Escherichia coli sequence type 1193 (ST1193) is an emerging multidrug-resistant pathogen. We performed longitudinal and cross-sectional surveillance for ST1193 among clinical and fecal E. coli isolates from Minneapolis Veterans Affairs Medical Center (VAMC) patients and their household members, other Minnesota centers, and national VAMCs and compared these ST1193 isolates with archival human and canine ST1193 isolates from Australia (2008).
KEYWORDS: Escherichia coli, molecular epidemiology, PCR, ST1193, antimicrobial resistance, fluoroquinolone resistance, intestinal colonization, surveillance, veterinary epidemiology, zoonotic infections
ABSTRACT
Escherichia coli sequence type 1193 (ST1193) is an emerging multidrug-resistant pathogen. We performed longitudinal and cross-sectional surveillance for ST1193 among clinical and fecal E. coli isolates from Minneapolis Veterans Affairs Medical Center (VAMC) patients and their household members, other Minnesota centers, and national VAMCs and compared these ST1193 isolates with archival human and canine ST1193 isolates from Australia (2008). We also developed and extensively validated a novel multiplex PCR assay for ST1193 and its characteristic fimH64 (type 1 fimbrial adhesin) allele. We found that ST1193-H64 (where “H64” refers to a phylogenetic subdivision within ST1193 that is characterized by the fimH64 allele), which was uniformly fluoroquinolone resistant, appeared to emerge in the United States in a geographically staggered fashion beginning around 2011. Its prevalence among clinical and fecal E. coli isolates at the Minneapolis VAMC rose rapidly beginning in 2013, peaked in early 2017, and then plateaued or declined. In comparison with other ST14 complex (STc14) isolates, ST1193-H64 isolates were more extensively multidrug resistant, whereas their virulence genotypes were less extensive but included (uniquely) K1 capsule and fimH64. Pulsed-field gel electrophoresis separated ST1193-H64 isolates from other STc14 isolates and showed genetic commonality between archival Australian versus recent U.S. isolates, fecal versus clinical isolates, and human versus canine isolates. Three main ST1193 pulsotypes differed significantly in resistance profiles and capsular types; emergent pulsotype 2123 was associated with trimethoprim-sulfamethoxazole resistance and K1 (versus K5) capsule. These findings clarify ST1193-H64’s temporal prevalence trends as a fluoroquinolone-resistant pathogen and commensal; document clonal subsets with distinctive geographic, temporal, resistance, and virulence gene associations; and establish a new laboratory tool for rapid and simple detection of ST1193-H64.
INTRODUCTION
Escherichia coli is an important extraintestinal pathogen with emerging antimicrobial resistance (1, 2). The emerging resistance is due mainly to a few widely disseminated clones, most notably the H30R subclone of sequence type 131 (ST131-H30R; associated with allele 30 of fimH [type 1 fimbrial adhesin]), which derives from virulence-associated phylogroup B2 (3–6). Recent reports from diverse locales describe a possible new such clone, ST1193, which likewise is from phylogroup B2 but, unlike ST131, represents sequence type complex 14 (STc14) and is associated with fimH allele 64, i.e., ST1193-H64 (7–12).
The earliest report regarding ST1193 documented its important contribution to the fluoroquinolone (FQ)-resistant (FQ-R) clinical E. coli population among humans in eastern Australia in 2008, with spillover into dogs (7). The Australian ST1193 isolates were all type O75, lactose negative, and FQ-R; most were coresistant to piperacillin, gentamicin, tetracycline, and trimethoprim-sulfamethoxazole. Their consensus virulence genotype included the F10 papA allele (P fimbria structural-subunit variant) without other pap elements, iha (adhesin-siderophore receptor), fimH (allele 64), sat (secreted autotransporter toxin), vat (vacuolating autotransporter toxin), fyuA (yersiniabactin receptor), iutA (aerobactin receptor), kpsM II (group 2 capsule), usp (uropathogenic specific protein), ompT (outer membrane protease), and malX (pathogenicity island marker). The only virulence gene variation involved the K1 versus K5 capsule variants, which segregated with different pulsed-field gel electrophoresis (PFGE) types (pulsotypes).
Subsequent reports from China (9, 10), Korea (11), Norway (12), Germany (13), and the United States (8) documented ST1193 as a progressively emerging FQ-R human pathogen in these regions, often second only to ST131-H30R among FQ-R isolates and sometimes associated with CTX-M extended-spectrum beta-lactamases (ESBLs) (9, 12). These isolates’ virulence genotype, O type, and lactose-negative phenotype, when reported, have resembled those of the 2008 Australian isolates, suggesting global clonal dissemination.
Gut colonization with multiresistant clones, such as ST131-H30 and ST1193-H64, may underlie these clones’ epidemic spread and ability to cause extraintestinal disease (14, 15). As such, the intestinal reservoir deserves attention, along with clinical isolates. To date, however, gut colonization with ST1193 has been reported only in an abstract describing a point-prevalence survey of newly admitted Norwegian inpatients (12).
Accordingly, we sought to clarify the prevalence trends and characteristics of ST1193-H64 in the United States among clinical and fecal surveillance isolates and to facilitate future global molecular epidemiological studies of this emerging lineage. For this, we devised and validated extensively a rapid and simple PCR assay for ST1193-H64 and then used it to screen for ST1193-H64 both cross-sectionally and longitudinally among clinical and fecal surveillance isolates from the Veterans Affairs Medical Center (VAMC) in Minneapolis, MN (MVAMC), other centers in Minnesota, and other VAMCs across the United States. We also compared the resulting isolates molecularly and phenotypically with historical human and canine ST1193-H64 isolates from Australia and with concurrent non-ST1193 isolates from STc14.
MATERIALS AND METHODS
Isolates.
The study isolates were derived from multiple published and unpublished sources, including collections of clinical E. coli isolates from diverse institutions and an ongoing prospective MVAMC-based fecal surveillance study. The clinical isolates included 6,569 E. coli isolates from the MVAMC microbiology laboratory, collected prospectively from November 2010 through May 2011 (here labeled 2011) and April 2012 through November 2018 (reference 16 and J. R. Johnson, unpublished data) (the 10-month hiatus was for logistical reasons). They also included (i) temporally matched sets of concurrent FQ-R and FQ-susceptible (FQ-S) E. coli isolates from 24 nationally distributed VAMCs in 2011 (approximately 20 each; total, 472) (17); (ii) all E. coli isolates from clinical laboratories in Olmsted County, MN, in 2013 (total, 299) (18); and (iii) all E. coli clinical isolates from the University of Minnesota Medical Center (UMMC) in July and August 2013 (total, 233) (19). These clinical isolates were predominantly (65% to 85%, by collection) from urine, followed by miscellaneous sources (e.g., abdominal, wound, drainage, tissue, and respiratory specimens); approximately 5% were from blood. Positive and negative controls for development and validation of an ST1193-H64-specific PCR assay (described below) included isolates for which the ST and the fimH allele had been determined previously, as selected from the E. coli Reference (ECOR) collection (20), a urinary-source bacteremia isolate collection (21), the above-mentioned national VAMC clinical-isolate collection (17), and a collection of Australian ST1193 isolates (7).
Fecal isolates were recovered from self-collected fecal swabs provided by veterans at the MVAMC and their household members (totals, 1,459 humans and 150 pets) from 2014 through November 2018. The veterans were recruited by sending invitations for study participation to all newly discharged MVAMC inpatients and randomly selected outpatients. Veterans who agreed to participate were encouraged to refer all available household members. Consenting veterans and household members collected fecal swabs according to an Institutional Review Board-approved protocol and mailed them at room temperature in commercial transport medium to the research laboratory. There, swabs were plated to Gram-negative selective media with and without ciprofloxacin (4 mg/liter) for overnight incubation at 37°C. Indole-positive, citrate-negative colonies with a characteristic E. coli morphology were regarded presumptively as E. coli.
Molecular typing.
Established multiplex PCR-based assays were used to identify E. coli phylogroups (22), nine clonal subsets (including STc14) within group B2 (23), ST131 and its H30R subclone (5), 50 E. coli virulence genes (24), and the O75 and O18 antigen-encoding rfb regions (25). Clonal typing was done for all FQ-R isolates (clinical and fecal), all FQ-S isolates from the University of Minnesota and Olmsted County, a random 25% subsample of non-ST131 group B2 FQ-S MVAMC clinical isolates through April 2017 and all such isolates thereafter, and selected FQ-S MVAMC fecal surveillance isolates. All PCR testing was done in duplicate using boiled lysates as template DNA, in parallel with standard positive and negative controls. Sequencing-based ST determination was done selectively using full or partial seven-locus multilocus sequence typing (MLST) (26) or two-locus fumC-fimH (CH) typing (27), with or without sequencing of additional MLST loci (28).
Genomic profiling was done using XbaI PFGE according to the PulseNet protocol (29). Pulsotypes were defined at 94% profile similarity based on computer-assisted Dice coefficient analysis of banding patterns within BioNumerics (Applied Maths) (30). Newly determined profiles were compared digitally with an existing large private profile library (currently, 6,901 profiles and 2,379 pulsotypes) (30).
Novel multiplex ST1193-H64 PCR assay.
A multiplex PCR assay for ST1193-H64 that included primers for icd, fimH, and fumC was developed. Allele 200 of the housekeeping gene icd, which distinguishes ST1193 from other STs within STc14, is specific to ST1193, plus several rare single-locus variants (based on an 18 June 2018 search of Enterobase [http://enterobase.warwick.ac.uk/species/index/ecoli]) (31). In silico analysis of 933 aligned icd allele sequences from Enterobase identified a distinctive icd-200-specific single-nucleotide polymorphism (SNP) (G→A) at bp 551 of the whole gene. The novel forward primer 1193icdF.21 (5′-ATTCCTGCGTGAAGAGATGGA-3′) was designed with this SNP at the 3′ end. The combination of this novel primer with the published reverse primer icdgpVII.r (5′-CAATTAAATCAGCCGCTTCG-3′), which is specific for the STc14-associated clonal subset VII within phylogroup B2 (23), yielded a unique icd-200-specific primer pair that produced a 600-bp PCR amplicon with ST1193 control strains.
The distinctive fimH allele that characterizes ST1193 is fimH64 (32). In silico analysis of a 448-allele fimH library (27) identified two SNPs (bp 80, C→T; bp 311, C→T) that occur jointly only in fimH64. Incorporation of these two SNPs into the respective 3′ ends of novel primers, i.e., forward primer fimH64F.18 (5′-TGTAAACCTTGCGCCCGT-3′) and reverse primer fimH64R (5′-TCAAATAAAGCGCCACCA-3′), yielded a novel fimH64-specific primer pair that produced a 266-bp amplicon with ST1193-fimH64 control strains.
fumC, a housekeeping gene used in MLST, was added as an internal amplification and species control by using published primers (fumC for, 5′-TCACAGGTCGCCAGCGCTTC-3′, and fumC rev, 5′-GTACGCAGCGAAAAAGATTC-3′) (26), which gave an 802-bp amplicon. After multiplex PCR, gel electrophoresis separates this amplicon well from the amplicons for icd-200 (600 bp) and fimH64 (266 bp).
After empirical optimization of PCR conditions and screening of multiple primer pairs with different calculated annealing temperatures, the best-performing primer combination and conditions were used in a single multiplex reaction for validation experiments. For each PCR, 1.6 μl sample DNA was combined with 15 μl of the following amplification master mix: 0.75 U GoTaq hot start polymerase (Promega), 1× GoTaq Flexi Buffer (Promega), 2.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates (dNTPs), 16 pmol per icd-200 primer, 7 pmol per fimH64 primer, 0.85 pmol per fumC control primer, and H2O to 15 μl. The cycling conditions were as follows: denaturation at 95°C for 2 min, 30 amplification cycles of 94°C for 20 s and 66°C for 1 min, extension at 72°C for 2 min, and then holding at 12°C.
A nonblinded operator did assay validation experiments in duplicate using 10 positive- and 150 negative-control strains. The negative controls represented 71 STs other than ST1193, distributed by E. coli phylogroup as follows: A (15 STs, 28 isolates), B1 (13 STs, 17 isolates), B2 (20 STs, 67 isolates, including 3 non-1193 STs from STc14), C (3 STs, 8 isolates), D (12 STs, 13 isolates), E (3 STs, 4 isolates), and F (5 STs, 13 isolates). The control strains for which fimH sequence was known represented 45 fimH alleles other than fimH64.
Antimicrobial susceptibility testing.
For the MVAMC clinical isolates, broth microdilution susceptibility results (from a bioMérieux Vitek instrument) were available from the MVAMC clinical laboratory for 12 antibiotics, i.e., ampicillin, ampicillin-sulbactam, cefazolin, ceftazidime, ceftriaxone, ciprofloxacin, ertapenem, imipenem, gentamicin, nitrofurantoin, piperacillin-tazobactam, and trimethoprim-sulfamethoxazole. For a subset of these isolates and selected others, susceptibility to 20 antibiotics was determined by a standardized disk diffusion method, using Clinical and Laboratory Standards Institute-approved procedures, reference strains, and interpretive criteria (33). Drugs included the 12 mentioned above plus an additional 8 drugs, i.e., amikacin, aztreonam, chloramphenicol, nalidixic acid, piperacillin, streptomycin, tetracycline, and trimethoprim. Intermediate isolates were analyzed as resistant.
Statistical methods.
Comparisons of proportions were tested using a two-tailed Fisher’s exact test or a chi-squared test, as appropriate. A chi-squared test for linear trend (http://epitools.ausvet.com.au/content.php?page=trend) was used to assess for prevalence shifts over time. Prevalence curve segments for statistical comparison were selected based on inspection of the curves for seeming inflection points. Differences in slope between adjacent segments of the prevalence curves were assessed for statistical significance by using a logistic regression model that included three variables as predictors of resistance prevalence: time, curve segment, and their interaction term (which reflected the slope change between segments). Two-group comparisons involving host age were tested using a two-tailed t test. The criterion for statistical significance was a P value of <0.05.
RESULTS
Validation of the ST1193-H64 PCR assay.
The novel multiplex PCR assay for ST1193-H64 exhibited 100% sensitivity (95% confidence interval [CI], 69% to 100%) with the 10 ST1193 positive-control isolates, all of which yielded both expected ST1193-specific bands (for icd200 and fimH64), and 100% specificity (95% CI, 98% to 100%) with 150 non-ST1193 negative-control isolates, none of which yielded either ST1193-specific band. The assay’s fumC internal-control band appeared with all 160 validation set isolates, whether ST1193 or non-ST1193, except ECOR strain 6 (ST77, phylogroup A). The assay confirmed all FQ-R ST1193 isolates identified in this study (as described below) as ST1193-H64 (not shown).
Emergence of E. coli ST1193-H64 as an FQ-R pathogen at MVAMC.
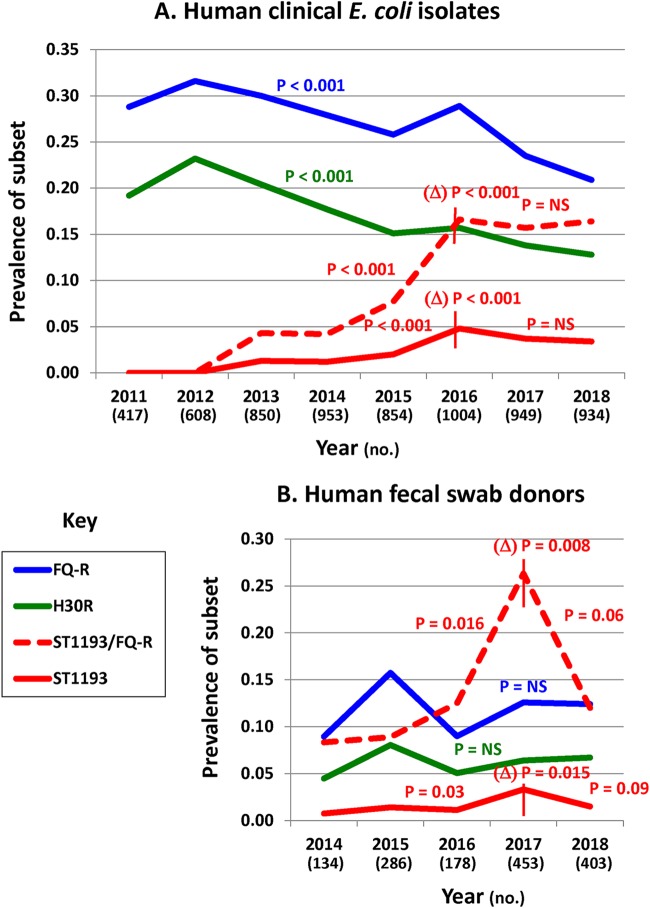
Longitudinal surveillance of MVAMC E. coli clinical isolates (October 2010 to November 2018: n = 6,569) showed a statistically significant overall prevalence decline for both the FQ-R subset and its main clonal component, ST131-H30R (for both, P < 0.001 for linear trend) (Fig. 1A). In contrast, ST1193-H64 (as identified by screening all non-ST131 FQ-R group B2 isolates for nine group B2 subtypes and then all STc14 isolates for ST1193-H64) appeared first in 2013 and rose rapidly in prevalence (P < 0.001 for linear trend). The prevalence of ST1193-H64 peaked in 2016, both among E. coli isolates overall (4.8%) and within the FQ-R subset (16.6%). These peaks were followed by statistically significant downward slope deflections (P < 0.001 for each), with downward-trending (overall) or stable (within the FQ-R subset) prevalences in 2017 and 2018 (slopes not significantly different from zero).
FIG 1.
Prevalence of FQ-R E. coli variants among clinical isolates and human volunteers. (A) Clinical isolates (6,569) at the MVAMC. (B) Fecal surveillance subjects (MVAMC veterans and their household members). H30R, prevalence of the ST131-H30R subclone; ST1193, prevalence of ST1193-H64; ST1193/FQ-R, prevalence of ST1194-H64 among FQ-R E. coli clinical isolates (A) or FQ-R E. coli-colonized subjects (B). P values not preceded by Δ are for the respective curve or curve segment’s fit to a chi-squared test for linear trend; those preceded by Δ are for the change in slope at the inflection point, according to multivariable logistic regression analysis. The vertical lines mark inflection points used to define curve segment boundaries (i.e., ST1193 and ST1193/FQ-R) for analyses of segmental slope and change in slope. Curves without a vertical line (i.e., FQ-R and H30R) were analyzed in total. P values are shown where P is less than 0.10. NS, nonsignificant (P ≥ 0.10). Numbers of isolates (top) or subjects (bottom) per year are shown below the x axis.
Emergence of ST1193-H64 as a gut commensal at MVAMC (2014 to 2018).
Prospective fecal surveillance of MVAMC patient volunteers and their human household members (May 2014 to November 2018: 1,459 total subjects) identified prevalence trends for colonization with FQ-R E. coli, ST131-H30, and ST1193-H64 that largely paralleled those observed among MVAMC clinical isolates (Fig. 1B). Specifically, for both FQ-R E. coli and ST131-H30, the prevalence curves trended downward after 2015, although not statistically significantly so. In contrast, for ST1193-H64, overall prevalence rose nearly 5-fold from 2014 through 2017, i.e., from 0.7% to 3.3% (P = 0.03 for linear trend), and fractional prevalence within the FQ-R subset rose over 4-fold, from 8.3% (2014) to 26.3% (2017) (P = 0.016 for linear trend). Thereafter, in 2017, both prevalence curves deflected significantly downward (for change in slope, overall, P = 0.015; within the FQ-R subset, P = 0.008), leading to borderline significantly negative slopes from 2017 to 2018 (overall, P = 0.09; within the FQ-R subset, P = 0.06).
The MVAMV-based household fecal surveillance also involved 150 pets, mainly dogs (Table 1). Colonization with FQ-R E. coli was significantly less common among pets than among humans and among pets was limited to dogs and involved only ST1193-H64 and other non-ST131-H30 FQ-R strains.
TABLE 1.
Prevalence of colonization with FQ-R E. coli in relation to host group among veterans and their human household members and pets
| Host group | No. of subjects with samples | No. (% within row) of subjects with specific culture result |
||
|---|---|---|---|---|
| FQ-R E. coli | ST131-H30R | ST1193 | ||
| Human | ||||
| Total | 1,459 | 181 (11.8)a,b,c | 93 (6.0)d,e,f | 28 (2.1)g |
| Veterans | 924 | 115 (11.8) | 62 (6.0) | 20 (2.5) |
| Household members | 535 | 66 (11.9) | 31 (6.0) | 8 (1.5) |
| Animals | ||||
| Total | 150 | 4 (2.5)a | 0 (0)d | 1 (0.6)g |
| Dogs | 93 | 4 (4.4)b | 0 (0)e | 1 (1.1)g |
| Other animals | 57 | 0 (0)c | 0 (0)f | 0 (0)g |
Humans versus all animals, P < 0.001.
Humans versus dogs, P = 0.02.
Humans versus other animals, P = 0.005.
Humans versus all animals, P = 0.001.
Humans versus dogs, P = 0.01.
Humans versus other animals, P = 0.049.
Humans versus other groups, all comparisons, P > 0.05.
Host age.
For MVAMC clinical isolates, the mean source patient age was 4.4 years higher in association with FQ-R isolates than with FQ-S isolates (71.7 versus 67.3 years; P < 0.001) but, for FQ-R isolates, was similar for ST1193-H64 and ST131-H30R isolates (71.0 versus 71.7 years). Similarly, for human fecal swab donors, the mean subject age was 6.0 years higher among those with FQ-R E. coli versus those with FQ-S E. coli only, non-E. coli, or no growth (68.9 versus 62.9 years: P < 0.001) but, among those with FQ-R E. coli, was similar for those with ST1193-H64 versus ST131-H30R (70.1 versus 68.4 years).
Historical survey.
For comparison with the MVAMC clinical isolates, we screened for ST1193-H64 within three relevant historical E. coli collections (two from 2011 and one from 2013) (Table 2). First, among 472 isolates collected systematically in 2011 from 22 nationally distributed VAMCs, ST1193-H64 accounted for 4 (1.7%) of 236 consecutive FQ-R isolates versus 0% of 236 concurrent FQ-S isolates (P = 0.12). Adjustment for the mean prevalence of FQ-R E. coli at the participating VAMCs (29%) yielded an estimated overall ST1193-H64 prevalence of 0.5%. The four identified ST1193-H64 isolates were derived from widely separated VAMCs in San Diego, CA (n = 2); Salt Lake City, UT (n = 1); and Miami, FL (n = 1). Second, among 299 consecutive clinical isolates collected concurrently in 2011 from clinical laboratories in Olmsted County, MN, ST1193-H64 accounted for 0.7% of isolates overall (n = 2), including 2.3% of 88 FQ-R isolates, versus 0% of 211 FQ-S isolates (P = 0.09). Third, among 233 consecutive clinical isolates collected 2 years later (2013) at the UMMC, ST1193-H64 accounted for 1.7% of isolates overall (n = 4), including 6% of 65 FQ-R isolates, versus 0% of 168 FQ-S isolates (P = 0.006).
TABLE 2.
Prevalence of ST1193 within collections of clinical E. coli isolates from multiple centers in 2011 and 2013
| Population | Location | Yr | Total no. of isolates | ST1193 [no. (% of total)] | FQ-R isolates |
FQ-S isolates |
P value, ST1193 in FQ-R vs. FQ-S | ||
|---|---|---|---|---|---|---|---|---|---|
| No. (% of total) | ST1193, no. (% of FQ-R) | No. (% of total) | ST1193, no. (% of FQ-S) | ||||||
| Veterans | 24 VAMCs | 2011 | 472 | 4 (0.5a ) | 236 (29a ) | 4 (1.7) | 236 (71a ) | 0 (0) | 0.12 |
| General | Olmsted County, MNb | 2011 | 299 | 2 (0.7) | 88 (29) | 2 (2.3) | 211 (71) | 0 (0) | 0.09 |
| Minneapolis, MNc | 2013 | 233 | 4 (1.7) | 65 (28) | 4 (6) | 168 (72) | 0 (0) | 0.006 | |
| Any of the above | All of the above | 2011–2013 | 1,004 | NAd | 389 (NA) | 10 (2.6) | 615 (NA) | 0 (0) | <0.001 |
Because the analyzed VAMC isolates were selected artificially to be 50% FQ-R and 50% FQ-S, the true total prevalence of ST1193 among VAMC isolates was estimated by adjusting the observed prevalence within the FQ-R and FQ-S subsets by the prevalence of those subsets at the contributing VAMCs, which is shown here as “% of total” for FQ-R and FQ-S isolates.
Olmsted County Medical Center and Mayo Clinic, Rochester, MN.
University of Minnesota Medical Center (East Bank facility, West Bank facility, and Children’s Hospital).
NA, percentage was not applicable (due to deliberate selection of FQ-R and FQ-S isolates for the VAMC collection).
In the three collections combined, ST1193-H64 was significantly more prevalent among FQ-R isolates than FQ-S isolates (10/389 [2.6%] versus 0/615 [0%]: P < 0.001) and among FQ-R isolates was significantly more prevalent in 2013 (4/65 [6%]) than in 2011 (6/324 [2%]: P = 0.045). In contrast, for the MVAMC clinical isolates, none of 312 total FQ-R isolates from either 2010 and 2011 (n = 120) or 2012 (n = 192) were ST1193-H64 (Fig. 1A), a significant difference versus the above-mentioned 2011 non-MVAMC isolates (P = 0.03).
Pulsotypes.
According to XbaI PFGE analysis, the present 95 ST1193-H64 isolates, plus 26 historical ST1193-H64 isolates from humans and dogs in Australia (2008), represented 13 total pulsotypes (94% profile similarity level) (Table 3; see Fig. S1 in the supplemental material). Three major pulsotypes (1297, 1298, and 2123) accounted for 14% to 45% of the isolates each (collectively, 78% of the isolates); the remaining 10 pulsotypes accounted for ≤7% of the isolates each.
TABLE 3.
Distribution of pulsotypes by locale and source among 133 isolates of E. coli ST1193
| Pulsotypea | No. of isolates |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (n = 133) | United States |
Australian clinicalc |
|||||||
| Fecalb MVAMC, 2014–2018 |
Clinicalb,c |
||||||||
| Human (n = 28) | Dog (n = 1) | MVAMC, 2010–2018 (n = 68) | 24 VAMCs, 2011 (n = 4) | Olmsted County, MN, 2011 (n = 2) | UMMC, 2013 (n = 4) | Human, 2008 (n = 24) | Dog, 2008 (n = 2) | ||
| 1297 | 56 | 14 | 1 | 26 | 2 | 2 | 3 | 8 | 0 |
| 2123 | 29 | 6 | 0 | 23 | 0 | 0 | 0 | 0 | 0 |
| 1298 | 18 | 1 | 0 | 2 | 0 | 0 | 0 | 14 | 1 |
| 2361 | 10 | 3 | 0 | 5 | 0 | 0 | 1 | 0 | 1 |
| 2359 | 4 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| 2364 | 4 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 2362 | 3 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 |
| 2365 | 3 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 |
Of 13 total ST1193 pulsotypes, 8 (shown in the table) accounted for ≥3 isolates each (127 total isolates; 95% of 133). The remaining 5 ST1193 pulsotypes accounted for either 2 isolates (pulsotype 2366; 2 MVAMC clinical isolates) or 1 isolate (3 MVAMC clinical isolates, 1 Australian human isolate) each.
Among U.S. isolates, all prevalence comparisons between fecal and clinical isolates for individual pulsotypes, P > 0.50 (Fisher’s exact test).
Among clinical isolates, the U.S. and Australian isolates differed significantly for the prevalence of pulsotypes 2123 (23/68 [28%] [United States] versus 0/26 [0%] [Australian]; P < 0.001) and 1298 (2/68 [3.0%] [United States] versus 15/26 [58%] [Australian]; P < 0.001). For all other pulsotypes, P was >0.05.
Two of the major pulsotypes were significantly distributed geographically: pulsotype 2123 was associated with U.S. isolates and pulsotype 1298 with Australian isolates (Table 3). Furthermore, among the U.S. clinical isolates, pulsotype 2123 was distributed temporally, accounting for only 4% (1/28) of isolates before the 4th quarter of 2015 but for 53% (18/34) of isolates thereafter (P < 0.001) (data not shown). The MVAMC fecal isolates exhibited a similar but statistically nonsignificant trend (not shown). In contrast, the various pulsotypes were distributed indifferently by clinical versus fecal origin and host species (Table 3).
In comparison, the 21 remaining STc14 isolates from these collections were more diverse by PFGE, representing 14 total pulsotypes, each accounting for only 1 to 4 (5% to 19%) isolates (see Fig. S2 in the supplemental material). In the similarity dendrogram, 12 of 13 ST1193-H64-associated index profiles (i.e., all but pulsotype 2123) clustered together, separately from the other profiles (see Fig. S2).
Antimicrobial resistance and lactose fermentation.
Overall, the ST1193-H64 isolates were extensively antimicrobial resistant, exhibiting a >50% resistance prevalence for 9 of 20 tested agents (ampicillin, piperacillin, cephalothin, streptomycin, trimethoprim, trimethoprim-sulfamethoxazole, nalidixic acid, ciprofloxacin, and tetracycline) (Table 4). Within STc14, compared with other STc14 isolates, the ST1193-H64 isolates had a significantly higher resistance prevalence for nine agents (gentamicin plus all of the above-mentioned agents except trimethoprim-sulfamethoxazole) and a significantly lower prevalence for none. They also were nearly all (99%) lactose negative versus only 41% of the other STc14 isolates (P < 0.001).
TABLE 4.
Antimicrobial resistance of E. coli isolates of ST1193-H64, its main pulsotypes (PFGEs), and other STc14 STs
| Antimicrobial agenta | Prevalence of resistance by ST1193-H64 status within STc14 (% within column) |
2-Group P valued | Prevalence of resistance by pulsotype within ST1193-H64 (% within column) |
3-Group P valued | |||
|---|---|---|---|---|---|---|---|
| Other STc14 isolates (n = 29) | ST1193-H64 (n = 117b or 77c) | PFGE 1297 (n = 55b or 31c) | PFGE 1298 (n = 17b or 16c) | PFGE 2123 (n = 23b or 9c) | |||
| Penicillin | |||||||
| AMP | 56 | 83 | 0.01 | 71f | 100e | 96 | 0.004 |
| SAM | 45 | 33 | 0.28 | 29 | 24 | 57f | 0.04 |
| PRL | 52 | 79 | 0.008 | 61f | 100e | 89 | 0.008 |
| Cephalosporin | |||||||
| KZ | 21 | 56 | 0.001 | 40g | 94g | 39 | <0.001 |
| Aminoglycoside | |||||||
| GEN | 0 | 21 | 0.004 | 11f | 77g | 0f | <0.001 |
| STR | 21 | 81 | <0.001 | 65f | 88 | 100 | 0.04 |
| AMK | 0 | 7 | 0.32 | 0 | 19 | 0 | 0.02 |
| Folate inhibitor | |||||||
| TMP | 3 | 68 | <0.001 | 58 | 88 | 100e | 0.007 |
| SXT | 45 | 63 | 0.09 | 49f | 82 | 93g | <0.001 |
| Quinolone | |||||||
| NA | 10 | 99 | <0.001 | 100 | 100 | 89 | 0.07 |
| CIP | 0 | 100 | <0.001 | 100 | 100 | 100 | 1.0 |
| Tetracycline | |||||||
| TE | 10 | 61 | <0.001 | 42f | 88e | 100f | <0.001 |
AMK, amikacin; AMP, ampicillin; CIP, ciprofloxacin; GEN, gentamicin; KZ, cefazolin; NA, nalidixic acid; PRL, piperacillin; SAM, ampicillin-sulbactam; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TE, tetracycline; TMP, trimethoprim. The antimicrobial classes and agents shown are those for which resistance was significantly clonally distributed. Additional studied agents, for which resistance was not significantly clonally distributed, included (range of resistance prevalence by clonal subset) piperacillin-tazobactam (TZP) (0% to 22%), ceftriaxone (CRO) (0% to 18%), ceftazidime (CAZ) (0% to 6%), ertapenem (ETP) (0% to 1%), imipenem (IMP) (0% to 3%), aztreonam (ATM) (0% to 10%), nitrofurantoin (NF) (0% to 12%), and chloramphenicol (C) (0% to 13%).
A larger set of ST1193-H64 isolates was tested against only AMP, CAZ, CRO, ETP, GEN, IMP, KZ, NF, SAM, and SXT.
A smaller set of ST1193-H64 isolates was also tested against AMK, ATM, C, NA, PRL, STR, TE, TMP, and TZP.
Two-group P values (by Fisher’s exact test, two-tailed) and 3-group P values (by 3-group Pearson χ2) are shown in boldface; P < 0.05.
P < 0.05 (Fisher’s exact test, two-tailed) for comparisons of each pulsotype versus all other ST1193 isolates.
P ≤ 0.01 (Fisher’s exact test, two-tailed) for comparisons of each pulsotype versus all other ST1193 isolates.
P ≤ 0.001 (Fisher’s exact test, two-tailed) for comparisons of each pulsotype versus all other ST1193 isolates.
Within ST1193-H64, the three main pulsotypes differed significantly from one another for resistance prevalences to 10 of the 20 tested agents (Table 4). Pulsotype 1297 had the lowest resistance prevalence for nine agents (i.e., all but gentamicin), whereas pulsotype 1298 had the highest resistance prevalence for four agents (penicillin, piperacillin, cefazolin, and gentamicin), and pulsotype 2123 had the highest resistance prevalence for five agents (ampicillin-sulbactam, streptomycin, trimethoprim, trimethoprim-sulfamethoxazole, and tetracycline).
Virulence genotype.
Overall, the ST1193-H64 isolates exhibited extensive virulence gene profiles, including a >85% prevalence for 14 genes: adhesins (papA [F10 allele], iha, fimH, and yfcV), toxins (sat and vat), siderophores (fyuA, iutA, and chuA), protectins (kpsM II and rfb [O75 variant]), and miscellaneous traits (usp, ompT, and malX) (see Table S1 in the supplemental material). Within STc14, non-ST1193 and ST1193-H64 isolates differed significantly in the prevalences of 16 genes: ST1193-H64 isolates had a higher prevalence of 4 (F10 papA allele, sat, K1, and O75 rfb variant) and a lower prevalence of the other 12. In contrast, within ST1193-H64, the virulence gene profiles of the three main pulsotypes differed significantly only for the prevalence of capsule variants K1 (100%, pulsotypes 1297 and 2123; 0%, pulsotype 1298: P < 0.001) and K5 (100%, pulsotype 1298; 0%, pulsotypes 1297 and 2123: P < 0.001).
DISCUSSION
Our longitudinal and cross-sectional survey for ST1193-H64 within multiple collections of clinical and fecal E. coli isolates (from Minnesota, national VAMCs, and Australia) and our characterization of the resulting isolates support five main conclusions. First, at MVAMC, ST1193-H64 emerged rapidly as an FQ-R pathogen beginning in 2013, significantly later than at VAMCs nationally or at other Minnesota centers, but appears now to have plateaued. Second, the prevalence of fecal colonization with ST1193-H64 among MVAMC patients and their household members has largely paralleled clinical prevalence, consistent with the fecal reservoir (and, possibly, within-household transmission) driving clinical emergence, as also supported by the similarity of clinical and fecal ST1193-H64 isolates. Third, ST1193-H64 appears more extensively antimicrobial resistant than other STc14 isolates, conceivably contributing to its emergence. In contrast, it has similarly or less extensive virulence gene profiles, which nonetheless uniquely include the K1 capsule variant, another possible contributor to clonal emergence by enhancing fitness in the clinical (21, 34) and possibly the commensal (35) niches. Fourth, ST1193-H64 exhibits three main PFGE subtypes, which segregate geographically and temporally and correspond to resistance profiles and capsular types. Fifth, our novel ST1193-H64 PCR-based assay was simple and accurate and so should facilitate the molecular epidemiological studies needed to further assess the prevalence, ecology, and clinical significance of ST1193-H64.
Regarding clonal emergence, several recent reports have documented ST1193-H64 as an emerging contributor to the FQ-R E. coli population (7–13), usually second only to ST131-H30, which in contrast emerged globally beginning nearly 2 decades ago (6, 24, 36–40). Our findings suggest a staggered timing for the emergence of ST1193-H64 at different centers, with historical isolates from Australia dating to 2008; a low prevalence in 2011 in Olmsted County, MN, and at VAMCs nationally in the United States; and a later appearance at MVAMC (2013), followed by a rapid rise and then a plateau and even a possible decline (fecal colonization).
The basis for such clonal prevalence fluctuations is largely unknown, although shifts in population level antibiotic use may contribute. Local clonal emergence, followed by decline, has been documented for other prominent multiresistant E. coli clones, including “clonal group A” (ST69) (41) and the O15:K52:H1 clonal group (STc31) (42). Our longitudinal data from MVAMC provide what, to our knowledge, is the first evidence in any locale of a statistically significant prevalence decline for either ST131-H30 or ST1193-H64. Whether these recent trends at MVAMC also apply to other locales and populations, and whether they will continue even at MVAMC, will be important to determine for public health, infection prevention, and rapid diagnostics purposes. The needed studies can now be done readily by using our novel PCR-based ST1193-H64 assay.
We identified at MVAMC a substantial fecal reservoir of ST1193-H64, which exhibited prevalence trends largely paralleling those seen among clinical isolates. These parallel clinical-versus-fecal temporal trends and the extensive similarity of ST1193-H64 fecal and clinical isolates support the notions that the fecal reservoir gives rise to clinical isolates (14, 43, 44) and that prevalence changes within the fecal reservoir may drive clinical prevalence (45–47). As such, identification of host factors that favor colonization and infection with specific organisms may help in understanding and mitigating clonal emergence.
In that regard, host age, although greater in association with FQ-R than with FQ-S isolates, did not differ between the two principal FQ-R subsets, ST1193-H64 and ST131-H30R. This conflicts with a recent study involving mainly non-VAMC clinical isolates in which source hosts were significantly younger for ST1193-H64 than for ST131-H30R (8), possibly due to population differences, as MVAMC patients are mainly elderly men with multiple comorbidities (16).
Identification of ST1193-H64 among canine clinical and fecal isolates suggests that dogs participate in the ecology of ST1193, possibly contributing to dissemination (7). Dogs are unlikely to be the primary reservoir, however, given the significant prevalence gradient from humans to dogs. Bidirectional transmission is likely, potentially making ST1193-H64 infection both a zoonosis and an anthroponosis.
Against the background of overall genetic homogeneity within ST1193-H64 across locales, sources, and hosts, we observed subtle molecular variation, some of which segregated geographically and/or temporally, e.g., the associations of pulsotype 1298 with K5 capsule and Australian isolates from 2008 and of pulsotype 2123 with K1 capsule and recent isolates from the United States. Likewise, among recent U.S. isolates, resistance profiles differed between the three main ST1193-H64 pulsotypes, with emergent pulsotype 2123 exhibiting the highest resistance prevalence for the most drugs, including (widely used) trimethoprim-sulfamethoxazole, which together with this pulsotype’s K1 capsule might have favored its epidemiologic success.
Our findings indicate a need for more extensive molecular epidemiological surveys and phylogenomic analyses of ST1193-H64. Such studies ideally would include isolates from diverse geographic locations, ecological contexts, hosts, and time periods; would incorporate epidemiological data regarding host characteristics and exposures; and would use whole-genome sequencing to more definitively resolve clonal sublineages and their accessory traits, as has been done for ST131-H30R (37–40, 48) and, preliminarily, ST1193-H64 and STc14 (49). Notably, within ST131-H30R, certain important subclones are associated with distinctive geographic locations and ESBL variants (37, 50, 51); whether this also occurs within ST1193-H64 awaits assessment.
The needed studies should be facilitated by our novel ST1193-H64 PCR assay, which was highly sensitive and specific. The assay detects both ST1193 per se and its distinctive fimH64 allele; this doubly confirms ST1193 status and screens for possible new molecular variants at either locus. Currently, fimH64 is the only fimH allele known to be associated with ST1193 (8), making its detection seemingly redundant. If, however, new ST1193 sublineages should emerge that carry alternate fimH alleles, as has occurred within ST131 (36), the fimH64 allele may acquire sub-ST taxonomic significance analogous to that of ST131’s fimH30 allele (27, 48).
Both lactose negativity and type O75 proved to be highly sensitive markers for ST1193-H64 but are known to lack specificity. That is, multiple other E. coli lineages are lactose negative, including ST648 (52), certain O78:H10 strains within ST10 (53), certain O15:K52:H1 strains within STc31 (54), and (as shown here) many non-ST1193 STc14 strains. Likewise, most non-ST1193 STc14 isolates are type O75. An important implication of ST1193-H64’s lactose negativity is that reliance on the lactose phenotype to identify E. coli will predictably create bias against detection of ST1193-H64.
Study limitations include the scant clinical and epidemiological data, focus on veterans and their household members, narrow geographical scope, post hoc (opportunistic) selection of time points for trend analyses, and absence of whole-genome data. Study strengths include the large sample size, inclusion of both fecal and clinical isolates from multiple institutions and diverse locales from 2008 through November 2018, longitudinal analysis of prevalence trends, extensive molecular and phenotypic typing, and development and validation of a novel ST1193-H64 PCR assay, which should facilitate future epidemiologic analyses.
In summary, we documented recent rise-and-fall prevalence trends for ST1193-H64 among clinical and fecal isolates from the MVAMC, detected ST1193-H64 at multiple other VAMCs and non-VA centers in diverse U.S. locales, showed genomic commonality between these isolates and archival Australian ST1193-H64 isolates, and identified differences in resistance and virulence gene profiles between ST1193-H64 and other STc14 isolates—and between the main ST1193-H64 pulsotypes—that might underlie clonal expansion and be clinically relevant. We also devised and extensively validated a rapid and simple multiplex PCR assay for ST1193-H64. Our findings suggest a need for additional studies of the molecular epidemiology, prevalence trends, and ecology of ST1193-H64, which our novel PCR assay should facilitate.
Supplementary Material
ACKNOWLEDGMENTS
We thank the participating clinical microbiology laboratories and study subjects. Veronika Tchesnokova provided a library of numbered fumC and fimH alleles to facilitate CH typing and information regarding the correspondence of CH types with sequence types.
This work was supported in part by the Office of Research and Development, Department of Veterans Affairs, grants 1 I01 CX000920-01 and 2I01CX000920-04 (to J.R.J.); the National Institute of Allergy and Infectious Diseases of the NIH (Antibacterial Resistance Leadership Group, award number UM1AI104681) (to J.R.J.); and Australian Research Council Linkage grant LP130100736 (to D.J.T. and J.M.).
J.R.J. has received grants or consultancies from Achaogen, Allergan, Janssen/Crucell, Melinta, Merck, Shionogi, and Tetraphase and has a patent application for tests to detect specific E. coli strains. We report no other financial conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01664-18.
REFERENCES
- 1.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 2.Review on Antimicrobial Resistance. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Wellcome Trust, London, United Kingdom: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf. [Google Scholar]
- 3.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 4.Croxall G, Hale J, Weston V, Manning G, Cheetham P, Achtman M, McNally A. 2011. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J Antimicrob Chemother 66:2501–2508. doi: 10.1093/jac/dkr349. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JR, Porter S, Thuras P, Castanheira M. 2017. The pandemic H30 subclone of sequence type 131 (ST131) is the leading cause of multidrug-resistant Escherichia coli infections in the United States (2011–2012). Open Forum Infect Dis 4:ofx089. doi: 10.1093/ofid/ofx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine M, Bertrand X, Madec J-Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platell J, Trott D, Johnson J, Heisig P, Heisig A, Clabots C, Johnston B, Cobbold R. 2012. Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob Agents Chemother 56:3898–3904. doi: 10.1128/AAC.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tchesnokova VL, Rechkina E, Larson L, Ferrier K, Weaver JL, Schroeder DW, She R, Butler-Wu SM, Aguero-Rosenfeld ME, Zerr D, Fang FC, Ralston J, Riddell K, Scholes D, Weissman S, Parker K, Spellberg B, Johnson JR, Sokurenko EV. 2019. Rapid and extensive expansion in the U.S. of a new multidrug-resistant Escherichia coli clonal group, sequence type ST1193. Clin Infect Dis 68:334–337. doi: 10.1093/cid/ciy525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia L, Liu Y, Xia S, Kudinha T, Xiao SN, Zhong NS, Ren GS, Zhuo C. 2017. Prevalence of ST1193 clone and IncI1/ST16 plasmid in E. coli isolates carrying blaCTX-M-55 gene from urinary tract infections patients in China. Sci Rep 7:44866. doi: 10.1038/srep44866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Lan F, Lu Y, He Q, Li B. 2017. Molecular characteristics of ST1193 clone among phylogenetic group B2 non-ST131 fluoroquinolone-resistant Escherichia coli. Front Microbiol 8:2294. doi: 10.3389/fmicb.2017.02294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YS, Oh T, Nam YS, Cho SY, Lee HJ. 2017. Prevalence of ST131 and ST1193 among bloodstream isolates of Escherichia coli not susceptible to ciprofloxacin in a tertiary care university hospital in Korea, 2013-2014. Clin Lab 63:1541–1543. doi: 10.7754/Clin.Lab.2017.170319. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen SB, Sunde M, Fladberg OA, Leegaard TM, Berg ES, Steinbakk M. 2017. Fluoroquinolone resistant Escherichia coli ST1193—another successful clone? ECCMID, ESCMID, Madrid, Spain. [Google Scholar]
- 13.Valenza G, Werner M, Eisenberger D, Nickel S, Lehner-Reindl V, Höller C, Bogdan C. 22 January 2019. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J Glob Antimicrob Resist doi: 10.1016/j.jgar.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JR, Davis G, Clabots C, Johnston BD, Porter S, Debroy C, Pomputius W, Ender PT, Cooperstock M, Slater BS, Banerjee R, Miller S, Kisiela D, Sokurenko E, Aziz M, Price LB. 2016. Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole genome sequence analysis. Open Forum Infect Dis 3:ofw129. doi: 10.1093/ofid/ofw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overdevest I, Haverkate M, Veenemans J, Hendriks Y, Verhulst C, Mulders A, Couprie W, Bootsma M, Johnson J, Kluytmans J. 2016. Prolonged colonization with Escherichia coli O25:ST131 versus other extended-spectrum β-lactamase-producing E. coli in a long-term care facility with a high endemic level of rectal colonization, the Netherlands, 2013–2014. Eurosurveillance 21 doi: 10.2807/1560-7917.ES.2016.2821.2842.30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drekonja DM, Kuskowski MA, Anway R, Johnston BD, Johnson JR. 2016. The niche for Escherichia coli sequence type 131 among veterans: urinary tract abnormalities and long-term care facilities. Open Forum Infect Dis 3:ofw138. doi: 10.1093/ofid/ofw138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators. 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. 2013. Escherichia coli sequence type ST131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol 34:361–369. doi: 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drawz S, Porter S, Kuskowski M, Johnston B, Clabots C, Kline S, Ferrieri P, Johnson J. 2015. Variation in resistance traits, phylogenetic background, and virulence genotypes among Escherichia coli clinical isolates from adjacent hospital campuses serving distinct patient populations. Antimicrob Agents Chemother 59:5331–5339. doi: 10.1128/AAC.00048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clermont O, Gordon D, Denamur E. 2015. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 161:980–988. doi: 10.1099/mic.0.000063. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J, Johnston B, Porter S, Thuras P, Aziz M, Price LB. 2019. Accessory traits and phylogenetic background predict Escherichia coli extraintestinal virulence better than does ecological source. J Infect Dis 219:121–132. doi: 10.1093/infdis/jiy459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 23.Clermont O, Christenson JK, Daubie A, Gordon DM, Denamur E. 2014. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J Microbiol Methods 101:24–27. doi: 10.1016/j.mimet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JR, Porter S, Thuras P, Castanheira M. 2017. Epidemic emergence in the United States of Escherichia coli sequence type 131-H30, 2000 to 2009. Antimicrob Agents Chemother 61:e00732-17. doi: 10.1128/AAC.00732-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn Microbiol Infect Dis 57:129–136. doi: 10.1016/j.diagmicrobio.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manges AR, Mende K, Murray CK, Johnston BD, Sokurenko E, Tchesnokova V, Johnson JR. 2017. Clonal distribution and associated characteristics of Escherichia coli clinical and surveillance isolates from a military medical center. Diagn Microbiol Infect Dis 87:382–385. doi: 10.1016/j.diagmicrobio.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 30.Johnson JR, Nicolas-Chanoine M-H, DebRoy C, Castanheira M, Robiscek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA, the MASTER Investigators. 2012. Comparison of Escherichia coli sequence type ST131 pulsotypes by epidemiologic traits, 1967-2009. Emerg Infect Dis 18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. 2018. A genomic overview of the population structure of Salmonella. PLoS Genet 14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchesnokova V, Riddell K, Scholes D, Johnson J, Sokurenko E. 2019. The uropathogenic Escherichia coli subclone ST131-H30 is responsible for most antibiotic prescription errors at an urgent care clinic. Clin Infect Dis 68:781–787. doi: 10.1093/cid/ciy523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2017. M100-S27: Performance standards for antimicrobial susceptibility testing. 27th informational supplement document. CLSI, Wayne, PA. [Google Scholar]
- 34.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4:80–128. doi: 10.1128/CMR.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowrouzian F, Adlerberth I, Wold AE. 2001. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol Infect 126:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddel K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine M, Debroy C, Robicsek A, Hansen G, Urban C, Platell JL, Trott DJ, Zhanel G, Weissman SJ, Cookson B, Fang F, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko E. 2013. Abrupt emergence of a single dominant multi-drug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoesser N, Sheppard AE, Pankhurst L, de Maio N, Moore C, Sebra R, Turner P, Anson L, Kasarkis A, Batty E, Kos V, Wilson D, Phetsouvanh R, Wyllie D, Sokurenko E, Manges A, Johnson TJ, Price LB, Peto T, Johnson J, Didelot X, Walker AS, Crook D, the Modernizing Medical Microbiology Informatics Group (MMMIG). 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan M-D, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JDD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben Zakour N, Alsheich-Hussain A, Ashcroft M, Nhu N, Roberts L, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNally A, Oren Y, Kelly D, Pascoe B, Dunn S, Sreecharan T, Vehkala M, Välimäki N, Prentice M, Ashour A, Avram O, Pupko T, Dobrindt U, Literak I, Guenther S, Schaufler K, Wieler LH, Zhiyong Z, Sheppard SK, McInerney JO, Corander J. 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12:e1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith SP, Manges AR, Riley LW. 2008. Temporal changes in the prevalence of community-acquired antimicrobial-resistant urinary tract infection affected by Escherichia coli clonal group composition. Clin Infect Dis 46:689–695. doi: 10.1086/527386. [DOI] [PubMed] [Google Scholar]
- 42.O’Neill PM, Talboys CA, Roberts AP, Azadian BS. 1990. The rise and fall of Escherichia coli O15 in a London teaching hospital. J Med Microbiol 33:23–27. doi: 10.1099/00222615-33-1-23. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 157:1127–1129. doi: 10.1016/S0022-5347(01)65154-1. [DOI] [PubMed] [Google Scholar]
- 44.Grüneberg RN. 1969. Relationship of infecting urinary organism to the faecal flora in patients with symptomatic urinary infection. Lancet i:766–768. doi: 10.1016/S0140-6736(69)90478-4. [DOI] [PubMed] [Google Scholar]
- 45.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Escherichia coli sequence type 131 (ST131) as a prominent cause of antimicrobial resistance among clinical and fecal E. coli isolates from reproductive-age women. J Clin Microbiol 51:3270–3276. doi: 10.1128/JCM.01315-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin Microbiol Infect 19:E173–E180. doi: 10.1111/1469-0691.12123. [DOI] [PubMed] [Google Scholar]
- 47.Kudinha T, Johnson JR, Andrew SC, Kong F, Anderson P, Gilbert GL. 2013. Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr Infect Dis J 32:543–548. doi: 10.1097/INF.0b013e31828ba3f1. [DOI] [PubMed] [Google Scholar]
- 48.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of ESBL-producing Escherichia coli ST131 is driven by a single highly virulent subclone, H30-Rx. mBio 6:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson TJ, Elnekave E, Miller EA, Monoz-Aguayo J, Flores-Figueroa C, Johnston B, Nielson DW, Logue CM, Johnson JR. 2019. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. mBio 63:e01913-18. doi: 10.1128/AAC.01913-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumura Y, Pitout JDD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradford PA, Motyl MR, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka K, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 52.Johnson JR, Johnston BD, Gordon DM. 2017. Rapid and specific detection of the Escherichia coli sequence type 648 complex within phylogroup F. J Clin Microbiol 55:1116–1121. doi: 10.1128/JCM.01949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olesen B, Scheutz F, Andersen RL, Menard M, Boisen N, Johnston B, Hansen DS, Krogfelt KA, Nataro JP, Johnson JR. 2012. Enteroaggregative Escherichia coli O78:H10, the cause of an outbreak of urinary tract infection. J Clin Microbiol 50:3703–3711. doi: 10.1128/JCM.01909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prats G, Navarro F, Mirelis B, Dalmau D, Margall N, Coll P, Stell A, Johnson JR. 2000. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J Clin Microbiol 38:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.