Abstract
The gene mutated in individuals with Huntington's disease (HD) encodes the 348-kDa huntingtin (HTT) protein. Pathogenic HD CAG-expansion mutations create a polyglutamine (polyQ) tract at the N terminus of HTT that expands above a critical threshold of ∼35 glutamine residues. The effect of these HD mutations on HTT is not well understood, in part because it is difficult to carry out biochemical, biophysical, and structural studies of this large protein. To facilitate such studies, here we have generated expression constructs for the scalable production of HTT in multiple eukaryotic expression systems. Our set of HTT expression clones comprised both N- and C-terminally FLAG-tagged HTT constructs with polyQ lengths representative of the general population, HD patients, and juvenile HD patients, as well as the more extreme polyQ expansions used in some HD tissue and animal models. Our expression system yielded milligram quantities of pure recombinant HTT protein, including many of the previously mapped post-translational modifications. We characterized both apo and HTT–HTT-associated protein 40 (HAP40) complex samples produced with this HD resource, demonstrating that this toolkit can be used to generate physiologically meaningful HTT complexes. We further demonstrate that these resources can produce sufficient material for protein-intensive experiments, such as small-angle X-ray scattering, providing biochemical insight into full-length HTT protein structure. The work outlined and the tools generated here lay a foundation for further biochemical and structural work on the HTT protein and for studying its functional interactions with other biomolecules.
Keywords: Huntington disease, neurodegeneration, protein expression, protein purification, biophysics, small-angle X-ray scattering (SAXS), molecular dynamics
Introduction
Huntington's disease (HD)6 is a devastating inherited neurodegenerative disorder that causes a range of progressive behavioral, cognitive, and physical symptoms. Incidence of HD varies in different parts of the world, but HD is thought to affect between 0.42 to 17.2 per 100,000 of the population (1). There are currently no disease-modifying therapies available for patients (2). HD is hallmarked by an expansion of a CAG-trinucleotide repeat tract in exon 1 of the HTT gene above a critical threshold of ∼35 CAG triplets (3, 4), translating to a polyglutamine (polyQ) expansion in the extreme N terminus of the huntingtin (HTT) protein. PolyQ-expanded HTT is thought to be responsible for the wide-ranging biochemical dysfunction observed in HD models and patients, including proteostasis network impairment (5), transcription dysregulation (6), mitochondrial toxicity (7, 8), cellular energy imbalance (9), synaptic dysfunction (10), and axonal transport impairment (8). Although it is thought that HTT is likely a scaffold protein (11, 12), the function of HTT, WT, or polyQ expanded, is still incompletely understood.
Biochemical investigation of the role of HTT, in either the WT or the disease state, is often dependent on obtaining large amounts of pure HTT protein of different polyQ lengths. The HTT protein is 3144 amino acids long (assuming a polyQ stretch of 23 residues, NCBI reference sequence NP_002102.4), a potentially daunting prospect for expression and purification given its size. A number of groups have published tools and methods by which full-length HTT might be expressed and purified from either insect (13–15) or mammalian cells (16). However, the tools produced and shared with the wider community are often limited by the number of different polyQ lengths, the position of an affinity tag, or their tractability for large-scale production for biochemical studies. To date, the published literature reporting experiments with purified HTT protein samples remains limited. Therefore, tools and detailed methods that will enable biochemical and biophysical studies of HTT by a larger number of researchers should accelerate our understanding of the function of this elusive protein.
Toward this end, we have cloned 28 HTT constructs that allow expression of HTT protein through transient transfection of mammalian cells or viral transduction of insect or mammalian cells. Constructs have either N- or C-terminal FLAG tags to assist in purification and yields of WT and polyQ-expanded HTT protein using these systems are up to 1 mg/liter of suspension culture of either insect or mammalian cells. The protein samples obtained from a simple two-step protocol are highly pure (>90% purity) and amenable to numerous downstream analyses and assays. Our constructs permit production of HTT in complex with the HTT-binding protein HAP40, as well as in its apo-form, and we have characterized these HTT protein samples. This includes mapping post-translational modifications (PTMs) of the proteins derived from both insect and mammalian cells, revealing similar modification motifs to those previously reported in the literature (17–19). Both apo and HTT–HAP40 complex samples are folded, as judged by reasonable thermal melting transitions of protein samples in solution. HTT and HTT–HAP40 samples were also assessed for monodispersity using size-exclusion chromatography in tandem with multiangle light scattering (SEC-MALS). We demonstrate how using these resources to generate large amounts of purified HTT protein sample enables protein-intensive experiments such as SAXS. We analyzed SAXS data for apo HTT samples and the HAP40 complex, providing initial insight to the complex structure in solution.
Results
Cloning of HTT expression constructs
Ligase-independent cloning (LIC) was used to clone the full-length HTT gene into the baculovirus transfer vector pBMDEL (Fig. 1A), for expression of proteins in insect cells as well as in mammalian cells. In addition to the sites for LIC, the vector contains a “stuffer” fragment that includes the SacB gene, allowing negative selection on 5% (w/v) sucrose, and a truncated VSVG fragment for pseudotyping of the baculovirus. As described previously for other HTT clones, a 15-bp repetitive element containing a mix of CAG and CAA codons was used to encode the polyQ expansion, in an effort to help maintain stability and integrity of the DNA sequence through various generations of vector propagation (20).
Figure 1.
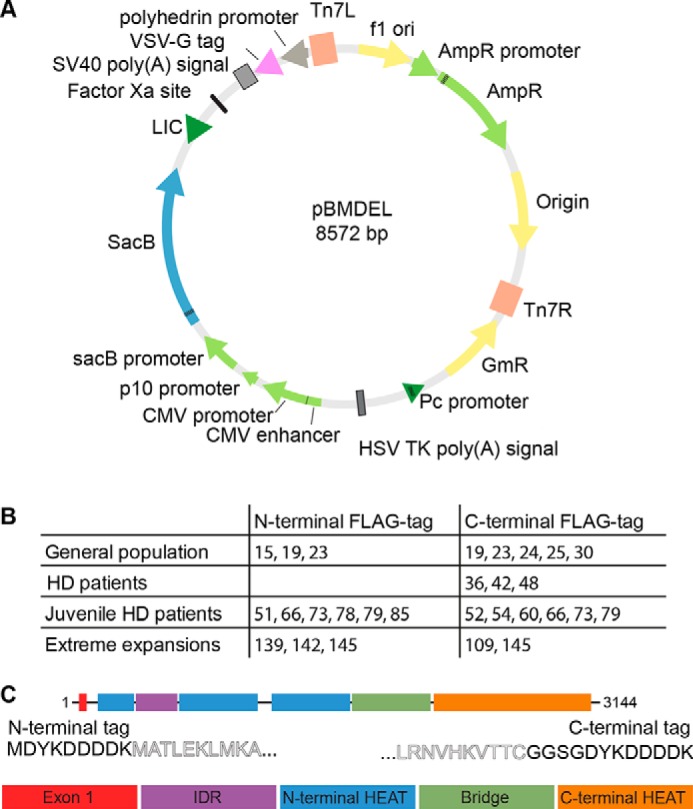
A, pBMDEL vector map. B, 28 HTT expression constructs with different polyQ lengths were generated with either N- or C-terminal FLAG tags. C, FLAG tags are appended to either end of the full-length HTT expression construct (comprising exon1, the N-terminal HEAT domains, the IDR, the Bridge domain, and the C-terminal HEAT domain) with minimal additional sequence.
As an ∼10-kb gene with multiple repetitive sequence elements, HTT is nontrivial to subclone between different vectors. We first generated N- and C-terminally FLAG-tagged pBMDEL–HTT constructs lacking part of the exon 1 sequence. Using different polyQ lengths encoding exon 1 PCR-generated cDNAs, our LIC-cloning protocol generated a variety of different polyQ-encoding HTT constructs due to the error-prone nature of the recombination step. By sequencing multiple colonies, we identified HTT clones with a variety of polyQ lengths with both N- and C-terminal FLAG tags (Fig. 1B). These entry vectors serve as valuable reagents to allow future generation of even more polyQ length HTT constructs. Additionally, by using a repetitive mix of codons for the polyQ expansion (CAG CAA CAG CAA CAA)n, we expect improved polyQ stability over generations of plasmid, bacmid, and baculovirus propagation compared with repetitive CAG codon tracts (20).
The resulting HTT open reading frames encoded within this series of constructs have either an N-terminal FLAG-octapeptide between the START methionine and the N-terminal methionine of the HTT amino acid sequence or have the FLAG-octapeptide linked to the extreme C terminus of HTT via a Gly–Gly–Ser–Gly linker (Fig. 1C). As subtle changes to the exon 1 amino acid sequence of the HTT protein have been shown to give rise to changes to biophysical properties of the protein (21, 22), the C-terminally FLAG-tagged constructs allow expression of a “clean” exon 1 sequence.
Expression of HTT variants in insect cells or mammalian cells yields functional proteins
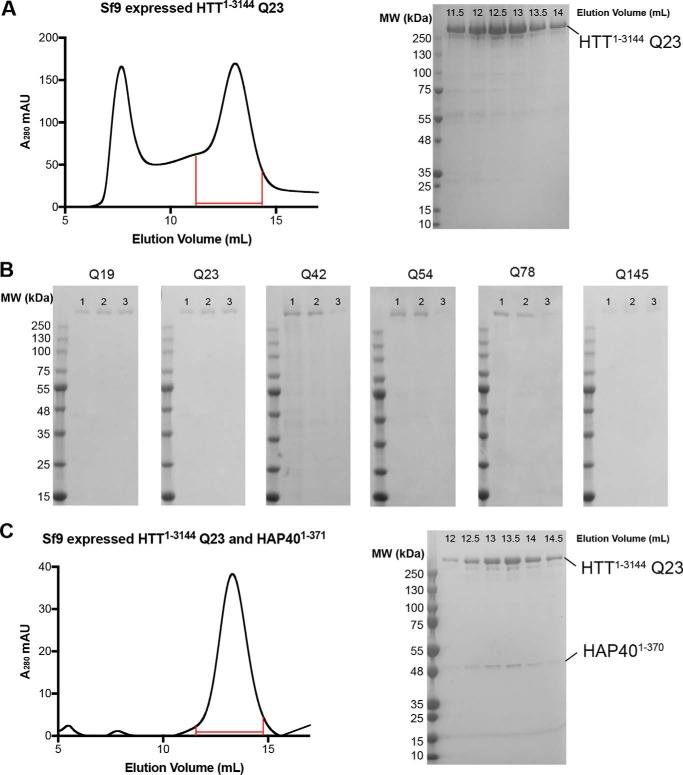
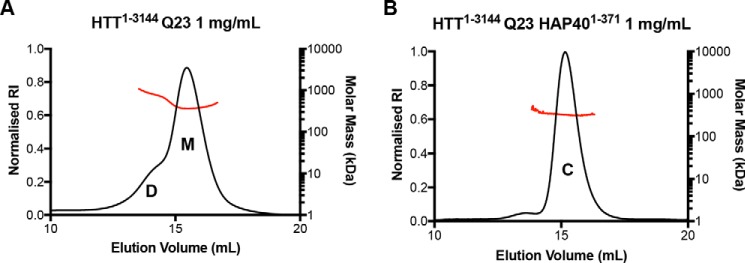
The HTT pBMDEL expression constructs we have developed allow the expression of HTT protein by three different methods: baculovirus-mediated expression in insect cells; transient transfection in mammalian cells; or transduction in mammalian cells (Fig. 2). All three methods allow cell growth in suspension culture permitting facile scaling of the culture volumes and thus scaling of the protein production as needed. Irrespective of the expression system, HTT protein could be purified in a two-step protocol, as described previously (14), from cell lysates in a Tris-salt buffer system comprising first a FLAG pulldown step and followed by size-exclusion chromatography using a Superose6 resin column (Fig. 2A). Similar to the HTT purification efforts of other research groups, multiple peaks are present in the size-exclusion chromatography profile, likely indicating the presence of a range of different oligomeric and/or aggregated states.
Figure 2.
A, Superose6 10/300 GL column size-exclusion chromatography profile of HTT1–3144 Q23 expressed in Sf9 cells and purified using the C-terminal FLAG tag. Coomassie-stained 4–20% SDS-PAGE analysis of size-exclusion chromatography fractions (0.5 ml volume) spanning 11.5–14 ml as marked on the elution profile. B, Coomassie-stained 4–20% SDS-PAGE analysis of C-terminally FLAG-tagged samples of HTT of different polyQ lengths from the following: lane 1, baculovirus-mediated expression in Sf9 insect cells; lane 2, transient transfection in mammalian EXPI293F cells; or lane 3, transduction in mammalian EXPI293F cells. C, Superose6 10/300 GL column size-exclusion chromatography profile of HTT1–3144 Q23 co-expressed with HAP401–371 in Sf9 cells and purified using the HTT construct C-terminal FLAG tag and HAP40 N-terminal His tag. Coomassie-stained 4–20% SDS-PAGE analysis of size-exclusion chromatography fractions (0.5 ml volume) spanning 12–14.5 ml is as marked on the elution profile.
Yields of the WT (Q23) purified HTT protein samples by the three expression methods can be as high as ∼1.6 mg/liter production in Sf9 insect cell culture to ∼1 mg/liter in transduced EXPI293F mammalian cells and ∼0.4 mg/liter in transiently transfected EXPI293F mammalian cells when measured after FLAG pulldown. Comparisons of preparations of HTT Q23 samples with either N- or C-terminal FLAG tag did not show significant difference in yield. In contrast, comparison of the yields of purified HTT with different polyQ expansions showed a trend of decreasing yield with increasing polyQ length. For example, in insect cells HTT Q42 yielded ∼0.5 mg/liter, whereas Q145 gives yields of <0.1 mg/liter. Longer polyQ lengths were also generally found to be more variable in yield between productions. For all constructs in each expression system, the two-step purification protocol yielded a protein sample that is >90% pure by Coomassie-stained SDS-PAGE analysis (Fig. 2B). Samples were analyzed by Western blotting using anti-HTT antibodies that revealed a discrete band of the expected molecular weight for each sample (Fig. S1).
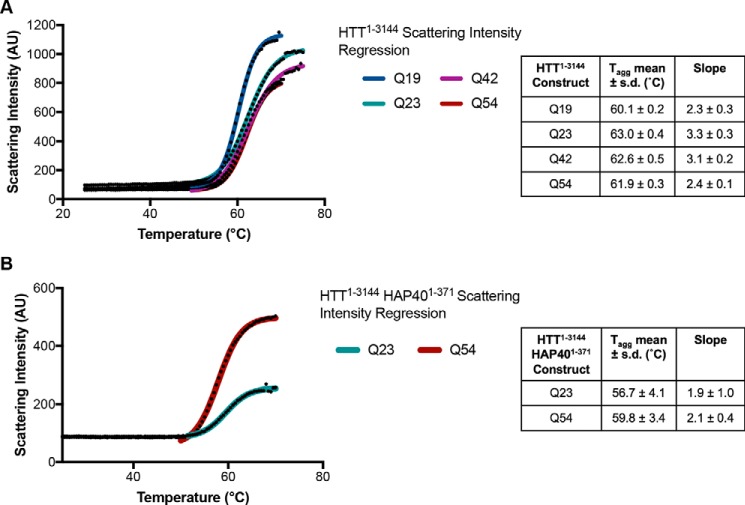
To assess whether these protein samples were folded, the C-terminal FLAG-tagged HTT samples of different polyQ lengths, purified from Sf9 cells, were analyzed by DSLS over a temperature gradient from 25 to 85 °C to assess thermal stability and propensity to aggregate under increasing temperatures. HTT samples were stable up to ∼55 °C with sigmoidal thermal melting curves reflective of a folded globular protein (Fig. 3A). Interestingly, irrespective of polyQ length, the temperature of aggregation (Tagg) values (35) for all HTT samples were similar at ∼60–63 °C, indicating that the polyQ repeats did not significantly affect protein thermal stability. This suggests that polyQ may not be interacting with the folded globular part of the protein.
Figure 3.
A, DSLS profiles of HTT1–3144 Q19, Q23, Q42, and Q54 (left) and the calculated thermal aggregation temperatures that show high stability indicating folded protein samples. B, similar results are seen for HTT1–3144–HAP401–371 for both Q23 and Q54 samples.
Purification of the HTT–HAP40 complex, previously reported from an adherent mammalian cell-expression system (23), could be achieved through a 3:1 viral titer ratio of HTT1–3144 Q23 in a C-terminally FLAG-tagged pBMDEL expression construct and HAP401–371 in an N-terminally His6-tagged pFBOH–MHL insect cell expression vector (Fig. 2C). The purification protocol was modified so that an additional Ni-affinity chromatography step was included following the FLAG pulldown. The final size-exclusion chromatography step reveals that the HTT–HAP40 complex is a monodisperse sample, indicating increased protein stability and conformational homogeneity compared with apo HTT. Formation of this complex by HTT produced in insect cells indicates that the protein expressed is correctly folded and functional with respect to formation of an important protein interaction.
HTT–HAP40 complexes with WT (Q23) and polyQ expanded (Q54) were also analyzed by DSLS (Fig. 3B) yielding similar thermal aggregation profiles (∼57–60 °C) again, suggesting a lack of interaction between the polyQ repeat and the globular portion of the protein complex.
HTT expressed in Sf9 insect cells retains reported phosphorylation PTMs
PTM of HTT is well-described for protein derived from various mammalian cell systems and in some detail for HTT extracted from post-mortem brain tissue (17–19). However, it is not known whether these PTMs are conserved in HTT expressed in Sf9 insect cells. Purified HTT Q23 and Q54 from Sf9 and EXPI293F were subjected to bottom-up proteomics (24, 25). PTMs were mapped for HTT expressed in Sf9 and EXPI293F cells and compared with published PTMs of mammalian-derived HTT (Tables 1–4 detail results for HTT Q23 samples from Sf9 and EXPI293F production, and complete data can be found on PRIDE (accession PXD010865) and in Zenodo).
Table 1.
Phosphorylation motifs identified for Sf9-expressed Q23 HTT1–3144 in references to literature
Modifications that have been discovered in proteomics studies, but not published, were retrieved from PhosphoSitePlus (17). Some modifications have not been described before. To illustrate the likelihood of these being physiologically relevant modifications, NetPhos 3.1 predictions for the putative enzyme and likelihood score are included (31). Only modifications with at least three peptide spectrum matches for at least one peptide containing the modification are listed in the table. All data are available via PRIDE (accession PXD010865) with summaries in Zenodo.
| Site | No. of PSMsa | Reports or predictions | HD patient/control tissue samples | In vitro or animal models |
|---|---|---|---|---|
| Ser-421 | 9 (9) | Reported in Refs. 16, 18, 26, 54–61, 61–79 | Yes | Yes |
| Ser-421/434 | 10 (10) | Both reported | Yes/no | Yes/yes |
| Ser-432 | 3 (7) | Reported in Refs. 16, 18, 75, 80, 81 | No | Yes |
| Ser-434 | 3 (5) | Reported in Refs. 16, 18, 54, 56, 57, 63–65, 68, 71–76, 79, 81–89 | No | Yes |
| Ser-622 | 3 (3) | Reported in Ref. 90 | No | Yes |
| Ser-780 | 3 (4) | Predicted NetPhos 3.1 (CDK1, 0.514) | No | No |
| Thr-816 | 3 (3) | Predicted NetPhos 3.1 (unspecified, 0.957) | No | No |
| Ser-1181 | 3 (5) | Reported in Refs. 16, 18, 26, 57, 72, 88, 90–92 | Yes | Yes |
| Ser-1201 | 27 (37) | Reported in Refs. 16, 18, 26, 54, 55, 57, 68, 69, 74, 75, 79, 85, 86, 88–97 | No | Yes |
| Ser-1469 | 6 (6) | Predicted NetPhos 3.1 (CDK1, 0.513) | No | No |
| Thr-1557 | 4 (6) | Predicted NetPhos 3.1 (unspecified, 0.777) | No | No |
| Ser-1864 | 4 (4) | Reported in Refs. 16, 18, 55, 98 | Yes | Yes |
| Ser-1866 | 5 (8) | Reported in Refs. 16, 18, 54, 55 | No | Yes |
| Ser-1867 | 3 (5) | Predicted NetPhos 3.1 (unspecified, 0.975) | No | No |
| Ser-1876 | 4 (4) | Reported in Refs. 16, 18, 54, 55, 57, 68, 70, 74, 75, 79, 83–90, 94–103 | Yes | Yes |
| Tyr-2102 | 3 (3) | Predicted NetPhos 3.1 (unspecified, 0.921) | No | No |
| Ser-2550 | 3 (3) | Reported in Refs. 16 | No | Yes |
| Ser-2775 | 3 (4) | Predicted NetPhos 3.1 (PKC, 0.491) | No | No |
| Ser-2776 | 7 (8) | Predicted NetPhos 3.1 (unspecified, 0.989) | No | No |
| Thr-2800 | 10 (13) | Predicted NetPhos 3.1 (CaM-II, 0.456) | No | No |
| Ser-2913 | 7 (10) | Predicted NetPhos 3.1 (unspecified, 0.990) | No | No |
| Thr-3133 | 10 (12) | Predicted NetPhos 3.1 (PKC, 0.813) | No | No |
a PSMs are reported as the number of peptide spectrum matches for the most abundant peptide containing the modification described with the total number of peptide spectra for all peptides containing this modification motif in parentheses.
Table 2.
Arginine and lysine monomethylation modifications identified for Sf9-expressed Q23 HTT1–3144 in references to literature
Methylation of huntingtin has not been previously described or reported. Only modifications with at least three peptide spectrum matches for at least one peptide containing the modification are listed in the table. All data are available via PRIDE (accession PXD010865) with summaries in Zenodo.
| Site | No. of PSMsa | Site | No. of PSMsa | Site | No. of PSMsa |
|---|---|---|---|---|---|
| Arg-221 | 4 (4) | Arg-1461 | 3 (5) | Lys-2802 | 5 (5) |
| Lys-700 | 3 (5) | Arg-1555 | 4 (4) | Arg-2908 | 4 (6) |
| Arg-908 | 7 (9) | Arg-1947 | 3 (5) | Arg-3112 | 3 (4) |
| Lys-1264 | 6 (6) | Lys-2163 | 3 (5) | Arg-3130 | 10 (10) |
| Lys-1448 | 3 (4) | Arg-2774 | 7 (15) |
a PSMs are reported as the number of peptide spectrum matches for the most abundant peptide containing the modification described with the total number of peptide spectra for all peptides containing this modification motif in parentheses.
Table 3.
Phosphorylation motifs identified for EXPI293F-expressed Q23 HTT1–3144 in references to literature
Modifications that have been discovered in proteomics studies, but not published, were retrieved from PhosphoSitePlus (17). Some modifications have not been described before. To illustrate the likelihood of these being physiologically relevant modifications, NetPhos 3.1 predictions for the putative enzyme and likelihood score are included (31). All data are available via PRIDE (accession PXD010865) with summaries in Zenodo.
| Site | No. of PSMsa | Previously reported | HD patient/control tissue samples | In vitro or animal models |
|---|---|---|---|---|
| Tyr-173 | 1 (1) | Predicted NetPhos 3.1 (unspecified, 0.587) | No | No |
| Ser-419 | 1 (1) | Reported in Refs. 16, 18, 60, 70, 75 | No | Yes |
| Ser-421 | 3 (3) | Reported in Refs. 16, 18, 26, 54–61, 61–79 | Yes | Yes |
| Ser-421/431 | 1 (1) | Both reported | Yes/no | Yes/yes |
| Ser-421/434 | 5 (5) | Both reported | Yes/no | Yes/yes |
| Ser-431 | 1 (1) | Reported in Refs. 16, 18, 55, 56, 63, 75, 80, 104 | No | Yes |
| Ser-432 | 2 (4) | Reported in Refs. 16, 18, 75, 80, 81 | No | Yes |
| Ser-434 | 5 (7) | Reported in Refs. 16, 18, 54, 56, 57, 63–65, 68, 71–76, 79, 81–89 | No | Yes |
| Ser-622 | 1 (1) | Reported in Ref. 90 | No | Yes |
| Ser-623 | 3 (4) | Reported in Ref. 105 | No | Yes |
| Ser-1063 | 1 (1) | Predicted NetPhos 3.1 (unspecified, 0.989) | No | No |
| Ser-1106 | 8 (8) | Reported in Refs. 16, 90 and Zhou (2011) PhosphoSitePlus dataset (https://www.phosphosite.org/curatedInfoAction.action?record=22693604) | No | Yes |
| Ser-1181 | 6 (8) | Reported in Refs. 16, 18, 26, 54–61, 61–79 | Yes | Yes |
| Ser-1197 | 2 (2) | Reported in Refs. 16, 89 | No | Yes |
| Ser-1201 | 12 (38) | Reported in Refs. 16, 18, 26, 54, 55, 57, 68, 69, 74, 75, 79, 85, 86, 88–97 | No | Yes |
| Thr-1262 | 1 (1) | Predicted NetPhos 3.1 (CDK1, 0.495) | No | No |
| Thr-1411 | 1 (1) | Reported in Refs. 16, 90 and Guo (2007) PhosphoSitePlus dataset (https://www.phosphosite.org/curatedInfoAction.action?record=25582702) | No | Yes |
| Thr-1859 | 1 (1) | Reported in Refs. 106 | No | Yes |
| Ser-1864 | 4 (5) | Reported in Refs. 16, 18, 55, 98 | Yes | Yes |
| Ser-1866 | 3 (5) | Reported in Refs. 16, 18, 54, 55 | No | Yes |
| Thr-1868 | 1 (1) | Reported in Refs 16 | No | Yes |
| Ser-2550 | 1 (1) | Reported in Ref. 16 | No | Yes |
| Ser-2690 | 2 (2) | Predicted NetPhos 3.1 (unspecified, 0.994) | No | No |
| Thr-2748 | 1 (1) | Predicted NetPhos 3.1 (PKC, 0.529) | No | No |
| Thr-3098/Tyr-3101 | 1 (1) | Predicted NetPhos 3.1 (GSK3, 0.446)/predicted NetPhos 3.1 (EGFR, 0.449) | No/no | No/no |
a PSMs are reported as the number of peptide spectrum matches for the most abundant peptide containing the modification described with the total number of peptide spectra for all peptides containing this modification motif in parentheses.
Table 4.
Other post-translational modifications identified for EXPI293F-expressed Q23 HTT1–3144 in references to literature
Methylation of huntingtin has not been previously described or reported. Only modifications with at least three peptide spectrum matches for at least one peptide containing the modification are listed in the table. All data are available via PRIDE (accession PXD010865) with summaries in Zenodo.
| Site | No. of PSMsa | Modification |
|---|---|---|
| Lys-826 | 1 (1) | Acetylation (Lys) |
| Arg-2053 | 1 (1) | Dimethylation (KR) |
| Arg-2781 | 4 (4) | Methylation (KR) |
| His-2786 | 6 (14) | Methylation (His) |
| Lys-2932 | 1 (1) | Acetylation (Lys) |
a PSMs are reported as the number of peptide spectrum matches for the most abundant peptide containing the modification described with the total number of peptide spectra for all peptides containing this modification motif in parentheses.
To map the PTMs on HTT Q23 produced in Sf9 cells as completely as possible, this sample was digested with five proteases having complementary and nonspecific cleavage specificity: trypsin; lysargiNase (27); pepsin; WT α-lytic protease (WaLP); and M190A α-lytic protease (MaLP) (28). Trypsin and lysargiNase cleave at the C and N termini, respectively, of lysine and arginine residues thus yielding complementary (or mirror-image) peptides. WaLP and MaLP preferentially cleave at aliphatic amino acids, whereas pepsin at pH >2 cleaves at Phe, Tyr, Trp, and Leu in position P1 or P1′ (29). MaLP, WaLP, and pepsin were selected to probe the Lys- and Arg-poor regions of the protein. Digestion of the other HTT samples was performed with trypsin alone.
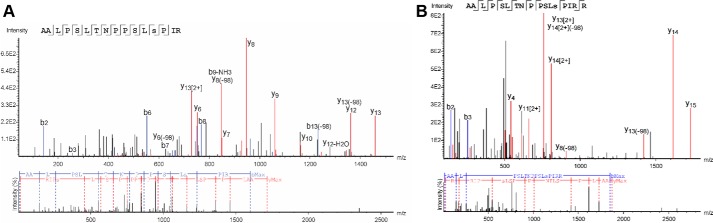
When LC/MS data from the five proteolysis reactions of Q23 HTT from Sf9 are searched together, at least 90% sequence coverage was observed, whereas trypsin digestion of the other HTT samples yielded at least 50% sequence coverage (Figs. S2 and S3). As we were able to digest such large overall amounts (hundreds of micrograms) of HTT protein in multiple rounds of MS experiments, due to the high levels of production from our expression systems, this also increased the overall peptide coverage and allowed us to map PTMs with lower incidence in the samples. Because of the large size of the HTT protein, routine MS protein identification experiments of intact sample are not feasible. Instead, we used peptide mapping analysis to confirm that the purified sample was indeed the HTT protein. As expected for the high purity of the samples indicated by SDS-PAGE analysis, HTT sequence peptides were the highest abundance proteins detected, although some contaminating proteins were detected. Details of these contaminants for the HTT Q23 samples from EXPI293F cells (Table S1) show that most of these proteins are unlikely to be true HTT interactors as they have high CRAPome scores (30) or are of very low abundance. Modifications were detected for all samples, with well-described phosphorylation motifs being present in HTT samples from both Sf9 and EXPI293F production methods (Fig. 4).
Figure 4.
Spectra of S1181 phosphorylation motifs detected from samples of HTT1–3144 Q23 derived from Sf9 (A) and EXPI293F (B) production. This modification has been detected in human brain samples, indicating that many of the detected phosphorylation motifs detected in the purified HTT samples are physiologically relevant.
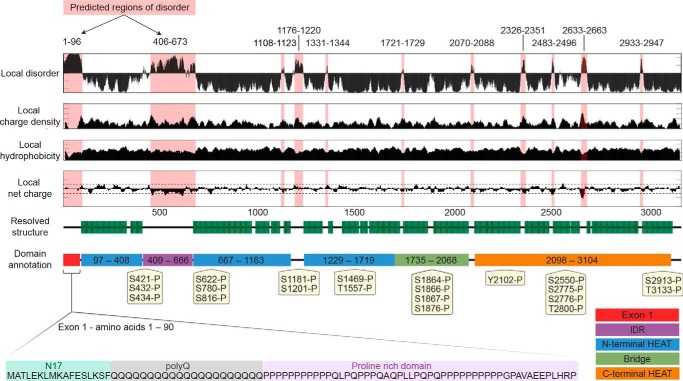
By employing multiple enzymes, sequences in regions containing sparse Lys and Arg residues, for example a 20-amino acid-long peptide within exon 1 (Fig. S4), were detected. Peptide–spectrum matches (PSMs) were used to prioritize the confident phosphorylation sites. HTT expressed in Sf9 cells retains many of the highly-observed phosphorylation sites described in the literature for mammalian cell lines and post-mortem tissue (Tables 1 and 2 and Figs. S5 and S6). A total of 22 phosphorylation events with at least three PSMs were mapped in Sf9-expressed Q23 HTT, 11 of which have been reported in at least one instance in the literature. Mapping the remaining modifications to the cryo-EM HTT–HAP40 model shows that most would-be surface-exposed residues in the context of apo HTT (Fig. S7) and their respective physiological likelihood and probably kinase, as determined by NetPhos analysis (31), are detailed in Table 1. Monomethylation of some lysine and arginine residues was also detected (Table 2). Sequence analysis of HTT using CIDER (32) and IUPred (33) in conjunction with analysis of the recently published near-atomic resolution cryo-EM structure of HTT in complex with HAP40 (Fig. 5) (23) reveals that most of the phosphorylation sites are within disordered regions of the protein structure as described previously (18). Although some of these previously unreported modifications may be artifacts of the Sf9 expression system, they appear to have a minimal effect on global huntingtin function, as seen by the ability of this sample to form a complex with HAP40.
Figure 5.
Sequence analysis of HTT complemented by structural data reveals a large modular protein connected by flexible unstructured linkers with numerous short unstructured loops protruding from the ordered domains. Local disorder, charge density, hydrophobicity, and net charge are based on sequence analysis, whereas the resolved structure and domain annotations are from cryo-EM (23). Of particular note, both exon1 and the IDR are not resolved in the current cryo-EM model. Phosphorylation motifs of insect cell–derived HTT Q23 mapped by peptide MS and found in at least three peptide spectrum matches are annotated.
A total of 25 phosphorylation motifs with at least three PSMs were mapped for HTT Q23 expressed in EXPI293F cells, 19 of which have been described previously in the literature (Tables 3 and 4 and Fig. S5). Interestingly, we also observed acetylation (Lys-826 and Lys-2932), monomethylation (Arg-2781 and His-2786), and dimethylation (Arg-2053) of our samples, none of which have been previously described in the literature. Acetylation of HTT at other sites has been previously described, and methylation motifs are observed in MS data of post-mortem human brain tissue samples of HTT (18), indicating that HTT protein methylation is a physiological modification.
Characterization of HTT and HTT–HAP40 protein sample monodispersity
Size-exclusion chromatography of HTT protein derived from insect Sf9 or mammalian EXPI293F cells using a Superose6 10/300 GL column gives a characteristic elution profile (Fig. 2A), with a void-aggregate peak followed by peaks previously attributed as being dimer and monomer species of HTT based on column standards (14–16). The recent cryo-EM structure of HTT in complex with HAP40 reveals a bi-lobed structure of HTT in which the N-HEAT and C-HEAT domains wrap around HAP40 yielding a more compact and globular structure (23). Furthermore, HAP40 was described as being critical for producing a conformationally homogeneous HTT sample amenable to cryo-EM structure determination. Our purified samples of apo HTT therefore lack a binding partner such as HAP40, which may account for the broad and overlapping elution peaks observed in gel-filtration analyses as well as the tendency for self-association when HTT samples are analyzed at higher protein concentrations.
To further understand this tendency for self-association and sample heterogeneity, a C-terminal FLAG-tagged HTT Q23 sample taken from the “monomer” peak of the Superose6 elution profile was analyzed by SEC-MALS using the same specification Superose6 column, which allows calculation of the in-solution protein mass. This analysis revealed a peak with a shoulder with approximate mass calculations indicating that this sample is a mixture of both HTT monomer and dimer (Fig. 6A). In contrast, the HTT–HAP40 complex sample run on the same SEC-MALS set up at the same total protein concentration is monodisperse, and the mass calculated across the peak is stable indicating the sample is homogeneous and not self-associating (Fig. 6B). Long-term storage and freeze-thaw of HTT–HAP40 samples had minimal effect on the peak profile, whereas apo HTT samples had a tendency to redistribute from monomer peak to a peak profile similar to that observed during purification. Taken together, these results suggest HAP40 binding reduces homotypic HTT interaction, possibly by competing for an interaction interface or through a linkage effect (34).
Figure 6.
A, analytical Superose6 10/300 GL gel filtration in tandem with SEC-MALS profiles of HTT1–3144 Q23 showing distinct dimer (D) and monomer (M) species as determined by the approximate in-solution mass values. B, SEC-MALS data for HTT1–3144–HAP401–371 reveals that HTT–HAP40 complex (C) is more monodisperse and homogeneous due to the stable mass calculated across the elution peak.
SAXS analysis of the HTT–HAP40 complex in solution
The cryo-EM structure of HTT–HAP40 (23) has laid a tremendous foundation for our understanding of the structure of the huntingtin protein with respect to its global architecture, HEAT repeat organization, and complex formation with the HAP40-binding partner protein. However, technical limitations of cryo-EM combined with sample limitations from the conformational flexibility and heterogeneity of HTT limit our current understanding of certain structural details. The Guo et al. (23) cryo-EM structure was resolved at a resolution of 4–5 Å and is missing several regions of the HTT protein. Roughly 25% of the huntingtin protein, including many functionally important elements such as exon1 and the highly modified 400–650 amino acid intrinsically disordered region (IDR), are not resolved in the cryo-EM maps, presumably due to the fact these regions are intrinsically disordered (Fig. 5).
To further understand the structure of the entire HTT protein, we conducted SAXS analysis of both the Q23 isoforms of HTT (isolated monomer peak) and the HTT–HAP40 complex in solution. Similar to other biophysical and structural analysis techniques, SAXS requires large (milligram) quantities of protein. Our toolkit of HTT reagents permits production of sufficient sample for structural analyses, expediting further investigation of the HTT protein by such methodologies.
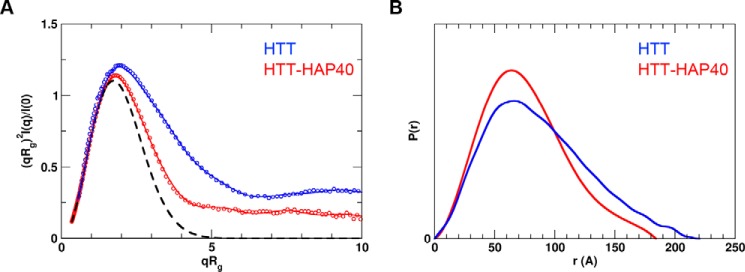
For both HTT and HTT–HAP40 sample data, Rg-based Kratky plots of the experimental curve do not fit the expected data for a generic globular protein of similar mass (Fig. 7). The experimentally calculated radius of gyration (Rg) for both samples was also much larger than that expected on the basis of the resolved residues of the cryo-EM structure (Table 5). This indicates that there is a degree of flexibility or disorder in both samples. This is not unexpected due to the large regions of the HTT protein sequence with predicted disorder, which are not present in the Guo et al. (23) cryo-EM HTT–HAP40 model. Interestingly, the normalized pair distance distribution function P(r) of HTT–HAP40 shows a narrower range of atomic radii compared with the apo HTT sample, consistent with the HAP40 complex being more compact. However, taking into account the high propensity of HTT self-association observed in our analytical size-exclusion chromatography profiles, apo HTT SAXS analysis may also be confounded by self-association of molecules in the concentrated solutions used for SAXS data collection. This assertion is corroborated by the higher molecular weight estimated from SAXS data (Table 5) for apo HTT compared with HTT–HAP40. Therefore, further SAXS analysis of the apoprotein was not pursued.
Figure 7.
Experimental SAXS data. A, Rg-based Kratky plots of experimental SAXS data for HTT–HAP40 complex (red) and apo HTT (blue). The experimental data are displayed as empty circles. The solid lines show regularized experimental curves determined using GNOM. The theoretical curve expected for a spherical protein of similar size is shown by a dashed line. B, normalized pair distance distribution function P(r) calculated from experimental SAXS data with GNOM.
Table 5.
SAXS parameters for data validation and interpretation
| Protein sample | Rga | Rgb | Dmaxc | Massd | Masse |
|---|---|---|---|---|---|
| Å | Å (real) | Å | kDa | kDa | |
| HTT/HAP40 | 63.3 + 2.1 | 57.7 + 0.4 | 179 | 386 (390) | 391 |
| HTT | 69.9 + 2.2 | 67.7 + 0.8 | 220 | 422 (350) | 428 |
a Radius of gyration (Rg) was calculated the using Guinier fit in the q range 0.015 < q < 0.025 Å−1 and 0.012< q < 0.019 Å−1 for HTT/HAP40 and HTT, respectively.
b Radius of gyration was calculated using GNOM.
c Maximum distance between atoms was calculated using GNOM.
d Molecular mass was estimated using SAXSMoW (47). The mass expected from the sequence is shown in the parentheses.
e Molecular mass was estimated from SAXS data based on volume of correlation (Vc) (48).
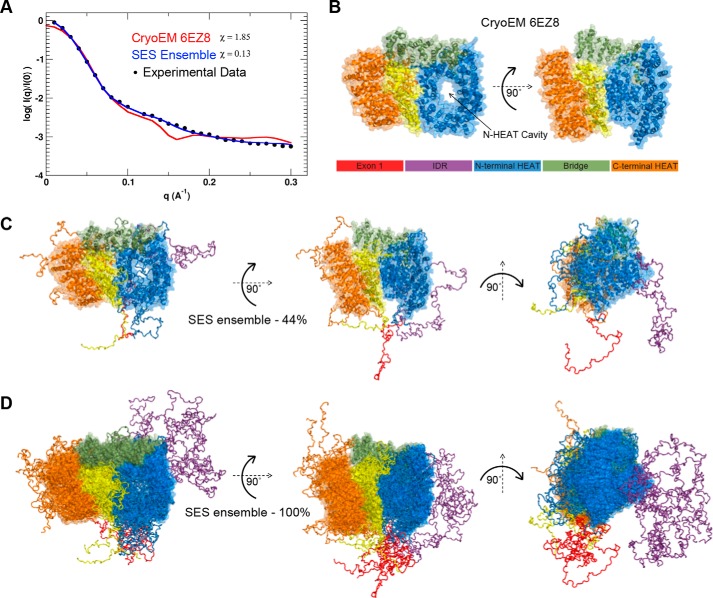
To better understand the HTT–HAP40 structure, including the disordered/missing regions of the cryo-EM model, we performed coarse-grained molecular dynamics (MD) simulations, and calculated an ensemble of conformations that best fits the solution SAXS data for HTT–HAP40. This modeling approach assumed that the residues with known coordinates in the cryo-EM model form a quasi-rigid complex, whereas the residues with missing coordinates are flexible. Predicted SAXS scattering curves were averaged over an ensemble of MD-simulated structures using the optimal weights for each ensemble member obtained with the sparse ensemble selection (SES) method (35). The resulting average “theoretical” scattering curve for the SES weighted ensemble of structures gave a much better fit to the experimental scattering data than that of the cryo-EM structure (Fig. 8A).
Figure 8.
Fitting SAXS data to structural models of HTT–HAP40 complex. A, experimental SAXS profiles (black circles) plotted with the theoretical profile for the EM structure of the HTT–HAP40 complex (red line) and the predicted profile averaged over the optimal SES ensemble (blue line). Surface representation of the EM structure (B) as well as the most populated (∼44%) solution model (C) and complete ensemble of the HTT–HAP40 complex (D) are shown with surface representation of the globular regions and backbone trace of the flexible regions (color key as in B).
The most populated model (44%) in this ensemble (Fig. 8C) shows extensive protruding regions of disorder extending out from the rigid complex core indicating that the overall envelope of the protein is likely to be larger than that calculated from the cryo-EM structure (Fig. 8B). For many of the HEAT repeats, the disordered regions of the connecting sequence are not seen in the cryo-EM structure, but the molecular modeling we have completed allows us to visualize how these might be arranged with respect to the rigid HEAT repeat core structure. The complete SES ensemble (Fig. 8D) further indicates how, in particular, the IDR and exon1 region of huntingtin are probably very structurally heterogeneous and dynamic in their conformation and are able to extend away from the more rigid core of the structure in many different arrangements due to their inherent flexibility. The extension of both exon1 and the IDR away from the core HTT–HAP40 complex is consistent with the accessibility of these domains to enzymes capable of post-translational modification.
A key feature of the cryo-EM structure is the large cavity that extends through the N-terminal HEAT repeat domain. This cavity is approximately the same diameter as a dsDNA helix, and it is tempting to envision a potential nucleic acid–binding role of this region of the HTT protein, especially given the functional links between HTT and DNA damage repair (36, 37). At the current resolution of the structural information, it is also difficult to analyze potential surface charge or “greasy” surface residue hotspots that could indicate interaction sites. However, our SES ensemble model indicates how this cavity could be capped by certain conformational states of disordered loop regions, not resolved in the cryo-EM structure. These loops could act as gatekeepers to any binding partner, nucleic acid, or protein by accessing this cavity. Similarly, an apparent cavity on the side of the N-terminal HEAT repeat domain in the cryo-EM model may also be capped by a flexible protein chain. Expansion of the polyQ region seems unlikely to affect the global structure of huntingtin given that exon1 is distal from the rigid HEAT repeat domains. Therefore, the mechanism by which the polyQ expansion affects huntingtin protein structure and function remains a question for future structural studies.
Discussion
We have generated a resource of 28 different HTT expression constructs that allow the generation of purified HTT samples of different polyQ lengths and affinity tags from three different expression systems. All constructs are available through Addgene, including the entry vector that will allow other researchers to make additional CAG expansion forms of the HTT gene should they require them. Our expression constructs permit facile scaling of culture volumes to enable the purification of milligram quantities of WT HTT protein from both insect and mammalian cells as well as substantial production of various polyQ-expanded HTT species.
These HTT proteins from different expression systems are modified with PTMs previously described in the literature as well as additional modifications of unknown physiological relevance but that do not seem to alter HTT function in its ability to form a complex with HAP40. We describe HTT protein methylation for the first time, a modification conferred by various protein methyltransferases, many with links to neurodegeneration (38). Further characterization of these modifications and their function could open up novel avenues for understanding HTT protein structure–function. The constructs cloned may also be used in future studies for co-expression with modifying enzymes to make highly site-specific PTMs on the sample as well as to test how certain enzymes might function on HTT.
Our work characterizing the biophysical properties of HTT confirms that the protein is not monodisperse or homogeneous when purified in its apo-form. Co-expression and purification with HAP40 allow purification of a more monodisperse protein sample, rendering it amenable to more detailed structural analysis, as performed previously by cryo-EM (23). It is unclear whether HAP40 is a constitutive binder of HTT in physiological settings, although its effects on the biophysical characteristics of the HTT protein are clearly significant. Interestingly, very few HTT protein–protein interaction studies have identified HAP40 as an interacting protein of HTT, and it is only identified in the published literature in a small number of articles (23, 39) compared with the multiple extensive HTT interaction network publications (41, 42).
Because of the high yield of both HTT and HTT–HAP40 proteins from our toolkit of resources, we were able to conduct preliminary biophysical analyses of these samples. Our SAXS analysis in tandem with molecular dynamics simulations permitted the generation of an SES ensemble, representing a possible solution structure of the HTT–HAP40 complex. This model gives insight into how HTT is post-translationally modified at flexible and accessible regions of sequence and suggests potential regulatory mechanisms such as steric capping of binding regions of the protein. Our SAXS model serves as an important resource for understanding the complete HTT–HAP40 complex, and it should help prevent misinterpretation of certain features of the cryo-EM structure that lacks ∼26% of the protein molecule. In particular, the exon1 region of HTT is distal to the complex core, and polyQ expansion does not affect HTT thermal stability as shown by DSLS. Therefore, it is likely that the effect of polyQ expansion on HTT structure–function is more nuanced. Both exon1 and the IDR have many of the hallmarks of interacting domains observed for intrinsically disordered protein regions (43), as they are heavily modified by PTMs, are conformationally flexible, and contain regions of charged amino acids (i.e. nearly 20% of residues in the IDR are negatively charged). The purported role of huntingtin as a scaffold protein could be explained through dynamic protein–protein interactions mediated through these regions of the structure (40, 41).
The precise molecular function of unexpanded HTT remains elusive, so it is unclear how the polyQ expansion may alter the HTT protein sufficiently to give rise to the wide-ranging biochemical dysfunction observed in HD models and patients. These reagents and the accompanying methods and validation for the production of HTT protein will provide an enabling framework for future research requiring purified HTT and its complexes for a wide range of polyQ lengths.
Experimental procedures
Cloning of HTT and HAP40 expression constructs
HTT expression constructs were assembled in two steps into the mammalian/insect cell vector pBMDEL, an unencumbered vector created for open distribution of these reagents. First, entry vectors for N-terminal FLAG-tagged and C-terminal FLAG-tagged HTT (amino acids 1–3144) were constructed without the polyQ regions, amino acids 7–85. PCR products encoding WT HTT were amplified from cDNA (Kazusa clone FHC15881) using primers N_int_FWD (ttaagaaggagatatactATGGACTACAAAGACGATGACGACAAGATGGCGACCCTGGAAAAGCgctGACCTTAGTCGCTAAcctgcaGGAGCCGCTGCACCGACCAAAG)/N_int_REV (gattggaagtagaggttttaGCAGGTGGTGACCTTGTGGAC) for the N-terminal FLAG-tagged HTT and C_int_FWD (ttaagaaggagatatactATGGCGACCCTGGAAAAGCgctGACCTTAGTCGCTAAcctgcaGGAGCCGCTGCACCGACCAAAG)/C_int_REV (gattggaagtagaggttttaCTTGTCGTCATCGTCTTTGTAGTCaccgcttccaccGCAGGTGGTGACCTTGTGGAC) for the C-terminal FLAG-tagged HTT. All PCR products were inserted using the In-Fusion cloning kit (Clontech) into the pBMDEL that had been linearized with BfuAI. Second, synthetic polyQ regions were inserted into the intermediate plasmids using the In-Fusion cloning kit. The polyQ regions were PCR-amplified using the primers polyQP_Fwd (ATGGCGACCCTGGAAAAGCTG)/polyQP_Rev (TGGTCGGTGCAGCGGCTCCTC) from template plasmids CH00007 (Q23), CH00008 (Q73), and CH00065 (Q145) (all from Coriell Institute Biorepository). PCR products were inserted into the intermediate vectors that had been linearized with AfeI and SbfI. Upon screening the assembled HTT expression constructs, we found that our cloning method generated a range of polyQ lengths. We selected constructs with polyQ lengths from Q15 to Q145. The HTT-coding sequences of intermediate and final expression constructs were confirmed by DNA sequencing. The sequences were confirmed by Addgene where these reagents have been deposited. HAP40 cDNA corresponding to amino acids 1–371 was subcloned into pFBOH–MHL expression vector using ligase-independent cloning.
HTT and HTT–HAP40 protein expression
The recombinant transfer vectors HTT pBMDEL and HAP40 pFBOH–MHL were transformed into DH10Bac Escherichia coli cells (Invitrogen, Bac-to-Bac System) to generate recombinant Bacmid DNA. Sf9 cells (Invitrogen) were transfected with Bacmid DNA using jetPRIME® transfection reagent (PolyPlus transfection, catalog no. 89129-924), and recombinant baculovirus particles were recovered. The recombinant virus titer was sequentially amplified from P1 to P3 virus stocks for protein production in the Sf9 insect cells and EXPI293F mammalian cells.
Baculovirus-mediated expression of HTT in insect cells
Sf9 cells at a density of ∼4.5 million cells/ml were infected with 8 ml of P3 recombinant baculovirus and grown at 130 rpm and 27 °C. HyQ SFX insect serum medium containing 10 μg/ml gentamicin was used as the culture medium. Infected cells were harvested when viability dropped to 80–85%, normally after ∼72 h post-infection. For HTT–HAP40 complex production, the same general protocol was followed but with a 3:1 ratio of HTT/HAP40 P3 recombinant baculovirus infection step.
Transduction of HTT in mammalian cells
EXPI293F cells (ThermoFisher Scientific, catalog no. A14527) were maintained in EXPI293 expression medium (ThermoFisher Scientific, catalog no. A1435102) in a humidified 8% CO2 incubator at 37 °C and 125 rpm. Cells were transduced at a density of 2–3 million cells/ml of culture. The transduction used recombinant baculoviruses of HTT constructs generated by transfecting Sf9 cells using Transfer vector pBMDEL and JetPRIME® transfection reagent (catalog no. 89129-924). The volume of the virus added into the cells was at ratio 6% of the total volume of the production. Infected cells were harvested after 7–10 days post-transduction depending on cell viability.
Transient transfection of HTT in mammalian cells
EXPI293F cells (ThermoFisher Scientific, catalog no. A14527) were maintained in EXPI293 expression medium (ThermoFisher Scientific, catalog no. A1435102) in a humidified 8% CO2 incubator at 37 °C and 125 rpm. Cells were transfected at a density of 2–3 million cells/ml of culture. FectoPRO® transfection reagent (VWR, catalog no. 116-001) and plasmid pBMDEL–HTT DNA were separately diluted in serum-free OptiMEM complexation medium (ThermoFisher Scientific, catalog no. 31985062) at 10% of the total production volume in a ratio of 1 μg of DNA to 1.2 μl of FectoPro to 0.5 μl of Booster/ml of cell culture. After 5 min of incubation at room temperature, the transfection mixtures were combined and incubated for an additional 20 min. The FectoPRO® transfection reagent/DNA/OptiMEM mixture was then added to the cells with an addition of FectoPRO® Booster in a ratio of 1 μg of DNA to 1.2 μl of FectoPro to 0.5 μl of Booster/ml of cell culture. The transfected cultures of EXPI293 cells were harvested after 72–96 h post-transfection–dependent cell density and viability.
HTT and HTT–HAP40 protein purification
The same protocol was used to purify HTT from insect and mammalian cell culture, adapted from Huang et al. (16) and Guo et al. (23). Cell cultures were harvested by centrifugation at 4000 rpm, 20 min, 4 °C (Beckman JLA 8.1000), washed in pre-chilled PBS, resuspended in 20 cell paste volumes of preparation buffer (50 mm Tris, pH 8, 500 mm NaCl), and stored at −80 °C prior to purification. Cell suspensions were thawed and diluted to at least 50 times the cell paste volumes with prechilled buffer and supplemented with 1 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine-HCl, and 20 units/ml benzonase. Note: two freeze–thaw cycles of cell suspensions were found to be sufficient for cell lysis. The lysate was clarified by centrifugation at 14,000 rpm, 1 h, 4 °C (Beckman JLA 16.2500), and then bound to 0.1 cell paste volumes of anti-FLAG resin (Sigma M2) at 4 °C with rocking for 2 h. Anti-FLAG resin was washed twice with the 100 cell paste volumes of buffer. HTT protein was eluted with 1 cell paste volume of buffer supplemented with 250 μg/ml 3×FLAG peptide (Chempep) run twice over the anti-FLAG resin. Residual HTT protein was washed from the beads with 0.5 cell paste volume of buffer. The sample was spin-concentrated with molecular weight cutoff of 100,000. Depending on the protein yield, the sample was run as one or more sample runs on Superose 6 10/300 GL column in size-exclusion chromatography buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 5% (v/v) glycerol, 1 mm TCEP) at 0.4 ml/min ensuring no more than 2 mg of protein was applied per run to minimize protein aggregation. For HTT–HAP40, the same protocol was followed except for using preparation buffer with just 300 mm NaCl, and an additional step where FLAG elution was rocked with 2 ml of equilibrated nickel-nitrilotriacetic acid resin for 30 min before washing in preparation buffer and then elution with buffer supplemented with 300 mm imidazole prior to the size-exclusion chromatography step.
SDS-PAGE and Western blot analysis
SDS-PAGE and Western blot analysis were performed according to standard protocols. In brief, purified proteins were denatured in sample buffer (50 mm Tris-HCl, 0.1 m DTT, 2% SDS, 0.1% bromphenol blue, and 10% glycerol, pH 6.8) at 98 °C for 5 min, followed by SDS-PAGE. After electrophoresis, the gel was either directly stained with Coomassie Blue or subjected to Western blot analysis. For Western blot analysis, proteins were transferred onto polyvinylidene difluoride membranes. The primary antibody used was anti-HTT (Abcam, ab109115, 1:5000), and the secondary antibody used was IRDye® 800CW anti-rabbit IgG (LI-COR, 926-32211, 1:5000). Membranes were visualized on an Odyssey® CLx Imaging System (LI-COR).
HTT MS
All data were acquired on an Agilent 1260 capillary HPLC system coupled to an Agilent Q-TOF 6545 mass spectrometer via the Dual Agilent Jetstream ion source.
Bottom-up proteomics for sequence coverage and PTM analysis
Proteins were processed according to established protocols (42). Briefly, proteins were reduced with DTT (10 mm final concentration) for 30 min at room temperature, alkylated with iodoacetamide (55 mm final concentration) for 30 min at room temperature, and incubated with trypsin (6 μl, 0.2 mg/ml) overnight at 37 °C. The digests acidified to pH 2 in hydrochloric acid and were desalted on-column (by diverting the first 2 min to waste), before analysis. Peptides were separated on a C18 Advance BioPeptide column (2.1 × 150 mm 2.7-μm particles) at a flow rate of 400 μl/min and an operating pressure of 4700 p.s.i. Peptides were eluted using a gradient from 100% solvent A (98:2 H2O/ACN with 1% formic acid) to 50% B (96:4 ACN/H2O with 1% formic acid) for 80 min. Mass spectra were acquired from m/z 300 to 1700 at a rate of eight spectra/s. The tandem mass spectra were acquired in automated MS/MS mode from m/z 100 to 1500 with an acquisition rate of three spectra/s. The top 10 precursors were selected and sorted by abundance only. Collision-induced dissociation was done using all ions at [4·(m/z)/100] − 1 and − 5.
Data analysis
Raw data were processed using PEAKS Studio 8.5 (build 20171002) and the reference complete human proteome FASTA file (Uniprot). Cysteine carbamidomethylation was selected as a fixed modification, and methionine oxidation and Asn/Gln deamidation as variable modifications. A minimum peptide length of five, a maximum of three missed cleavage sites, and a maximum of three labeled amino acids per peptide were employed.
HTT sequence disorder prediction
Disorder prediction was performed using IUPred (33, 43). A threshold of 0.5 was used to define disordered or ordered regions, with predicted disordered regions shaded in light red (Fig. 5). Further sequence analysis of HTT was performed using the sequence analysis tool local CIDER (32). Hydrophobicity was calculated using a normalized Kyte-Doolittle scale (43, 44). Resolved structure and domain annotations were based on the solved cryo-EM structure of HTT in complex with HAP40 (23).
DSLS
The thermal stability of HTT1–3144 Q19, Q23, Q42, and Q54 as well as HTT1–3144–HAP401–371 Q23 and Q54 samples were analyzed by DSLS using Stargazer. With four repeats per sample, HTT and HTT–HAP40 proteins at 1 mg/ml in 20 mm HEPES, pH 7.5, 300 mm NaCl, 5% (v/v) glycerol, 1 mm TCEP were heated from 20 to 85 °C. Protein aggregation was monitored using a CCD camera. The temperature of aggregation (Tagg) was analyzed and fitted as described previously (35).
SEC-MALS
The absolute molar masses and mass distributions of purified protein samples of HTT1–3144 Q23 and HTT1–3144 Q23–HAP401–371 complex with C-terminal FLAG tag at 1 mg/ml were determined using SEC-MALS. Samples were injected through a Superose 6 10/300 GL column (GE Healthcare) equilibrated in 20 mm HEPES, pH 7.5, 300 mm NaCl, 5% (v/v) glycerol, 1 mm TCEP followed in-line by a Dawn Heleos-II light scattering detector (Wyatt Technologies) and a 2414 refractive index detector (Waters). Molecular mass calculations were performed using ASTRA 6.1.1.17 (Wyatt Technologies) assuming a dn/dc value of 0.185 ml/g.
SAXS data collection and analysis
SAXS measurements were carried out at the beamline 12-ID-B of the Advanced Photon Source, Argonne National Laboratory. The energy of the X-ray beam was 14 keV (wavelength λ = 0. 8856 Å), and two setups (small- and wide-angle X-ray scattering) were used simultaneously to cover scattering q ranges of 0.006 < q < 2.6 Å−1, where q = (4π/λ)sinθ, and 2θ is the scattering angle. Thirty two-dimensional images were recorded for each buffer or sample solutions using a flow cell, with the accumulated exposure time of 0.8–2 s to reduce radiation damage and obtain good statistics. No radiation damage was observed as confirmed by the absence of systematic signal changes in sequentially collected X-ray scattering images. The 2D images were corrected and reduced to 1D scattering profiles using the Matlab software package at the beamlines. The 1D SAXS profiles were grouped by sample and averaged.
Concentration-series measurements for each sample were carried out at 300 K. We used HTT concentrations of 1.0, 2.0, and 4.0 mg/ml and complex HTT/HAP40 concentrations of 0.5, 1.0, and 2.0 mg/ml, respectively, in 20 mm HEPES, pH 7.5, 300 mm NaCl, 5% (v/v) glycerol, 1 mm TCEP. The data were processed and analyzed with ATSAS program package (45). The scattering profile of the protein was calculated by subtracting the background buffer contribution from the sample-buffer profile, and the difference data were extrapolated to zero solute concentration by standard procedures. Guinier analysis and the experimental radius of gyration (Rg) estimation from the data of infinite dilution were performed using PRIMUS. The pair distance distribution function, P(r), and the maximum dimension of the protein, Dmax, in real space was calculated with the indirect Fourier transform using program GNOM (46). The molecular weights were estimated separately based on volume calculated by SAXMoW (47), and volume of correlation (Vc) (48) was calculated by DATVC in q range of 0 < q < 0.3 Å−1. The theoretical scattering intensity of a structural model was calculated and fitted to the experimental scattering intensity using CRYSOL (49) and FoXS (50) programs.
Fitting structural ensemble to SAXS data
The SAXS data indicate that the HTT/HAP40 complex possesses some degree of flexibility. The known EM structure of the complex (Protein Data Bank code 6ez8) is missing ∼26% of the residues, and it does not fit the SAXS data. We assume that the residues with known coordinates form a quasi-rigid part of the complex, although the residues with missing coordinates are flexible. We performed coarse-grained MD simulations to generate the initial ensemble of possible conformations of the complex. The MD trajectory of 1000 ns was generated at 300 K, and the theoretical scattering profiles in the q range 0 < q < 0.3 Å−1 for 5000 frames taken from the trajectory were calculated using FoXS. The calculated scattering curves were averaged over the entire ensemble of structures using the optimal weights for each ensemble member obtained with the SES method (35), and this average profile was compared with the experimental scattering data.
Coarse-grained molecular dynamics simulations
We used a coarse-grained model of HTT/HAP40 protein complex to enhance the sampling efficiency in the conformational space of the complex. In this model, amino acid residues in the proteins are represented as single beads located at their Cα positions and interacting via appropriate bonding, bending, torsion angle, and nonbonding potential. A Gö-like model of Clementi et al. (51) was employed to maintain the structured globular domains as quasi-rigid in the simulation. For flexible regions, we adopted a simple model in which adjacent amino acids beads are joined together into a polymer chain by means of virtual bond and angle interactions with a quadratic potential as shown in Equation 1,
| (Eq. 1) |
with the constants Kb = 50 kcal/mol and Kα = 1.75 kcal/mol and the equilibrium values b0 = 3.8 Å and α0 = 112° for bonds and angles, respectively. The excluded volume between nonbonded beads was treated with pure repulsive potential as shown in Equation 2,
| (Eq. 2) |
where rij is the inter-bead distance; σR = 4 Å, and ϵR = 2.0 kcal/mol.
The interaction between quasi-rigid domains is modeled with the residue-specific pair interaction potentials that combine short-range interactions with the long-range electrostatics as described (52, 53). The short-range interaction is given by a Lennard-Jones 12–10−6–type potential, and simple Debye-Hückel–type potential is used for the electrostatic interactions (53). In this study, we used the dielectric constant of 80 and the Debye screening length of 10 Å, which corresponds to a salt concentration of about 100 mm. In-house software was developed and used to carry out constant temperature molecular dynamics simulations of the coarse-grained model described above.
Author contributions
R. J. H. conceptualization; R. J. H., L. T.-S., and C. H. A. supervision; R. J. H. and C. H. A. funding acquisition; R. J. H., P. L., S. A., A. L., A. H., B. H., A. S. H., J. C. H., L. F., and A. S. investigation; R. J. H., P. L., S. A., A. L., A. S., and C. H. A. methodology; R. J. H., P. L., S. A., A. L., A. S. H., J. C. H., and A. S. writing-original draft; R. J. H. and C. H. A. project administration; R. J. H., P. L., S. A., A. L., A. H., B. H., A. S. H., J. C. H., L. F., L. T.-S., A. S., and C. H. A. writing-review and editing; C. H. A. resources.
Supplementary Material
Acknowledgments
We thank Dr. Xiaobing Zuo (Argonne National Laboratory) for expert support with SAXS measurements, and we acknowledge the use of the SAXS Core Facility of the Center for Cancer Research (CCR), NCI, National Institutes of Health. This research used 12-ID-B beamline of the Advanced Photon Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We acknowledge Dr. Pravin Mahajan who constructed the pBMDEL vector. We thank Shili Duan for cloning the HAP40 expression construct.
This work was supported in part by National Institutes of Health, NCI, under Contract HHSN26120080001E, by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research, a Huntington's Disease Society of America Berman/Topper Family Career Development Fellowship (to R. J. H.), the Natural Sciences and Engineering Research Council of Canada (to C. H. A.), Huntington Society of Canada Grant RGPIN-2015-05939 (to C. H. A.), the CHDI Foundation (to L. T. S. and C. H. A.), the SGC, a registered charity (No. 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute Grant OGI-055, Innovative Medicines Initiative (EU/EFPIA) ULTRA-DD Grant 115766, Janssen, Merck KGaA, Darmstadt, Germany, MSD, Novartis Pharma AG, Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, São Paulo Research Foundation (FAPESP), Takeda, and The Wellcome Trust. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7, Table S1 and supporting Refs. 1 and 2.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD010865.
All contributing data and analysis files for this manuscript can be found under Creative Commons Attribution 4.0 International license in Zenodo: https://doi.org/10.5281/zenodo.2591677.
- HD
- Huntington's disease
- HTT
- huntingtin
- polyQ
- polyglutamine
- SAXS
- small-angle X-ray scattering
- SEC-MALS
- size-exclusion chromatography in tandem with multiangle light scattering
- TCEP
- tris(2-carboxyethyl)phosphine
- IDR
- intrinsically disordered region
- PTM
- post-translational modification
- LIC
- ligase-independent cloning
- PSM
- peptide spectrum match
- MD
- molecular dynamics
- SES
- sparse ensemble selection
- ACN
- acetonitrile
- DSLS
- differential static light scattering.
References
- 1. Kay C., Hayden M. R., and Leavitt B. R. (2017) Epidemiology of Huntington disease. Handb. Clin. Neurol. 144, 31–46 10.1016/B978-0-12-801893-4.00003-1 [DOI] [PubMed] [Google Scholar]
- 2. Ross C. A., Aylward E. H., Wild E. J., Langbehn D. R., Long J. D., Warner J. H., Scahill R. I., Leavitt B. R., Stout J. C., Paulsen J. S., Reilmann R., Unschuld P. G., Wexler A., Margolis R. L., and Tabrizi S. J. (2014) Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 10, 204–216 10.1038/nrneurol.2014.24 [DOI] [PubMed] [Google Scholar]
- 3. Kay C., Fisher E. R., and Hayden M. R. (2014) Huntington's Disease Bates G. P., Tabrizi S. J., and Jones L., eds) 4th Ed., Chapter 7, pp. 131–164, Oxford University Press, New York [Google Scholar]
- 4. The Huntington's Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- 5. Kim M., Lee H. S., LaForet G., McIntyre C., Martin E. J., Chang P., Kim T. W., Williams M., Reddy P. H., Tagle D., Boyce F. M., Won L., Heller A., Aronin N., and DiFiglia M. (1999) Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J. Neurosci. 19, 964–973 10.1523/JNEUROSCI.19-03-00964.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seredenina T., and Luthi-Carter R. (2012) What have we learned from gene expression profiles in Huntington's disease? Neurobiol. Dis. 45, 83–98 10.1016/j.nbd.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 7. Johri A., Chandra A., and Flint Beal M. (2013) PGC-1α, mitochondrial dysfunction, and Huntington's disease. Free Radic. Biol. Med. 62, 37–46 10.1016/j.freeradbiomed.2013.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reddy P. H., and Shirendeb U. P. (2012) Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington's disease. Biochim. Biophys. Acta 1822, 101–110 10.1016/j.bbadis.2011.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ju T.-C., Lin Y.-S., and Chern Y. (2012) Energy dysfunction in Huntington's disease: insights from PGC-1α, AMPK, and CKB. Cell. Mol. Life Sci. 69, 4107–4120 10.1007/s00018-012-1025-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nithianantharajah J., and Hannan A. J. (2013) Dysregulation of synaptic proteins, dendritic spine abnormalities and pathological plasticity of synapses as experience-dependent mediators of cognitive and psychiatric symptoms in Huntington's disease. Neuroscience 251, 66–74 10.1016/j.neuroscience.2012.05.043 [DOI] [PubMed] [Google Scholar]
- 11. Zheng Z., and Diamond M. I. (2012) Huntington disease and the huntingtin protein. Prog. Mol. Biol. Transl. Sci. 107, 189–214 10.1016/B978-0-12-385883-2.00010-2 [DOI] [PubMed] [Google Scholar]
- 12. Rui Y.-N., Xu Z., Patel B., Chen Z., Chen D., Tito A., David G., Sun Y., Stimming E. F., Bellen H. J., Cuervo A. M., and Zhang S. (2015) Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 17, 262–275 10.1038/ncb3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vijayvargia R., Epand R., Leitner A., Jung T.-Y., Shin B., Jung R., Lloret A., Singh Atwal R., Lee H., Lee J.-M., Aebersold R., Hebert H., Song J.-J., and Seong I. S. (2016) Huntingtin's spherical solenoid structure enables polyglutamine tract-dependent modulation of its structure and function. Elife 5, e11184 10.7554/eLife.11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seong I. S., Woda J. M., Song J.-J., Lloret A., Abeyrathne P. D., Woo C. J., Gregory G., Lee J.-M., Wheeler V. C., Walz T., Kingston R. E., Gusella J. F., Conlon R. A., and MacDonald M. E. (2010) Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 19, 573–583 10.1093/hmg/ddp524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W., Serpell L. C., Carter W. J., Rubinsztein D. C., and Huntington J. A. (2006) Expression and characterization of full-length human Huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 281, 15916–15922 [DOI] [PubMed] [Google Scholar]
- 16. Huang B., Lucas T., Kueppers C., Dong X., Krause M., Bepperling A., Buchner J., Voshol H., Weiss A., Gerrits B., and Kochanek S. (2015) Scalable production in human cells and biochemical characterization of full-length normal and mutant Huntingtin. PLoS One 10, e0121055 10.1371/journal.pone.0121055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ratovitski T., O'Meally R. N., Jiang M., Chaerkady R., Chighladze E., Stewart J. C., Wang X., Arbez N., Roby E., Alexandris A., Duan W., Vijayvargia R., Seong I. S., Lavery D. J., Cole R. N., and Ross C. A. (2017) Post-translational modifications (PTMs), identified on endogenous Huntingtin, cluster within proteolytic domains between HEAT repeats. J. Proteome Res. 16, 2692–2708 10.1021/acs.jproteome.6b00991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saudou F., and Humbert S. (2016) The biology of huntingtin. Neuron 89, 910–926 10.1016/j.neuron.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 20. Michalik A., Kazantsev A., and Van Broeckhoven C. (2001) Method to introduce stable, expanded, polyglutamine-encoding CAG/CAA trinucleotide repeats into CAG repeat-containing genes. BioTechniques 31, 250–252, 254 10.2144/01312bm02 [DOI] [PubMed] [Google Scholar]
- 21. Cariulo C., Azzollini L., Verani M., Martufi P., Boggio R., Chiki A., Deguire S. M., Cherubini M., Gines S., Marsh J. L., Conforti P., Cattaneo E., Santimone I., Squitieri F., Lashuel H. A., et al. (2017) Phosphorylation of huntingtin at residue T3 is decreased in Huntington's disease and modulates mutant huntingtin protein conformation. Proc. Natl. Acad. Sci. U.S.A. 114, E10809–E10818 10.1073/pnas.1705372114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vieweg S., Ansaloni A., Wang Z.-M., Warner J. B., and Lashuel H. A. (2016) An intein-based strategy for the production of tag-free Huntingtin exon 1 proteins enables new insights into the polyglutamine dependence of Httex1 aggregation and fibril formation. J. Biol. Chem. 291, 12074–12086 10.1074/jbc.M116.713982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo Q., Bin Huang, Cheng J., Seefelder M., Engler T., Pfeifer G., Oeckl P., Otto M., Moser F., Maurer M., Pautsch A., Baumeister W., Fernández-Busnadiego R., and Kochanek S. (2018) The cryo-electron microscopy structure of huntingtin. Nature 555, 117–120 10.1038/nature25502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jünger M. A., and Aebersold R. (2014) Mass spectrometry-driven phosphoproteomics: patterning the systems biology mosaic. Wiley Interdiscip. Rev. Dev. Biol. 3, 83–112 10.1002/wdev.121 [DOI] [PubMed] [Google Scholar]
- 25. Trevisiol S., Ayoub D., Lesur A., Ancheva L., Gallien S., and Domon B. (2016) The use of proteases complementary to trypsin to probe isoforms and modifications. Proteomics 16, 715–728 10.1002/pmic.201500379 [DOI] [PubMed] [Google Scholar]
- 26. Schilling B., Gafni J., Torcassi C., Cong X., Row R. H., LaFevre-Bernt M. A., Cusack M. P., Ratovitski T., Hirschhorn R., Ross C. A., Gibson B. W., and Ellerby L. M. (2006) Huntingtin phosphorylation sites mapped by mass spectrometry modulation of cleavage and toxicity. J. Biol. Chem. 281, 23686–23697 10.1074/jbc.M513507200 [DOI] [PubMed] [Google Scholar]
- 27. Huesgen P. F., Lange P. F., Rogers L. D., Solis N., Eckhard U., Kleifeld O., Goulas T., Gomis-Rüth F. X., and Overall C. M. (2015) LysargiNase mirrors trypsin for protein C-terminal and methylation-site identification. Nat. Methods 12, 55–58 10.1038/nmeth.3177 [DOI] [PubMed] [Google Scholar]
- 28. Meyer J. G., Kim S., Maltby D. A., Ghassemian M., Bandeira N., and Komives E. A. (2014) Expanding proteome coverage with orthogonal-specificity α-lytic proteases. Mol. Cell. Proteomics 13, 823–835 10.1074/mcp.M113.034710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keil B. (1992) Specificity of Proteolysis, Chapter 5, pp. 43–228, Springer-Verlag Berlin Heidelberg, Germany: 10.1007/978-3-642-48380-6 [DOI] [Google Scholar]
- 30. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J.-P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods 10, 730–736 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blom N., Gammeltoft S., and Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 10.1006/jmbi.1999.3310 [DOI] [PubMed] [Google Scholar]
- 32. Holehouse A. S., Das R. K., Ahad J. N., Richardson M. O., and Pappu R. V. (2017) CIDER: resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 112, 16–21 10.1016/j.bpj.2016.11.3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dosztányi Z., Csizmok V., Tompa P., and Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- 34. Posey A. E., Ruff K. M., Harmon T. S., Crick S. L., Li A., Diamond M. I., and Pappu R. V. (2018) Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 293, 3734–3746 10.1074/jbc.RA117.000357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berlin K., Castañeda C. A., Schneidman-Duhovny D., Sali A., Nava-Tudela A., and Fushman D. (2013) Recovering a representative conformational ensemble from underdetermined macromolecular structural data. J. Am. Chem. Soc. 135, 16595–16609 10.1021/ja4083717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ayala-Peña S. (2013) Role of oxidative DNA damage in mitochondrial dysfunction and Huntington's disease pathogenesis. Free Radic. Biol. Med. 62, 102–110 10.1016/j.freeradbiomed.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maiuri T., Mocle A. J., Hung C. L., Xia J., van Roon-Mom W. M., and Truant R. (2017) Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum. Mol. Genet. 26, 395–406 [DOI] [PubMed] [Google Scholar]
- 38. Ackloo S., Brown P. J., and Müller S. (2017) Chemical probes targeting epigenetic proteins: applications beyond oncology. Epigenetics 12, 378–400 10.1080/15592294.2017.1279371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters M. F., and Ross C. A. (2001) Isolation of a 40-kDa Huntingtin-associated protein. J. Biol. Chem. 276, 3188–3194 10.1074/jbc.M008099200 [DOI] [PubMed] [Google Scholar]
- 40. Ratovitski T., Chighladze E., Arbez N., Boronina T., Herbrich S., Cole R. N., and Ross C. A. (2012) Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11, 2006–2021 10.4161/cc.20423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shirasaki D. I., Greiner E. R., Al-Ramahi I., Gray M., Boontheung P., Geschwind D. H., Botas J., Coppola G., Horvath S., Loo J. A., and Yang X. W. (2012) Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75, 41–57 10.1016/j.neuron.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gundry R. L., White M. Y., Murray C. I., Kane L. A., Fu Q., Stanley B. A., and Van Eyk J. E. (2009) Preparation of proteins and peptides for mass spectrometry analysis in a bottom-up proteomics workflow. Curr. Protoc. Mol. Biol., 2009, Chapter 10, Unit 10.25 10.1002/0471142727.mb1025s88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dosztányi Z., Csizmók V., Tompa P., and Simon I. (2005) The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347, 827–839 10.1016/j.jmb.2005.01.071 [DOI] [PubMed] [Google Scholar]
- 44. Kyte J., and Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 45. Konarev P. V., Volkov V. V., Petoukhov M. V., and Svergun D. I. (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 10.1107/S0021889806004699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feigin L. A., and Svergun D. I. (1987) Structure Analysis by Small-angle X-Ray and Neutron Scattering, Chapter 3, pp. 94–104, Springer-Verlag Inc., New York: 10.1007/978-1-4757-6624-0 [DOI] [Google Scholar]
- 47. Fischer H., Neto O., De M., Napolitano H. B., Polikarpov I., and Craievich A. F. (2010) Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43, 101–109 10.1107/S0021889809043076 [DOI] [Google Scholar]
- 48. Rambo R. P., and Tainer J. A. (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 10.1038/nature12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 10.1107/S0021889895007047 [DOI] [Google Scholar]
- 50. Schneidman-Duhovny D., Hammel M., and Sali A. (2010) FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 38, W540–W544 10.1093/nar/gkq461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clementi C., Nymeyer H., and Onuchic J. N. (2000) Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 298, 937–953 10.1006/jmbi.2000.3693 [DOI] [PubMed] [Google Scholar]
- 52. Kim Y. C., and Hummer G. (2008) Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J. Mol. Biol. 375, 1416–1433 10.1016/j.jmb.2007.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim Y. C., Tang C., Clore G. M., and Hummer G. (2008) Replica exchange simulations of transient encounter complexes in protein–protein association. Proc. Natl. Acad. Sci. U.S.A. 105, 12855–12860 10.1073/pnas.0802460105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., et al. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J., Levine D. A., Townsend R., Erdmann-Gilmore P., Snider J. E., Davies S. R., et al. (2014) Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics 13, 1690–1704 10.1074/mcp.M113.036392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carrier M., Joint M., Lutzing R., Page A., and Rochette-Egly C. (2016) Phosphoproteome and transcriptome of RA-responsive and RA-resistant breast cancer cell lines. PloS One 11, e0157290 10.1371/journal.pone.0157290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sharma K., D'Souza R. C., Tyanova S., Schaab C., Wiśniewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 10.1016/j.celrep.2014.07.036 [DOI] [PubMed] [Google Scholar]
- 58. Humbert S., Bryson E. A., Cordelières F. P., Connors N. C., Datta S. R., Finkbeiner S., Greenberg M. E., and Saudou F. (2002) The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell 2, 831–837 10.1016/S1534-5807(02)00188-0 [DOI] [PubMed] [Google Scholar]
- 59. Warby S. C., Chan E. Y., Metzler M., Gan L., Singaraja R. R., Crocker S. F., Robertson H. A., and Hayden M. R. (2005) Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum. Mol. Genet. 14, 1569–1577 10.1093/hmg/ddi165 [DOI] [PubMed] [Google Scholar]
- 60. Moritz A., Li Y., Guo A., Villén J., Wang Y., MacNeill J., Kornhauser J., Sprott K., Zhou J., Possemato A., Ren J. M., Hornbeck P., Cantley L. C., Gygi S. P., Rush J., and Comb M. J. (2010) Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 3, ra64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pardo R., Colin E., Régulier E., Aebischer P., Déglon N., Humbert S., and Saudou F. (2006) Inhibition of calcineurin by FK506 protects against polyglutamine-huntingtin toxicity through an increase of huntingtin phosphorylation at S421. J. Neurosci. 26, 1635–1645 10.1523/JNEUROSCI.3706-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rangone H., Poizat G., Troncoso J., Ross C. A., MacDonald M. E., Saudou F., and Humbert S. (2004) The serum- and glucocorticoid-induced kinase SGK inhibits mutant huntingtin-induced toxicity by phosphorylating serine 421 of huntingtin. Eur. J. Neurosci. 19, 273–279 10.1111/j.0953-816X.2003.03131.x [DOI] [PubMed] [Google Scholar]
- 63. Watkin E. E., Arbez N., Waldron-Roby E., O'Meally R., Ratovitski T., Cole R. N., and Ross C. A. (2014) Phosphorylation of mutant huntingtin at serine 116 modulates neuronal toxicity. PLoS One 9, e88284 10.1371/journal.pone.0088284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beausoleil S. A., Villén J., Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- 65. Zahedi R. P., Lewandrowski U., Wiesner J., Wortelkamp S., Moebius J., Schütz C., Walter U., Gambaryan S., and Sickmann A. (2008) Phosphoproteome of resting human platelets. J. Proteome Res. 7, 526–534 10.1021/pr0704130 [DOI] [PubMed] [Google Scholar]
- 66. Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.-J., Saudou F., and Humbert S. (2008) Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27, 2124–2134 10.1038/emboj.2008.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zala D., Colin E., Rangone H., Liot G., Humbert S., and Saudou F. (2008) Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum. Mol. Genet. 17, 3837–3846 10.1093/hmg/ddn281 [DOI] [PubMed] [Google Scholar]
- 68. Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., and Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 10.1021/ac9004309 [DOI] [PubMed] [Google Scholar]
- 69. Pan C., Olsen J. V., Daub H., and Mann M. (2009) Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics 8, 2796–2808 10.1074/mcp.M900285-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Han G., Ye M., Liu H., Song C., Sun D., Wu Y., Jiang X., Chen R., Wang C., Wang L., and Zou H. (2010) Phosphoproteome analysis of human liver tissue by long-gradient nanoflow LC coupled with multiple stage MS analysis. Electrophoresis 31, 1080–1089 [DOI] [PubMed] [Google Scholar]
- 71. Christensen G. L., Kelstrup C. D., Lyngsø C., Sarwar U., Bøgebo R., Sheikh S. P., Gammeltoft S., Olsen J. V., and Hansen J. L. (2010) Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol. Cell. Proteomics 9, 1540–1553 10.1074/mcp.M900550-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., and Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 10.1126/science.1199498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weber C., Schreiber T. B., and Daub H. (2012) Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J. Proteomics 75, 1343–1356 10.1016/j.jprot.2011.11.004 [DOI] [PubMed] [Google Scholar]
- 74. Klammer M., Kaminski M., Zedler A., Oppermann F., Blencke S., Marx S., Müller S., Tebbe A., Godl K., and Schaab C. (2012) Phosphosignature predicts dasatinib response in non-small cell lung cancer. Mol. Cell. Proteomics 11, 651–668 10.1074/mcp.M111.016410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schweppe D. K., Rigas J. R., and Gerber S. A. (2013) Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J. Proteomics 91, 286–296 10.1016/j.jprot.2013.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Britton D., Zen Y., Quaglia A., Selzer S., Mitra V., Lössner C., Jung S., Böhm G., Schmid P., Prefot P., Hoehle C., Koncarevic S., Gee J., Nicholson R., Ward M., et al. (2014) Quantification of pancreatic cancer proteome and phosphorylome: indicates molecular events likely contributing to cancer and activity of drug targets. PLoS One 9, e90948 10.1371/journal.pone.0090948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Marion S., Urs N. M., Peterson S. M., Sotnikova T. D., Beaulieu J.-M., Gainetdinov R. R., and Caron M. G. (2014) Dopamine D2 receptor relies upon PPM/PP2C protein phosphatases to dephosphorylate huntingtin protein. J. Biol. Chem. 289, 11715–11724 10.1074/jbc.M113.544312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Naia L., Ferreira I. L., Cunha-Oliveira T., Duarte A. I., Ribeiro M., Rosenstock T. R., Laço M. N., Ribeiro M. J., Oliveira C. R., Saudou F., Humbert S., and Rego A. C. (2015) Activation of IGF-1 and insulin signaling pathways ameliorate mitochondrial function and energy metabolism in Huntington's disease human lymphoblasts. Mol. Neurobiol. 51, 331–348 10.1007/s12035-014-8735-4 [DOI] [PubMed] [Google Scholar]
- 79. Yi T., Zhai B., Yu Y., Kiyotsugu Y., Raschle T., Etzkorn M., Seo H.-C., Nagiec M., Luna R. E., Reinherz E. L., Blenis J., Gygi S. P., and Wagner G. (2014) Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, E2182–E2190 10.1073/pnas.1404943111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dong G., Callegari E., Gloeckner C. J., Ueffing M., and Wang H. (2012) Mass spectrometric identification of novel post-translational modification sites in Huntingtin. Proteomics 12, 2060–2064 10.1002/pmic.201100380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xiao K., Sun J., Kim J., Rajagopal S., Zhai B., Villén J., Haas W., Kovacs J. J., Shukla A. K., Hara M. R., Hernandez M., Lachmann A., Zhao S., Lin Y., Cheng Y., et al. (2010) Global phosphorylation analysis of β-arrestin–mediated signaling downstream of a seven transmembrane receptor (7TMR). Proc. Natl. Acad. Sci. U.S.A. 107, 15299–15304 10.1073/pnas.1008461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Luo S., Vacher C., Davies J. E., and Rubinsztein D. C. (2005) Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J. Cell Biol. 169, 647–656 10.1083/jcb.200412071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mayya V., Lundgren D. H., Hwang S.-I., Rezaul K., Wu L., Eng J. K., Rodionov V., and Han D. K. (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein–protein interactions. Sci. Signal. 2, ra46 [DOI] [PubMed] [Google Scholar]
- 85. Beli P., Lukashchuk N., Wagner S. A., Weinert B. T., Olsen J. V., Baskcomb L., Mann M., Jackson S. P., and Choudhary C. (2012) Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225 10.1016/j.molcel.2012.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., and Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 10.1038/nmeth.2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bian Y., Song C., Cheng K., Dong M., Wang F., Huang J., Sun D., Wang L., Ye M., and Zou H. (2014) An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics 96, 253–262 10.1016/j.jprot.2013.11.014 [DOI] [PubMed] [Google Scholar]
- 88. Stuart S. A., Houel S., Lee T., Wang N., Old W. M., and Ahn N. G. (2015) A phosphoproteomic comparison of B-RAFV600E and MKK1/2 inhibitors in melanoma cells. Mol. Cell. Proteomics 14, 1599–1615 10.1074/mcp.M114.047233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Boeing S., Williamson L., Encheva V., Gori I., Saunders R. E., Instrell R., Aygün O., Rodriguez-Martinez M., Weems J. C., Kelly G. P., Conaway J. W., Conaway R. C., Stewart A., Howell M., Snijders A. P., and Svejstrup J. Q. (2016) Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 15, 1597–1610 10.1016/j.celrep.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., and Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Anne S. L., Saudou F., and Humbert S. (2007) Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J. Neurosci. 27, 7318–7328 10.1523/JNEUROSCI.1831-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., and Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 10.1016/j.molcel.2008.07.007 [DOI] [PubMed] [Google Scholar]
- 93. Brill L. M., Xiong W., Lee K.-B., Ficarro S. B., Crain A., Xu Y., Terskikh A., Snyder E. Y., and Ding S. (2009) Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5, 204–213 10.1016/j.stem.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schreiber T. B., Mäusbacher N., Kéri G., Cox J., and Daub H. (2010) An integrated phosphoproteomics work flow reveals extensive network regulation in early lysophosphatidic acid signaling. Mol. Cell. Proteomics 9, 1047–1062 10.1074/mcp.M900486-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., and Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3 [DOI] [PubMed] [Google Scholar]
- 96. Zhou H., Di Palma S., Preisinger C., Peng M., Polat A. N., Heck A. J., and Mohammed S. (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271 10.1021/pr300630k [DOI] [PubMed] [Google Scholar]
- 97. Luerman G. C., Nguyen C., Samaroo H., Loos P., Xi H., Hurtado-Lorenzo A., Needle E., Stephen Noell G., Galatsis P., Dunlop J., Geoghegan K. F., and Hirst W. D. (2014) Phosphoproteomic evaluation of pharmacological inhibition of leucine-rich repeat kinase 2 reveals significant off-target effects of LRRK-2-IN-1. J. Neurochem. 128, 561–576 10.1111/jnc.12483 [DOI] [PubMed] [Google Scholar]
- 98. Franz-Wachtel M., Eisler S. A., Krug K., Wahl S., Carpy A., Nordheim A., Pfizenmaier K., Hausser A., and Macek B. (2012) Global detection of protein kinase D-dependent phosphorylation events in nocodazole-treated human cells. Mol. Cell. Proteomics 11, 160–170 10.1074/mcp.M111.016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang R., Ferraris J. D., Izumi Y., Dmitrieva N., Ramkissoon K., Wang G., Gucek M., and Burg M. B. (2014) Global discovery of high-NaCl-induced changes of protein phosphorylation. Am. J. Physiol. Cell Physiol. 307, C442–C454 10.1152/ajpcell.00379.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rolland D., Basrur V., Conlon K., Wolfe T., Fermin D., Nesvizhskii A. I., Lim M. S., and Elenitoba-Johnson K. S. (2014) Global phosphoproteomic profiling reveals distinct signatures in B-cell non-Hodgkin lymphomas. Am. J. Pathol. 184, 1331–1342 10.1016/j.ajpath.2014.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Raijmakers R., Kraiczek K., de Jong A. P., Mohammed S., and Heck A. J. (2010) Exploring the human leukocyte phosphoproteome using a microfluidic reversed-phase-TiO2-reversed-phase high-performance liquid chromatography phosphochip coupled to a quadrupole time-of-flight mass spectrometer. Anal. Chem. 82, 824–832 10.1021/ac901764g [DOI] [PubMed] [Google Scholar]
- 102. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 [DOI] [PubMed] [Google Scholar]
- 103. Xia Q., Cheng D., Duong D. M., Gearing M., Lah J. J., Levey A. I., and Peng J. (2008) Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J. Proteome Res. 7, 2845–2851 10.1021/pr8000496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Imami K., Sugiyama N., Imamura H., Wakabayashi M., Tomita M., Taniguchi M., Ueno T., Toi M., and Ishihama Y. (2012) Temporal profiling of lapatinib-suppressed phosphorylation signals in EGFR/HER2 pathways. Mol. Cell. Proteomics 11, 1741–1757 10.1074/mcp.M112.019919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 10.1016/j.cmet.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wiśniewski J. R., Nagaraj N., Zougman A., Gnad F., and Mann M. (2010) Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 9, 3280–3289 10.1021/pr1002214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.