Abstract
Recent progresses in clinical diagnostic analyses have demonstrated the decisive influence of host gut microbiota on the status of metabolic disorders. Short chain fatty acids (SCFAs) produced by gut microbiota, in particular, are considered as a key biomarker, both of communication between gut microbiota and the host, and of impact on host metabolic homeostasis. Microbiota modulation and concomitant anti-obesity effects of probiotics have been reported by different researchers. However, the underlying modulatory functions of probiotics on gut microbiota towards host metabolic homeostasis are still not fully understood. In this study, the impact of Lactobacillus sakei CJLS03 (isolated from Korean kimchi) on obesity-related biomarkers was investigated using a diet-induced obese mouse model. Body weight increase, SCFAs, the gut microbiota and various obesity-associated biomarkers were significantly and beneficially influenced by L. sakei CJLS03 administration compared to the control groups. Analytical data on faecal samples support the role of the colonic microbial population in SCFA production. The composition of the latter may be influenced by modulation of the distal gastro-intestinal microbiota by putative probiotics such as L. sakei CJLS03.
Subject terms: Biochemistry, Applied microbiology
Introduction
Obesity has become a major social issue associated with health and quality of life. Approximately one-third of the world’s population is reported as obese or overweight, and this phenomenon is expected to increase further1,2. Currently, several drugs are being used under approval of Food and Drug Administration (FDA) around world for the purpose of weight loss with known mechanisms that promote fat metabolism, inhibit absorption of consumed fat, or suppress appetite. However, such pharmacological approach may cause serious side effects, resulting in a problem in continuous administration3. Accordingly, it is necessary to consider a new therapeutic approach that is safer and improves the harmony of the overall metabolism in the body.
Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer beneficial effects on the host”4. Combinations of antibiotics, probiotics and prebiotics have been suggested as a basis for novel therapeutic approaches for the beneficial modulation of commensal microbiota4. Proven probiotics have a relatively recent history of safe use and reported beneficial effects. Various papers have reported that the resulting beneficial effects include amelioration of obesity and of associated immuno-metabolic diseases4–8. Most of the studies (both in vivo and clinical trials) reported that consumption of probiotics resulted in reduced fat accumulation and relief in the level of biomarkers related to metabolic disorders such as blood glucose and triglycerides. Administration of L. sakei strain OK67 isolated from kimchi ameliorated HFD induced body and epididymal fat weight gains and reduced expression of various pro-inflammatory cytokines and serum endotoxin in a HFD induced obesity mouse model9. An earlier study has reported on the reduction of body weight, body mass index and visceral and subcutaneous fat by application of L. gasseri SBT2055 (in fermented milk) in a double-blind randomized, placebo-controlled trial over 12 weeks10. Despite the potent beneficial effects against metabolic syndrome, the mechanisms underlying these functional probiotic attributes are thus far only partially understood11. Novel insights in the complex relationship between gut microbiota and host health suggest that beneficial effects may be linked to the ‘normalisation’ of the host gut microbiota1,11. Thus, an imbalance (dysbiosis) of intestinal microbiota may lead to obesity and related metabolic disorders12,13. A ‘dysbiotic’ or imbalanced condition reflects disruption and negative shifts in abundance, diversity and relative distribution of the gut microbiota, and, as for metabolic syndrome, may also be associated with inflammatory bowel diseases (IBD), cancer, neurological disorders and type 1 and type 2 diabetes12,14–16.
The key role of the gut microbiota in host metabolism is reflected by a higher utilization of indigestible carbohydrates in obese people17 for which increased numbers of indigestible carbohydrate utilizing microbiota have been reported18. With caecal short chain fatty acids (SCFAs) providing 10% of the daily absorbable energy, changes in the gastrointestinal tract (GIT) microbiota will strongly impact host energy metabolism19, as is also reflected in the higher SCFA concentrations in faecal samples of overweight and obese individuals20. However, an increasing number of reports describe beneficial (including anti-obesogenic) functions of SCFAs; these are confirmed by a recent study in which germ-free mice received transplanted faecal material of discordant obese and lean twins21. Despite the well-recognized role of SCFAs as extra energy source for de novo lipogenesis, accumulating literature reports explaining a new role of SCFA’s such as G protein coupled receptor 41 and 43 (GPR41/GPR43) mediated satiety and insulin sensitivity regulation22–26. The importance of SCFA production by commensal microbiota is also emphasised by other host health benefits such as the production of vitamins27.
In this study we administered L. sakei strain CJLS03 (isolated from Korean kimchi) to a diet-induced obese C57BL/6 murine model and analysed both faecal microbiota modulation and the SCFA level of the serum and faeces by gas chromatography (GC/FID) while the expression level of various obesity related genes was determined in the liver and epididymal adipose tissue.
Results
Impact of L. sakei CJLS03 on weight gain and obesity associated biomarkers
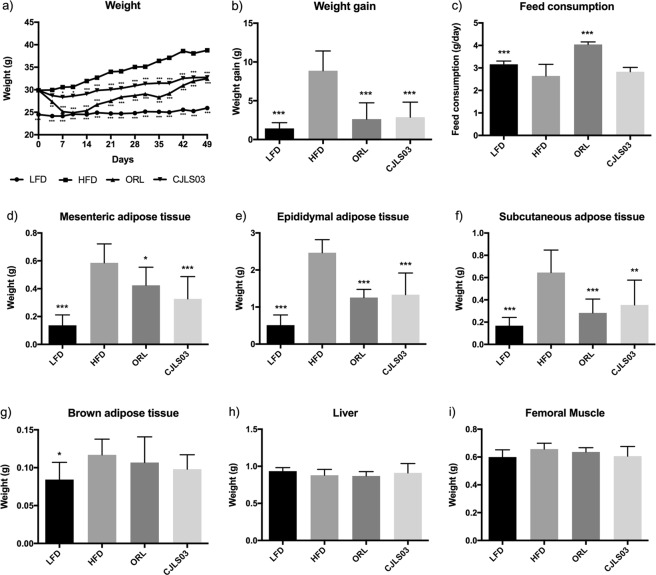
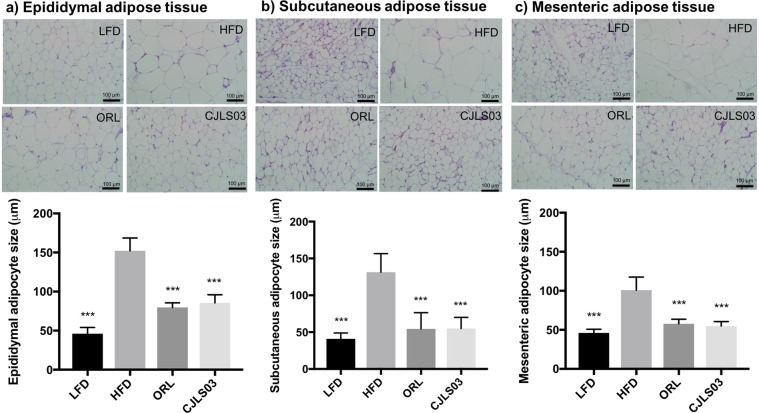
Our previous experiments involving three different L. sakei strains have shown the most promising anti-obesity effects for L. sakei CJLS0328, but prompted further research on related biomarkers. In this investigation we used orlistat as control, this drug being the commercially available anti-obesity drug in various countries such as the United States, the European Union, Australia and Japan; in some countries it is available over the counter when used at a dose of under 60 mg. It was reported to show a distinct weight loss effect with minimal systemic absorption29,30. Mice on a high-fat diet (HFD) showed significantly increased body weight gain and decreased feed consumption after 7 weeks compared to low-fat diet (LFD) fed controls (Fig. 1a–c). Weights of epididymal, mesenteric, subcutaneous and interscapular brown adipose tissues were markedly increased by HFD feeding, whereas weights of the liver and femoral muscle were not altered compared to the LFD control group (Fig. 1d–i). Administration of L. sakei CJLS03 resulted in a significantly lower weight gain of the experimental group, compared to the HFD-fed control, but with no change in feed consumption (Fig. 1a–c). The L. sakei CJLS03 treated group showed a significant weight reduction which was also reflected in the mesenteric, epididymal and subcutaneous adipose tissues, but without any significant reduction in the brown adipose tissue compared to the HFD control group (Fig. 1d–g). Liver and femoral muscle weight did not show any significant differences among the groups when compared to the HFD control (Fig. 1h,i). Body and adipose tissue weights were reduced, and feed consumption was significantly increased in the orlistat (ORL)-treated group compared to the HFD group. Haematoxylin and eosin microscopy showed that L. sakei CJLS03 supplementation significantly reduced the average size of epididymal, subcutaneous and mesenteric adipocytes in comparison to the HFD control group (Fig. 2). Subcutaneous and mesenteric adipose tissue sizes were reduced to the level of the ORL and LFD groups with L. sakei CJLS03 administration (Fig. 2b,c). Moreover, the L. sakei CJLS03 treatment was less effective than for the ORL and LFD groups in the reduction of epididymal adipose tissue (Fig. 2a).
Figure 1.
Anti-obesity effect of L. sakei CJLS03 determined in a diet-induced obesity (DIO) mouse model. The level of significance was measured using ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.05 (*), 0.01 (**) and <0.001 (***) compared to the HFD group. Feeding groups: LFD: low-fat diet plus PBS; HFD: high-fat diet plus PBS; ORL: high-fat diet plus orlistat; CJLS03: high-fat diet plus L. sakei CJLS03.
Figure 2.
Adipocyte size of each group. ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.05 (*), 0.01 (**) and <0.001 (***) compared to the HFD group. Feeding groups: LFD: low-fat diet plus PBS; HFD: high-fat diet and PBS; ORL: high-fat diet plus orlistat; CJLS03 high-fat diet plus L. sakei CJLS03.
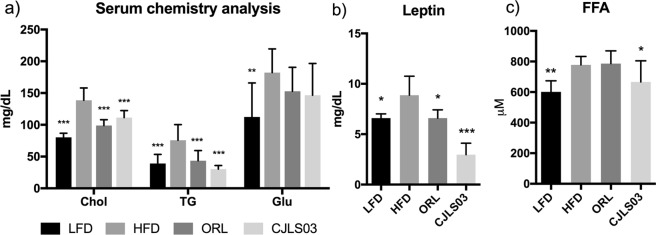
Obesity associated serum biomarkers such as cholesterol, triglycerides and glucose were analysed. Compared to the HFD group, serum cholesterol and triglycerides (TG) were significantly reduced in the L. sakei CJLS03 and ORL feeding groups (Fig. 3a), and (including glucose) also in the LFD control group. Serum leptin was significantly lower in the LFD, ORL and L. sakei CJLS03 groups while those receiving L. sakei CJLS03 showed the highest level of significance (Fig. 3b). Concentrations of free fatty acids (FFA) in the serum decreased significantly when L. sakei CJLS03 administration was compared to the HFD control group (Fig. 3c).
Figure 3.
Serum biomarkers determined after administration of L. sakei CJLS03 in the DIO mouse model using ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.05 (*), 0.01 (**) and <0.001 (***) compared to the HFD group. Feeding groups: LFD: low-fat diet plus PBS; HFD: high-fat diet and PBS; ORL: high-fat diet plus orlistat; CJLS03 high-fat diet plus L. sakei CJLS03. FFA: Free fatty acids.
Influence of L. sakei CJLS03 administration on biomarkers in the epididymal adipose tissue
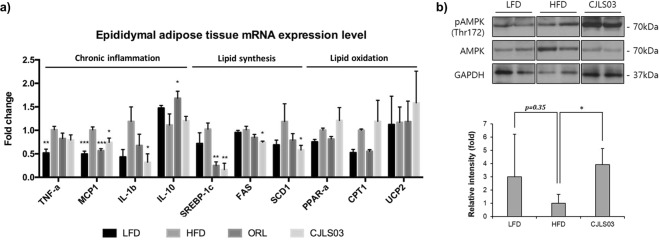
Various biomarkers in the epididymal adipose tissue (EAT) were significantly influenced by L. sakei CJLS03 supplementation when compared to the HFD control group. Low-grade chronic inflammation in adipose tissue is one of the major causes for the development of obesity-induced insulin resistance and metabolic impairments31. HFD feeding resulted in significantly increased gene expression of tumour necrosis factor alpha (TNF-a) and monocyte chemotactic protein 1 (MCP1) compared to the LFD control group (Fig. 4a). The L. sakei CJLS03 feeding group showed significant reduction in some pro-inflammatory cytokines in the EAT, such as MCP1 and interleukin 1 beta (IL-1b) in comparison with the HFD control group. Compared to the HFD group, MCP1 was also significantly reduced in the ORL group while interleukin 10 (IL-10), an anti-inflammatory cytokine, was significantly increased (Fig. 4a). Consistent with the decrease of adipose tissue weight (Fig. 1e), the mRNA levels of fatty acid synthesis gene expression were reduced in the HFD group receiving L. sakei CJLS03, including the sterol regulatory element-binding protein 1 (SREBP-1c), fatty acid synthase (FAS) and stearoyl-CoA desaturase-1 (SCD1). ORL group showed significantly reduced gene expression levels of SREBP-1c compared to HFD control group. Activation of AMP-activated kinase (AMPK), a central regulator of cellular energy homeostasis, triggers catalytic processes to generate ATP while inhibiting anabolic pathways32. In comparison to the LFD and HFD groups, the protein level of phosphorylated AMPK was significantly increased in the L. sakei CJLS03 feeding group (Fig. 4b) (see also Supplementary Figure S1).
Figure 4.
Host obesity and immune associated biomarkers in epididymal adipose tissue after L. sakei CJLS03 application in the DIO mouse model using (a) mRNA based qRT-PCR method and (b) Western blot. Full-length Western blots are shown Supplementary Figure S1. The mRNA expression levels and protein loadings were normalized to the expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.05 (*), 0.01 (**) and <0.001 (***) compared to the HFD group. Feeding groups: LFD: low-fat diet plus PBS; HFD: high-fat diet and PBS; ORL: high-fat diet plus orlistat; CJLS03 high-fat diet plus L. sakei CJLS03.
Modulation of intestinal microbiota and short chain fatty acid composition by L. sakei CJLS03 treatment
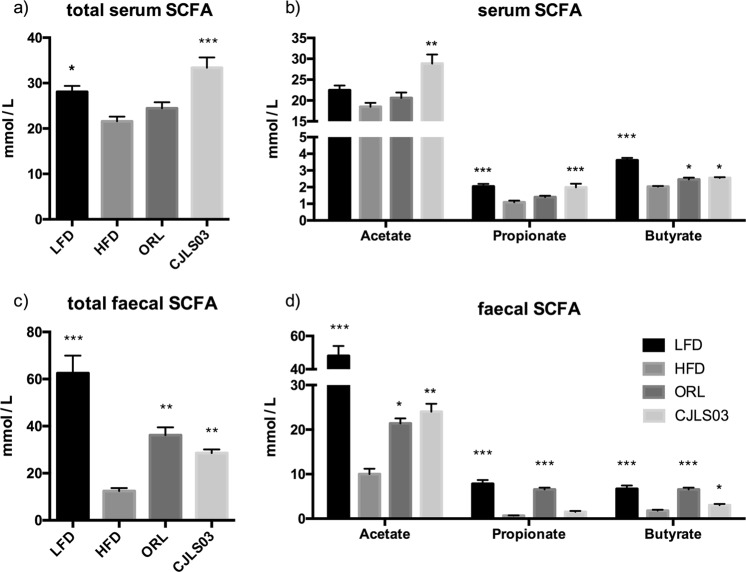
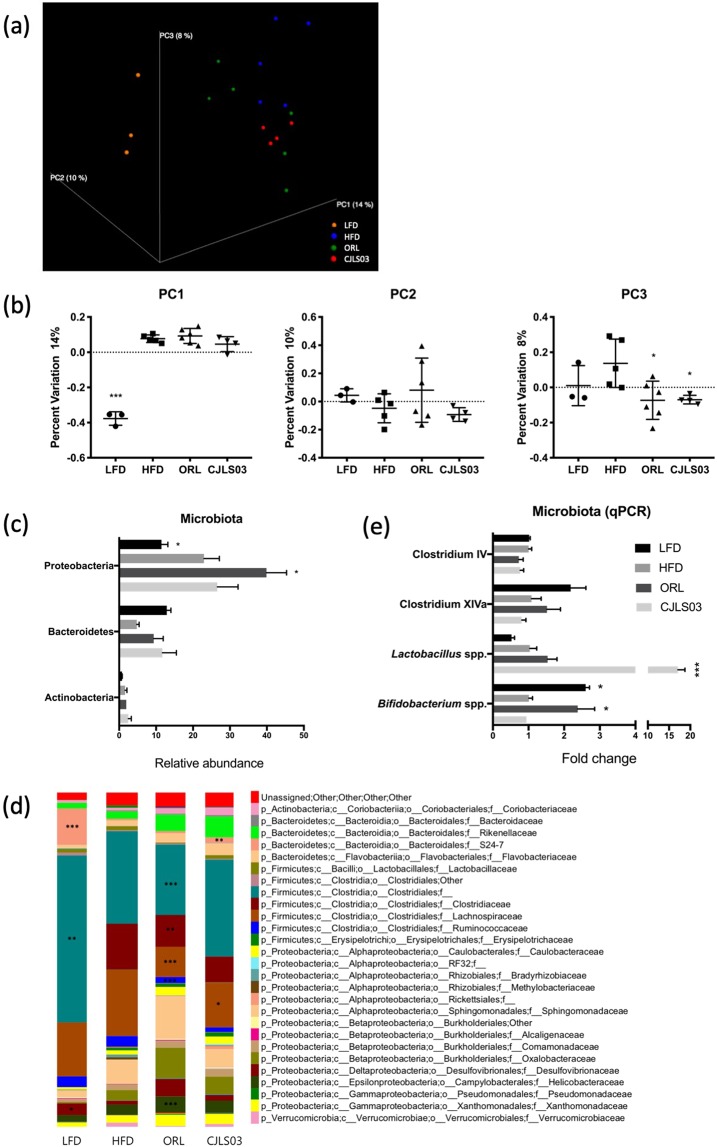
The L. sakei CJLS03 feeding group showed significantly higher SCFA levels in the serum and faeces compared to the HFD control group and (only for the serum) the ORL group (Fig. 5a,c). Yet, total SCFA levels in the faecal samples were lower than for the LFD control and ORL groups. Levels of acetate and propionate were significantly higher in the serum of the L. sakei CJLS03 group than in the HFD and ORL groups, but with significantly higher butyrate levels in the LFD group. Significantly elevated levels only of acetate were detected in the faecal samples both of the L. sakei CJLS03 and LFD control feeding groups, while propionate and butyrate levels were significantly higher only in the LFD and ORL groups, when compared to the HFD and CJLS03 treatment (Fig. 5b,d). A noticeable taxonomic shift in the faecal microbiota was detected when using Qiime analysis (Fig. 6a). The PCoA plots strongly differed for all the high-fat diet feeding groups (beta diversity), with a higher percent variation towards PC1, while both Orlistat and L. sakei CJLS03 feeding induced a shift towards PC3 (Fig. 6b). L. sakei CJLS03 treatment showed no dramatic modulation of the major phyla, although, compared to the LFD, Orlistat resulted in a significant increase in the Proteobacteria (Fig. 6c). At the family level interesting diferences were detected when the HFD and both treatments (ORL and CJLS03) were compared with the LFD, with a strong reduction in the Bacteroidales Family S24-7, and increases in the Rikenellaceae (especially for the two treatments), the Clostridiaceae, the Sphingomomonadaceae and Oxalobacteraceae (Fig. 6d). Compared to the HFD control group, significantly elevated numbers of Lactobacillus spp. were detected in the CJLS03 group, while Orlistat treatment resulted in levels of Bfidobacterium spp. similar to those for the LFD control (Fig. 6e). Administration of L. sakei CJLS03 with the HFD had no effect on the Gram-positive groups Clostridium IV and XIVa (Firmicutes) and Bifidobacterium spp. (Actinobacteria) when compared to the HFD control, except for a highly significant (more than 15-fold) increase of the Lactobacillus population in the L. sakei CJLS03 group (Fig. 6e). On the other hand, Orlistat treatment with the HFD resulted in levels of Bfidobacterium spp. similar to those of the LFD control group (Fig. 6e).
Figure 5.
Results from gas chromatographic analysis of short chain fatty acids from serum (a,b) and faecal (c,d) samples after application of L. sakei CJLS03 in a DIO mouse model. ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.05 (*), 0.01 (**) and <0.001 (***) compared to the HFD group. Feeding groups: LFD: low-fat diet plus PBS; HFD: high-fat diet and PBS; ORL: high-fat diet plus orlistat; CJLS03 high-fat diet plus L. sakei CJLS03.
Figure 6.
Microbiota analyses of faecal samples. Summary of taxonomic identification by a three dimensional PCoA plot of LFD, HFD, ORL and L. sakei CJLS03 treatment groups using unweighted unifrac (a), showing percentage variation (beta diversity) for each of PC1, PC2, PC3 (b), taxonomic summary of the phylum level (c), the family level (d), and some of the major groups/genera within the Firmicutes (the Clostridium clusters IV and XIVa and Lactobacillus) and the genus Bifdobacterium (Actinobacteria) as measured by qPCR (e). ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.05 (*), 0.01 (**) and <0.001 (***) compared to the HFD group. Feeding groups: LFD: low-fat diet plus PBS; HFD: high-fat diet and PBS; ORL: high-fat diet plus orlistat; CJLS03 high-fat diet plus L. sakei CJLS03.
Discussion
We have investigated the anti-obesity effect of L. sakei CJLS03 using a diet-induced obese murine model and found that L. sakei CJLS03 administration resulted in a strong increase in SCFAs concomitantly with microbiota modulation. Compared to the HFD control group plus placebo the impact of this putative probiotic strain was comparable to that of the ORL group as was reflected in a significant weight and (epididymal and subcutaneous) adipose tissue reduction. Feed consumption of the LFD and ORL groups was significantly higher than the CJLS03 and HFD groups between which no changes could be detected. An overall probiotic derived anti-obesity effect was influenced by reduced fatty acid synthesis gene expression and up-regulated beta-oxidation in the EAT. A probiotic associated anti-obesity effect and related phenomena in HFD-fed mice were described by Wang and colleagues7. They consider probiotic induced glucose-insulin homeostasis in the EAT and microbiome modulation a major cause of weight gain attenuation. Yet, specific metabolites associated with and derived from the resident microbiota were not fully defined by these workers. Administration of L. sakei strain OK67 isolated from kimchi ameliorated HFD induced body and epididymal fat weight gains and reduced expression of various pro-inflammatory cytokines and serum endotoxin in a HFD induced obesity mouse model9. An earlier study has reported on the reduction of body weight, body mass index and visceral and subcutaneous fat by application of L. gasseri SBT2055 (in fermented milk) in a double-blind randomized, placebo-controlled trial over 12 weeks10. Increased numbers of indigestible carbohydrate utilizing microbiota, including members of the family Ruminococcaceae have been reported for obese individuals18. Since the caecal concentration of short chain fatty acids (SCFAs) produced by commensal microbial fermentation constitutes 10% of the daily absorbable energy, changes in the gastrointestinal tract (GIT) microbiota should therefore strongly impact host energy metabolism19. Faecal samples of overweight and obese individuals contained higher SCFA concentrations as compared to lean counterparts20. Riduara and colleagues reported that germ-free mice receiving (transplanted) faecal material of discordant obese and lean twins21, showed elevated levels of SCFAs in the caecum of those mice receiving faecal transplants of the lean twin concomitantly with a lean phenotype in the recipient mice20. Data from our study are pointing to SCFAs as possible anti-obesity metabolites of the (modulated) gut microbiota. In this study we have detected significantly elevated SCFA levels in faecal and serum samples of the L. sakei CJLS03 feeding group concomitantly with modulation of the microbiota. The role of butyrate in glucose-insulin homeostasis was described by Gao and colleagues33, showing the potential of dietary butyrate supplementation for prevention and treatment of diet-induced insulin resistance in mice. Butyrate has induced an increase in beta-cell proliferation and function in juvenile rats and also significantly decreased hyperglycemia in induced beta-cell apoptosis34, while it reduced insulin resistance in type-2 diabetic rats35. The underlying mechanism was proposed to be linked to energy expenditure and induction of mitochondria function and resulted from increased insulin expression34. Thus, direct feeding of butyrate resulted in reduced fat levels without reduction in food intake. Increased levels of AMPK activity and up-regulation of beta-oxidation genes were found in the butyrate feeding group, thus supporting the results recorded in this study. Moreover, it has been shown that a reduced lactate supply (e.g., by reduced lactic acid producing population) may lead to a depletion of butyrate producing taxa, resulting in a negative impact on the host immune system36,37. Not only butyrate but all major SCFAs, including acetate and propionate, have been reported to exert a beneficial role in host health, thereby providing a mechanistic basis for explaining the potential impact of microbiota modulation on host patho-physiological status also involving metabolic syndrome27,38. Den Besten and coworkers39 have shown the major three SCFAs (acetate, propionate and butyrate) to reduce lipogenesis and increase beta-oxidation in adipose tissue. Consequently, gut microbiota modulation resulting in SCFA increase may serve as an indication of a ‘healthy’ microbiota23,40. Still, SCFA production in the host gut is a highly complex process of interactive metabolic pathways, details of which are still mostly unveiled. As a lactic acid bacterium, L. sakei CJLS03 is not a butyrate producer per se; however, by producing lactate it can influence the gastrointestinal environment, modulate the gut microbial population (as, e.g., in the case of lactic acid bacteria such as enterococci)41, and provide a carbon source that can be further utilised for production of SCFAs37. This was reflected in the elevated levels of total SFCAs in the serum of the CJLS03 group (Fig. 4a), and suggested by the 15-fold higher abundance of Lactobacillus spp. in the CJLS03 group (Fig. 6e). Even when some lactic acid bacterial strains have been claimed to produce SCFAs (apart from acetate and lactate), increased SCFA concentrations have only been produced in interaction with complex gut microbial populations, e.g., in a simulated chicken caecum42, by the use of selected synbiotics in a model system of the human colon43, and in an in vitro (SHIME) simulator of the human gut44,45. Based on formerly published data, caecal microbiota may be stronger modulated by probiotic administration, thus resulting in a higher correlation of microbiota with major SCFA levels, typical of the distal small intestine and proximal large intestine46.
Compared to the HFD control, L. sakei CJLS03 feeding has influenced the host faecal microbiota in a different way than either the LFD control or the ORL treatment. Interestingly, the bifidobacteria of the ORL group (as detected by qRT-PCR) were “restored” to approximately the level of the LFD control, suggesting a beneficial effect of Orlistat on the Bifidobacterium population (but not the Actinobacteria as a whole) when administered to the group receiving the highfat diet (Fig. 6c and e). An assessment of the possible effects of Orlistat and other treatments on the Bifidobacterium population should, however, take into consideration the limitations given by the primers chosen to amplify the V1–V3 variable 16S rRNA regions. Specific sample preparation procedures include mechanical disruption (by bead-beating) for DNA extraction, and appear more reliable when applied in conjunction with “optimised universal PCR primers”47. Moreover, when using the MiSeq platform for sequencing of the V1–V3 region of the 16S rRNA gene the removing of low-frequency sequences may reduce error rates48. A specific ‘indicator’ bacterial group resulting from the L. sakei CJLS03 administration could not be detected in this study, except for a noticeable increase of the phylum Bacteroidetes, a significant increase of Lactobacillus species and also an increase in the abundance of the family Rikenellaceae (Fig. 6).
In conclusion, the administration of L. sakei CJLS03 in a diet-induced obese mouse model showed significant reduction of adipose tissue and an attenuation of various obesity associated biomarkers including serum cholesterol and triglyceride without reduction in feed consumption. These parameters were well correlated with increasing amounts of SCFAs, both in faecal and serum samples concomitantly with modulation of the gut microbiota. It has indeed been shown that obesity-enhancing changes in the mouse gut microbiota may be partially reversed by cessation of the high fat diet49. It appears that elevated SCFA levels may be an indication of a ‘healthier’ microbiota when compared to the HFD control group; this was most probably the result of an increased abundance in the gut (faecal) microbiota upon administration of strain CJLS03. Serum chemical analysis indicated no sign of toxicity while reduced free fatty acid and leptin levels were detected in the L. sakei CJLS03 feeding group. Overall, particular probiotics such as L. sakei CJLS03 may confer host health benefits by gut microbiota modulation and stimulation of SCFA production. The positive role of SCFAs as key bacterial metabolites has been recently highlighted in an extensive summary by Koh and colleagues40.
Changes in traditional lifestyle to that typical of industrialized countries seems to be associated with shifts in the gut microbiota50. L. sakei CJLS03 appears to be a promising probiotic candidate for combating the growing incidence of metabolic syndrome and obesity probably resulting from a shift in the gut microbiota. On the other hand, in underdeveloped countries, sufficient energy harvesting via the diet constitutes a major issue that may justify a different approach in probiotic development with a focus on the enhancement of energy conversion.
Methods
All experiments were performed in accordance with relevant guidelines and regulations.
Animal experiments
The animal study was approved by the ethical committee of Handong Global University, Pohang, South Korea. Four-week-old, specific pathogen free (SPF) male C57BL/6 mice were purchased from Saeron Bio, South Korea. High-fat diet (Research Diets D12492) (HFD), low-fat diet (Research diets D12450) (LFD) and composition of diet is described in Supplementary Table 1. Autoclaved tap water was provided ad libitum, while the animals were housed at 23 °C and 55 ± 10% humidity, in a 12 h light/dark cycle. All mice were separated into four different groups each receiving different treatments (Table 1).
Table 1.
Study design and animal treatments based on a high-fat (HFD) and low-fat diet (LFD) in a murine model.
| Group (n = 7) | Feed type | Treatment |
|---|---|---|
| LFD | LFD | 200 μL PBS (non-obese control) |
| HFD | HFD | 200 μL PBS (obese control) |
| ORL | HFD | Orlistat 40 mpk |
| CJLS03 | HFD | 1 × 109 CFU/day of L. sakei CJLS03 suspended in 200 μL PBS |
The experiment comprised three weeks of an adaptation period followed by eight weeks of oral administration of substrates listed in Table 1. On the last day of the experiment, the mice were sacrificed by dislocation of the cervical vertebrata. The organs, e.g., small intestine, liver, epididymal adipose tissue and mesenteric adipose tissue were collected, weighed, and kept at −80 °C until analysis. One part of adipose tissues of each group were kept in 10%v/v formalin/PBS, and then embedded in paraffin for staining with haematoxylin and eosin for microscopy. Adipocyte size measuring has been accepted as an indication of obesity. Average of three different randomly chosen adipocytes from five mice per each group was analysed to measure the size of adipocyte. Blood, acquired by cardiac puncture, was centrifuged for 20 min at 2500 g to isolate serum.
Bacterial strains and culture conditions for the animal study
L. sakei CJLS03 was isolated from Korean fermented kimchi. This strain was grown in MRS (Difco Laboratories INC., USA) and prepared daily for feeding. It was grown for seven hours to reach late log phase and collected (16,000 g, 5 min, 4 °C) and washed twice with PBS. The strain was prepared in an approximate number of 1 × 109 CFU/mL according to a pre-optimised standard curve (data not shown) using optical density by SPECTROstar Nano (BMG Labtech, USA). The prepared suspension (1 × 109 CFU/mL) of the strain was suspended in 100 µL of PBS to be administered to each mouse by oral gavage daily.
Extraction of microbial genomic DNA from faecal samples
Genomic DNA (gDNA) was extracted from mouse faeces using the ReliaPrep gDNA Tissue Miniprep System (Promega, USA), after mechanical disruption of microbial cell walls. Briefly, 50 mg of faecal samples were suspended in 720 μL of lysis buffer solution (320 μL of PBS + 400 μL of CLD) in a screw cap micro-tube (Sarstedt, Germany) together with 0.3 g of 0.1 mm zirconium/silica beads (Biospec, USA), followed by disruption in a mini-beadbeater-16 (Biospec, USA) for 2 min and centrifuged at 14,000 × g for 3 min at room temperature. The supernatant was thoroughly mixed with 250 μL of Binding Buffer (BBA) and placed on a binding column. For further cleaning and eluting the manufacturer’s instructions were followed.
Gut microbiota analysis and bioinformatics
Gut microbial metagenome analysis was performed with the Roche 454 GS FLX plus system (AtoGen, Korea) using genomic DNA (gDNA). Briefly, primers were designed based on the V1–V3 variable region of 16S rDNA sequence (forward, 8f: 5′-AGAGTTTGATCMTGGCTCAG-3′; reverse 518r: 5′-ATTACCGCGGCTGCTGG-3′) and tagging of 10 bp unique barcode labels; the adaptor sequence was applied to monitor multiple samples in a single sequencing plate. Flow pattern B raw data from the 454 GS FLX plus system (Roche, Switzerland) was denoised and filtered using FlowClus51 and further analysed using MacQIIME52 1.8.0 pipeline. Chimeras were eliminated using Usearch 6.1 and sequences were clustered into operational taxonomic units (OTU) at 99% sequence similarity, and taxonomically assigned using RDP database. Composition and inter-community beta diversity of taxonomically assigned microbiota were interpreted using three-dimensional (3D) principal coordinates analysis (PCoA) using unweighted UniFrac distance, analyzed and visualized by MacQIIME 1.9.1. Each PC1, PC2 and PC3 is presented separately as column scatter graph with mean and standard deviation, thereby showing the differences in taxonomic abundance profiles between the respective treatment groups (see Fig. 6b). Further clusters and species levels were measured using qRT-PCR as described by Ji et al.13, using specific primers for Clostridium cluster IV (forward-GCACAAGCAGTGGAGT, reverse-CTTCCTCCGTTTTGTCAA), Clostridium cluster XIVa (forward-AAATGACGGTACCTGACTAA, reverse-CTTTGAGTTTCATTCTTGCGAA), Lactobacillus species (forward-AGCAGTAGGGAATCTTCCA, reverse-CACCGCTACACATGGAG), and Bifidobacterium species (forward-GCGTGCTTAACACATGCAAGTC, reverse-CACCCGTTTCCAGGAGCTATT).
Short chain fatty acid analysis
SCFA analysis was performed according to the method of Schwiertz et al.20. Briefly, 50 mg of deep-frozen faeces were mixed with 500 μL of extraction solution (comprising 0.1 mol oxalic acid /L and 40 mmol sodium azide /L), incubated in a horizontal shaker for an hour at room temperature, and centrifuged at 16000 × g for 10 min. 100ul of serum samples were prepared without the extraction step, but centrifuged briefly at 2000 g for 15 min without the extraction step. The supernatant was filtered through a 0.45 μm Minisart RC 4 syringe filter (Sartorius Stedim Biotech, Germany) and transferred to a Clear gas chromatography vial (Shimadzu, USA) and tightly sealed using Ribbed blue screw vial cap with bonded silicone (Shimadzu, USA) until analysis. A GC-2010 (Shimadzu, Japan) and HP-Innowax 30 m × 0.32 mm × 0.25 μm column (Agilent, USA) were used for detection; N2 gas served as carrier gas. 1 μL of each sample was injected by Shimadzu Auto-sampler AOC-20is (Shimadzu, Japan) at 260 °C and detected by flame ionised detector (FID). The column temperature was increased from 100 °C up to 180 °C at a rate of 25 °C/min. Volatile free acid standard mix (Supelco, USA) was used as analytical standard of C2 through C5.
Blood analysis
50 µL of blood serum sample of each mouse were diluted into 450 µl of PBS and glucose, triglycerides, and cholesterol levels measured using an automated biochemical analyser BS-200 (Mindray, China) in Pohang Technopark (South Korea). The concentration of leptin in the serum was determined by the enzyme-linked immunosorbent assay (ELISA) method using ELISA Kit, pink-ONE (Koma, Korea) as described in the manual. Free fatty acids (FFA) were analysed using EnzyChrom Free Fatty Acid Assay Kit (BioAssay Systems, USA) according to the manufacturer’s manual.
Measurement of host immune and obesity biomarkers
Extraction of mRNA from adipose tissue followed the specific protocol of RNeasy RNA tissue miniprep system (Promega, USA). Briefly, each organ sample was homogenised by a hand-held homogeniser (IKA, Germany) in lysis buffer and centrifuged. The supernatant was mixed with isopropanol and passed through a column provided with the kit. After several washing and DNase treatments, the purity and concentration of the eluted RNA were measured by SPECTROstar (BMG LABTECH, Germany). 2–3 µg of complement DNA (cDNA) were prepared using GoScript™ Reverse Transcription System (Promega, USA) through a Verity 96-well thermal cycler (ABI research, USA) after 10 min of incubation with oligodT primer at 70 °C. Various obesity related gene expression levels were monitored by the qRT-PCR method and calculated using the Delta Delta C(t) method normalised by GAPDH. Primer information is as follows: TNF-a (forward- ACTGCCAGAAGAGGCACTCC, reverse- CGATCACCCCGAAGTTCA), MCP-1 (forward - CGGAACCAAATGAGATCAGAA, reverse – TGTGGAAAAGGTAGTGGATGC), IL-1b (forward-GACCTTCCAGGATGAGGACA, reverse-AGCTCATATGGGTCCGACAG), IL-10 (forward-GGTTGCCAAGCCTTATCGGA, reverse-ACCTGCTCCACTGCCTTGCT), SREBP-1c (forward-AGCAGCCCCTAGAACAAACAC, reverse-CAGCAGTGAGTCTGCCTTGAT), FAS (forward- CTGGACTCGCTCATGGGTG, reverse-CATTTCCTGAAGTTTCCGCAG), SCD1 (forward-TCAACTTCACCACGTTCTTCA, reverse-CTCCCGTCTCCAGTTCTCTT), PPAR-a (forward-GTACGGTGTGTATGAAGCCATCTT, reverse-GCCGTACGCGATCAGCAT), CPT1 (forward-AAGCCTTTGGGTGGATATGTGA, reverse-ATGGAACTGGTGGCCAATGA), UCP2 (forward-CTACAAGACCATTGCACGAGAGG, reverse-AGCTGCTCATAGGTGACAAACAT). Proteins were extracted from EAT using PRO-PREP buffer (iNtRON Biotechnology Inc., Korea). Samples were analysed by SDS-PAGE-immunoblotting assay. Antibodies (Cell signalling technology, Beverly, MA) against phospho-AMPK (Thr172), total AMPK and GAPDH were used as primary antibodies, followed by anti-rabbit IgG-HRP conjugated secondary antibody.
Statistical analysis
All values were expressed in average and standard deviation (SD) and experimental determinants were confirmed with at least triplicate biological samples. ANOVA, Dunnett’s multiple comparisons test was used to distinguish the level of significance based on probability of 0.033 (*), 0.002 (**) and <0.001 (***).
Supplementary information
Acknowledgements
This work was supported by the CJ CheilJedang Corporation, Seoul, Republic of Korea. This is also to gratefully acknowledge support by the Bio & Medical Technology Development Program of the National Research Foundation (NRF), funded by the Ministry of Science, ICT and Future Planning (No. 2016M2A9A5923160 and 2018M3A9F3021964).
Author Contributions
Y.J., B.J.K., C.-K.H. and W.H.H. contributed to developing the study concept, design, and supervision. Y.J., S.P., Y.C., B.K., H.P., E.H., D.J. and H.Y.J. conducted the experiments. All authors contributed to the writing and reviewing of the manuscript and approved the final manuscript before submission. W.H.H. did the final check and submitted the manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
This project has been conducted as part of a scientific collaboration between Handong Global University and CJ CheilJedang Corporation. YC, DJ and BJK are employees of CJ CheilJedang Corporation, South Korea. Other authors declare that there are no potential conflicts of interests with respect to the authorship and/or publication of this article.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43092-y.
References
- 1.Alexandraki, V., Tsakalidou, E., Papadimitriou, K. & Holzapfel, W. Status and trends of the conservation and sustainable use of microorganisms in food processes. FAO Background Study Paper No. 65, Rome, 160 pp (2013).
- 2.Ng M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. The Lancet. 2014;384(9945):766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessesen DH, Van Gaal LF. Progress and challenges in anti-obesity pharmacotherapy. Lancet Diabetes Endo. 2017;6:237–248. doi: 10.1016/S2213-8587(17)30236-X. [DOI] [PubMed] [Google Scholar]
- 4.Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat. Rev. Drug. Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 5.Ji YS, et al. Modulation of the murine microbiome with a concomitant anti-obesity effect by Lactobacillus rhamnosus GG and Lactobacillus sakei NR28. Benef. Microbes. 2012;3:13–22. doi: 10.3920/BM2011.0046. [DOI] [PubMed] [Google Scholar]
- 6.Kadooka Y, et al. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur. J. Clin. Nutr. 2010;64:636–643. doi: 10.1038/ejcn.2010.19. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high-fat diet-fed mice. ISME J. 2015;9:1–15. doi: 10.1038/ismej.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodard GA, et al. Probiotics improve outcomes after Roux-en-Y gastric bypass surgery: a prospective randomized trial. J. Gastrointest. Surg. 2009;13:1198–1204. doi: 10.1007/s11605-009-0891-x. [DOI] [PubMed] [Google Scholar]
- 9.Lim, et al. Lactobacillus sakei OK67 ameliorates high-fat diet–induced blood glucose intolerance and obesity in mice by inhibiting gut microbiota lipopolysaccharide production and inducing colon tight junction protein expression. Nutr. Res. 2016;36:337–348. doi: 10.1016/j.nutres.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Kadooka Y, et al. Effect of Lactobacillus gasseri SBT2055 in fermented milk on abdominal adiposity in adults in a randomised controlled trial. Br. J. Nutr. 2013;110:1696–1703. doi: 10.1017/S0007114513001037. [DOI] [PubMed] [Google Scholar]
- 11.Patterson E, et al. Gut microbiota, obesity and diabetes. Postgrad. Med. J. 2016;92:286–300. doi: 10.1136/postgradmedj-2015-133285. [DOI] [PubMed] [Google Scholar]
- 12.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozupone CA, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepage P, et al. A metagenomic insight into our gut’s microbiome. Gut. 2013;62:146–158. doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 15.Clarke G, et al. Minireview: Gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paun A, Danska JS. Modulation of type 1 and type 2 diabetes risk by the intestinal microbiome. Pediatr. Diabetes. 2016;17:469–477. doi: 10.1111/pedi.12424. [DOI] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomicanalysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint HJ, et al. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3(4):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conterno L, Fava F, Viola R, Tuohy KM. Obesity and the gut microbiota: does upregulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011;6:241–260. doi: 10.1007/s12263-011-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwiertz A, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 21.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rios-Covian D, et al. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015;7:2839–2849. doi: 10.3390/nu7042839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topping DL. Targeted delivery of short-chain fatty acids to the human large bowel. Am. J. Clin. Nutr. 2016;104:1–2. doi: 10.3945/ajcn.116.137711. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Shimizu Y, Kimura I. Gut microbial metabolite short-chain fatty acids and obesity. Biosci. Microbiota, Food and Health. 2017;36:135–140. doi: 10.12938/bmfh.17-010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBlanc JG, et al. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell. Fact. 2017;16:79. doi: 10.1186/s12934-017-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji Y, et al. Dose dependent anti-obesity effect of three different Lactobacillus sakei strains using a diet induced obese murine model. PeerJ Preprints. 2018;6:e26959v1. doi: 10.7717/peerj.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vickers SP, Jackson HC, Cheetham SC. The utility of animal models to evaluate novel anti-obesity agents. Br. J. Pharmacol. 2011;164(4):1248–1262. doi: 10.1111/j.1476-5381.2011.01245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patil SH, et al. A comparative study of efficacy of argyreia speciose and orlistat for their anti-obesity action in high fat diet induced obese rat. Int. J. Basic & Clin. Pharmacol. 2017;6(3):613–617. doi: 10.18203/2319-2003.ijbcp20170823. [DOI] [Google Scholar]
- 31.Thomas D, Apovian CM. Macrophage functions in lean and obese adipose tissue. Metabolism. 2017;72:120–143. doi: 10.1016/j.metabol.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Day EA, Ford RJ, Steinberg GR. AMPK as a therapeutic target for treating metabolic diseases. Trends Endocrinol. Metab. 2017;28:545–560. doi: 10.1016/j.tem.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Gao Z, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan S, Jena GB. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: Study in juvenile diabetic rat. Chem.-Biol. Interact. 2014;213:1–12. doi: 10.1016/j.cbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Khan S, Jena GB. Sodium butyrate reduces insulin-resistance, fat accumulation anddyslipidemia in type-2 diabetic rat: A comparative study with metformin. Chem.-Biol. Interact. 2016;254:124–134. doi: 10.1016/j.cbi.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 36.De Goffau MC, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 37.Heeney et al. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opinion Biotechnol. 49, 140–147 (2017). [DOI] [PMC free article] [PubMed]
- 38.Samuel BS, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.den Besten G, et al. Short-Chain Fatty Acids Protect Against High-Fat Diet–Induced Obesity via a PPARγ-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes. 2015;64:2398–2408. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 40.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 41.Carasi et al. Enterococus durans EP1, a promising anti-inflammatory probiotic able to stimulate SIgA and to increase Faecalibacterium prausnitzii abundance. Front. Immunol. 8, 88, PMC5300979 (2017). [DOI] [PMC free article] [PubMed]
- 42.Meimandipour A, et al. Selected microbial groups and short-chain fatty acids profile in a simulated chicken cecum supplemented with two strains of Lactobacillus. Poultry Sci. 2010;89:470–476. doi: 10.3382/ps.2009-00495. [DOI] [PubMed] [Google Scholar]
- 43.Van Zanten GC, et al. The effect of selected synbiotics on microbial composition and short-chain fatty acid production in a model system of the human colon. PloS One. 2012;7:e47212. doi: 10.1371/journal.pone.0047212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan A, et al. Survival and metabolic activity of selected strains of Propionibacterium freudenreichii in the gastrointestinal tract of human microbiota-associated rats. Br. J. Nutr. 2007;97(4):714–724. doi: 10.1017/S0007114507433001. [DOI] [PubMed] [Google Scholar]
- 45.Sivieri K, et al. Lactobacillus acidophilus CRL 1014 improved “gut health” in the SHIME® reactor. BMC Gastroenterol. 2013;13:100. doi: 10.1186/1471-230X-13-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.den Besten G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker AW, et al. 16S rRNA gene based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3(1):26. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen HK, et al. Pipeline for amplifying and analyzing amplicons of the V1–V3 region of the 16S rRNA gene. BMC Res. Notes. 2016;9:380. doi: 10.1186/s13104-016-2172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shang, et al. Short term high fat diet induces obesity-enhancing changes in mouse gut microbiota that are partially reversed by cessation of the high fat diet. Lipids. 2017;52:499–511. doi: 10.1007/s11745-017-4253-2. [DOI] [PubMed] [Google Scholar]
- 50.de la Cuesta-Zuluaga, et al. Gut microbiota is associated with obesity and cardiometabolic disease in a population in the midst of Westernization. Sci. Rep. 2018;8:1356. doi: 10.1038/s41598-018-19850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaspar JM, Thomas WK. FlowClus: efficiently filtering and denoising pyrosequenced amplicons. BMC Bioinformatics. 2015;16:105. doi: 10.1186/s12859-015-0532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.