A simple pipeline is introduced for de novo protein structure determination by calcium SAD phasing using an in-house diffractometer equipped with a chromium rotating anode.
Keywords: calcium phasing, chromium rotating anode, single-wavelength anomalous diffraction, calcium-binding proteins
Abstract
With better tools for data processing and with synchrotron beamlines that are capable of collecting data at longer wavelengths, sulfur-based native single-wavelength anomalous dispersion (SAD) phasing has become the ‘first-choice’ method for de novo protein structure determination. However, for many proteins native SAD phasing can be simplified by taking advantage of their interactions with natural metal cofactors that are stronger anomalous scatterers than sulfur. This is demonstrated here for four unique domains of a 1.5 MDa calcium-dependent adhesion protein using the anomalous diffraction of the chelated calcium ions. In all cases, low anomalous multiplicity X-ray data were collected on a home-source diffractometer equipped with a chromium rotating anode (λ = 2.2909 Å). In all but one case, calcium SAD phasing alone was sufficient to allow automated model building and refinement of the protein model after the calcium substructure had been determined. Given that Ca atoms will be present in a significant percentage of proteins that remain uncharacterized, many aspects of the data-collection and processing methods described here could be broadly applied for routine de novo structure elucidation.
1. Introduction
To produce three-dimensional images of the electron density surrounding atoms within crystallized proteins, phase information that cannot be recorded during X-ray diffraction data collection must be estimated by other methods. These include direct methods (Usón & Sheldrick, 1999 ▸), molecular replacement (Taylor, 2003 ▸), multiple isomorphous replacement, and multi-wavelength anomalous diffraction (MAD) and single-wavelength anomalous diffraction (SAD) methods (Hendrickson, 1991 ▸; Ealick, 2000 ▸). The vast majority of crystal structures deposited in the Protein Data Bank (PDB; http://www.rcsb.org) were determined at synchrotrons using MAD or SAD, and involve selenomethionine (SeMet) derivatives of the native protein (Hendrickson, 2014 ▸). However, SeMet incorporation is not without its challenges, a major one being its negative impact on expression yields of the labeled protein. As an alternative, SAD phasing using the weak anomalous scattering power of sulfur from native cysteine and methionine residues (S-SAD) is an advantageous strategy for de novo structure elucidation and can succeed with diffraction data obtained using the longer wavelength X-rays of home X-ray sources with copper (Cu; λ = 1.5418 Å) or chromium (Cr; λ = 2.2909 Å) anodes. Unfortunately, this method can be frustrated by its reliance on highly redundant diffraction data, which are not easily obtained for samples that are sensitive to radiation damage. It is therefore not surprising that relatively few de novo S-SAD structures have been deposited in the PDB (∼150; Weinert et al., 2015 ▸).
Metal ions that are naturally present in the protein of interest can provide excellent alternative anomalous scatterers to sulfur. It is estimated that one third of all proteins contain at least one metal cofactor, most commonly Ca2+, Fe3+, Mg2+, Zn2+ or Cu2+ (Shi & Chance, 2008 ▸). These bind to specific sites to perform catalytic, regulatory and structural functions in diverse classes of proteins (Degtyarenko, 2000 ▸). For proteins that bind Fe atoms, structure determination by native SAD or MAD is often straightforward because the scattering power is proportional to the square of the atomic number Z (Z = 26 for iron; Piontek et al., 2010 ▸) and the iron absorption edge (7.1120 keV, λ = 1.7433) is accessible at synchrotron beamlines and home sources. Structures determined with Mn2+ (Z = 25) and Zn2+ (Z = 30) are also represented in the PDB (Stevenson et al., 2004 ▸; Shi et al., 2005 ▸). If the experimenter is careful to mitigate or eliminate potential contributors of background noise, or factors that may mask anomalous signal detection, native SAD phasing is also possible by recording the weak anomalous scattering signal from light metal ions (Z ≤ 20) that are present in the crystallized sample, such as Ca2+ (Z = 20). Of the 130 990 crystal structures presently in the PDB, 10 068 or 7.7% have Ca2+ bound, and many of these have multiple Ca atoms present (Yáñez et al., 2012 ▸).
The properties of Ca2+ make it ideal for coordination by proteins, both directly to side-chain O atoms or backbone carbonyl groups, as well as indirectly through waters bound to the protein. As a result, many different classes of calcium-binding proteins (CaBPs) have evolved (Kawasaki & Kretsinger, 2017 ▸). These include EF-hand-containing proteins such as the calmodulin family, C2-domain-containing proteins such as phospholipase C and synaptotagmin, calreticulin, calsequestrin and annexins that are found inside the cell, where Ca2+ levels can be submicromolar (Yáñez et al., 2012 ▸; Heizmann & Hunziker, 1991 ▸). Outside the cell, where Ca2+ levels are typically in the low-millimolar range, there are also EF-hand proteins, C-type lectins, cadherins, RTX proteins and proteins with EGF-like and gamma-carboxyl glutamic acid domains, to name just a few (Permyakov & Kretsinger, 2010 ▸).
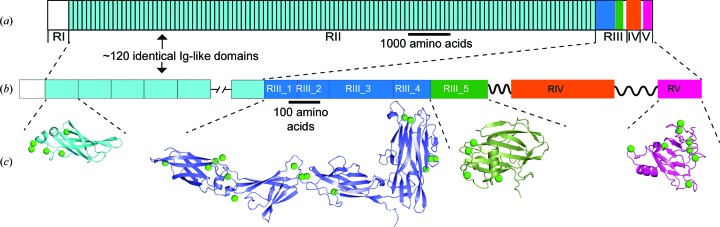
Here, we describe the crystallization conditions, data-collection strategies and structure-determination methods that were used on five sections that together make up >90% of a 1.5 MDa ice-binding adhesin protein, MpIBP (Fig. 1 ▸). This enormous adhesin is expressed on the exterior surface of the Gram-negative marine bacterium Marinomonas primoryensis, where it contributes to the ability of the bacterium to bind surface ice in an Antarctic lake for better access to oxygen and nutrients. It is hypothesized that this surface protein uses >600 Ca2+ ions to help to rigidify the chain formed by one of its domains repeated ∼120 times in tandem in region II (RII) and to aid in the proper folding and function of domains in the other regions (RI, RIII, RIV and RV) (Guo et al., 2012 ▸, 2013 ▸). Here, the natural incorporation of Ca2+ within the protein scaffold, as occurs in an abundance of proteins, presented a straightforward path to obtain the phasing information needed for de novo protein structure determination.
Figure 1.
X-ray crystal structures solved by Ca-SAD phasing. (a) Linear domain map of MpIBP drawn to scale. The RII repeats are colored cyan, the RIII_1–4 domains are colored blue and RIII_5 is colored green, while RV is colored magenta. (b) Expanded view of the linear domain map colored as in (a). (c) Structures of the RII monomer (cyan), RIII_1–4 (blue), RIII_5 (green) and RV (magenta) are shown in cartoon representation with the anomalous substructure Ca2+ atoms depicted as green spheres.
2. Materials and methods
The RII monomer and regions RIII_1–2, RIII_1–4, RIII_5 and RV of MpIBP (Fig. 1 ▸) were expressed in Escherichia coli BL21 (DE3) cells using pET-24a and pET-28a vectors, purified using affinity and size-exclusion chromatography, and then concentrated to 5–100 mg ml−1. All proteins were crystallized using the microbatch-under-oil method by mixing equal volumes of the concentrated protein with a precipitant solution. The RII monomer crystallized in the presence of a precipitant solution consisting of 0.2 M calcium acetate, 0.1 M sodium acetate pH 4.5, 30%(v/v) polyethylene glycol 400. RIII_1–2 crystallized in the presence of 0.2 M calcium chloride, 0.1 M MES pH 6, 20%(v/v) polyethylene glycol 3350. RIII_5 crystallized in the presence of 0.2 M calcium chloride, 0.1 M HEPES pH 7, 20%(v/v) polyethylene glycol 3350, 12%(w/v) myo-inositol. Crystals of RIII_1–4 grew in the presence of 0.1 M calcium chloride, 0.1 M sodium acetate pH 4.6, 30%(w/v) PEG 400, and RV crystallized in the presence of 0.1 M calcium chloride, 0.1 M sodium acetate pH 4.6, 30%(v/v) polyethylene glycol 400. Prior to data collection, all crystals were individually flash-cooled in a cryosolution consisting of 80%(v/v) precipitant solution and 20%(v/v) glycerol, with the exception of RIII_1–4, which was flash-cooled directly in the precipitant solution.
All diffraction data were collected in-house at 100 K using a Rigaku MicroMax-007 HF rotating-anode X-ray generator equipped with a chromium target and an R-AXIS IV++ image-plate detector. The detector was equipped with a helium-flushed beam path to reduce the air absorption of long-wavelength X-rays, and the crystal-to-detector distance was set to 105 mm.
Diffraction images for the RII monomer, RIII_5 and RV crystals were indexed and integrated with iMosflm (Battye et al., 2011 ▸) and then scaled with AIMLESS in CCP4 (Evans, 2006 ▸). For all three of these proteins, AutoSol from the PHENIX suite (Terwilliger et al., 2009 ▸) was used to identify Ca-atom sites and calculate phases. Uncertain of how many Ca atoms there were in each protein, we input a search number that was approximately 5% of the number of residues in the protein as a starting point. AutoBuild (Terwilliger et al., 2008 ▸; Adams et al., 2010 ▸) was then used to build and refine the initial models.
The diffraction images for the RIII_1–2 and RIII_1–4 crystals were indexed and integrated with XDS (Kabsch, 2010 ▸) and then scaled with AIMLESS (Evans & Murshudov, 2013 ▸). For RIII_1–2, CRANK2 from the CCP4 suite (Ness et al., 2004 ▸) was used to identify Ca-atom sites, calculate phases and build the preliminary model. The structure of RIII_1–4 was solved using a combined molecular-replacement/SAD (MR-SAD) method in CRANK2 (Skubák & Pannu, 2013 ▸) using RIII_1–2 as the search model and calcium as the heavy atom to obtain phase information and the initial model.
All initial protein models were retraced in the experimentally phased electron-density maps with Buccaneer (Cowtan, 2006 ▸) and were then corrected for remaining errors with Coot v.0.7 (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸). Final model refinement was performed in REFMAC5 (Murshudov et al., 2011 ▸; Vagin et al., 2004 ▸). These structures were used as molecular-replacement models for higher resolution native data sets collected at synchrotron sites, the coordinates of which have been deposited in the PDB with the following accession codes: 4kdw (RII monomer), 5j6y (RIII_5), 5k8g (RIII_1–4) and 5juh (RV) (Guo et al., 2013 ▸, 2017 ▸).
Illustrations of protein structures and electron density were created using PyMOL. Electron-density and anomalous difference map coefficients were calculated using phenix.refine.
3. Results
3.1. Crystallization conditions and data-collection procedure for Ca-SAD phasing
Three of the five regions of the 1.5 MDa Ca2+-dependent ice-binding adhesin MpIBP were dissected into five protein constructs with molecular weights ranging from 10 to 57 kDa (Fig. 1 ▸). BLAST searches (Altschul et al., 1990 ▸) of their sequences against the PDB did not reveal usable homology models, negating structure determination by molecular replacement alone. Instead, we postulated that the requirement of these proteins for calcium for correct folding could be exploited as a source of anomalous scatterers if their occupancy and orderliness in the crystals were high (Garnham et al., 2008 ▸; Gilbert et al., 2005 ▸). Therefore, the calcium concentration in the crystallization and cryocooling solutions was maintained within the range 0.1–0.2 M for all proteins.
Diffraction data were collected from single crystals of each of the five proteins using an X-ray home source equipped with a chromium anode (X-ray energy = 5.41 keV). At this energy, the anomalous term f′′ for calcium is 2.51 e−. This is significantly higher than the f′′ value of 1.29 e− for calcium when using a copper anode (calculated using the graph from http://skuld.bmsc.washington.edu/scatter/AS_index.html). For further comparison, the f′′ values for sulfur are 1.15 and 0.56 e− at the Cr Kα and Cu Kα wavelengths, respectively (Ibers & Hamilton, 1974 ▸; Yang et al., 2003 ▸). In order to measure the small Bijvoet differences as accurately as possible, this system included Osmic Confocal MaxFlux optics optimized for Cr Kα radiation and a helium-filled path to the R-AXIS IV++ image-plate-based area detector to minimize air scattering. To minimize radiation damage by the longer wavelength Cr Kα radiation (2.2909 Å), X-ray exposure times were limited to 2–5 min per frame and 1° oscillations. Using this approach, ≤500° of data yielded sufficient anomalous multiplicity values (3.8-fold to 14-fold) to discern the anomalous signal from Ca atoms. This information, and all other diffraction statistics, are presented in Table 1 ▸. The statistics for model refinement are shown in Table 2 ▸. The experimental phasing and model-building approaches for each protein construct are described in the following subsections.
Table 1. Statistics for diffraction data.
Values in parentheses are for the outer shell.
| Protein | RII monomer | RIII_1–2 | RIII_1–4 | RIII_5 | RV |
|---|---|---|---|---|---|
| No. of molecules in asymmetric unit | 1 | 1 | 1 | 1 | 1 |
| No. of amino acids in asymmetric unit | 102 | 190 | 511 | 188 | 137 |
| Anomalous scatterers | Ca [5] | Ca [9] | Ca [15] | Ca [8] | Ca [8] |
| Space group | P21 | P22121 | P1 | P212121 | P212121 |
| Unit-cell parameters | |||||
| a (Å) | 28.69 | 29.04 | 50.29 | 45.13 | 34.87 |
| b (Å) | 43.02 | 45.93 | 61.34 | 50.62 | 43.66 |
| c (Å) | 32.26 | 156.84 | 63.64 | 79.52 | 78.16 |
| Wavelength (Å)/energy (keV) | 2.2909/5.41 | 2.2909/5.41 | 2.2909/5.41 | 2.2909/5.41 | 2.2909/5.41 |
| Resolution range (Å) | 32.03–2.12 (2.26–2.12) | 78.42–2.43 (2.49–2.43) | 47.83–2.45 (2.55–2.45) | 42.7–2.44 (2.58–2.44) | 43.66–2.43 (2.52–2.43) |
| Total rotation range (°) | 360 | 750 | 666 | 475 | 258 |
| Anomalous multiplicity | 3.8 | 14.3 | 3.6 | 8.1 | 5 |
| Multiplicity | 7.1 | 25.8 | 7.2 | 14.9 | 9.1 |
| Completeness (%) | 89.5 (69.6) | 98.4 (90.7) | 92.5 (83.2) | 99.3 (94.4) | 99.4 (97.1) |
| 〈I/σ(I)〉 | 17.0 (3.2) | 25.59 (10.6) | 5.8 (1.1) | 33.4 (28.6) | 13.31 (7.2) |
| R merge | 0.081 | 0.083 | 0.344 | 0.07 | 0.1 |
| CC1/2 | 99.3 (98.9) | 99.9 (99.3) | 97 (30.9) | 99.8 (99.7) | 99.5 (98.2) |
Table 2. Statistics for structure refinement.
Values in parentheses are for the outer shell.
| Protein | RII monomer | RIII_1–2 | RIII_1–4 | RIII_5 | RV |
|---|---|---|---|---|---|
| Resolution (Å) | 25.7–2.42 (2.48–2.42) | 78.42–2.43 (2.49–2.43) | 47.83–2.45 (2.55–2.45) | 42.70–2.44 (2.50–2.44) | 39.08–2.43 (2.58–2.43) |
| No. of reflections | 2605 | 7976 | 20610 | 6825 | 22197 |
| R work/R free (%) | 14.4/23.4 | 32.8/36.3 | 19.2/23.2 | 18.56/26.27 | 18.8/26.5 |
| B factors (Å2) | |||||
| Protein | 21.6 | 36.8 | 37.7 | 6.2 | 17.41 |
| Ligand | 30.1 | 44.4 | 35.7 | 3.5 | 21.39 |
| Water | 23.1 | 23.5 | 41.5 | 5.0 | 21.70 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.012 | 0.0143 | 0.0145 | 0.017 | 0.0194 |
| Bond angles (°) | 1.54 | 2.07 | 1.71 | 1.7 | 2.22 |
| Ramachandran statistics | |||||
| Favored | 98 | 92 | 94.8 | 95 | 97.1 |
| Allowed | 2 | 8 | 4.9 | 5 | 2.9 |
| Outliers | 0 | 0 | 0.2 | 0 | 0 |
3.2. Determination of the heavy-atom substructure and protein model building
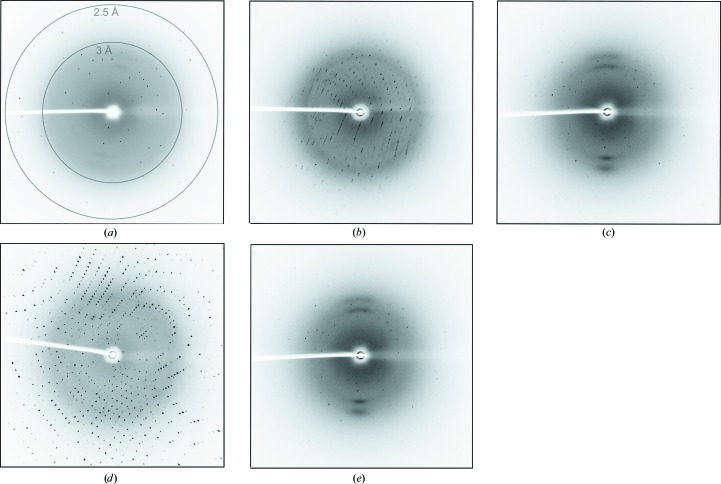
The RII monomer, RIII_5 and RV domains proved to be the most straightforward cases for automated structure determination using Ca-SAD phasing. The RII monomer (10 kDa) crystallized in space group P21, with one molecule in the asymmetric unit, and diffracted to 2.12 Å resolution (Fig. 2 ▸ a). The RIII_5 domain (20 kDa) crystallized in space group P212121, with one molecule in the asymmetric unit, and diffracted to 2.44 Å resolution (Fig. 2 ▸ d). The RV domain (14 kDa) also crystallized in space group P212121 and contained one molecule in the asymmetric unit. It diffracted to 2.43 Å resolution (Fig. 2 ▸ e). Diffraction images of the RII monomer, RIII_5 and RV domains were integrated with iMosflm (Battye et al., 2011 ▸) and then scaled with AIMLESS in CCP4 (Evans, 2006 ▸). To determine the Ca-atom substructure and calculate experimental phases for each domain, the anomalous data, protein sequence and X-ray wavelength were input into the AutoSol routine in PHENIX with ‘Ca’ selected as the anomalously scattering atom (Adams et al., 2010 ▸). Uncertain of how many Ca atoms were coordinated by each protein, we input a search number that was approximately 5% of the number of residues in each protein as a starting point. In the cases of the RII monomer, RIII_5 and RV, Ca-atom substructure determination was straightforward using the HySS program within PHENIX AutoSol (Fig. 3 ▸), and the phases were sufficient, with figure-of-merit (FOM) values of near 0.3 or above, for iterative automated model building by AutoBuild within PHENIX (Terwilliger et al., 2008 ▸; Adams et al., 2010 ▸; Table 3 ▸).
Figure 2.
Examples of X-ray diffraction images. (a) Representative diffraction image of the RII monomer with resolution rings indicated at 2.5 and 3 Å. (b) Representative diffraction image of the RIII_1–2 protein. (c) Representative diffraction image of the RIII_1–4 protein. (d) Representative diffraction image of the RIII_5 protein. (e) Representative diffraction image of RV. The images in (b)–(e) were collected at the same wavelength and detector position as that in (a).
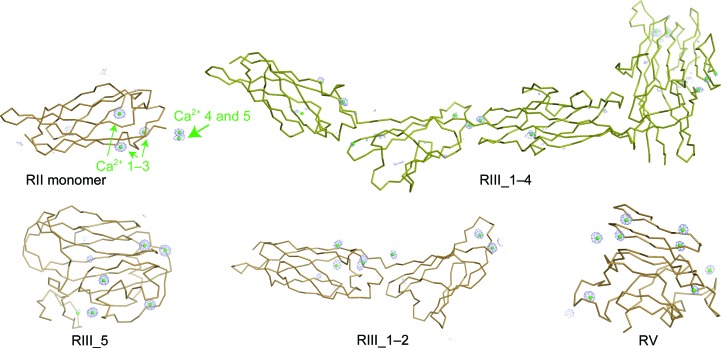
Figure 3.
The five solved structures of MpIBP domains shown in ribbon representation. Protein backbone is colored bronze. Electron density for the anomalous scatterers (Ca2+) is shown as blue mesh, with the anomalous difference map contoured at 3σ.
Table 3. Statistics of phase quality determined by the HySS program from AutoSol in PHENIX .
| Protein | RII monomer | RIII_5 | RV |
|---|---|---|---|
| BAYES-CC | 10.8 ± 25.3 | 13.7 ± 26.5 | 12.4 ± 25.9 |
| Z-score | 3.027 | 0.590 | 1.436 |
| FOM | 0.51 | 0.44 | 0.293 |
The model of the RII monomer built from experimental phases comprised 102 residues, and the electron-density map showed a very good final correlation coefficient and connectivity (Fig. 4 ▸ a). The RIII_5 and RV domain models comprised 188 residues and 137 residues, respectively, and also showed good map coverage (Figs. 4 ▸ d and 4 ▸ e). Each of these initial models were rebuilt with Buccaneer (Cowtan, 2006 ▸) using the maps produced by AutoBuild, and then manually corrected in Coot v.0.7 (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸). Each structure was then refined with REFMAC5 (Murshudov et al., 2011 ▸; Vagin et al., 2004 ▸), and when the models were complete there were five Ca2+ ions bound to the RII monomer and eight Ca2+ ions bound to both the RIII_5 and RV domains (Table 1 ▸). Notably, not all bound calcium ions appear to be integral to the structure of these proteins. Using the RII monomer as an example, Ca2+ ions 1–3 appear to be needed for stability of the protein (Fig. 3 ▸ a), but Ca2+ ions 4 and 5 are present as a result of crystal packing and have slightly lower occupancies (Table 4 ▸).
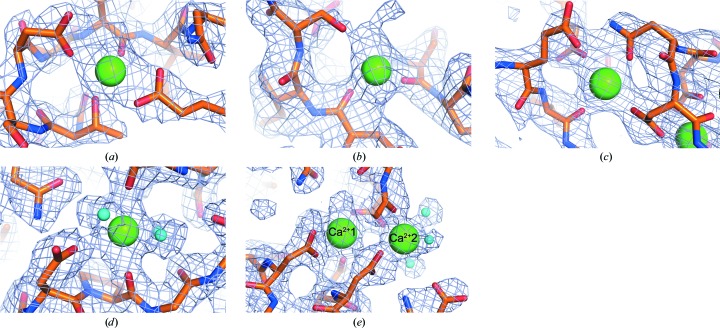
Figure 4.
Sample electron densities showing calcium-binding sites and surrounding amino acids. Electron density is shown as blue mesh with the 2F o − F c map contoured at 1σ. Polypeptide chains are shown in stick representation with the carbon backbone colored orange, O atoms colored red and N atoms colored blue. Ca2+ ions are shown as large green spheres, while water molecules bound to Ca2+ are indicated as small cyan spheres. (a) RII monomer. (b) RIII_1–2. (c) RIII_1–4. (d) RIII_5. (e) RV.
Table 4. Occupancies and B factors of individual Ca2+ ions in the RII monomer structure.
| Ca2+ in RII monomer | Occupancy | B factor (Å2) |
|---|---|---|
| Ca2+ 1 | 1 | 30.4 |
| Ca2+ 2 | 1 | 20.4 |
| Ca2+ 3 | 0.9 | 17.7 |
| Ca2+ 4 | 0.7 | 32.3 |
| Ca2+ 5 | 0.8 | 26.8 |
The RIII_1–2 and RIII_1–4 domain structures were slightly more challenging to solve using Ca-SAD phasing. RIII_1–2 (20 kDa) crystallized in space group P22121, with one molecule in the asymmetric unit, and diffracted to 2.43 Å resolution (Fig. 2 ▸ b). We found that indexing and integrating the diffraction images with XDS (Kabsch, 2010 ▸), followed by scaling with AIMLESS (Evans & Murshudov, 2013 ▸), was needed to account for the lower quality of these data. The structure of RIII_1–2 was solved using Ca-SAD methods in CRANK2 to obtain phase information and to perform automated building of the initial 190-residue model. The overall quality of the resulting electron-density map was sufficient to rebuild the RIII_1–2 model with Buccaneer and Coot v.0.7; the model, and the positions of its nine bound Ca2+ ions, was then refined with REFMAC5.
RIII_1–4 (57 kDa) crystallized in space group P1, with one molecule in the asymmetric unit, and diffracted weakly to 2.4 Å resolution (Fig. 2 ▸ c). Diffraction images were indexed and integrated with XDS and were scaled with AIMLESS. With an anomalous multiplicity of only 3.6 (Table 1 ▸), RIII_1–4 was the most difficult case for Ca-SAD phasing and required the data to be truncated at 2.45 Å resolution. The data set had a high R merge of 0.344 and it was not possible to solve the structure with AutoSol in PHENIX using SAD methods alone. We resorted to using an MR-SAD method in CRANK2 to obtain phase information and build the initial model. In this case, the structure of RIII_1–2 was used as a partial search model for molecular replacement and calcium was used as the heavy atom in the combined MR-SAD approach. This gave experimentally phased electron-density maps that were suitable for model building and structure completion by Buccaneer and Coot v.0.7, respectively. Refinement with REFMAC5 gave an accurate RIII_1–4 model composed of 511 residues that fitted well into the electron-density map (Fig. 4 ▸ c) and showed the positions of 15 bound Ca2+ ions.
In general, our approach was to collect a large amount of data for all crystals in order to achieve a high anomalous multiplicity. We found that the proteins with higher molecular weight, such as RIII_1–2 (190 amino acids) and RIII_1–4 (511 amino acids), required a higher anomalous multiplicity for phasing than those of low molecular weight (for example the RII monomer, 102 amino acids). To determine whether sufficient data were available for Ca-SAD phasing, anomalous signal indicators such as SigAno were tracked as the data were being collected. Although we did not encounter appreciable evidence of radiation damage during data collection, we were careful to monitor this and other data-collection issues that can degrade the anomalous signal. In such cases, we would examine and truncate the data as needed to provide the necessary information for phasing. Therefore, the optimal data-collection strategy must be determined empirically for each crystal.
3.3. Model building and refinement
For the five calcium-binding proteins that were studied, we found that XDS typically gave a more robust performance than iMosflm for data of poorer quality, and we found that CRANK2 tended to be faster than AutoSol in calculating calcium-substructure maps (Fig. 3 ▸). The well coordinated Ca2+ ions (those with six or more ligands from protein or water) usually had occupancies of one or close to one, and their sites could typically be identified early on during substructure determination using AutoSol in PHENIX or CRANK in CCP4. Those Ca2+ ions with fewer ligands (typically more solvent-accessible) have lower occupancies of 0.3 or 0.4. The initial maps obtained from Ca-SAD phasing typically had R factors in the region of 30%, and their relatively high resolution allowed most aspects of model building to be automated (Fig. 4 ▸). These structures solved at home provided sufficient models to solve higher resolution structures by molecular replacement alone or by MR-SAD when synchrotron data were available. The crystal structure of the RII monomer was solved to a resolution of 1.35 Å by single-wavelength anomalous dispersion and molecular-replacement methods using Ca2+ as the heavy atom to obtain phase information (PDB entry 4kdw; Guo et al., 2013 ▸). High-resolution structures (from data sets obtained at synchrotron facilities) of RIII_5 (PDB entry 5j6y), RIII_1–4 (PDB entry 5k8g) and RV (PDB entry 5juh) were determined by molecular replacement with Phaser in CCP4 using their respective low-resolution structures solved by in-house Ca2+ phasing as search models (Guo et al., 2017 ▸).
4. Discussion
The successful use of Ca-SAD phasing to provide the first view of over 90% of the massive ice-binding adhesion protein MpIBP demonstrates that the weak anomalous differences produced by this common natural cofactor can be extracted reliably from data collected from a single crystal. For other proteins known to chelate calcium, we recommend using native Ca-SAD as a first attempt at phasing, particularly if the crystals diffract well and are tolerant of normal X-ray exposure times, as it avoids the need to unnaturally derivatize the protein. This strategy could also be used in cases where structure determination by molecular replacement is possible but model bias is a concern.
Many of the calcium-binding proteins within the cytoplasm of the cell are involved in calcium signaling and can exist in both calcium-bound and calcium-free forms. A comparison of the structures of these two forms can provide valuable information about the mechanism of action of the protein. One such example is from the cysteine protease family of calpains (Campbell & Davies, 2012 ▸). The main calpain isoforms calpain-1 and calpain-2 are normally in the calcium-free (apo) state (Hosfield et al., 1999 ▸; Strobl et al., 2000 ▸) and become transiently activated on cooperatively binding up to ten Ca2+ ions: eight of these ions bind to paired EF-hand motifs and two bind to atypical binding sites in the protease core (Moldoveanu et al., 2008 ▸; Hanna et al., 2008 ▸). This metal-ion binding causes a conformational change to the protease core that forms the active-site cleft with the correct juxtaposition of the catalytic triad residues for proteolysis to occur. When the calcium signal dissipates, calpain returns to the inactive, calcium-free state. Related roles of other calcium-binding proteins in the cytoplasm include calcium buffering, storage and transport, where again the metal ion tends to serve as a ligand.
In environments outside the cell, such as blood, lymph and seawater, where calcium levels are in the low-millimolar range, there is an even greater range of functions for calcium-binding proteins (listed in Section 1). Here we also see an increased use of calcium as an integral structural component of the protein required for proper folding, rather than just a ligand. The RTX adhesin, from which the five structures featured here were derived, is a good example. As with other proteins exported from Gram-negative bacteria through the type 1 secretion system (T1SS), the adhesin must exit the cell through a narrow pore as an extended polypeptide chain, C-terminus first, and then be folded on the outside as the domains emerge into a calcium-rich environment (Thomas et al., 2014 ▸). Bacterial adhesins appear to be involved in the initial stages of biofilm formation, and are gaining in importance as targets for blocking bacterial infection and colonization (Spaulding et al., 2017 ▸). The RTX adhesin featured here has calcium-dependent ligand-binding domains for affinity for ice, polysaccharides and peptides that determines its biofilm location (sea ice) and binding partners (diatoms) (Guo et al., 2017 ▸). Determining the structures and binding partners of ligand-binding domains in the adhesins of bacterial pathogens will provide additional strategies for blocking infections.
With the help of the tremendous advances in diffraction hardware, crystallographic software, data-collection methods and strategies, and the use of data statistics that are now available, our results also show that the anomalous signal of lighter metal atoms can be collected quickly and that the lower X-ray energy (longer wavelength) of Cr Kα radiation from an in-house rotating-anode X-ray generator can provide sufficient phasing power from calcium anomalous signals to routinely phase protein diffraction data. This method provides a welcome example of how to obtain structural information on new proteins without waiting for synchrotron beam time. We also encourage the use of Cr Kα radiation to improve the strength of anomalous scattering of many other intrinsic elements in macromolecules, such as zinc, iron, phosphorus and sulfur, because of the increased f′′ (Yang et al., 2003 ▸; Roeser et al., 2005 ▸). Research groups that are considering the acquisition of a home diffractometer are encouraged to investigate chromium systems as an option. Those seeking to upgrade older home sources could also investigate the relatively modest modifications required to convert a copper X-ray system to chromium, as the instrument can still be useful for native data collections. Moreover, the relative ease of phasing with calcium, iron, manganese, zinc or copper on a home-source diffractometer is a valuable enough method to consider adding metal-binding sites to the target protein produced by recombinant methods. Thus, the inclusion of minimal EF-hand or zinc-finger motifs might provide sufficient sites for phasing.
Acknowledgments
We thank Mrs Sherry Gauthier, Dr Qilu Ye, Dr Sarathy Karunan Partha, Tony He and Rebecca Lang for their assistance with cloning and crystallization trials.
Funding Statement
This work was funded by Natural Sciences and Engineering Research Council of Canada grants RGPIN 2016-04810 and RGPIN 2013-356025. Canadian Institutes of Health Research, Institute of Genetics grant .
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol. 215, 403–410. [DOI] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Campbell, R. L. & Davies, P. L. (2012). Biochem. J. 447, 335–351. [DOI] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Degtyarenko, K. (2000). Bioinformatics, 16, 851–864. [DOI] [PubMed]
- Ealick, S. E. (2000). Curr. Opin. Chem. Biol. 4, 495–499. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Garnham, C. P., Gilbert, J. A., Hartman, C. P., Campbell, R. L., Laybourn-Parry, J. & Davies, P. L. (2008). Biochem. J. 411, 171–180. [DOI] [PubMed]
- Gilbert, J. A., Davies, P. L. & Laybourn-Parry, J. (2005). FEMS Microbiol. Lett. 245, 67–72. [DOI] [PubMed]
- Guo, S., Garnham, C. P., Karunan Partha, S., Campbell, R. L., Allingham, J. S. & Davies, P. L. (2013). FEBS J. 280, 5919–5932. [DOI] [PubMed]
- Guo, S., Garnham, C. P., Whitney, J. C., Graham, L. A. & Davies, P. L. (2012). PLoS One, 7, e48805. [DOI] [PMC free article] [PubMed]
- Guo, S., Stevens, C. A., Vance, T. D. R., Olijve, L. L. C., Graham, L. A., Campbell, R. L., Yazdi, S. R., Escobedo, C., Bar-Dolev, M., Yashunsky, V., Braslavsky, I., Langelaan, D. N., Smith, S. P., Allingham, J. S., Voets, I. K. & Davies, P. L. (2017). Sci. Adv. 3, e1701440. [DOI] [PMC free article] [PubMed]
- Hanna, R. A., Campbell, R. L. & Davies, P. L. (2008). Nature (London), 456, 409–412. [DOI] [PubMed]
- Heizmann, C. W. & Hunziker, W. (1991). Trends Biochem. Sci. 16, 98–103. [DOI] [PubMed]
- Hendrickson, W. A. (1991). Science, 254, 51–58. [DOI] [PubMed]
- Hendrickson, W. A. (2014). Q. Rev. Biophys. 47, 49–93. [DOI] [PMC free article] [PubMed]
- Hosfield, C. M., Elce, J. S., Davies, P. L. & Jia, Z. (1999). EMBO J. 18, 6880–6889. [DOI] [PMC free article] [PubMed]
- Ibers, J. A. & Hamilton, W. C. (1974). Editors. International Tables for X-ray Crystallography, Vol. IV, p. 149. Birmingham: Kynoch Press.
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kawasaki, H. & Kretsinger, R. H. (2017). Protein Sci. 26, 1898–1920. [DOI] [PMC free article] [PubMed]
- Moldoveanu, T., Gehring, K. & Green, D. R. (2008). Nature (London), 456, 404–408. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ness, S. R., de Graaff, R. A., Abrahams, J. P. & Pannu, N. S. (2004). Structure, 12, 1753–1761. [DOI] [PubMed]
- Permyakov, E. A. & Kretsinger, R. H. (2010). Calcium Binding Proteins, pp. 361–397. Hoboken: John Wiley & Sons.
- Piontek, K., Ullrich, R., Liers, C., Diederichs, K., Plattner, D. A. & Hofrichter, M. (2010). Acta Cryst. F66, 693–698. [DOI] [PMC free article] [PubMed]
- Roeser, D., Dickmanns, A., Gasow, K. & Rudolph, M. G. (2005). Acta Cryst. D61, 1057–1066. [DOI] [PubMed]
- Shi, W. & Chance, M. R. (2008). Cell. Mol. Life Sci. 65, 3040–3048. [DOI] [PMC free article] [PubMed]
- Shi, W., Zhan, C., Ignatov, A., Manjasetty, B. A., Marinkovic, N., Sullivan, M., Huang, R. & Chance, M. R. (2005). Structure, 13, 1473–1486. [DOI] [PubMed]
- Skubák, P. & Pannu, N. S. (2013). Nature Commun. 4, 2777. [DOI] [PMC free article] [PubMed]
- Spaulding, C. N., Klein, R. D., Ruer, S., Kau, A. L., Schreiber, H. L., Cusumano, Z. T., Dodson, K. W., Pinkner, J. S., Fremont, D. H., Janetka, J. W., Remaut, H., Gordon, J. I. & Hultgren, S. J. (2017). Nature (London), 546, 528–532. [DOI] [PMC free article] [PubMed]
- Stevenson, C. E. M., Tanner, A., Bowater, L., Bornemann, S. & Lawson, D. M. (2004). Acta Cryst. D60, 2403–2406. [DOI] [PubMed]
- Strobl, S., Fernandez-Catalan, C., Braun, M., Huber, R., Masumoto, H., Nakagawa, K., Irie, A., Sorimachi, H., Bourenkow, G., Bartunik, H., Suzuki, K. & Bode, W. (2000). Proc. Natl Acad. Sci. USA, 97, 588–592. [DOI] [PMC free article] [PubMed]
- Taylor, G. (2003). Acta Cryst. D59, 1881–1890. [DOI] [PubMed]
- Terwilliger, T. C., Adams, P. D., Read, R. J., McCoy, A. J., Moriarty, N. W., Grosse-Kunstleve, R. W., Afonine, P. V., Zwart, P. H. & Hung, L.-W. (2009). Acta Cryst. D65, 582–601. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Thomas, S., Holland, I. B. & Schmitt, L. (2014). Biochim. Biophys. Acta, 1843, 1629–1641. [DOI] [PubMed]
- Usón, I. & Sheldrick, G. M. (1999). Curr. Opin. Struct. Biol. 9, 643–648. [DOI] [PubMed]
- Vagin, A. A., Steiner, R. A., Lebedev, A. A., Potterton, L., McNicholas, S., Long, F. & Murshudov, G. N. (2004). Acta Cryst. D60, 2184–2195. [DOI] [PubMed]
- Weinert, T., Olieric, V., Waltersperger, S., Panepucci, E., Chen, L., Zhang, H., Zhou, D., Rose, J., Ebihara, A., Kuramitsu, S., Li, D., Howe, N., Schnapp, G., Pautsch, A., Bargsten, K., Prota, A. E., Surana, P., Kottur, J., Nair, D. T., Basilico, F., Cecatiello, V., Pasqualato, S., Boland, A., Weichenrieder, O., Wang, B.-C., Steinmetz, M. O., Caffrey, M. & Wang, M. (2015). Nature Methods, 12, 131–133. [DOI] [PubMed]
- Yáñez, M., Gil-Longo, J. & Campos-Toimil, M. (2012). Adv. Exp. Med. Biol. 740, 461–482. [DOI] [PubMed]
- Yang, C., Pflugrath, J. W., Courville, D. A., Stence, C. N. & Ferrara, J. D. (2003). Acta Cryst. D59, 1943–1957. [DOI] [PubMed]