Abstract
Sequential processing of the amyloid precursor protein (APP) by β- and γ-secretases generates the Aβ peptide, a major constituent of the senile plaques observed in Alzheimer's disease. The cleavage by γ-secretase also results in the cytoplasmic release of a 59- or 57-residue-long C-terminal fragment (Cγ). This processing resembles regulated intramembrane proteolysis of transmembrane proteins such as Notch, where the released cytoplasmic fragments enter the nucleus and modulate gene expression. Here, we examined whether the analogous Cγ fragments of APP also exert effects in the nucleus. We find that ectopically expressed Cγ is present both in the cytoplasm and in the nucleus. Interestingly, expression of Cγ59 causes disappearance of PAT1, a protein that interacts with the APP cytoplasmic domain, from the nucleus and induces its proteosomal degradation. Treatment of cells with lactacystin prevents PAT1 degradation and retains its nuclear localization. By contrast, Cγ57, a minor product of γ-cleavage, is only marginally effective in PAT1 degradation. Furthermore, Cγ59 but not Cγ57 potently represses retinoic acid-responsive gene expression. Thus, our studies provide the evidence that, as predicted by the regulated intramembrane proteolysis mechanism, Cγ seems to function in the nucleus.
Amyloid precursor protein (APP) is a ubiquitously expressed type I membrane protein with a large external domain, a single transmembrane domain, and a short cytoplasmic tail (1, 2). APP undergoes a series of proteolytic events near and within its membrane domain (3–5), which results in the shedding of the large N-terminal domain, generation of the extracellular 40- or 42-residue-long Aβ peptides, and release of the resultant 59- or 57-residue-long C-terminal fragments (Cγ; cleaved by γ-secretase) in the cytoplasm. The processing of APP is similar to that of other proteins that undergo regulated intramembrane proteolysis (RIP; refs. 6–8). One of the best-characterized examples of RIP is the processing of Notch (8, 9), which is remarkably similar to that of APP insofar as the same activity (γ-secretase/presenilin 1) cuts within their transmembrane segments (10–12). Upon release, the Notch intracellular domain (NICD) enters the nucleus, interacts with CSL proteins, and regulates gene expression (9). This raises the possibility that Cγ, the analogous fragment released from APP, may regulate events in the nucleus.
We previously identified a protein termed PAT1 in a yeast two-hybrid system by using the 11-aa residue peptide (amino acid sequence KKKQYTSIHHG) from the juxtamembrane region of the APP cytoplasmic domain, which constitutes a part of Cγ (13). Here, we report that PAT1 also is present in the nucleus and that Cγ causes selective degradation of PAT1 and represses retinoic acid-responsive gene expression.
Methods
Cloning and Generation of Inducible Cell Lines.
Cytomegalovirus promoter-driven and tetracycline-regulated mammalian expression plasmids of FLAG-PAT11–585, FLAG-PAT11–411, FLAGPAT11–270, GFP-Cγ59, GFP-Cγ57, and GFP-C31 were constructed in pcDNA4/TO (Invitrogen) by standard techniques, using the full-length PAT1 that has been described (13). We were not able to observe the expression of the untagged version of Cγ constructs presumably because of high turnover and used only the green fluorescent protein (GFP)-tagged versions throughout these studies. To prepare the stable cell lines, MDCK (Madin–Darby canine kidney) cells were transfected with pcDNA6/TR (Invitrogen) and selected in Blasticidin, and an individual clone (designated MDCKTR) exhibiting high induction (>30-fold) was isolated. MDCKTR cells were transfected with PAT1 or its truncation mutants and selected in Zeocin. Single-cell clones were screened by indirect immunofluorescence and Western blot analysis.
Cell Culture, Transfection, and Drug Treatment.
MDCK clones were maintained in MEM with 10% FCS, 2 mM glutamine, and 150 μg/ml each of penicillin and streptomycin. To induce the expression of proteins, cells were incubated for 15–18 h with 1 μg/ml tetracycline (Invitrogen) and 2 mM sodium butyrate. In transient transfections, cells were induced with tetracycline alone. Staurosporine (Sigma) treatment was usually for 4 h after 15 h of induction. When used, lactacystin was added 15 min before the addition of staurosporine and was present during staurosporine treatment. Transient transfection was performed by using FuGENE6 according to the manufacturer's instructions (Roche Diagnostics).
Immunofluorescence.
Cells were plated on glass coverslips for up to 18 h, induced for 15 h, and fixed in methanol at −20°C for 7 min. In some experiments, the cells were fixed in 4% paraformaldehyde (Electron Microscopy Science) for 30 min at room temperature and permeabilized with 0.5% Triton X-100 for 4 min. The cells were blocked in PBS containing 10% FCS and 0.025% saponin (Sigma). Antibody incubation was performed for 30–45 min at 37°C. The Cγ constructs were visualized by using 369 antibody against the cytoplasmic domain of APP because the GFP signal in GFP-Cγ fusion proteins was extremely weak and became undetectable after methanol fixation.
Luciferase Assay.
CV-1 or MDCKTR cells were plated in 12-well plates 24 h before transfection by using FuGENE6 (Roche Diagnostics). Each well received reporter pRARE-TK-luc, cmv-β-gal, a pRAR plasmid DNA (200 ng, 100 ng, and 5 ng, respectively), with an additional one or two plasmids as indicated. All-trans-retinoic acid (RA; 1 μM) was added after 24 h, and cells were harvested 48 h after transfection and assayed for luciferase (Promega luciferase assay kit) and β-galactosidase activities (Sigma). The luciferase activity was normalized by the β-galactosidase activity. Values shown are averages of transfection carried out in duplicate or triplicate and repeated at least three times. MDCKTR cells showed similar results.
Western Blotting.
Western blotting was performed essentially as described (13). Antibodies used were anti-FLAG M5 (Sigma), anti-ER (Calbiochem), and antibody 369 (gift of S. Gandy, New York University).
Results
PAT1 Is a Nucleocytoplasmic Protein.
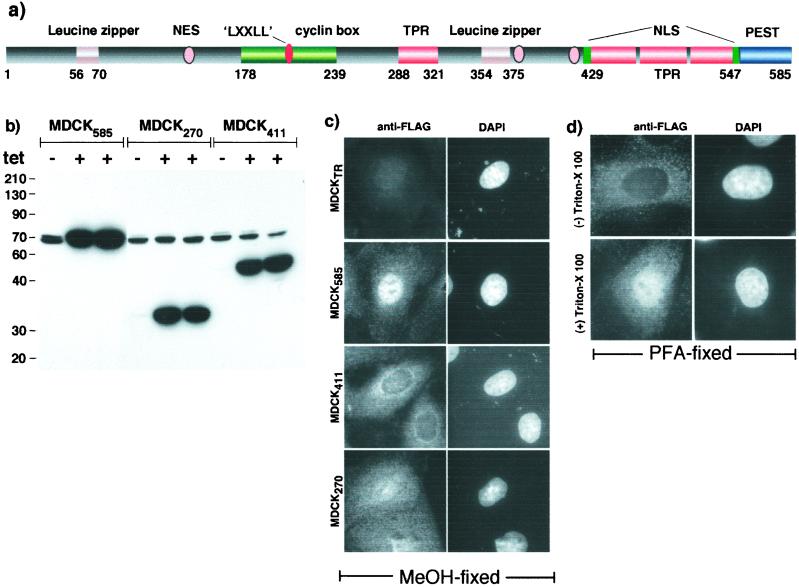
PAT1 is a soluble cytoplasmic protein related to kinesin light chain. Sequence analysis predicts the presence of several protein–protein-interacting motifs (Fig. 1a), including two leucine zipper motifs generally involved in dimerization and also in binding DNA (14), a putative cyclin box found in many cyclin-dependent kinases, a “LXXLL” motif (NR-Box) that is present in transcription coregulators (15), four TPR domains also present in kinesin light chain (16), and a PEST protein-degradation domain (17) at the extreme C terminus that is common in regulatory proteins with high turnover rates. Also present are multiple putative nuclear localization signals and leucine-rich nuclear export signals (NESs) (18). To study how PAT1 modulates APP function, we made several truncation mutants of PAT1. However, efforts to generate stable cell lines expressing any of the mutant proteins were unsuccessful, suggesting that such changes were lethal. We, therefore, generated MDCK cell lines that express FLAG-tagged PAT1 constructs in a tetracycline-inducible manner. Fig. 1b shows that various MDCK clones expressed little or no ectopic PAT1 in the absence of 1 μg/ml tetracycline but showed robust expression in its presence and that the expression levels were comparable in these clones.
Figure 1.
PAT1 and its intracellular localization. (a) Schematic representation of putative motifs present in PAT1. See text for details. (b) Inducible expression of PAT1 and deletion mutants. MDCK cells stably transfected with PAT11–585, PAT11–270, or PAT11–411 were induced (+) or not (−) with tetracycline and analyzed by Western blotting with anti-PAT1 antibody (mAb37). The endogenous PAT1 shows as a doublet at 70 kDa. (c) MDCK cells were fixed in methanol and stained with anti-FLAG antibody (Left) and counterstained with 4′,6-diamidino-2-phenylindole (Right). (d) MDCK585 cells were fixed in 4% paraformaldehyde and treated without (Upper) or with (Lower) 0.5% Triton X-100 before staining with anti-FLAG antibody (×63 objective).
To study the intracellular distribution of PAT1, MDCK clones were induced for 15 h, fixed in methanol at −20°C, and subjected to immunofluorescence microscopy using anti-FLAG antibody. Whereas the parental cell line (MDCKTR) expressing only the tet repressor showed no signal, MDCK585 cells expressing full-length PAT1 (PAT11–585) showed the protein to be present in both cytoplasm and nucleus, with the nuclear signal being much more prominent than the cytoplasmic staining (Fig. 1c). Interestingly, PAT11–411 was excluded from the nucleus, whereas PAT11–270 was distributed equally in the two compartments. Previously, we failed to detected PAT1 signal in the nucleus presumably because the tyramide signal-amplification system did not gain access to the nuclear compartment (13). The nuclear presence of PAT1 was not an artifact of methanol fixation; PAT1–585 cells that were fixed in paraformaldehyde showed nuclear staining of PAT1 when permeabilized with Triton X-100 (Fig. 1d). We also found the full-length FLAG-PAT1 to be present in the nucleus and FLAG-PAT1–411 in the cytoplasm when transiently expressed in M17 neuroblastoma, COS1, HeLa, HEK293, and MCF-7 cell lines (not shown).
Cγ Localizes to Both Cytoplasm and Nucleus.
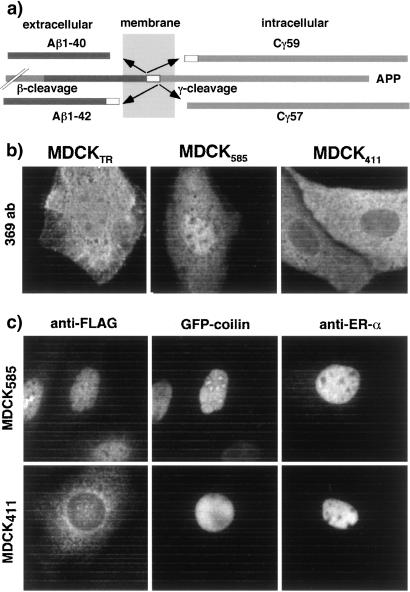
Although the Cγ fragment of APP is generated in the cytoplasm, it is also found in the nucleus (refs. 19–21; see also Fig. 2b). To learn whether PAT1 could be responsible for the presence of Cγ in the nucleus, we asked whether Cγ might accumulate in the nucleus in MDCK585 cells but remain in the cytoplasm in MDCK411 cells. The levels of the endogenous Cγ, like those of other fragments produced by RIP (see ref. 6), including the NICD (22), are too low to be detected reliably by conventional immunodetection techniques. We, therefore, transiently expressed the GFP fusions of Cγ59, Cγ57, and C31 of the APP cytoplasmic domain in MDCKTR, MDCK585, and MDCK411 cells. We were unable to observe expression of the untagged version of Cγ fragments presumably because they were turned over rapidly in MDCK cells and their steady-state levels were below the detection limits. The Cγ59 and Cγ57 fragments (Fig. 2a) would be produced by γ-secretase cleavage together with Aβ-1–40 and Aβ-1–42, respectively; C31 is produced by caspase cleavage (23) of the cytoplasmic domain of APP and lacks the PAT1-interacting region. Fig. 2b shows that Cγ59 was distributed rather evenly in the cytoplasm and the nucleus of MDCKTR (Fig. 2b Left) and MDCK585 cells and that very few cells showed slight accumulation of Cγ59 in the nucleus (Fig. 2b Center). However, Cγ59 was absent from the nucleus of MDCK411 cells, suggesting that in these cells PAT11–411 largely prevented Cγ59 from entering the nucleus. To examine the specificity of this effect, we monitored the distribution of nuclear proteins GFP-coilin and estrogen receptor-α (ER-α) in transiently transfected MDCK585 and MDCK411 cells. As expected, both these proteins were present in the nucleus of MDCK585 cells (Fig. 2c Upper) and also were found in the nucleus of MDCK411 cells (Fig. 2c Lower). Similarly, endogenous corepressor SMRT was found in the nucleus of MDCK411 cells (not shown). Cγ57 was present in both compartments in MDCK585 cells but was mostly cytoplasmic in MDCK411 cells, whereas C31 was nuclear and cytoplasmic in both MDCK585 and MDCK411 (not shown). Thus, Cγ is selectively accumulated in the cytoplasm of MDCK411.
Figure 2.
Cγ59 is selectively retained in the cytoplasm in MDCK411 cells. (a) Processing of APP. APP, initially cleaved by β-secretase close to the external surface of the membrane, can be cleaved by γ-secretase at two sites within the plane of the membrane. The major cleavage site (90–95%) produces Aβ-1–40 plus Cγ59, whereas the minor site yields minor Aβ-1–42 plus Cγ57. (b) Cells transiently transfected with GFP-Cγ59 were stained with antibody 369 that recognizes the cytoplasmic tail portion of APP. Note that Cγ59 is present in the nucleus as well as cytoplasm in MDCKTR and MDCK585 cells and mostly cytoplasmic in MDCK411 cells. (c) MDCK411 cells do not accumulate other nuclear proteins in the cytoplasm. MDCK585 (Upper) or MDCK411 cells (Lower) were transiently transfected with GFP-coilin or ER-α and stained with anti-FLAG antibody (Left and Center) or anti-ER-α antibody (Right). Left and Center show a view of the same field through rhodamine channel (Left) or observed for GFP signal (Center; ×63 objective).
Overexpression of Cγ59 Results in Down-Regulation of PAT1.
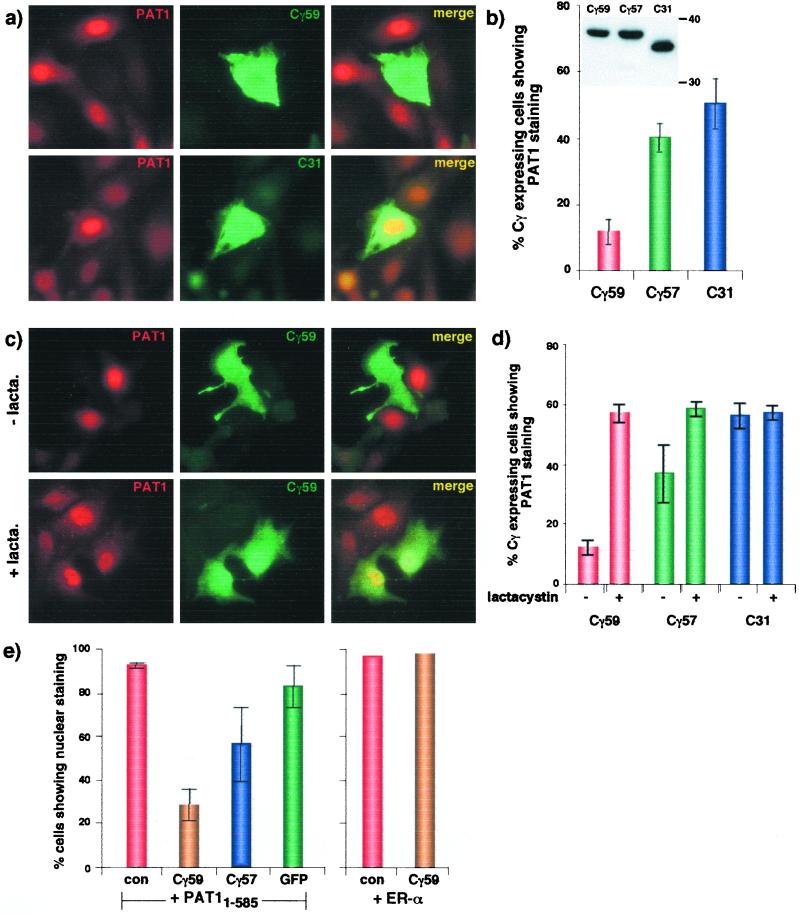
To study further the interaction between Cγ and PAT1, we transiently transfected the MDCK585 cells to express Cγ59 and double-labeled them with antibodies to localize both PAT1 and Cγ. Surprisingly, we observed that the PAT1 signal was almost completely absent from the cells that expressed Cγ59 (Fig. 3a Upper). By contrast, a large fraction of MDCK585 cells that expressed C31 also expressed PAT1 (Fig. 3a Lower). Quantification of these data shows that although only 10% of Cγ59-expressing cells showed the presence of PAT1, nearly 40% of Cγ57-expressing cells and up to 60% of C31-expressing cells stained positive for PAT1 (Fig. 3b; at steady state, only 60–70% of MDCK585 cells show the presence of PAT1). Because Cγ59, Cγ57, and C31 were expressed at similar levels (Fig. 3b Inset), we conclude that expression of Cγ59 significantly down-regulated PAT1 levels, Cγ57 was much less effective, and C31 had a minimal or no effect. We also tested the effect of C47, which lacks the membrane-embedded 12 residues but contains the PAT1-interacting region, and observed no significant effects on PAT1 down-regulation (not shown). Expression of Cγ59 had no effect on the nuclear localization of ER-α, which was 100% nuclear in MDCK585 cells expressing or not expressing Cγ59 (not shown). Expression of Cγ59 in MDCK411 cells did not cause the disappearance of PAT11–411 (which lacks the PEST domain), suggesting that the C-terminal region of PAT1 or its nuclear localization is essential for PAT1 down-regulation (not shown). To examine whether the loss of PAT1 signal was because of proteosomal degradation, we treated the transfected cells with lactacystin for 4 h before fixation. In the presence of lactacystin, PAT1 was present in Cγ59-expressing cells (Fig. 3c). Quantification of the data shows that lactacystin increased the number of cells that coexpress both Cγ59 and PAT11–585 to the same level as those coexpressing C31 and PAT11–585. Lactacystin treatment had a moderate effect on Cγ57-expressing cells and had no effect on C31-expressing cells (Fig. 3d).
Figure 3.
Expression of Cγ59 causes disappearance of PAT1 from the nucleus. (a) MDCK585 cells transiently transfected with GFP-Cγ59 (Upper) or GFP-C31 (Lower) were labeled with anti-FLAG antibody followed by rhodamine-anti-mouse IgG (Left) or antibody 369 followed by FITC-anti-rabbit IgG (Center). (b) Quantification of data is shown on the right (means ± SD of at least three independent experiments). Inset shows that these three proteins were expressed at similar levels. (c) Lactacystin treatment prevents the disappearance of PAT1 in Cγ59-expressing MDCK585 cells. Details are as above. (d) Quantification of the data is shown on the right (means ± SD of at least three experiments). (e) MDCKTR cells were transiently cotransfected with PAT11–585 or ER-α together with other plasmids as indicated and labeled as above. con, Control (empty vector). Only those cells that showed signal for both proteins were quantified for nuclear vs. cytoplasmic localization of PAT1. Cγ59 did not affect the nuclear localization of ER-α.
We verified these observations by cotransfecting PAT11–585 with Cγ59, Cγ57, or GFP in MDCKTR (Fig. 3e). In more than 90% of cells expressing PAT1 with control plasmid, the nuclear PAT1 signal was much stronger than the cytoplasmic signal (nuclear > cytoplasmic). However, when coexpressed with Cγ59, less than 30% cells showed nuclear > cytoplasmic signal and the rest of the cells displayed predominantly cytoplasmic signal (cytoplasmic > nuclear; these data most certainly underestimate the effect of Cγ on down-regulation of PAT1 because the cells that were positive for Cγ59 but negative for PAT1 were excluded to avoid counting the singly transfected cells). Coexpression with GFP alone did not change the predominant nuclear concentration of PAT1, whereas Cγ57 had an intermediate effect. As observed in MDCK585 cells, coexpression of Cγ59 had no effect on nuclear localization of ER-α. Similar observations were made when these proteins were transiently coexpressed in COS1 cells (not shown). Taken together, these results show that Cγ59 selectively causes cytoplasmic relocation and proteosomal degradation of PAT1.
Cγ59 Represses RA-Responsive Gene Transcription.
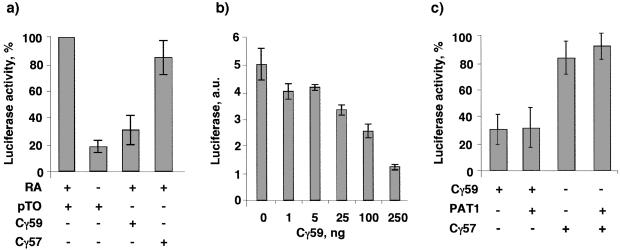
We next determined whether Cγ59 and Cγ57, like the analogous fragments produced by Notch and SREBP, also regulate transcription by examining their effects on RA-responsive transcription. We chose to study the RA-responsive gene expression because previous studies have indicated that the C-terminal region of APP influences the RA pathway (24, 25) and because PAT1 contains NR-box motifs (LXXLL) that mediate interaction of transcriptional coregulators with nuclear hormone receptors, including the RA receptor (RAR). We cotransfected CV1 cells with the reporter plasmid RARE-TK-Luc (containing luciferase gene downstream of the RA response element) with plasmids expressing Cγ59 or Cγ57. The parental plasmid (pTO) in which Cγ constructs were cloned was used as a negative control. Cells were transfected with a constitutive β-galactosidase expression plasmid to normalize the transfection efficiency. Cells were treated with or without 1 μM retinoic acid for 24 h and lysed, luciferase activity was measured, and normalized luciferase activity observed in cells cotransfected with pTO vector in the presence of RA is expressed as 100%. Fig. 4a shows that, in the absence of RA, only a basal level of transactivation is observed, which is stimulated 5- to 6-fold by the addition of RA. However, cotransfection of Cγ59 with the reporter plasmid greatly repressed transactivation almost to the basal level. By contrast, cotransfection with Cγ57 or with C47 (not shown) resulted in little or no repression. Cotransfection of the reporter plasmid with PAT11–585 also had no significant effect on RA-responsive gene expression (not shown). The repression by Cγ59 was dose-dependent (Fig. 4b), with maximal repression observed with 250 ng of plasmid DNA per well. Higher amounts of DNA did not cause further repression. Because Cγ59 also down-regulated PAT1 in the nucleus, we next examined whether the inhibition could be prevented by overexpressing PAT1. The reporter plasmid was cotransfected with Cγ59 and PAT11–585 or pTO plasmid DNA in CV1 cells. Fig. 4c shows that expression of PAT1 did not prevent repression by Cγ59, nor did it have any effect on Cγ57. Thus, overexpression of PAT11–585 is not sufficient to overcome the repression, which suggests that repression of transcription by Cγ59 may be independent of its deleterious effects on PAT1. Alternately, additional protein factors may be required to restore the transcription.
Figure 4.
Cγ59 potently represses RA-responsive transcription. (a) CV1 cells were cotransfected with a plasmid mixture of RARE-TK-Luc, RAR, and β-galactosidase (to normalize for transfection efficiency) together with the plasmids indicated below each bar. A 59-residue-long, γ-secretase-cleaved, C-terminal fragment of APP (Cγ59) but not the 57-residue-long fragment (Cγ57) represses transactivation. (b) Indicated amounts of GFP-Cγ59 plasmid DNA were cotransfected with the reporter plasmid DNA. The data show that Cγ59 represses transactivation in a dose-dependent manner. (c) Overexpression of PAT11–585 does not affect the repression by Cγ59. Cells were cotransfected with the reporter plasmid mix together with 250 ng each of Cγ and PAT11–585 or Cγ with pTO (parental plasmid vector). The normalized luciferase activity obtained with pTO plasmid is expressed as 100%. Bars = means ± SD of at least three experiments performed in duplicate except in b, which shows values from a single, representative experiment repeated twice.
Discussion
APP and RIP.
These data show that the Cγ fragment has at least two biological effects in the nucleus—it down-regulates PAT1 and represses RA-responsive transcription. The notion that APP may follow RIP was prompted by the remarkable similarity of APP processing to that of Notch, which, in turn, suggested that Cγ, like NICD, may function in the nucleus (3). Also, a biological function for Cγ in the nucleus could explain the physiological necessity of APP processing. On the other hand, the Cγ fragment is extremely small compared with the NICD (and the analogous fragments from other proteins) and lacks motifs commonly found in transcriptional regulators, suggesting that Cγ may not necessarily function in signaling in the way NICD functions. Our findings show that the ectopically expressed Cγ fragment does exhibit two biological activities—induction of proteosomal degradation and transcriptional modulation. Whether the endogenous Cγ at physiological concentrations shows similar effects remains to be seen, but identification of the RA-responsive genes as potential downstream targets should provide a sensitive functional parameter to address this issue.
Because Cγ down-regulates nuclear PAT1 and causes its proteosomal degradation, our studies provide an indication that the γ-secretase-released product of APP can regulate events in the nucleus. Importantly, because Cγ59, the major product of the γ-cleavage, is much more potent than Cγ57 in down-regulating nuclear PAT1, these findings suggest a physiological role for the γ-cleavage of APP at the major site, namely, that of signal transduction. These findings are consistent with a RIP model in which Cγ released from APP (perhaps as a result of an as-yet-unidentified signal) enters the nucleus and functions in an APP-signaling pathway by down-regulating PAT1. The consequences of PAT1 degradation by Cγ remain to be established. We have observed that the absence of nuclear PAT1 (as observed in MDCK411 cells or achieved by treatment with staurosporine) results in apoptosis and makes cells proapoptotic (unpublished results). In this respect, it is noteworthy that several APP cytoplasmic fragments (APP-CTFs), including the Cγ peptides (26), have been shown to cause apoptosis and/or to make the cells proapoptotic (27–29). However, direct evidence that the apoptotic effects of Cγ or APP-CTFs are mediated by PAT1 is not yet available.
APP, Retinoid Pathway, and Alzheimer's Disease Neuropathology.
The second finding of our studies suggests that Cγ may function as a repressor of RA-responsive gene expression. Two lines of observations indicate that this repression could be physiologically relevant. Studies have shown that RA treatment increases the expression of APP (30–34) and BACE (35), an enzyme that cleaves APP and makes it a substrate for the γ-cleavage. Thus, repression of RA-responsive genes by Cγ simply could be a part of the autoregulatory, feedback mechanism. Second, overexpression of APP (24) or its C-terminal fragment (25) has been shown to antagonize the RA-mediated responses, and it has been shown that the APP-CTFs interfere with the neuronal differentiation induced by RA treatment of mouse embryonic P19 stem cells (24, 36) but not with the myocytic differentiation of the same cells induced by DMSO (36).
Indeed, a closer scrutiny of the possible connection between RA and APP-CTF/Cγ reveals interesting parallels between the effects of RA and APP in memory and cognition. The RA-signaling pathway has been proposed to play a role in adult neuronal plasticity, and gene knockout studies show that deficiency of RAR-β virtually eliminates hippocampal CA1 long-term potentiation (LTP) and causes deficits in spatial learning and memory tasks (37). The transgenic mice overexpressing the “Swedish-mutant” APP695SWE (38) or overexpressing the C-terminal fragment of APP also exhibit severely impaired CA1 LTP and diminished spatial memory (27) and show hippocampal degeneration (39). Moreover, administration of RA is shown to alleviate the deficit in the CA1 LTP and reverse the age-related down-regulation of RA-transcription and cognition deficits in aged mice in vivo (40). These observations are only correlative, but suggestive, and deserve further exploration. Also, although no phenotype has been reported in APP/APLP2 knockout mice, suggesting a possible role of Cγ59 in suppression of RA-responsive genes, it is intriguing that these mice (41), like the double-RAR knockout mice (42), display postnatal lethality. Finally, although direct genetic or biochemical proof to support a role for endogenous Cγ has been lacking, the effects ascribed to APP-CTF, a substrate for the γ-secretase (43, 44), could, in fact, be mediated by Cγ59.
While this manuscript was in preparation, a study appeared supporting the view that Cγ may function in gene expression (45). Chimeric proteins containing the APP or its C-terminal fragment and exogenous DNA-binding domain (Gal4- or LexA-) were shown to potently stimulate transcription in the presence but not in the absence of Fe65, an activator of transcription (46). Thus, Cγ, like other transcriptional modulators, may function both as a transcriptional repressor and as an activator. Alternately, the variations in results simply could be a result of the differences in the reporter-assay systems used to study the role of Cγ in transcriptional regulation. It should be noted that Fe65 recently has been shown to colocalize with APP, actin, and Mena and has been implicated in regulation of actin-based motility (47) and stimulate Aβ secretion (48).
In summary, our results suggest that Cγ is able to transduce signal to the nucleus and regulate gene expression, thus indicating that APP is a bona fide member of the family of regulatory proteins that follow the RIP. Furthermore, these results indicate that APP may repress RA-responsive genes and thereby modulate cell growth and differentiation (49).
Acknowledgments
We thank K. Herrup, B. Lamb, H.-Y. Kao, M. Montano, B. Katzenellenbogen, and R. Evans for comments on the manuscript or for reagents. This work was supported partly by a grant from the Human Frontier Science Program to S.W.P.
Abbreviations
- APP
amyloid precursor protein
- Cγ
C-terminal fragment cleaved by γ-secretase
- RIP
regulated intramembrane proteolysis
- NICD
Notch intracellular domain
- GFP
green fluorescent protein
- ER-α
estrogen receptor-α
- RA
retinoic acid
- RAR
RA receptor
- CTF
cytoplasmic fragment
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Price D L, Sisodia S S. Annu Rev Neurosci. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D J. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 3.De Strooper B, Annaert W. J Cell Sci. 2000;113:1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 4.Vassar R, Citron M. Neuron. 2000;27:419–422. doi: 10.1016/s0896-6273(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 5.Haass C, De Strooper B. Science. 1999;286:916–919. doi: 10.1126/science.286.5441.916. [DOI] [PubMed] [Google Scholar]
- 6.Brown M S, Ye J, Rawson R B, Goldstein J L. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 7.Rudner D Z, Fawcett P, Losick R. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niwa M, Sidrauski C, Kaufman R J, Walter P. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- 9.Mumm J S, Kopan R. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 11.Ray W J, Yao M, Mumm J, Schroeter E H, Saftig P, Wolfe M, Selkoe D J, Kopan R, Goate A M. J Biol Chem. 1999;274:36801–36807. doi: 10.1074/jbc.274.51.36801. [DOI] [PubMed] [Google Scholar]
- 12.Struhl G, Greenwald I. Nature (London) 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 13.Zheng P, Eastman J, Vande Pol S, Pimplikar S W. Proc Natl Acad Sci USA. 1998;95:14745–14750. doi: 10.1073/pnas.95.25.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alber T. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 15.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Gindhart J G, Jr, Goldstein L S. Trends Biochem Sci. 1996;21:52–53. [PubMed] [Google Scholar]
- 17.Rechsteiner M, Rogers S W. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 18.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 19.DeGiorgio L A, DeGiorgio N, Milner T A, Conti B, Volpe B T. Brain Res. 2000;874:137–146. doi: 10.1016/s0006-8993(00)02545-2. [DOI] [PubMed] [Google Scholar]
- 20.Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B. J Neurochem. 2001;78:1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- 21.Kimberly W T, Zheng J B, Guenette S, Selkoe D J. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- 22.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 23.Lu D C, Rabizadeh S, Chandra S, Shayya R F, Ellerby L M, Ye X, Salvesen G S, Koo E H, Bredesen D E. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- 24.Yoshikawa K, Aizawa T, Hayashi Y. Nature (London) 1992;359:64–67. doi: 10.1038/359064a0. [DOI] [PubMed] [Google Scholar]
- 25.Honda S, Itoh F, Yoshimoto M, Hinoda Y, Imai K. J Clin Lab Anal. 1998;12:172–178. doi: 10.1002/(SICI)1098-2825(1998)12:3<172::AID-JCLA8>3.0.CO;2-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passer B, Pellegrini L, Rousso C, Siegel R M, Lenardo M J, Schettini G, Bachmann M, Tabaton M, D'Adamio L. J Alzheimer Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 27.Nalbantoglu J, Tirado-Santiago G, Lahsaini A, Poirier J, Goncalves O, Verge G, Momoli F, Welner S A, Massicotte G, Julien J P, et al. Nature (London) 1997;387:500–505. doi: 10.1038/387500a0. [DOI] [PubMed] [Google Scholar]
- 28.Berger-Sweeney J, McPhie D L, Arters J A, Greenan J, Oster-Granite M L, Neve R L. Brain Res Mol Brain Res. 1999;66:150–162. doi: 10.1016/s0169-328x(99)00014-5. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto Y, Niikura T, Ito Y, Nishimoto I. J Biol Chem. 2000;275:34541–34551. doi: 10.1074/jbc.M005332200. [DOI] [PubMed] [Google Scholar]
- 30.Konig G, Masters C L, Beyreuther K. FEBS Lett. 1990;269:305–310. doi: 10.1016/0014-5793(90)81181-m. [DOI] [PubMed] [Google Scholar]
- 31.Fukuchi K, Deeb S S, Kamino K, Ogburn C E, Snow A D, Sekiguchi R T, Wight T N, Piussan H, Martin G M. J Neurochem. 1992;58:1863–1873. doi: 10.1111/j.1471-4159.1992.tb10063.x. [DOI] [PubMed] [Google Scholar]
- 32.Hung A Y, Koo E H, Haass C, Selkoe D J. Proc Natl Acad Sci USA. 1992;89:9439–9443. doi: 10.1073/pnas.89.20.9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lahiri D K, Nall C. Brain Res Mol Brain Res. 1995;32:233–240. doi: 10.1016/0169-328x(95)00078-7. [DOI] [PubMed] [Google Scholar]
- 34.Beckman M, Iverfeldt K. Neurosci Lett. 1997;221:73–76. doi: 10.1016/s0304-3940(96)13292-4. [DOI] [PubMed] [Google Scholar]
- 35.Satoh J, Kuroda Y. Neuropathology. 2000;20:289–296. doi: 10.1046/j.1440-1789.2000.00349.x. [DOI] [PubMed] [Google Scholar]
- 36.Fukuchi K, Sopher B, Furlong C E, Smith A C, Dang N, Martin G M. Neurosci Lett. 1993;154:145–148. doi: 10.1016/0304-3940(93)90192-n. [DOI] [PubMed] [Google Scholar]
- 37.Chiang M Y, Misner D, Kempermann G, Schikorski T, Giguere V, Sucov H M, Gage F H, Stevens C F, Evans R M. Neuron. 1998;21:1353–1361. doi: 10.1016/s0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 38.Chapman P F, White G L, Jones M W, Cooper-Blacketer D, Marshall V J, Irizarry M, Younkin L, Good M A, Bliss T V, Hyman B T, et al. Nat Neurosci. 1999;2:271–276. doi: 10.1038/6374. [DOI] [PubMed] [Google Scholar]
- 39.Oster-Granite M L, McPhie D L, Greenan J, Neve R L. J Neurosci. 1996;16:6732–6741. doi: 10.1523/JNEUROSCI.16-21-06732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Etchamendy N, Enderlin V, Marighetto A, Vouimba R M, Pallet V, Jaffard R, Higueret P. J Neurosci. 2001;21:6423–6429. doi: 10.1523/JNEUROSCI.21-16-06423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heber S, Herms J, Gajic V, Hainfellner J, Aguzzi A, Rulicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, et al. J Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross S A, McCaffery P J, Drager U C, De Luca L M. Physiol Rev. 2000;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 43.McLendon C, Xin T, Ziani-Cherif C, Murphy M P, Findlay K A, Lewis P A, Pinnix I, Sambamurti K, Wang R, Fauq A, et al. FASEB J. 2000;14:2383–2386. doi: 10.1096/fj.00-0286fje. [DOI] [PubMed] [Google Scholar]
- 44.Pinnix I, Musunuru U, Tun H, Sridharan A, Golde T, Eckman C, Ziani-Cherif C, Onstead L, Sambamurti K. J Biol Chem. 2001;276:481–487. doi: 10.1074/jbc.M005968200. [DOI] [PubMed] [Google Scholar]
- 45.Cao X, Sudhof T C. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 46.Duilio A, Zambrano N, Mogavero A R, Ammendola R, Cimino F, Russo T. Nucleic Acids Res. 1991;19:5269–5274. doi: 10.1093/nar/19.19.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabo S L, Ikin A F, Buxbaum J D, Greengard P. J Cell Biol. 2001;153:1403–1414. doi: 10.1083/jcb.153.7.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sabo S L, Lanier L M, Ikin A F, Khorkova O, Sahasrabudhe S, Greengard P, Buxbaum J D. J Biol Chem. 1999;274:7952–7957. doi: 10.1074/jbc.274.12.7952. [DOI] [PubMed] [Google Scholar]
- 49.Maden M. Proc Nutr Soc. 2000;59:65–73. doi: 10.1017/s0029665100000082. [DOI] [PubMed] [Google Scholar]