocity v (computed from Eq. 4). Cells alternate between crests with a slow drift to the left: vdrift ≈ v(τ+ − τ−)/(τ+ + τ−), where τ± is the reversal period in the right- (+) and left- (−) moving wave trains. The wavy line traces the orbit of a single material point (bacterium). Note that the left-going density, n−, is nearly constant at its mean value, whereas the amplitude of n+ is periodic and large.
The necessary condition for wave propagation is given by a parameter inequality:
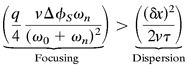 |
(see Eq. 14, which is published as supporting information on the PNAS web site www.pnas.org). This ensures that the focusing effect of density-dependent reversals that speed the cell to its refractory period can compensate for the drift to maintain the wave train. Here (δx)2 is the variance in position caused by the drift in velocity and phase.
Individual Cell Behavior in Unidirectional Waves.
Welch and Kaiser also tracked the paths of individuals participating in the ripple phase (see Fig. 4 and accompanying movies in ref. 10). We can imitate their single-cell tracking experiments by superimposing the trajectories of single bacteria, computed by using the stochastic Eqs. 1 and 2, onto the solution of the density Eq. 3. First, consider individuals from a train of right-moving waves; Movie 1 illustrates the motions dynamically. A static picture is shown in Fig. 2b, where the motion of a single cell is traced on a space–time plot superimposed on a unidirectional wave train. Refractory and sensitive periods are shown dashed and solid, respectively. The waves are generated by cells that oscillate back and forth, corresponding to right angle turns in Fig. 2b. In the crests, more cells are moving in the same direction as the crests (1→2), while in the troughs more cells are moving against the waves (2→3).
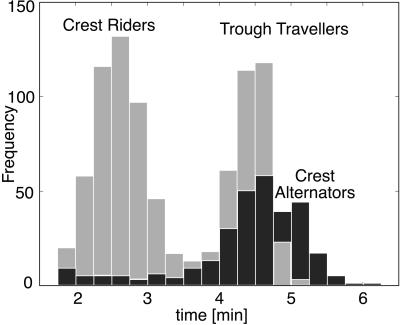
Figure 4.
The distribution of reversal frequencies (compare with figure 7 of ref. 10). The lower peak corresponds to cells that reverse in the crests (e.g., 1→2 in Fig. 2b); the larger peak corresponds to cells that traverse the troughs (e.g., 2→3 in Fig. 2b). There is also a third, somewhat longer, period when there are counterpropagating waves because of cells that alternate between the two wave trains, seen in the broader and higher second peak in the experimental histogram (see figure 7 of ref. 10).
When a cell reverses in a crest, it is refractory to signals from oncoming cells and can therefore penetrate the wave and glide against the wave velocity without turning. By the time the cell emerges from its refractory period, it is in the trough, where it encounters few reversal signals, and so it reaches the previous crest in the wave train about the time that its phase is close to its reversal point. Entering the wave, the cell encounters increasing interactions, which advances its phase, so it quickly reverses again, completing the cycle. Thus, wave crests constantly lose cells to reversals but gain new ones from the crest ahead.
Cells entering the wave crest and reversing go into their refractory period, where their phase velocity is at its minimum, which tends to populate the refractory sector, thus synchronizing the cells' phases. This synchronization maintains the ripple's stability (a quantitative version of this argument is presented in the supporting information on the PNAS web site). The slow drift of cells in the direction opposite the direction of the wave train (see Fig. 2b) can be understood from Fig. 2c, which plots the density of right- and left-going cells. In a unidirectional wave train, the concentration of left-going cells in the right-going crests is much larger than the concentration of right-going cells in the trough. Therefore, less C-signaling events are received while moving in the trough, so the average time a cell spends there is longer, resulting in a bimodal reversal frequency distribution, as shown in Fig. 4.
Interpenetrating Waves.
A feature of the ripples that distinguishes them from other developmental waves is that colliding waves do not annihilate but appear to pass through one another (Movie 2a which is published as supporting information on the PNAS web site). When two wave fronts collide, cells in their sensitive phase increase their reversal frequency because of increased collisions, whereas cells in their refractory phase continue unaffected. Consequently, the outgoing waves consist of a combination of individuals from both incoming waves, as shown in Fig. 3b. This give the appearance of waves passing through one another, analogous to soliton water waves (ref. 20; see also ref. 21). Movie 2b shows the computed behavior of individual bacteria superimposed on the wave pattern.
Figure 3.
(a) Space–time plot of two colliding wave trains. See Movie 3, which is published as supporting information on the PNAS web site. (b) Two colliding waves appear to pass through one another. Those cells in the incoming wave that are in their refractory period continue to pass through, whereas those in their sensitive period may reverse on collision with counter-moving cells. Therefore, the outgoing waves after a collision consist of individuals from both incoming waves. (c) Plot of the directional densities, n+ (solid) and n− (dashed), in counterpropagating wave trains moving at velocities v and −v, respectively.
Tracing individuals in two counterpropagating wave trains produces a different picture from unidirectional waves (see Movie 2c). Here most of the cells are caught in crests, moving with the right-going crest to the right and with the left-going crest to the left. Cells switch their allegiance from one crest to another when two crests collide, confirming the interpretation of Sager and Kaiser and of Welch and Kaiser that, although wave crests appear to pass through one another, most of the individual cells reverse (3, 10). There is an obvious symmetry between right- and left-going cells, and therefore no drift (net transport of bacteria) occurs in this situation. Fig. 3c plots the density of right- and left-going cells in a colliding wave train. Most of the time, collisions between oppositely moving cells in the troughs are spatially separated so that C-signaling levels are low. Therefore, even though most of the cells move in crests all the time, their reversal time distribution in Fig. 4 is centered near the “trough” peak of the unidirectional wave. A quantitative analysis demonstrating the stability of the two-dimensional waves is given in the supporting information on the PNAS web site. We will see below when we treat the two-dimensional case that the bimodal reversal frequency distribution measured in figure 7 of Welch and Kaiser (10) can be interpreted as an average of the reversal histograms in Fig. 4.
The Pattern of Cell Reversals.
Bi- and unidirectional waves show two distinct types of individual behaviors. Cells in bidirectional waves (“wave alternators”) tend to move within the crest all the time, switching wave trains when they collide. The cells in unidirectional waves tend either to move with the wave in the direction of wave propagation (“crest riders”) or to travel in a trough while going the opposite way (“trough travelers”). Trough travelers (Fig. 2c) are not greatly influenced by C-signaling, because they move in regions with few counterpropagating cells. Similarly, crest alternators (Fig. 3c) are not influenced much by C-signaling, because they spend most of their time “moving with the crowd,” and by the time they encounter a wave collision, they are nearly ready to reverse on their own. Therefore, for both trough travelers and wave alternators, the reversal time is close to τ∼π/ω0, the time that determines the wavelength (λ = 2vτ) for both uni- and bidirectional wave trains. The situation is different for crest riders (Fig. 2c), which are constantly advanced in phase as they collide with counterpropagating cells, so their reversal time is much smaller. In the reversal distributions and individual trajectories of Welch and Kaiser, all cell types are represented. Each cell can move for some time with one or the other wave and then commence alternating between two wave crests (10). The bimodal reversal distribution arises as follows. Even though both alternators and crest riders travel in the crests, the alternators arrive at the point of wave collision being ready to reverse, so they are not much affected by C-signaling, i.e., they reverse spontaneously and glide with the oppositely moving crest. However, the “crest riders” are probably not ready to reverse at the collision point, but the C-signaling dose they get during the collision significantly advances them in their cycle, resulting in shorter reversal times for crest riders.
Finally, it is worth noting that the phases of cells participating in a unidirectional wave train are not spatially synchronized: in any volume element, phases of the cells traveling in the same direction are uncorrelated. In contrast, cells participating in two colliding wave trains do have their phases spatially synchronized, so that a cohort traveling together in a crest will also have nearby phases.
Initiation of Waves.
When cell density exceeds a critical value, a uniform field becomes unstable: if the cells are aligned, small perturbations initiate an outward-propagating wave. Fig. 7, which is published as supporting information on the PNAS web site, shows how the initial outgoing wave pair triggers “echo” waves that maintain the source until eventually the field is filled with counterpropagating and colliding waves (Movies 3 a and b). In a two-dimensional field, this corresponds to a bulls-eye patterns (see figure 4 of ref. 3).
In the experiments of Welch and Kaiser, the waves appeared simultaneously around the entire periphery of the colony. Starting with a field of a small randomly distributed perturbations, several collision times are required to reach a steady-state wave train. Therefore, our numerical solution of the model equations predicts the existence of some transition period that probably took place before the waves became optically visible.
Two-Dimensional Waves.
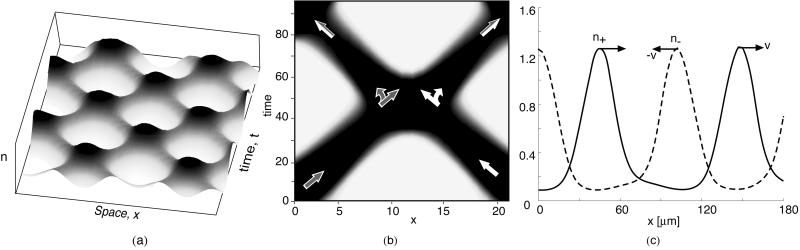
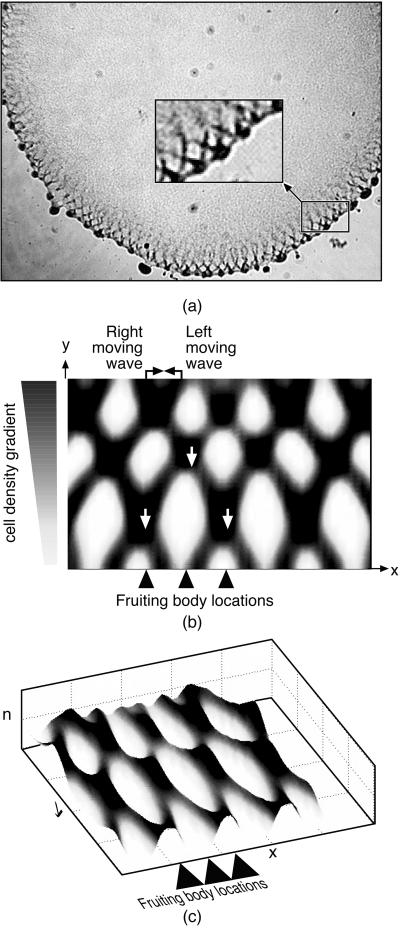
The ripples observed by Welch and Kaiser at the edge of their submerged agar culture consisted of a pair of obliquely oriented oppositely propagating wave trains (see Fig. 5a and figure 2 of ref. 10). Around the periphery of the population, incipient fruiting bodies are forming, spaced roughly one wavelength apart. The radial flux of cells out of colony into the developing fruiting bodies creates a cell density gradient in an annular ring around the colony periphery. If this density gradient is introduced into the two-dimensional model (Eqs. 3–5), it reproduces closely the observed wave pattern. This is shown in Fig. 5 b and c as cell density and perspective wave plots. Movies 2 a–c show the pattern of counterpropagating waves that appear to pass through one another as well as the tracks of individual cells.
Figure 5.
(a) During the early ripple phase, bacteria form a large disc-like population. Counter-rotating waves propagate along the rim of the colony as streams form to feed aggregation centers—precursors of fruiting bodies—distributed along the colony rim about a wavelength apart. The Inset shows the fruiting bodies being fed by the radial component of flow (frame from ref. 10). (b and c) Counterpropagating wave train computed from the model Eqs. 3 and 5 in contour and perspective plots. The tilt in the waves is caused by the density gradient between the proximal (Top) and peripheral (Bottom) regions of the colony, which causes a slow drift of individuals outward, bringing with it cells with retarded phases. This phase-coupling causes the waves to tilt: right-moving waves tilt right, and left-moving waves tilt left. Where the waves intersect, the population density is doubled. The intersection regions move downward to the colony periphery, where they seed the incipient fruiting body aggregates with a spacing about equal to the wavelength of the two wave trains.
Spacing of Fruiting Bodies.
The wave pattern generated by the model also shows how the spacing of fruiting bodies around the periphery evolves. Where the oblique wave trains intersect, the local bacterial population is almost twice that in the individual waves (see Fig. 5b and Movie 2a). As the waves counterpropagate, these intersection regions move radially outwards (downwards in Fig. 5b). Thus the colony edge is “pulsed” with extra bacteria each time an intersection hits the boundary. Because the intersections are spaced about a wavelength apart, the wavelength of the counterpropagating waves will be echoed by the spacing of the peripheral aggregations that form the incipient fruiting bodies. Fig. 5c shows a perspective view of how the wave intersections determine the fruiting body locations.
Discussion
There are many examples of pattern formation in microorganisms and a large literature on mathematical models describing them. A few notable examples, among many, include periodic patterns in colonies of Proteus mirabilis (22, 23), Salmonella typhimurium (24), Escherichia coli (25), Bacillus subtilis (26), and D. discoideum (27–29). The signature feature of these patterns is that they depend on diffusion-mediated chemotaxis and/or growth and death of the cells. This is quite different from the rippling pattern in myxobacteria, which depend on contact-mediated signaling and advective motion rather than diffusion. Although some aspects of swarming behavior of myxobacteria colonies were considered in (30–33), to the best of our knowledge, rippling behavior has not been successfully modeled before.
The mathematical model presented here has four critical ingredients: (i) each bacterium possesses an internal biochemical cycle whose progression controls the time between gliding direction reversals. (ii) Contact-mediated C-signaling alters the probability of reversal by modulating the rate of the biochemical cycle. (iii) Immediately after reversal, the C-signaling system enters a refractory phase, where it does not respond to collisions with countermoving cells. (iv) The response to C-signal depends nonlinearly on local cell density.
The model successfully reproduces the principal features of the ripple phase, both qualitatively and quantitatively. For example, the spacing of the waves obeys the predicted relationship (Eq. 6), the location of the nascent fruiting bodies, and the behavior of individual cells as they move with and against the wave directions. Most importantly, the model demonstrates that no diffusible signals are necessary: the ripple patterns can be generated by density dependent intercellular signals relayed by cell contact alone.
Indeed, the success of the model leads us to view the four assumptions on which the model is based as predictions that can be addressed experimentally: the existence of a refractory period, nonlinear density dependence, and wave tilt being determined by the transverse density gradient. A crucial prediction of the model—confirmed by the experiments of Welch and Kaiser (10)—is the correlation between the behavior of individual cells with the macroscopic properties of the waves.
The importance of rippling behavior for myxobacterial cell development is unknown. Although rippling precedes fruiting body formation, fruiting body formation can proceed without the ripple phase preamble. However, when rippling is present, the cells are able to distribute themselves with a nearly constant time-averaged spatial density that fosters the formation of equidistant fruiting bodies. The model shows how an even distribution arises in the experimental system of Welch and Kaiser (10). The cells also align in ripples, enhancing the formation of streams into nascent fruiting bodies. Changes in the quantitative characteristics of the C-signaling and gliding systems can trigger the transition from rippling to aggregation (11). Thus, rippling patterns are not robust in the same sense that the swarming and aggregation patterns are. Their importance to biologists may lie in their value as a sensitive spatiotemporal assay for cell signaling and motility. That is, rippling is a transient pattern that reveals important information (e.g., refractory period and cooperativity) about intercellular signaling.
Supplementary Material
Acknowledgments
We thank David Zusman, John Kirby, and Dave Astling for invaluable input during the development of this model. A special thanks to Philip Colella, who introduced us to the numerical methods used in the simulations. G.O. and D.K. were supported by National Institutes of Health grants. A.M. was supported by a National Science Foundation grant. O.I. was supported by a Howard Hughes Medical Institute predoctoral fellowship.
Footnotes
A model representing the biochemical cycle by a delay, or functional, kinetic equation yields similar results to the model presented here. However, the explicit representation of the C-signal biochemistry by a phase variable is simpler and makes contact with a substantial literature on phase-coupled systems (14).
In this model, the C-signaling system behaves in some respects analogous to an “integrate and fire” neuron (13).
Eq. 5 is a simple and convenient representation for the density dependence of the phase velocity. We will use it, because little is known about how the biochemistry of the reversal cycle is affected by C-signaling protein. In the supporting information on the PNAS web site (www.pnas.org), we discuss modifications of Eq. 5 to address the mutant dilution experiments of Sager and Kaiser (3). However, we show that the results of our simulations are not sensitive to this modification.
References
- 1.Dworkin M, Kaiser D. Science. 1985;230:18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- 2.Shimkets L. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sager B, Kaiser D. Genes Dev. 1994;8:2793–2804. doi: 10.1101/gad.8.23.2793. [DOI] [PubMed] [Google Scholar]
- 4.Monk P B, Othmer H G. Proc R Soc London. 1990;240:555–589. [Google Scholar]
- 5.Murray J D. Mathematical Biology. Berlin: Springer; 1993. [Google Scholar]
- 6.Kim S, Kaiser D, Kuspa A. Annu Rev Microbiol. 1992;46:117–139. doi: 10.1146/annurev.mi.46.100192.001001. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin M, Eide D. J Bacteriol. 1983;154:437–442. doi: 10.1128/jb.154.1.437-442.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jelsbak L, Søgaard-Andersen L. Proc Natl Acad Sci USA. 1999;96:5031–5036. doi: 10.1073/pnas.96.9.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi W, Ngok F, Zusman D. Proc Natl Acad Sci USA. 1996;93:4142–4146. doi: 10.1073/pnas.93.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch R, Kaiser D. Proc Natl Acad Sci USA. 2001;98:14907–14912. doi: 10.1073/pnas.261574598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jelsbak L, Søgaard-Andersen L. Curr Opin Microbiol. 2000;3:637–642. doi: 10.1016/s1369-5274(00)00153-3. [DOI] [PubMed] [Google Scholar]
- 12.Gronewold T, Kaiser D. Mol Microbiol. 2001;40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 13.Glass L, Mackey M C. From Clocks to Chaos: The Rhythms of Life. Princeton, NJ: Princeton Univ. Press; 1988. [Google Scholar]
- 14.Winfree A. The Geometry of Biological Time. Berlin: Springer; 2001. [Google Scholar]
- 15.Bassler B L. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 16.Oleskin A V, Botvinko I V, Tsavkelova E A. Mikrobiologiya. 2000;69:309–327. [PubMed] [Google Scholar]
- 17.Lomax H, Pulliam T H, Zingg D W. Fundamentals of Computational Fluid Dynamics. Berlin: Springer; 2001. [Google Scholar]
- 18.Press W H. Numerical Recipes in C: The Art of Scientific Computing. Cambridge, U.K.: Cambridge Univ. Press; 1997. [Google Scholar]
- 19.Van Leer B. J Comp Phys. 1977;23:276–299. [Google Scholar]
- 20.Filippov A T. The Versatile Soliton. Boston: Birkhäuser; 2000. [Google Scholar]
- 21.Meinhardt H, Prusinkiewicz P, Fowler D. The Algorithmic Beauty of Sea Shells. Berlin: Springer; 1998. [Google Scholar]
- 22.Esipov S, Shapiro J. J Math Biol. 1997;36:249–268. [Google Scholar]
- 23.Czirok, A., Matsushita, M. & Vicsek, T. (2001) Phys. Rev. E 63, article 031915. [DOI] [PubMed]
- 24.Woodward D, R, T, Myerscough M, Murray J D, Berg H C. Biophys J. 1995;68:2181–2189. doi: 10.1016/S0006-3495(95)80400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyson R, Lubkin S R, Murray J D. Proc R Soc London Ser B. 1999;266:299–304. doi: 10.1098/rspb.1999.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendelson N, Lega J. J Bacteriol. 1998;180:3285–3294. doi: 10.1128/jb.180.13.3285-3294.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palsson E, Othmer H. Proc Natl Acad Sci USA. 2000;97:10448–10453. doi: 10.1073/pnas.97.19.10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano S. Dev Growth Diff. 2000;42:541–550. doi: 10.1046/j.1440-169x.2000.00547.x. [DOI] [PubMed] [Google Scholar]
- 29.Maree A, Hogeweg P. Proc Natl Acad Sci USA. 2001;98:3879–3883. doi: 10.1073/pnas.061535198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfistner B, Alt W. In: Biological Motion. Alt W, Hoffmann G, editors. New York: Springer; 1989. pp. 556–563. [Google Scholar]
- 31.Stevens A. J Biol Syst. 1995;3:1059–1068. [Google Scholar]
- 32.Deutsch A. J Biol Syst. 1995;3:947–955. [Google Scholar]
- 33.Othmer H, Stevens A. Siam J Appl Math. 1997;57:1044–1081. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.