Abstract
Many bacteria, including Legionella pneumophila, rely on the type IV secretion system to translocate a repertoire of effector proteins into the hosts for their survival and growth. Type IV coupling protein (T4CP) is a hexameric ATPase that links translocating substrates to the transenvelope secretion conduit. Yet, how a large number of effector proteins are selectively recruited and processed by T4CPs remains enigmatic. DotL, the T4CP of L. pneumophila, contains an ATPase domain and a C-terminal extension whose function is unknown. Unlike T4CPs involved in plasmid DNA translocation, DotL appeared to function by forming a multiprotein complex with four other proteins. Here, we show that the C-terminal extension of DotL interacts with DotN, IcmS, IcmW and an additionally identified subunit LvgA, and that this pentameric assembly binds Legionella effector proteins. We determined the crystal structure of this assembly and built an architecture of the T4CP holocomplex by combining a homology model of the ATPase domain of DotL. The holocomplex is a hexamer of a bipartite structure composed of a membrane-proximal ATPase domain and a membrane-distal substrate-recognition assembly. The presented information demonstrates the architecture and functional dissection of the multiprotein T4CP complexes and provides important insights into their substrate recruitment and processing.
Many bacterial species are equipped with a type IV secretion system (T4SS) that delivers DNA–protein conjugates or virulence proteins across the cell envelope and into recipient cells to mediate horizontal gene transfer or to establish a survival niche in hosts1. The T4SS is mainly composed of two components: a transenvelope secretion conduit and a type IV coupling protein (T4CP)2. T4CP is an AAA+ type hexameric transmembrane ATPase that plays a dual role: recruiting substrates and conveying them to the secretion conduit3. A prototypic T4SS is the VirB/ VirD4 system, encoded by the Ti plasmid in Agrobacterium tumefaciens4, where VirD4 is the T4CP. It translocates the covalently linked transfer DNA–relaxase complex as well as three effector proteins (VirE2, VirE3 and VirF) into plant cells5,6. After being recruited to VirD4, the substrate has to be processed to pass through the narrow T4SS channel. In addition to VirD4, at least two other ATPases, VirB4 and VirB11, are required for substrate processing, which includes protein unfolding7,8.
Legionella pneumophila is a Gram-negative bacterium and a causative pathogen of Legionnaires’ disease. It is an intracellular pathogen that translocates about 300 bacterial proteins, termed effector proteins, into a phagocytic macrophage to subvert immune defense mechanisms and alter homeostasis of the host cell. Isolation of L. pneumophila mutants with severe defects in macrophage killing and intracellular multiplication led to the identification of the 26 Dot/Icm genes (Defective for Organelle Trafficking/IntraCellular Multiplication defect) located at two loci on the bacterial genome9–14. The Dot/Icm loci are related to the conjugal DNA transfer (Tra) region of the Shigella flexneri IncI ColIb-P9 plasmid. The proteins encoded by this region constitute a T4SS, called I-T4SS (after the conjugative plasmid IncI) or T4BSS (after type IVB)15,16. Of the 26 Dot/Icm proteins, DotC, DotD, DotF (IcmG), DotG (IcmE) and DotH (IcmK) constitute a core transmembrane complex that appears to function analogously to the T4SS secretory conduit of A. tumefaciens17. DotL (IcmO) has been identified as a T4CP based on sequence homology with other known T4CPs, including TrwB of Escherichia coli18. In comparison with TrwB, whose cytoplasmic part consists of a nucleotide-binding domain (NBD) and an all α-helical domain (AAD)19, DotL contains an additional ~200-residue segment following the N-terminal NBD. This C-terminal extension is unrelated to any functionally annotated proteins, while a similar or even longer extension is found in T4CPs from other pathogenic bacteria, including Coxiella, Yersinia and Pseudomonas species. Intriguingly, unlike the other characterized T4CPs, DotL appears to form a multiprotein T4CP complex with four other proteins, DotM (IcmP), DotN (IcmJ), IcmS and IcmW (refs 2,17,20). The functions of these four putative subunits are elusive, and how they interact with DotL is virtually unknown.
Here, we show that the C-terminal extension of DotL forms a pentameric complex together with DotN, IcmS, IcmW and a previously unknown subunit LvgA. Moreover, we show that this assembly interacts with Legionella effector proteins. We also present a pseudo-atomic model for the Dot/Icm T4CP holocomplex.
Results
Structure of IcmSW bound to DotL(656–783)
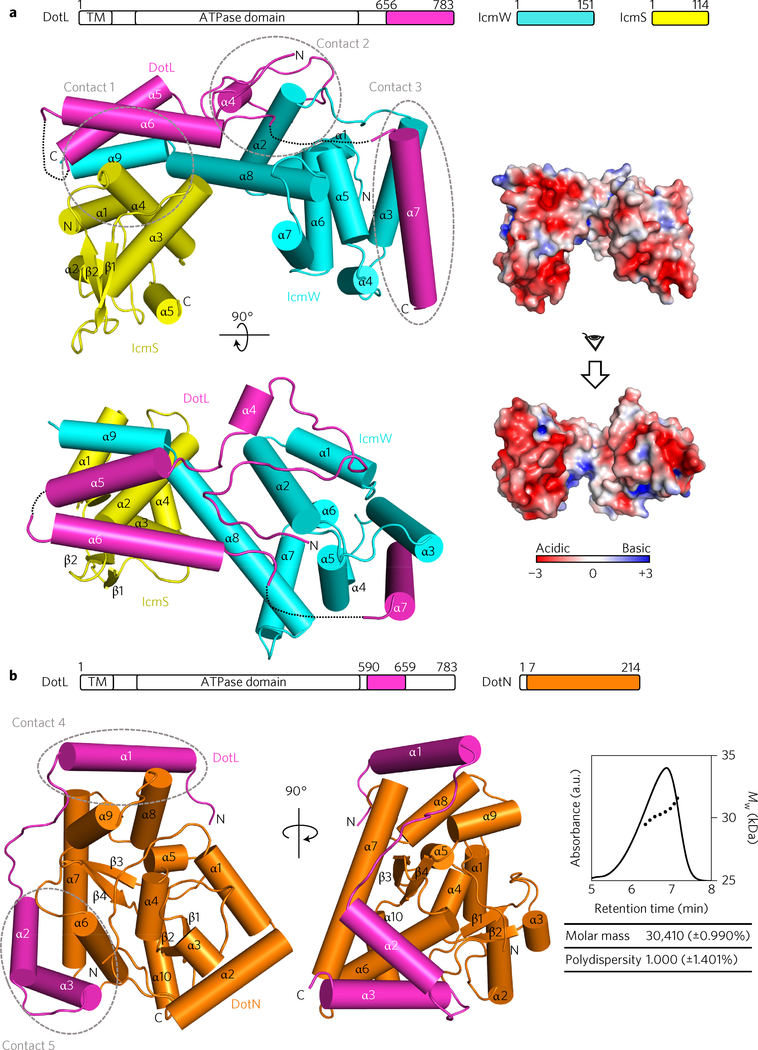
IcmS and IcmW are known to form a stable complex (designated as IcmSW)21. By coexpression, we obtained a triple complex composed of IcmS, IcmW and a C-terminal fragment of DotL, DotL(656–783), which has been shown previously to bind IcmS and IcmW (ref. 20). Subsequently, we determined the structure of this T4CP subcomplex, designated DotL(656–783)–IcmSW. The structure shows that IcmS and IcmW are composed entirely or mostly of α-helices and that the two C-terminal α-helices (α8, α9) of IcmW interact with IcmS to form a bilobed structure (Fig. 1a). The binding interfaces between IcmS and IcmW are quite extensive and are composed mainly of hydrophobic residues, explaining the tight interaction between the two proteins. One face of IcmSW is rather flat and the opposite face is predominantly concave. The overall surface of IcmSW is negatively charged (Fig. 1a), but the concave surface contains a number of exposed hydrophobic residues: Leu86, Ala97, Leu101, Ile102, Leu119 and Leu123 of IcmW. As describe in the following, these hydrophobic residues are involved in the binding of another subunit LvgA. DotL(656–783) forms an extended rope-like structure that spans the entire length of the flat surface of IcmSW and also a side of IcmW (Fig. 1a). Presumably, the observed structure of DotL(656–783) cannot be maintained without IcmSW, which is consistent with the role of IcmSW in stabilizing DotL (ref. 2). None of the three protein structures is meaningfully similar to known structures in the Protein Data Bank (PDB) according to the program Dali22.
Figure 1 |. Crystal structures of DotL(656–783)–IcmSW and DotL(590–659)–DotN.
Two perpendicular views are shown, with schematic drawings of the constructs at the top. a, DotL(656–783)–IcmSW. The three proteins are colour-coded. The first N-terminal α-helix of DotL(656–783) is named α4 to distinguish it from the first α-helix (α1) of DotL(590–659) in b. Dotted lines indicate disordered regions. The surface electrostatic potential was calculated using Adaptive Poisson-Boltzmann Solver (APBS)47 at ±3kT/e. b, DotL(590–659)-DotN. Two proteins are colour-coded. Right: estimate of the molecular mass of the complex by AF4-MALS, where particles elute in order of increasing size. Absorbance at 280 nm (solid line) and molecular weight (Mw, dotted line) are plotted. Intermolecular interactions of DotL at the five indicated regions (contacts 1–3 in a and 4 and 5 in b) are shown in detail in Supplementary Fig. 1b. TM, transmembrane.
Structure of isolated DotN
We next determined the structure of DotN with a truncation of six N-terminal residues. A sequence analysis by HHpred (ref. 23) indicated that a 62-residue sequence of DotN (residues 35–96) is highly similar to a number of the HNH superfamily nucleases containing four Zn2+-chelating cysteines (Supplementary Fig. 1a). Consistently, DotN crystals exhibited strong X-ray absorption at the Zn K-edge. Subsequently, the structure of DotN was determined by the single wavelength anomalous dispersion (SAD) method using a native DotN crystal. In the asymmetric unit of the crystal, 12 molecules formed six dimeric pairs, which are essentially the same as each other, indicating that DotN alone is likely to form a homodimer (Supplementary Fig. 1a). A single Zn atom was found coordinated by four cysteines (Cys52, Cys55, Cys84 and Cys87). This Zn cage is completely buried inside the protein, indicating that the zinc ion plays a structural role in protein folding and the stability of DotN (Supplementary Fig. 1a).
Structure of DotN bound to DotL(590–659)
We found that DotL (590–783) could be produced in a soluble form when E. coli cells expressing this construct were lysed together with cells expressing DotN or IcmSW. Furthermore, these four proteins formed a stable quaternary complex during protein purification. However, this complex was refractory to crystallization. By deletion analysis, we further found that DotL(590–659) forms a stable complex with DotN. Subsequently, the structure of this complex was determined by the zinc SAD method (Fig. 1b).
In this crystal structure, DotN forms a 1:1 complex with DotL (590–659), indicating that the homodimer of free DotN disassembles into monomers to bind DotL(590–659). Formation of the 1:1 complex was supported by an asymmetrical flow field-flow fractionation coupled with multi-angle light scattering (AF4-MALS), showing that the deduced molecular mass is close to the calculated molecular mass of 31.44 kDa (Fig. 1b). Conceivably, the DotN–DotL(590–659) interaction is tighter than the DotN–DotN interaction. DotL(590–659) interacts with DotN in a linear fashion, as if it wraps about halfway around DotN. On the surface of DotN, the N-terminal portion of DotL(590–659) forms an α-helix (α1), followed by an extended loop (α1–α2) and two C-terminal α-helices (α2 and α3) (Fig. 1b). The extended structure of this DotL segment would be disordered without DotL, which explains the known dependence of DotL stability on DotN (ref. 17).
LvgA is an integral component of the coupling complex
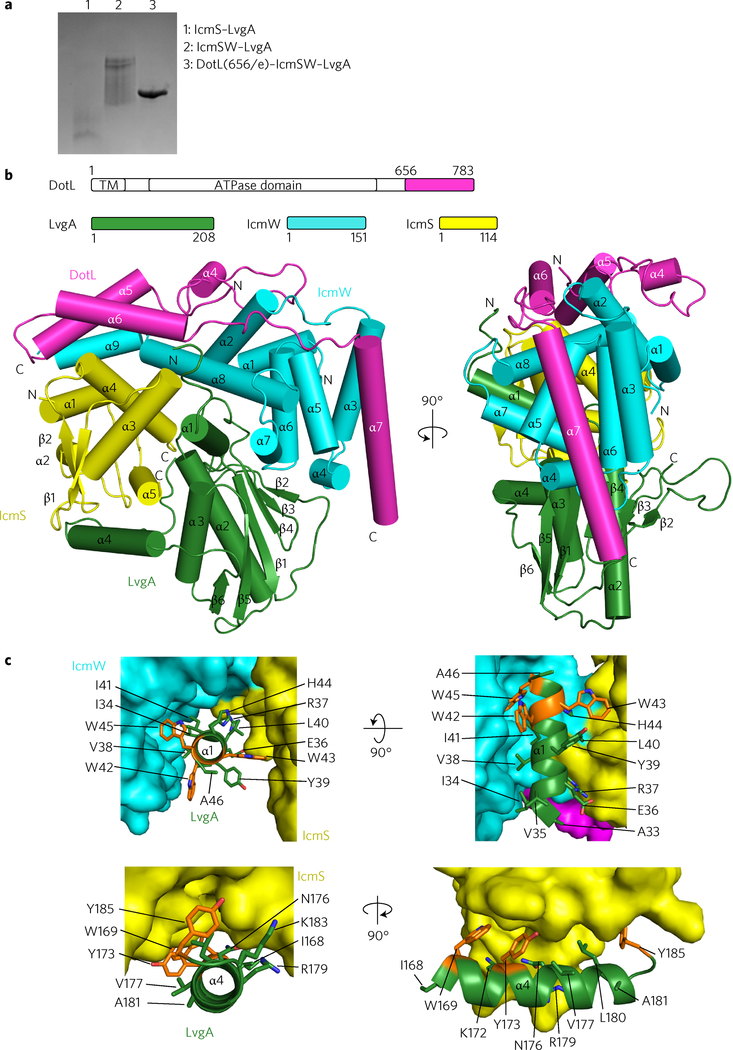
LvgA is an acidic protein that was identified as a virulence factor present in all strains of L. pneumophila24. The open reading frame of LvgA resides outside the Dot/Icm loci, but LvgA is known to interact directly with IcmS (ref. 25). We postulated that this protein might be an integral component of the coupling complex. Consistently, whereas LvgA alone was expressed in a completely insoluble form in E. coli, the protein co-expressed together with IcmS was soluble and the resulting heterodimer could be purified. However, this complex appeared heterogeneous, as judged from the elution profile from a gel-filtration column and a smeared band pattern on a native polyacrylamide gel (Fig. 2a). Coexpression of LvgA with IcmSW resulted in a ternary complex, IcmSW–LvgA, which could be purified, but again it exhibited signs of heterogeneity (Fig. 2a). In contrast, coexpression of LvgA with IcmSW and DotL(656–783) resulted in a homogeneous quaternary complex, DotL(656–783)–IcmSW–LvgA. This complex was maintained throughout purification and exhibited a well-defined single band on a native polyacrylamide gel (Fig. 2a). The dependence of LvgA folding on IcmS and its tight interaction with DotL(656–783)–IcmSW suggest that LvgA is an integral subunit of the coupling complex.
Figure 2 |. Crystal structure of DotL(656–783)–IcmSW–LvgA.
a, Native PAGE analysis. DotL(656–783)–IcmSW–LvgA is a homogeneous complex in comparison with IcmS–LvgA and IcmSW–LvgA. A representative image from three replicate experiments is shown. b, Two orthogonal views of the structure. Schematic drawings of the constructs are shown at the top. The four proteins are colour-coded. c, Highlighted hydrophobic interactions between LvgA and IcmSW. Three clustered tryptophan residues on α1 of LvgA interacting with IcmS (yellow) and IcmW (cyan) are shown as orange sticks (top). Three aromatic residues on α4 of LvgA interacting with IcmS are shown as orange sticks (bottom). Detailed residue–residue interactions are shown in Supplementary Fig. 2.
Structure of LvgA bound to DotL(656–783)–IcmSW
Subsequently, we determined the structure of the DotL(656–783)–IcmSW–LvgA complex at 2.8 Å. LvgA adopts a mixed α/β fold, in which two juxtaposed α2 and α3 helices sit on a rather flat β-sheet composed of six β-strands (Fig. 2b). According to the Dali server, the structure of LvgA is unrelated to any known proteins (highest Z-score of 4.6). While LvgA does not interact with DotL (656–783), it interacts with both IcmS and IcmW at the wide concave surface between the two proteins (Fig. 2b). This interaction is mediated mainly by the N-terminal α-helix (α1, residues 31–45) of LvgA. In addition, its C-terminal α-helix (α4, residues 167–184) and the following loop interact with IcmS (Fig. 2b). The interactions between these two α-helices of LvgA and IcmSW are mainly hydrophobic, involving Trp42, Trp43 and Trp45 on α1 and Trp169, Tyr173 and Tyr185 on α4 (Fig. 2c). By these interactions, LvgA shields a number of otherwise exposed hydrophobic patches of IcmSW. Notably, both α1 and α4 of LvgA extend out from the main body of LvgA, as if they do not belong to LvgA, but to IcmS or IcmW. These observations explain why LvgA alone is insoluble and depends on IcmS for its solubility.
Structure of DotL(590–783)–DotN–IcmSW–LvgA
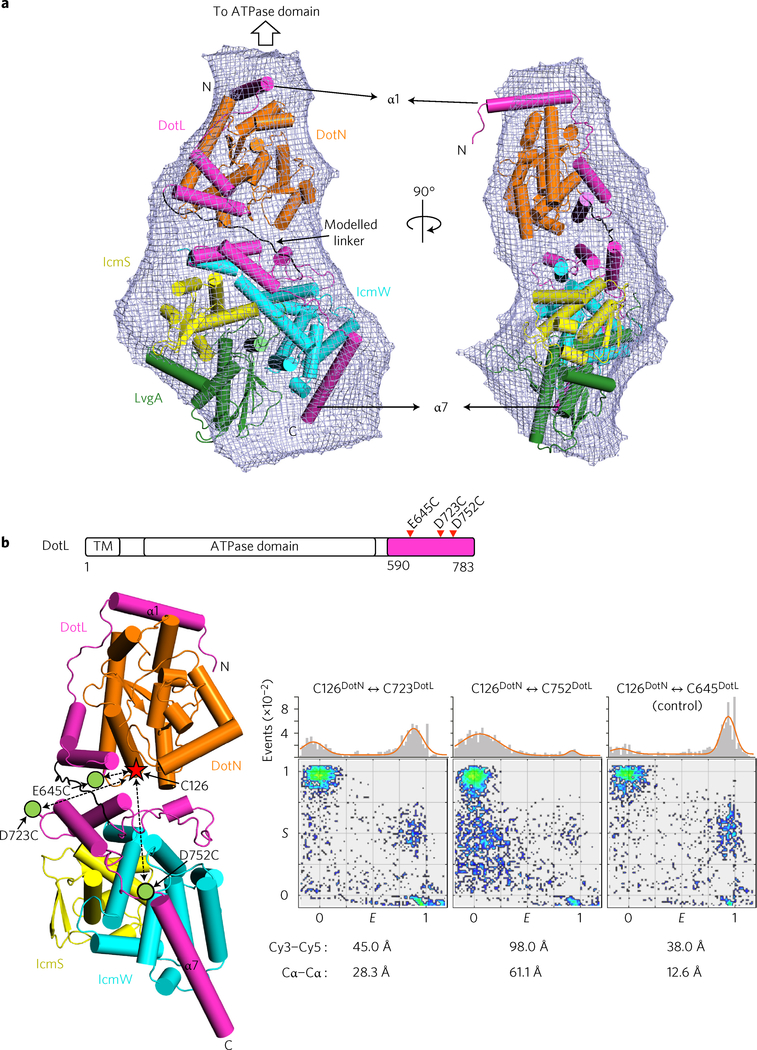
To obtain structural information about the entire C-terminal assembly of DotL, we produced a pentameric complex of DotL(590–783)–DotN–IcmSW–LvgA. This complex itself does not form a higher-order oligomer in solution, as judged based on an AF4-MALS analysis (Supplementary Fig. 3a). Crystals of this complex were obtained, but they diffracted poorly and resisted improvement. We next performed a small-angle X-ray scattering (SAXS) analysis of this complex. The molecular envelope derived from the analysis had an elongated shape with a slim end and a fat end (Fig. 3a). The DotL(590–659)–DotN structure (small) and the DotL (656–783)–IcmSW–LvgA structure (large) were fitted visually into the slim portion and fat portion, respectively. The two structures in this orientation were used as an input to calculate a homology model for DotL residues 660–673, which correspond to the linker segment between the DotN-binding region and the IcmSW-binding region. The program MODELLER26,27 modelled this segment as an extended loop structure between DotL(590–659)–DotN and DotL(656–783)–IcmSW–LvgA (Fig. 3a), which is consistent with a secondary structure prediction. The calculated scattering curve and distance distribution function P(r) are in good agreement with the experimental observations (Supplementary Fig. 3b,c). In this reconstituted, pseudo-atomic resolution structure of DotL(590–783)–DotN–IcmSW–LvgA, the first N-terminal α-helix of DotL(590–783) is at the top and the following polypeptide cascades down along the complex, with LvgA located at the bottom (Fig. 3a).
Figure 3 |. Structural reconstitution of DotL(590–783)–DotN–IcmSW–LvgA.
a, SAXS analysis of DotL(590–783)–DotN–IcmSW–LvgA. The crystal structures of DotL(590–659)–DotN and DotL(656–783)–IcmSW–LvgA were docked into the SAXS envelope (mesh). The computationally modelled DotL segment (residues 660–673) is shown in black. The first and last α helices of DotL(590–783) are labelled. SAXS curves and P(r) functions are shown in Supplementary Fig. 3b,c. b, ALEX-FRET analysis of DotL(590–783)–DotN–IcmSW. Cys126 of DotN is denoted with a star, and the positions of cysteine mutations (E645, D723 and D752 of DotL) by green dots. In the two-dimensional FRET efficiency E versus stoichiometry parameter S graph (see Methods), each dot denotes a single dye-labelled complex. The E and S values of each molecule were determined from its fluorescent signals. A molecule with the correct FRET pair has an S value of ~0.5 (middle region). The control pair (inter-Cα distance of 12.6 Å), generated by an E645C mutation in DotL, exhibited high FRET signals. Shown below the E versus S plots are the inter-Cα distances between the dye-labelled residues and the Cy3–Cy5 distances estimated from the FRET signals. A representative image from three replicate experiments is shown.
To elaborate further, we analysed the DotL(590–783)–DotN–IcmSW subcomplex by fluorescence resonance energy transfer (FRET). For cysteine-specific covalent attachment of the donor (Cy3) and acceptor (Cy5) dyes, three exposed cysteine residues (Cys14 of DotN, Cys136 of IcmW and Cys8 of IcmS) were replaced with alanine. One remaining exposed cysteine residue, Cys126 of DotN, was chosen to make a pair with an artificially introduced cysteine at the position of Asp723 or Asp752 of DotL (Fig. 3b). These two surface-exposed aspartate residues are 28.3 and 61.1 Å apart from Cys126 of DotN (inter Cα distance), respectively, in the reconstituted structure. Because of the stochastic nature of dye labelling, the alternating-laser excitation FRET (ALEX-FRET) method was used to discriminate the correct FRET pair (Cy3–Cy5) from the non-FRET pairs (Cy3–Cy3) and (Cy5–Cy5) in the sample. The complex containing the Cys126DotN–Cys723DotL pair exhibited high values of FRET efficiency E, and the complex containing the Cys126DotN–Cys752DotL pair exhibited low FRET signals (Fig. 3b). These data support the structural reconstitution based on the SAXS envelope and also indicate that the entire C-terminal assembly of DotL is rigid.
Interaction between the T4CP complex and effector proteins
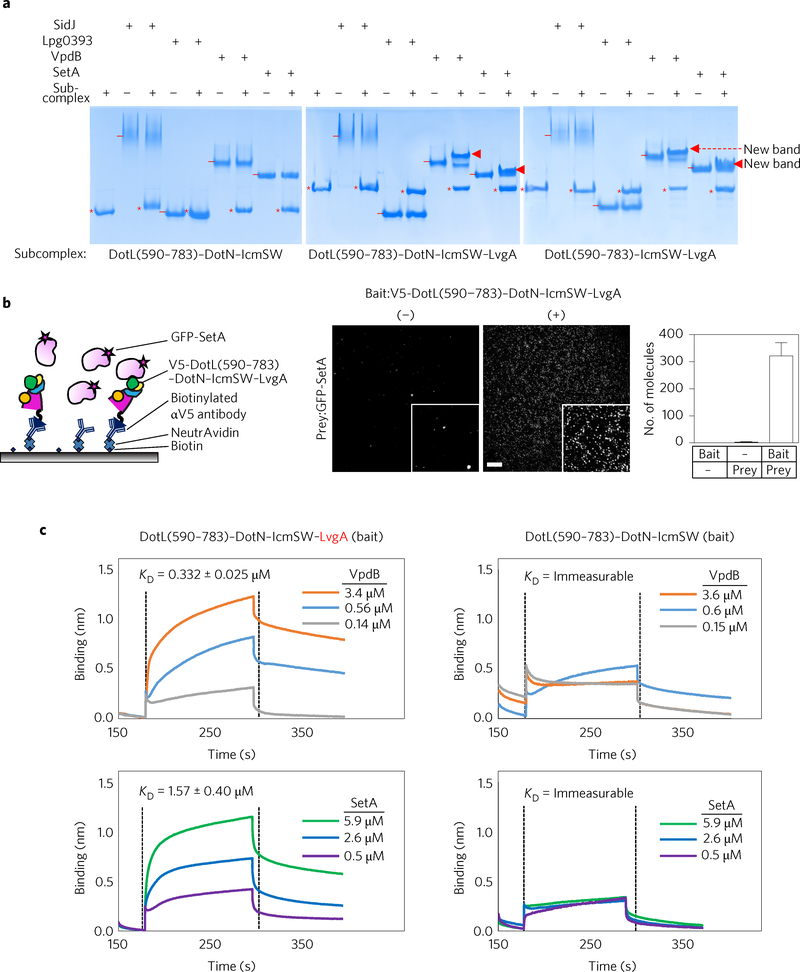
Because IcmSW was reported to interact with a number of Dot/Icm T4SS substrates20,21,28–31, we tested how different T4CP subcomplexes containing IcmSW would interact with effector proteins. Four different Legionella effector proteins (SidJ, Lpg0393, VpdB and SetA) were purified, and their potential binding to DotL(590–783)–DotN–IcmSW was examined. In a native PAGE-based protein-binding assay, the four effector proteins showed no notable interaction with this complex except for a slight tailing of effector protein bands (Fig. 4a, left). In contrast, DotL(590–783)–DotN–IcmSW–LvgA exhibited additional protein bands for VpdB and SetA that are likely to arise from a protein–protein interaction (Fig. 4a, middle). DotN was not involved in formation of the additional protein bands, because DotL(590–783)–IcmSW–LvgA lacking DotN exhibited similar additional protein bands with VpdB and SetA (Fig. 4a, right). The same assay was performed with IcmSW and IcmS–LvgA to find that IcmSW alone exhibited no notable interaction (Supplementary Fig. 4a) and that IcmS–LvgA exhibited merely detectable interactions with SetA and VpdB (Supplementary Fig. 4b).
Figure 4 |. Effector protein binding.
a, Native PAGE analysis. The four effector proteins (6 μM) were incubated with the indicated subcomplexes at a 1:1 molar ratio. The subcomplexes and effector proteins are indicated by ‘*’ and ‘−’, respectively. Newly formed protein bands are indicated by triangles. A representative image from four replicate experiments is shown. b, Detection by SiMPull. Schematic for SiMPull (left). Anti-V5 antibodies coated to a slide immobilized DotL(590–783)–DotN–IcmSW–LvgA with a V5 tag at the N terminus of DotL. GFP-tagged SetA was added and unbound protein was washed out. GFP signals detected by TIRF microscopy are shown, with the insets displaying magnified areas (middle). Scale bar, 5 μm. The average number of fluorophores per image is shown as a bar graph (right). Ten images for each recording were used to calculate standard deviations (2.2 and 48.7), indicated by error bars (zero for bait alone). A representative image from three replicate experiments is shown. c, Quantification. The indicated complexes were immobilized on a Ni-NTA biosensor via the (His)10-MBP tag fused to DotL(590–783). The interaction with VpdB or SetA at the three different concentration was analysed in triplicate by biolayer interferometry, and the deduced KD values are shown. The association and dissociation constants for VpdB were 1.18 × 104 ± 2.70 × 103 M−1 s−1 and 0.39 × 10−2 ± 5.50 × 10−3 s−1, respectively, and those for SetA were 1.31 × 104 ± 2.80 × 103 M−1 sx1 and 2.03 × 10×2 ± 5.70 × 10−3 s−1, respectively.
Using the SiMPull (single-molecule pull-down) assay32, the interaction between DotL(590–783)–DotN–IcmSW–LvgA and SetA was confirmed (Fig. 4b). We next quantified the interaction of DotL(590–783)–DotN–IcmSW–LvgA with VpdB and SetA by biolayer interferometry (BLItz) to find that they interact with each other with an apparent dissociation constant KD of ~332 nM and 1.6 μM, respectively (Fig. 4c, left). Consistent with the native PAGE analysis, interaction between DotL(590–783)–DotN–IcmSW lacking LvgA and VpdB or SetA was undetectable (Fig. 4c, right).
Whether translocation of VpdB and SetA is dependent on IcmSW is unknown. We purified recombinant PieA and a C-terminal fragment (residues 1830–2225) of SidH. These two proteins are known to depend on IcmSW for their translocation31,33. Both PieA and SidH(1830–2225) exhibited no detectable interaction with DotL(590–783)–DotN–IcmSW, but notable interactions with DotL (590–783)–DotN–IcmSW–LvgA and DotL(590–783)–IcmSW–LvgA (Supplementary Fig. 4c,d), as observed for VpdB and SetA.
Together, these data demonstrate that IcmSW and LvgA are required to form the multiprotein assembly with the C-terminal extension of DotL to interact with effector proteins and that LvgA appears to be a critical subunit in the T4CP complex for binding a subset of effector proteins.
A model for the architecture of the T4CP holocomplex
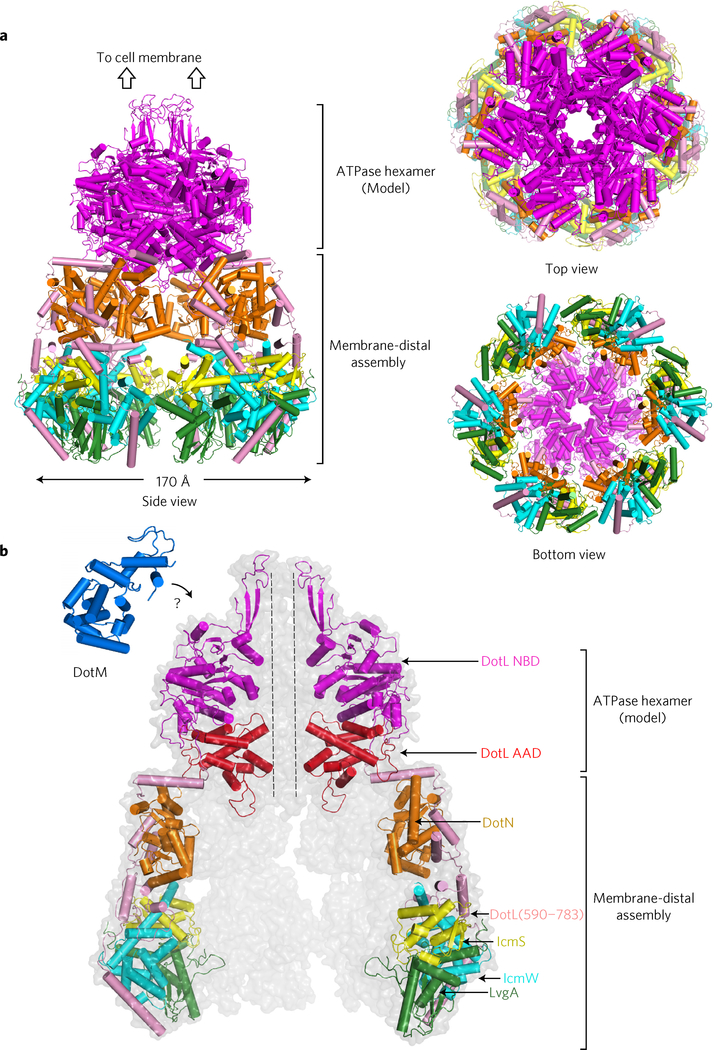
According to a sequence alignment by HHpred (ref. 23), the ATPase domain (NBD plus AAD) of DotL is composed of residues 118–585. Based on the reported homohexameric structure of TrwB lacking the N-terminal transmembrane region34, we built a hexameric model of the ATPase domain of DotL, which was then combined with the six copies of the structure of DotL(590–783)–DotN–IcmSW–LvgA (Fig. 5a). In the hexameric structure of TrwB, the C-terminal end is located most remotely from the excised membrane segment. The C-terminal end in the model for the DotL ATPase domain is located similarly and could be connected reasonably to the N-terminal end of DotL in the DotL(590–783)–DotN–IcmSW–LvgA structure. With the length constraint at the connection, the orientation of DotL (590–783)–DotN–IcmSW–LvgA was manually adjusted to align the six copies without a steric clash. This resulted in an elongated bell-shaped architecture composed of the membrane-proximal ATPase hexamer and the membrane-distal C-terminal assemblies of DotL (Fig. 5a). The holocomplex has a narrow central channel (diameter of ~20 Å) in the ATPase hexamer and a large chamberlike space surrounded by the six copies of the C-terminal assembly of DotL. Located at the bottom of the holocomplex, LvgA forms the bottom rim, indicating that an effector protein bound to LvgA would be positioned quite far from the membrane.
Figure 5 |. Model for the T4CP holocomplex.
a, Three different views. The holocomplex is an elongated bipartite structure. Each subunit or domain (in the case of DotL) is colour coded as in b. b, Enlarged view. Two monomeric units at the front are removed and two monomeric units at the back are faintly shown. The structure of DotM (161–371) is shown. The cytoplasmic domain of DotM might interact with the NBD of DotL (‘?’ symbol).
Both DotL and DotM subunits lack their transmembrane segment.
This model for the T4CP holocomplex does not include DotM, which is a transmembrane protein and is known to interact with DotL (ref. 2). We determined the structure of a cytoplasmic fragment of DotM (residues 161–371) at 1.8 Å resolution (Supplementary Fig. 5). DotM(161–371) is an α-helical globular protein. The structure is not similar to known protein structures according to aDali search (highest Z-score, 4.7). Because the size of the cytoplasmic domain of DotM is smaller than the NBD domain of DotL, it is presumed to interact with the top portion of DotL below the inner membrane of L. pneumophila (Fig. 5b).
Discussion
The DotL T4CP forms a multi-subunit T4CP complex through its C-terminal extension
The T4CP AAA+ ATPases commonly contain a transmembrane domain, an NBD and an AAD. Some T4CP ATPases, for example, TrwB of E. coli, are composed of these three domains only, but other T4CP ATPases, such as VirD4 of A. tumefaciens, contain an additional domain at the C terminus. This domain, which we designate as the C-terminal extension, varies greatly in length as well as in sequence among bacteria (Fig. 6). The functional roles of this extension are poorly understood. For the TraD T4CP in E. coli, the eight-residue tail of its C-terminal extension (~140 amino acids) has been shown to interact with the plasmid conjugation factor TraM (ref. 35), which binds cognate plasmid. Analogously, the C-terminal extension (~124 residues) of VirD4 has been demonstrated to be required for the translocation of the VirE2 effector protein, although a binding interaction between the two was not detectable36,37. In the case of TrwB, which lacks a C-terminal extension, the AAD plays an equivalent role, as this domain binds to DNA and cognate processing proteins38.
Figure 6 |. C-terminal extension of T4CPs.
Multiple T4CPs are classified into the two subtypes and their domain organizations are shown with different colours. The domain boundaries were determined by HHpred (ref. 23). The C-terminal extension is denoted CTE. Right: experimentally identified substrates are indicated by ‘O’, and ‘ND’ indicates ‘not determined’. E.c., Escherichia coli (R388 plasmid); S.f., Shigella flexneri 4c (1205p3 plasmid); P.s., Pseudomonas syringae (NCPPB880–40 plasmid); E.f., Enterococcus faecalis (CF10 plasmid); N.u., Nitrosomonas ureae; E.c., Escherichia coli (IncP-α RP4 plasmid); B.h., Bartonella henselae; A.t., Agrobacterium tumefaciens (Ti plasmid); E.c., Escherichia coli (O157_Sal plasmid); E.c., Escherichia coli (F plasmid); A.p., Anaplasma phagocytophilum; L.p., Legionella pneumophila; P.s., Piscirickettsia salmonis; R.g., Rickettsiella grylli; C.b., Coxiella burnetii; P.p., Pseudomonas putida; B.v., Burkholderia vietnamiensis; X.c., Xanthomonas campestris pv. vesicatoria str. 85–10; M.a., Micavibrio aeruginosavorus; A., Acidovorax sp. Root70; Y.p., Yersinia pseudotuberculosis; X.c., Xanthomonas citri. Genetic organizations of the associated T4ASSs or T4BSSs are provided in Supplementary Fig. 6.
The Dot/Icm T4CP is likely to require a more elaborate substrate-recognition mechanism than T4CPs mediating conjugative plasmid DNA transfer, because the former has to recognize a large number of effector proteins while the latter recognizes a DNA-relaxase adduct and additionally a few effectors as the substrate(s). Acquisition of the intricate C-terminal assembly of DotL during evolution, which would be absent in TrwB-like T4CPs lacking a C-terminal extension, might have been essential for DotL to recognize and process a large number of effector proteins. Notably, T4CPs associated with a T4BSS contain a C-terminal extension, which in general is longer than that of T4CPs associated with a T4ASS (Fig. 6). Whether the length of the C-terminal extension may reflect the complexity of the interacting proteins remains to be investigated further. Because the C-terminal extensions are heterogeneous in their length and sequence, the set of proteins interacting with a C-terminal extension differ from one pathogen to another. Consistently, DotN, IcmS, IcmW and LvgA homologues are found only in Legionellales. This diversification is perhaps a reflection of the great variations of the effectors translocated through T4SS among pathogens39.
Potential role of DotM
DotM is known to stabilize DotL, presumably through direct protein–protein interaction2. Although we determined the structure of DotM(161–371), no further clues for the functional roles of DotM were gained. Because DotM (161–371) is small and globular (Supplementary Fig. 5), the cytosolic domain of DotM could interact with the membraneproximal NBD domain of DotL (Fig. 5b), in addition to the reported DotL–DotM interaction through their transmembrane domains2. We sought to address this possibility, but our effort was hampered by the protein instability of all DotL constructs containing the ATPase domain.
In contrast to other non-ATPase subunits of the T4CP complex, DotM homologues are found in alpha-, beta-, gamma- and delta- Proteobacteria according to a sequence homology search. The wide distribution of DotM supports a possibility that this subunit may play a general role in substrate translocation by interacting with T4CP ATPase, for example, through regulation of the ATPase activity of T4CP or by linking T4CP to the secretory conduit.
Substrate recognition/recruitment
Both IcmSW and IcmS–LvgA were presumed to function as a cytosolic adaptor involved in substrate recognition by the Dot/Icm complex25,28,31, like secretion chaperones in the type III secretion system, which bind substrate proteins and maintain them in a secretion-competent state40–42. The data presented here support that they function as an integral component of the Dot/Icm T4CP complex rather than as a free heterodimer. Consistently, although a potential interaction between IcmSW or IcmS–LvgA and four tested effector proteins was undetectable or marginally detected (Supplementary Fig. 4a,b), the interactions of DotL(590–783)–IcmSW–LvgA with VpdB and SetA were clearly observed (Fig. 4).
So far, two different secretion signals have been identified in the Legionella effector proteins that depend on the Dot/Icm T4SS for their translocation: a glutamic acid stretch called E-block (EExxE) and several hydrophobic amino acids close to the C-terminal end43,44. SetA contains none of the identified translocation signals. VpdB has a C-terminal 585EEFNEQ590 sequence, which conforms to the E-block motif and a hydrophobic residue Leu595 at the C-terminal −4 position. Intriguingly, however, we observed that VpdB(11–485) lacking this sequence motif interacted with DotL (590–783)–DotN–IcmSW–LvgA indistinguishably from fulllength VpdB (Supplementary Fig. 4e). Thus, VpdB and SetA seem to belong to another group of Leginonella effectors that contain an unknown translocation signal45.
Of note, VpdB and SetA share no detectable sequence homology with each other and they exhibit considerably different binding affinities (0.33 versus 1.57 μM) for the substrate-recognition assembly. Given the large number of Legionella effectors, substrate recognition by the C-terminal assembly of the T4CP complex is expected to be promiscuous. Different surfaces of the substrate-recognition assembly, including that of DotN, might be used for interaction with different effector proteins. On the other hand, as yet unknown adaptor proteins may be involved in substrate recruitment, because DotL(590–783)–DotN–IcmSW–LvgA exhibited no interaction with SidJ and Lpg0393 that we tested. We also note that RalF, whose secretion is independent of IcmSW (refs 20,31), did not show a detectable interaction with DotL (590–783)–DotN–IcmSW–LvgA (Supplementary Fig. 4f).
Architecture of the Dot/Icm T4CP holocomplex and substrate processing
At this point, we do not know whether the substrate-recognition assembly in the T4CP holocomplex may form a proximity-induced hexamer forced by the hexamerization of the ATPase domain of DotL. In this case, the six substrate-recognition assemblies would form a chamber below the ATPase hexamer (Fig. 5b). Alternatively, they could undergo a hinge bending motion at the junction between the ATPase domain and the C-terminal extension of DotL. In either case, the substrate protein recruited to the T4CP holocomplex should be placed under the ATPase hexamer and then unfolded to pass through the central channel of the ATPase hexamer.
In L. pneumophila, the homologues of the three substrate-processing ATPases of A. tumefaciens VirD4, VirB4 and VirB11 are present as DotL, DotO and DotB, respectively. The functional role of the cytosolic DotB protein in the T4SS substrate export has been verified through genetic analysis46. Given the architectural view of the T4CP holocomplex (Fig. 5), it is tempting to speculate that hexameric DotB may stabilize or induce hexamerization of the substrate-recognition assemblies by binding them at the bottom of the T4CP holocomplex. In this scenario, the spacious chamber formed by the six substrate-recognition assemblies (Fig. 5) may serve as a substrate-unfolding chamber, where the ATPase activities of DotB and DotL provide energy for protein unfolding.
Conclusions
The presented study reveals how five different subunits assemble to form the Dot/Icm T4CP holocomplex, which probably represents the architectures of multi-subunit coupling complexes mediating effector protein translocation in other pathogenic bacteria. The membrane-distal part of the T4CP holocomplex, below the membrane-proximal ATPase hexamer, is identified as the interface for effector protein recognition. This work provides a strong foundation for addressing the enigmatic questions of how selective recognition of effector proteins and ATP-driven substrate processing are achieved by multi-subunit T4CP complexes. A detailed mechanistic understanding of these complexes would be valuable for therapeutic intervention in effector translocation into host cells, which is critical for the infectivity of many pathogenic bacteria.
Methods
Protein production
DotL(656–783)–IcmSW
Genomic DNA (NCBI accession no. AE017354.1) from L. pneumophila (Philadelphia-1) was purchased from ATCC. The genes encoding full-length IcmS (AAU26539.1) and IcmW (AAU28746.1) were cloned into the pRSFDuet-1 (Novagen) vector by standard polymerase chain reaction (PCR) cloning methods. IcmS with a C-terminal fusion of the cysteine protease domain (CPD) of the V. cholera MARTX toxin48 with a (His)10-tag and IcmW without a tag were coexpressed from this vector in E. coli BL21 (DE3) RIPL strain (Novagen) at 18 °C overnight after 200 μM isopropyl-β-D-thiogalactoside (IPTG) induction. A DNA fragment coding for residues 656–783 of DotL (AAU26543.1) was subcloned into the pET22b CPD-(His)10 vector to produce DotL(656–783) with C-terminal fusion of CPD-(His)10. The protein was expressed in the E.coli BL21 (DE3) RIPL strain at 18 °C overnight. E. coli cells expressing these proteins were co-sonicated and cleared lysate was applied onto a gravity flow column containing HisPur Cobalt Resin (Thermo Scientific). The column was washed with buffer A (20 mM Tris-HCl (pH 7.5), 100 mM NaCl and 3 mM 2-mercaptoethanol) and the CPD-(His)10 tag was autolytically cleaved off on this resin by incubation with 100 μM sodium phytate (Sigma Aldrich). The flow-through fraction was further purified using a HiTrap Q anion exchange column (GE Healthcare) and a HiLoad 26/60 Superdex 75 gel-filtration column (GE Healthcare) equilibrated with buffer A supplemented with 1 mM dithiothreitol (DTT) instead of 2-mercaptoethanol. Selenomethionine (SelMet)-labelled DotL(656–783)–IcmSW was obtained by using the E. coli B834 (DE3) RIL strain (Novagen) and purified according to the same procedure for the native protein.
DotN homodimer
DotN(7–214) (AAU26552.1) was cloned into pET22b CPD-(His)10 vector and expressed in E.coli BL21 (DE3) RIPL strain at 18 °C overnight. The cleared lysate was applied onto a gravity flow column with HisPur Cobalt Resin and washed with buffer A. DotN protein was eluted from the resin by treatment with 100 μM sodium phytate. The eluate was further purified with a HiTrapQ anion exchange column and a HiLoad 26/60 Superdex 75 column.
DotL(590–659)–DotN
The DotN binding region of DotL, residues 590–659, was cloned into pET22b CPD-(His)10 vector and expressed in E.coli BL21 (DE3) RIPL strain at 18 °C overnight. Cells expressing DotN(7–214) cloned into pET22b CPD-(His)10 were co-sonicated. The purification procedure was virtually identical to that of the DotN homodimer.
DotL(656–783)–IcmSW–LvgA
The gene coding for LvgA (AAU26621.1) was cloned into pMAL vector (AmpR) (New England Biolabs) to produce N-terminal (His)10-MBP tagged LvgA, and the genes coding for full-length IcmS and IcmW were cloned into pRSFDuet-1 vector (KanR). The E. coli BL21 (DE3) RIPL strain transformed with the two vectors expressed the three proteins. A DNA fragment coding for DotL(656–783) was cloned into the pET22b vector, and the protein was expressed from the E. coli BL21 (DE3) RIPL strain. Equal volumes of E. coli cultures were co-sonicated and cleared lysate was applied onto a gravity flow column containing HisPur Cobalt Resin. The column was washed with buffer A and the proteins were eluted with buffer A containing an additional 200 mM imidazole. After treatment with tobacco etch virus protease overnight, the complex was further purified using a HiTrap Q anion exchange column and a HiLoad 26/60 Superdex 75 gel-filtration column.
DotM(161–371)
DotM (AAU26542.1) was cloned into pET22b CPD-(His)10 vector and expressed in BL21 (DE3) RIPL strain at 18 °C overnight. The purification procedure was virtually identical to that for the DotN homodimer. SelMet-labelled DotM(161–371) was obtained using E. coli B834 (DE3) RIL strain (Novagen) and purified according to the same procedure used for the native protein.
DotL(590–783)–DotN–IcmSW, DotL(590–783)–DotN–IcmSW–LvgA and DotL(590–783)–IcmSW–LvgA
(His)10-MBP was N-terminally fused to DotL(590–783) and DotN(7–214). The expression and purification procedures for these three subcomplexes were virtually identical to that used for DotL(656–783)–IcmSW–LvgA.
SidJ(128–873), Lpg0393, VpdB, VpdB(11–485), SetA, PieA, SidH(1830–2225) and RalF
SidJ(128–873) (AOU14010), Lpg0393 (YP_094437), SetA (Q5ZU30) and PieA (YP_095979.1) were expressed as a fusion protein containing a C-terminal CPD-(His)10 tag, VpdB (WP_010946959) and VpdB (11–485) with an N-terminal (His)10-GST tag, SidH(1830–2225) (AAU28877.1) and RalF (WP_010947666) with an N-terminal (His)10-YFP tag. The procedures for expression and purification of these effector proteins were similar to those used for DotN and DotL(656–783)–IcmSW–LvgA.
Crystallization, structure determination and refinement
DotL(656–783)–IcmSW
The DotL(656–783)–IcmSW subcomplex (20 mg ml−1) was crystallized in a solution containing 25% (vol/vol) PEG 550 monoethylether, 100 mM 2-(N-morpholino) ethanesulfonic acid (pH 6.5), 10 mM zinc sulfate and 5 mM spermine tetrahydrochloride (Sigma Aldrich). Crystals appeared overnight and reached their maximum in ~3 days. The SelMet-substituted complex was crystallized under the same conditions. The crystallization solution served as the cryoprotectant in cryocooling of the crystals. A SAD data set was collected at the Se absorption peak. The Se positions were identified and the phases were calculated using PHENIX software49.
DotN homodimer
The DotN homodimer (10 mg ml−1) was crystallized in a solution containing 600 mM sodium citrate and 100 mM sodium cacodylate (pH 6.5). For cryoprotection, the crystals were soaked in the crystallization solution containing an additional 12% PEG400. X-ray fluorescence scans indicated the presence of Zn with the Zn K edge at 9.66 keV. A SAD data set was collected at a wavelength of 1.2828 Å. The Zn anomalous signal was sufficient for phase determination using PHENIX software49.
DotL(590–659)–DotN
The DotL(590–659)–DotN (15 mg ml−1) was crystallized in a solution containing 100 mM sodium malonate (pH 4.0), 10% (vol/vol) PEG3350 and 20% (vol/vol) glycerol. This solution also served as a cryoprotectant. The structure was determined by Zn SAD with PHENIX software49.
DotL(656–783)–IcmSW–LvgA
The DotL(656–783)–IcmSW–LvgA subcomplex (25 mg ml−1) was crystallized in a solution containing 100 mM Bis-Tris (pH 5.5), 15% (vol/vol) PEG3350 and 8 mM spermine tetrahydrochloride. Crystals appeared in about a month. For cryoprotection, the crystals were soaked in the crystallization solution containing additional 15% glycerol. Phases were determined by molecular replacement using the structure of DotL(656–783)–IcmSW as a search model with PHASER50.
DotM(161–371)
The DotM(161–371) (36 mg ml−1) was crystallized in a solution containing 100 mM Tris-HCl (pH 8.5), 17% (vol/vol) PEG3350 and 200 mM MgCl2. SelMet-substituted DotL(161–371) was crystallized under the same conditions, but required microcrystal seeds of the native protein. For cryoprotection, the crystals were soaked in crystallization solution containing additional 10% glycerol. The best X-ray data set was obtained with a SelMet-substituted crystal. Phase determination and auto model-building were performed using PHENIX49.
All crystals were obtained using the hanging-drop vapour diffusion method at 20 °C. All X-ray diffraction data were collected at beamline 5C at the Pohang Accelerator Laboratory. All diffraction data were processed with the HKL2000 suite51. All model building and structure refinement were carried out using COOT52 and CNS53. Crystallographic data statistics are summarized in Supplementary Table 1.
SAXS analysis
SAXS data were collected with the SX165 detector at a sample-to-detector distance of 1,000 or 4,000 mm on the 4C SAXS II beamline at Pohang Light Source II. All scattering measurements were carried out at 4 °C using a FP50-HL refrigerated circulator (JULABO). Samples of varying protein concentrations (2, 4 and 6 mg ml−1 DotL(590–783)–DotN–IcmSW–LvgA) were used to obtain triplicate measurements. For each sample, we recorded six successive frames of 5–10 s exposure at 0.734 Å wavelength. These frames, indicating no sign of radiation damage, were averaged for further analysis. Data were processed and analysed using software applications embedded in the ATSAS package (http://www.embl-hamburg.de/biosaxs/software.html). The one-dimensional scattering data I(q) as a function of q (q = 4πsinθ/λ, where 2θ is the scattering angle and λ is the wavelength) were obtained by radial averaging. Scattering intensities from the buffer solution were measured and used for background subtraction. The I(q) data of the samples were then extrapolated to zero concentration. The program GASBOR54 was used to reconstruct the molecular shapes. The SAXS curves and distance distribution function P(r) were calculated using CRYSOL55 and GNOM56, respectively.
ALEX-FRET analysis
The DotL(590–783)–DotN–IcmSW complex was labelled with Cy3- and Cy5-maleimide (GE Healthcare). For dual labelling, the two fluorescent dyes were added at a 1:2 (protein:dye) molar ratio. The mixture was incubated for 6 h at 4 °C. Unreacted dyes were removed by a PD minitrap G-25 size-exclusion column (GE healthcare). Dye labelling was confirmed by a fluorescent protein band on an SDS–PAGE gel. The ALEX-FRET method and data analysis have been described in previous works57,58. The laser intensities for donor and acceptor excitation were 80 and 30 μW, respectively. To detect single diffusing molecules in solution, the protein sample was diluted to ~50 pM in a buffer solution containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM NaN3, 1 mM DTT, 5% glycerol (vol/vol), 100 μg ml−1 bovine serum albumin and 1 mM β-mercaptoethanol. Labview software (National Instrument) was used to select fluorescent bursts induced by single molecules. The FRET efficiency E and stoichiometry parameter S were calculated as described previously57:
where (D, donor; A, acceptor) denotes the fluorescent emission of acceptor dye by donor excitation (FRET signal), the fluorescent emission of donor dye excited by the donor-excitation laser, the fluorescent emission of acceptor dye excited by the acceptor-excitation laser. The distance between Cy3 and Cy5 was estimated by the equation of R = R0(1/E − 1)1/6, with R0 = 6 nm for the Cy3–Cy5 pair59.
SiMPull
Slides and chambers were constructed as previously described60. All samples were diluted in T100 buffer (20 mM Tris-HCl (pH 7.5), 100 mM NaCl) and experiments were performed at 23 °C. Slide chambers were first blocked by flowing in 10% bovine serum albumin solution and incubating for 30 min. Slides were washed and then treated with NeutrAvidin (Thermo) followed by biotinylated V5 antibody (Genetex) diluted 1:100. After thorough washing, 40 μl V5 tagged DotL (590–783)–DotN–IcmSW–LvgA was added to slide chambers and allowed to bind for 10 min. Unbound complex was washed out of the chambers with T100 buffer. A 40 μl volume of 50 nM SetA protein tagged with green fluorescent protein (GFP) or mCherry at the N terminus was added, allowed to bind for 10 min, and unbound SetA was then washed out. Image acquisition was performed using a total internal reflection fluorescence (TIRF) microscope and an electron-multiplying charge-coupled device camera. Single-molecule spots were identified using IDL scripts and mean spot counts were calculated using MATLAB scripts.
Biolayer interferometry
The KD value for the protein–protein interaction was derived from measurement of the kinetic parameters using a BLItz instrument (ForteBio). T4CP subcomplexes containing a (His)10 tag were loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) biosensor for 120 s. A baseline was determined by incubating the sensor with 500 μl T100 buffer for 30 s. A 4 μl volume of VpdB and SetA in the T100 buffer was used to measure protein association until reaching steady state. The binding was used to measure protein dissociation (for a duration of 120 s). Binding kinetics were deduced using BLItz Pro software (ForteBio).
Homology modelling
The template-based structure modelling method GOAL was used to generate models for the monomeric structure of the DotL ATPase domain61. In GOAL, the two structures of TrwB (apo and ADP-bound) were used as templates, and its sequence was aligned with the target sequence using the multiple sequence-structure alignment protocol of MSACSA (ref. 62) followed by generation of structure models by the global optimization method of conformation space annealing63,64. These monomeric models were used to build the hexameric structure of the target by placing six copies of a monomer on the hexameric structures of TrwB using MODELLER (ref. 27). The final structure was selected from 100 models based on the MODELLER energy, the steric clash score and the discrete optimized protein energy score. Finally, side-chain remodelling was carried out using SCWRL4 (refs 65,66).
Data availability
The coordinates of the structures have been deposited in the Protein Data Bank under accession codes 5X1E for DotL(656–783)–IcmS–IcmW, 5X42 for DotL(590–659)–DotN, 5X90 for DotL(656–783)–IcmS–IcmW–LvgA, 5X1H for DotN and 5X1U for DotM(161–371). Other data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
This study made use of Beamlines 4C and 5C at Pohang Accelerator Laboratory, Korea, and was supported by the National Research Foundation of Korea (NRF) (grant no. 2008–00576) and KAIST Future Systems Healthcare Project, Ministry of Science, ICT and Future Planning. J.D.K. was supported by a TJ PARK postdoctoral fellowship from POSCO TJ PARK Foundation. The authors thank the Korea Institute for Advanced Study for providing computing resources (KIAS Center for Advanced Computation Linux Cluster).
Footnotes
Competing interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available for this paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wallden K, Rivera-Calzada A & Waksman G Microreview: type IV secretion systems: versatility and diversity in function. Cell. Microbiol 12, 1203–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent CD, Friedman JR, Jeong KC, Sutherland MC & Vogel JP Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol 85, 378–391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomis-Ruth FX, Sola M, de la Cruz F & Coll M Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des 10, 1551–1565 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Zechner EL, Lang S & Schildbach JF Assembly and mechanisms of bacterial type IV secretion machines. Phil. Trans. R. Soc. Lond. B 367, 1073–1087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vergunst AC et al. Virb/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jacome EJ & Hooykaas PJJ Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 31, 860–868 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atmakuri K, Cascales E & Christie PJ Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol 54, 1199–1211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakubowski SJ, Krishnamoorthy V, Cascales E & Christie PJ Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol 341, 961–977 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isberg RR, O’Connor TJ & Heidtman M The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol 7, 13–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finsel I & Hilbi H Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell. Microbiol 17, 935–950 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Berger KH & Isberg RR Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol 7, 7–19 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Marra A, Blander SJ, Horwitz MA & Shuman HA Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl Acad. Sci. USA 89, 9607–9611 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal G, Purcell M & Shuman HA Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl Acad. Sci. USA 95, 1669–1674 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel JP, Andrews HL, Wong SK & Isberg RR Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Juhas M, Crook DW & Hood DW Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol 10, 2377–2386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie PJ & Vogel JP Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8, 354–360 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent CD et al. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol 62, 1278–1291 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Buscher BA et al. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J. Bacteriol 187, 2927–2938 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomis-Ruth FX, Moncalian G, de la Cruz F & Coll M Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein—detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277, 7556–7566 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Sutherland MC, Nguyen TL, Tseng V & Vogel JP The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog. 8, e1002910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cambronne ED & Roy CR The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3, e188 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L & Rosenström P Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alva V, Nam SZ, Soding J & Lupas AN The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44, W410–W415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edelstein PH, Hu B, Higa F & Edelstein MAC lvgA, a novel Legionella pneumophila virulence factor. Infect. Immun 71, 2394–2403 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent CD & Vogel JP The Legionella pneumophila IcmS–LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol 61, 596–613 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Fiser A, Do RK & Sali A Modeling of loops in protein structures. Protein Sci. 9, 1753–1773 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb B & Sali A Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 5.6.1–5.6.37 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Bardill JP, Miller JL & Vogel JP IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol 56, 90–103 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Coers J et al. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol 38, 719–736 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Habyarimana F, Price CT, Santic M, Al-Khodor S & Kwaik YA Molecular characterization of the Dot/Icm-translocated AnkH and AnkJ eukaryotic-like effectors of Legionella pneumophila. Infect. Immun 78, 1123–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ninio S, Zuckman-Cholon DM, Cambronne ED & Roy CR The Legionella IcmS–IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol 55, 912–926 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Jain A et al. Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninio S, Celli J & Roy CR A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 5, e1000278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomis-Rüth FX et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Lu J et al. Structural basis of specific TraD–TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol 70, 89–99 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Atmakuri K, Ding ZY & Christie PJ Vire2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol 49, 1699–1713 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitaker N et al. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J. Bacteriol 198, 2701–2718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitaker N et al. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J. Bacteriol. 197, 2335–2349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voth DE, Broederdorf LJ & Graham JG Bacterial type IV secretion systems: versatile virulence machines. Future Microbiol. 7, 241–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett JCQ & Hughes C From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8, 202–204 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Parsot C, Hamiaux C & Page AL The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol 6, 7–14 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Stebbins CE & Galan JE Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414, 77–81 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Nagai H et al. A C-terminal translocation signal required for Dot/lcm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl Acad. Sci. USA 102, 826–831 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang L et al. The E block motif is associated with Legionella pneumophila translocated substrates. Cell. Microbiol. 13, 227–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lifshitz Z et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl Acad. Sci. USA 110, E707–E715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sexton JA, Yeo HJ & Vogel JP Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol 57, 70–84 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen A et al. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat. Chem. Biol 5, 469–478 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mccoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brünger AT et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Svergun DI, Petoukhov MV & Koch MHJ Determination of domain structure of proteins from X-ray solution scattering. Biophys. J 80, 2946–2953 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svergun D, Barberato C & Koch MHJ CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr 28, 768–773 (1995). [Google Scholar]
- 56.Semenyuk AV & Svergun DI GNOM—a program package for small-angle scattering data-processing. J. Appl. Crystallogr 24, 537–540 (1991). [Google Scholar]
- 57.Lee NK et al. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J 88, 2939–2953 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim C, Lee OC, Kim JY, Sung W & Lee NK Dynamic release of bending stress in short dsDNA by formation of a kink and forks. Angew. Chem. Int. Ed 54, 8943–8947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy R, Hohng S & Ha T A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain A, Liu R, Xiang YK & Ha T Single-molecule pull-down for studying protein interactions. Nat. Protoc 7, 445–452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joo K et al. Template based protein structure modeling by global optimization in CASP11. Proteins 84(Suppl. 1), 221–232 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Joo K, Lee J, Kim I, Lee SJ & Lee J Multiple sequence alignment by conformational space annealing. Biophys. J 95, 4813–4819 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J, Scheraga HA & Rackovsky S New optimization method for conformational energy calculations on polypeptides: conformational space annealing. J. Comput. Chem 18, 1222–1232 (1997). [Google Scholar]
- 64.Lee J, Lee IH & Lee J Unbiased global optimization of Lennard–Jones clusters for N < or = 201 using the conformational space annealing method. Phys. Rev. Lett 91, 080201 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Joo K et al. All-atom chain-building by optimizing MODELLER energy function using conformational space annealing. Proteins 75, 1010–1023 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Krivov GG, Shapovalov MV & Dunbrack RL Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the structures have been deposited in the Protein Data Bank under accession codes 5X1E for DotL(656–783)–IcmS–IcmW, 5X42 for DotL(590–659)–DotN, 5X90 for DotL(656–783)–IcmS–IcmW–LvgA, 5X1H for DotN and 5X1U for DotM(161–371). Other data that support the findings of this study are available from the corresponding author upon request.