Human papillomaviruses (HPVs) are highly infectious pathogens that commonly infect the oropharynx and uterine cervix. The idea that posttranslational modifications of viral proteins coordinates viral genome replication is less explored. We recently discovered that fibroblast growth factor receptor 3 (FGFR3) phosphorylates the viral E2 protein. The current study demonstrates that FGFR3 phosphorylates E2 at tyrosine 138, which inhibits association with the C-terminal peptide of Brd4. This study illustrates a novel regulatory mechanism of virus-host interaction and provides insight into the role of Brd4 in viral replication.
KEYWORDS: Brd4, HPV, kinase, tyrosine phosphorylation, viral replication
ABSTRACT
The papillomavirus (PV) E2 protein coordinates viral transcription and genome replication. Following a strategy to identify amino acids in E2 that are posttranslationally modified, we reported that tyrosine kinase fibroblast growth factor receptor 3 (FGFR3) complexes with and phosphorylates E2, which inhibits viral DNA replication. Here, we present several lines of evidence indicating that tyrosine (Y) 138 of HPV-31 E2 is a substrate of FGFR3. The active form of FGFR3 bound to and phosphorylated the region of amino acids (aa) 107 to 175 in HPV-31 E2. The E2 phenylalanine (F) mutant Y138F displayed reduced FGFR3-induced phosphotyrosine. A constitutive kinase-active FGFR3 inhibited wild-type (WT) E2-induced E1-dependent DNA replication, while the 138F mutant retained activity. The tyrosine to glutamic acid (E) mutant Y138E, which can mimic phosphotyrosine, failed to induce transient DNA replication, although it maintained the ability to bind and localize the viral DNA helicase E1 to the viral origin. The bromodomain-containing protein 4 (Brd4) binds to E2 and is necessary for initiation of viral DNA synthesis. Interestingly, the Y138E protein coimmunoprecipitated with full-length Brd4 but was defective for association with its C-terminal domain (CTD). These results imply that the activity of the FGFR3 kinase in the infected epithelial cell restricts the HPV replication program through phosphorylation of E2 at Y138, which interferes with E2 binding to the Brd4 CTD, and that this interaction is required for initiation of viral DNA synthesis.
IMPORTANCE Human papillomaviruses (HPVs) are highly infectious pathogens that commonly infect the oropharynx and uterine cervix. The idea that posttranslational modifications of viral proteins coordinates viral genome replication is less explored. We recently discovered that fibroblast growth factor receptor 3 (FGFR3) phosphorylates the viral E2 protein. The current study demonstrates that FGFR3 phosphorylates E2 at tyrosine 138, which inhibits association with the C-terminal peptide of Brd4. This study illustrates a novel regulatory mechanism of virus-host interaction and provides insight into the role of Brd4 in viral replication.
INTRODUCTION
Human papillomaviruses (HPVs) are double-stranded DNA viruses that cause diseases ranging from benign genital warts to malignant tumors of cervical, oral, and anal epithelia. The circular genome of approximately 8 kb encodes six early (E) proteins and two late (L) proteins (1). HPVs rely on host replication proteins for viral DNA synthesis. During the initial phase of replication, which occurs during the infection of epithelial basal cells, the E1 and E2 proteins are expressed and establish and maintain a low copy number of viral episomes (2). During epithelial differentiation in HPV-positive cells, upper-level keratinocytes are induced to reenter the synthesis (S) phase, during which the viral genome copy number increases, a process termed amplification (3). The ∼400-amino acid (aa) HPV E2 proteins are known to exercise many functions during the viral life cycle, including regulation of viral transcription and genome replication and partitioning (4). The E2 protein encompasses two domains separated by a flexible serine- and arginine-rich hinge region, the N-terminal transcription activation domain (TAD), which binds to cellular proteins, and the C terminus, which mediates DNA binding and dimerization (DBD) (5, 6). Several posttranslational modifications (PTM) of E2, such as phosphorylation, acetylation, and sumoylation, have been described (7–11). The regulatory effects of these PTMs during the course of viral replication are not well understood.
The first tyrosine (Y) phosphorylation was identified at position 102 in the TAD of the bovine papillomavirus (BPV) E2 protein (7). By comparing wild-type (WT) E2 to phosphodeficient and phosphomimetic mutants, we demonstrated that this phosphorylation restricted E1-dependent replication, as well as transcriptional activation. Setting out to identify the kinase responsible for this PTM, we screened a number of cellular receptors, as well as nonreceptor tyrosine kinases, for coimmunoprecipitation (co-ip) with E2 irrespective of the residue they phosphorylate. This led to the identification of fibroblast growth receptor 3 (FGFR3) as a candidate E2 interaction partner. However, our results indicated that BPV-1 Y102 was unlikely to be the target of the FGFR3 kinase, since Y102F-mediated transient replication was still susceptible to FGFR3 inactivation (12). These findings inferred that another Y residue(s) in PV E2 was phosphorylated by FGFR3. FGFR3 is activated by extracellular fibroblast growth factor (FGF) and results in downstream signaling, leading to mitogenesis, accompanied by signal transducer and activator of transcription (STAT) and phosphatidylinositol 3-kinase activation (13). FGFR3 signaling is also implicated in differentiation and apoptosis (14). FGFR3 is overexpressed in cervical, oral, and oropharyngeal squamous cell carcinomas and is abundant in skin (15–17).
To locate the tyrosine residue that is phosphorylated by FGFR3, we employed site-directed mutagenesis approaches at multiple conserved tyrosine residues of HPV E2 to generate the nonphosphorylatable phenylalanine (F), as well as potential phosphomimetic glutamic acid (E) mutants, and analyzed these for effects on E1-dependent DNA replication. Our experiments indicate that constitutively active FGFR3 phosphorylates HPV-31 E2 at Y138. Moreover, the phosphomimetic E2 Y138E is defective for induction of DNA replication, and this may be attributable to its inability to bind the C-terminal domain (CTD) of Brd4, a global chromatin regulator, transcription cofactor, and regulator of viral replication (18).
RESULTS
FGFR3 phosphorylates HPV-31 E2 at Y138.
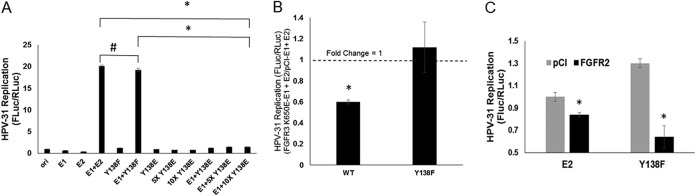
Previous experiments implied that FGFR3 phosphorylation of the PV E2 protein suppressed viral DNA replication (12). This initially led us to test the conserved tyrosine residues within the TAD of E2 as the substrates for phosphorylation in this assay. The tyrosine at position 138 in HPV-31 is conserved in many PV E2 proteins, including BPV-1, HPV-16, and HPV-18. We mutated HPV-31 E2 Y138 to the nonphosphorylatable residue phenylalanine (Y138F) or the phosphomimetic mutant glutamate (Y138E). Transient cotransfections of HPV-31 E1 and E2 together with an HPV ori-luciferase plasmid replicon (19) into C33A cells demonstrated that the activities of E2 Y138F and the WT were equal (Fig. 1A). Luciferase expression remained at baseline upon cotransfection of E1 and HPV-31 E2 Y138E, even at ten times the amount of transfected Y138E plasmid. If FGFR3 kinase-phosphorylated E2 at Y138 caused this inhibition, then the nonphosphorylatable residue phenylalanine mutant E2 Y138F should not show decreased transient replication. As shown in Fig. 1B, E1-dependent ori replication with E2 Y138F was not susceptible to constitutively active FGFR3 K650E (13). In contrast, another E2 interaction partner, FGFR2, reduced transient replication with Y138F (Fig. 1C) (20). We previously reported that coexpression of the constitutively kinase-active FGFR3 K650E suppresses HPV-31 E1- and E2-mediated transient replication, so the Y138E result was consistent with this phenotype (12). These observations suggested that Y138 is the target of FGFR3.
FIG 1.
E2- and E1-dependent transient DNA replication. C33A cells were transfected with HPV-31 E1 and HPV-31 Y138 mutants, along with pFLORI31 and pRL constructs. After 72 h, cells were lysed, and firefly and Renilla luciferase levels were measured using Dual-Glo luciferase reagent. Firefly luciferase levels were normalized to Renilla luciferase levels. Values are expressed as means ± SEM (n = 8). (A) P < 0.0001 by 1-way analysis of variance (ANOVA); #, P = 0.045 by two-tailed t test; *, P < 0.0001 by two-tailed t test. (B) Coexpression of constitutive kinase-active FGFR3 K650E. FGFR3 K650E suppressed WT (fold change < 1) but not Y138F replication (fold change = 1). Values are expressed as means ± SEM (n = 8). *, P < 0.01 by two-tailed t test comparing FGFR3 K650E to controls. (C) Coexpression of FGFR2 in 0.5% fetal bovine serum (FBS). Values are expressed as means ± SEM (n = 8). * P < 0.001 by two-tailed t test compared to pCI control groups.
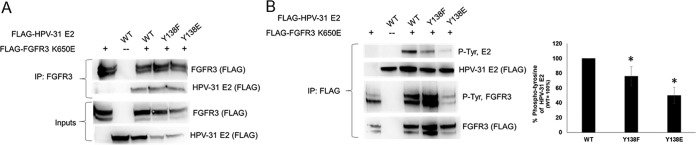
To further explore this possibility, HEK293TT cells were transfected with FLAG-HPV-31 E2 expression plasmids for WT, Y138F, and Y138E along with FLAG-FGFR3 K650E constructs. FGFR3 K650E coimmunoprecipitated with both WT and Y138 mutants (Fig. 2A). The phosphotyrosine signal was reduced in HPV-31 E2 Y138F (by 25%; Fig. 2B), suggesting that this residue is phosphorylated by FGFR3. Y138E had 50% reduced phosphotyrosine signal (Fig. 2B). We also observed decreased tyrosine phosphorylation on FGFR3, suggesting that Y138E may inhibit FGFR3 autophosphorylation.
FIG 2.
(A) HEK293TT cells were transfected with FLAG-FGFR3 K650E and FLAG-HPV-31 E2 mutants. Cells were lysed, and FLAG-FGFR3 K650E immunoprecipitated (IP) with FGFR3 antibodies. Complexes were blotted with FLAG antibodies. (B) HEK293TT cells were transfected with FLAG-FGFR3 K650E and FLAG-HPV-31 E2 mutants. Cells were lysed, and HPV-31 E2 immunoprecipitated with FLAG gel. Complexes were blotted with PY-1000 and FLAG antibodies. Densitometry from three independent experiments is shown. Phosphotyrosine E2 levels were normalized to total E2 levels.
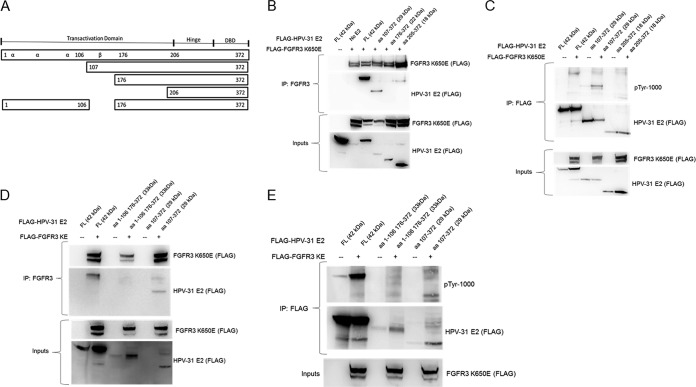
Next, we created a series of progressive N-terminal deletions of HPV-31 E2 fragments to identify the region of binding and phosphorylation by FGFR3 (Fig. 3A). The kinase-active form of FGFR3 bound the WT E2 in the region of aa 107 to 372 but did not bind aa 176 to 372 or the DBD aa 205 to 372 (Fig. 3B). These data coincide with the functional assays that suggested Y138 as the residue targeted by FGFR3. Immunoblots with phosphotyrosine antibody detected increased WT and aa 107 to 372 reactive proteins with coexpressed constitutively active FGFR3 (Fig. 3C). We also created an HPV-31 E2 truncated protein that expresses amino acids 1 to 106 and 176 to 372 to confirm that FGFR3 did not bind to these regions (Fig. 3D) and did not phosphorylate this truncated E2 protein (Fig. 3E). Nonetheless, tyrosine phosphorylation was not completely abolished, suggesting that FGFR3 phosphorylates HPV-31 E2 at Y138 and perhaps additional or alternative tyrosine residues within aa 107 to 175 of E2.
FIG 3.
(A) Illustration of E2 truncations and deletions. HEK293TT cells were transfected with FLAG-FGFR3 K650E and these FLAG-HPV-31 E2 constructs. (B) After 48 h, cells expressing aa 107 to 372 and aa 176 to 372 were treated with 20 μM MG132 for 6 h. Cells were lysed, and FLAG-FGFR3 K650E immunoprecipitated with FGFR3 antibodies. Complexes were blotted with FLAG antibodies. (C) FLAG-HPV-31 E2 and FLAG-FGFR3 K650E immunoprecipitated with FLAG antibodies. Complexes were blotted with PY-1000 and FLAG antibodies (D) FLAG-FGFR3 K650E was immunoprecipitated with FGFR3 antibodies. Complexes were blotted with FLAG antibodies. (E) FLAG-HPV-31 E2 and FLAG-FGFR3 K650E immunoprecipitated with FLAG antibodies. Complexes were blotted with PY-1000 and FLAG antibodies.
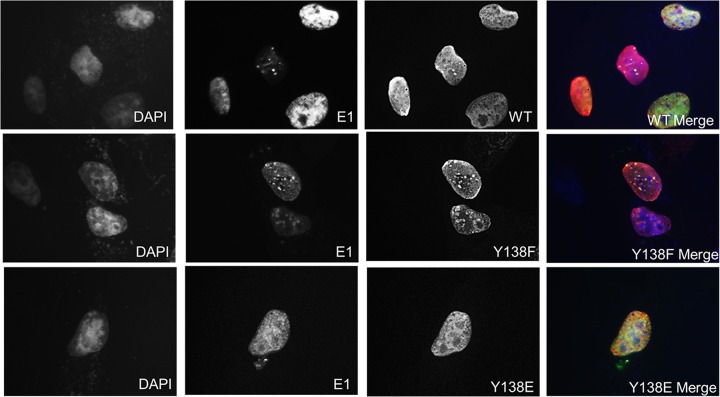
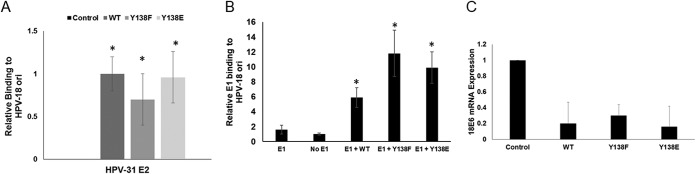
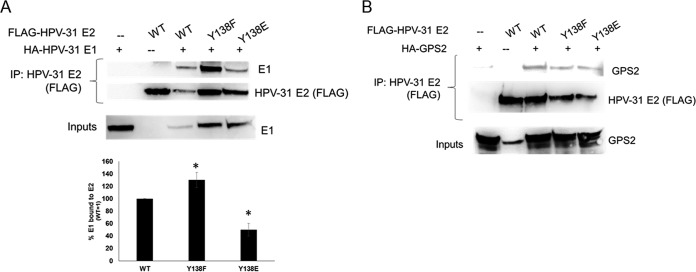
We next assessed the properties of E2 Y138 that might explain the inability to support E1-dependent PV DNA replication. We first tested nuclear localization. CV-1 cells were transfected with FLAG-HPV-31 E2 WT, Y138F, and Y138E constructs in the presence of HPV-31 E1 and the HPV-31 ori (Fig. 4). The HPV-31 E2 Y138 mutants were all localized to the nucleus; however, Y138E was not detected in discrete nuclear “replication” foci, in contrast to WT and Y138F E2 proteins. This led us to consider that the Y138 phospho-E2 protein might not be able to localize to the HPV-31 ori. E2 WT and Y138 mutants were transfected into HeLa cells, and chromatin immunoprecipitations (ChIP) demonstrated that the Y138F and Y138E mutants were present at the E2 palindromes present in the integrated HPV-18 genomes (Fig. 5A). We also performed E1 ChIPs and found that E2 WT, Y138F, and Y138E localized E1 to the viral origin (Fig. 5B). The expression of E2 represses the transcription of the HPV-18 E6 oncogene in HeLa cells, which have several hundred tandem copies of genomes integrated into cellular DNA at chromosomes 8 and 9 and abnormal chromosomes 5 and 22 (21). HeLa cells were transfected with HPV-31 E2 Y138 mutants, and HPV-18 E6 transcripts were measured by quantitative PCR (qPCR). As seen in Fig. 5C, Y138F and Y138E repressed transcription from the integrated viral genomes. Because chromatin accessibility and DNA binding activity remained intact, this implied that Y138E may be interfering with either viral or cellular replication/transcription partners. We therefore performed a series of co-ip experiments with E1, as well as with other E2 binding partners. HEK293TT cells were transfected with HPV-31 E2 Y138 mutants and HPV-31 E1 constructs. Mutation of tyrosine Y138 did not affect the ability to bind the viral helicase HPV-31 E1 (Fig. 6A). However, Y138E bound E1 at a lower efficiency (50%) and Y138F bound E1 at a higher efficiency (120%) than that of the WT. We previously reported that BPV-1 E2 Y138H bound to BPV-1 E1 but had reduced binding to GPS2 (AMF-1), which decreased the replication and transcriptional abilities of BPV-1 (22). To test GPS2 binding, HEK293TT cells were transfected with HPV-31 E2 Y138 mutants and GPS2 constructs. HPV-31 Y138F and HPV-31 Y138E were both able to coimmunoprecipitate GPS2 (Fig. 6B).
FIG 4.
E2 Y138 mutants are nuclear, but Y138E do not form replication foci. CV-1 cells were transfected with FLAG-HPV-31 E2 Y138 mutants, HA-HPV-31 E1, and HPV-31 ori. Immunofluorescence staining was carried out with FLAG (red) and E1 (green) antibodies at 100×; 4′,6-diamidino-2-phenylindole (DAPI; blue) was used to stain the nuclei.
FIG 5.
(A) HeLa cells were transfected with FLAG-WT and E2 Y138 mutants. After 48 h, ChIP assays were performed using the ChIP-IT express chromatin immunoprecipitation kit (Active Motif) with FLAG antibodies. HPV-18 LCR DNA was amplified using RT-PCR, and values were normalized to those of input DNA. Control = no FLAG E2 DNA; WT = 1. Values are expressed as means ± SEM. *, P ≤ 0.05 by two-tailed t test compared to control. (B) HeLa cells were transfected with FLAG-HPV-31 E1 and FLAG-WT or Y138 E2 mutants. After 48 h, the ChIP assays were performed with rat anti-E1 antibodies. HPV-18 LCR DNA was amplified using RT-PCR, and values were normalized to those of input DNA. No E1 = 1. Values are expressed as means ± SEM. *, P ≤ 0.05 by two-tailed t test compared to no E1 group. No statistically significant difference was found between WT and Y138 mutant groups. E1 only (n = 6), no E1 (n = 12), E1+ WT (n = 11), E1 + Y138F (n = 12), and E1 + Y138E (n = 12). (C) HeLa cells were transfected with WT and Y138 E2 mutants. After 48 h, RNA was isolated and cDNA was synthesized. Real-time PCR was used to measure HPV-18 E6 cDNA and normalized to actin cDNA.
FIG 6.
Y138E binds HPV-31 E1 and GPS-2. (A) HEK293TT cells were transfected with HA-HPV-31 E1 and FLAG-HPV-31 E2 Y138 mutants. FLAG pulldown was completed with M2 gel and immunoblotted with 16E1 and FLAG antibodies. Densitometry for four independent experiments is shown. E1 levels were normalized to E2 levels. (B) HEK293TT cells were transfected with HA-GPS2 and FLAG-HPV-31 E2 Y138 mutants. FLAG pulldown was completed with M2 gel and immunoblotted with GPS-2, HPV-16 E1, and FLAG antibodies.
HPV-31 E2 Y138E does not bind the C-terminal domain of Brd4.
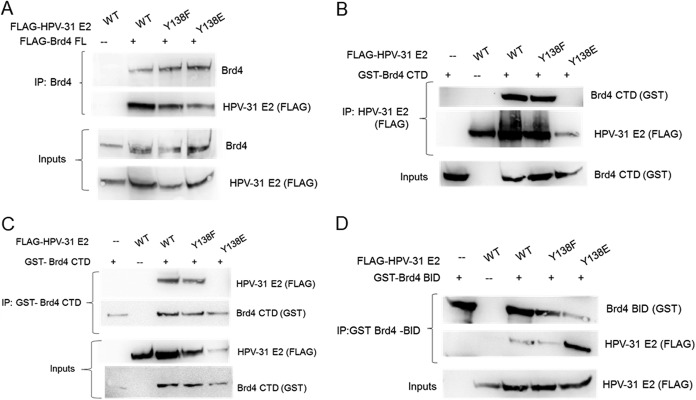
The E2 binding partner Brd4 has been previously shown to regulate several phases of the viral life cycle through binding of its C-terminal domain (CTD; aa 1222 to 1362) to the E2 transactivation domain (18, 23–28). The E2 DNA binding domain has also been reported to bind a basic residue-enriched interaction domain (BID) within amino acids 524 to 579 of Brd4 and to bind to the phosphorylation-dependent interaction domain (PDID) formed by amino acids 287 to 530 (18). The PDID region contains N-terminal CK2 phosphorylation sites (NPS) that mediate Brd4 binding to high-risk E2 proteins (18). Full-length Brd4 protein effectively coimmunoprecipitated with both HPV-31 E2 Y138F and Y138E (Fig. 7A). As expected, the Brd4-BID fragment bound Y138F and Y138E in co-ip experiments (Fig. 7D). However, while Y138F was able to bind to the CTD, Y138E did not (Fig. 7B and C). Previous studies observed that expression of the CTD increased the stability of HPV E2 proteins (26, 29). In the presence of the Brd4 CTD, we consistently observed decreased Y138E protein levels compared to those of WT and Y138F E2 proteins.
FIG 7.
Y138E does not bind to the Brd4 C-terminal domain (CTD). (A) HEK293TT cells were transfected with FLAG-Brd4 full-length and FLAG-HPV-31 E2 Y138 mutants. Brd4 pulldown with Brd4 antibodies and immunoblotted with Brd4 and FLAG antibodies. (B) HEK293TT cells were transfected with glutathione S-transferase (GST)-CTD and FLAG-HPV-31 E2 Y138 mutants. FLAG pulldown with M2 gel and immunoblotted with GST and FLAG antibodies. (C) HEK293TT cells were transfected with GST-CTD and FLAG-HPV-31 E2 Y138 mutants. GST pulldown with glutathione Sepharose beads and immunoblotted with GST and FLAG antibodies. (D) HEK293TT cells were transfected with GST-BID and FLAG-HPV-31 E2 Y138 mutants. GST pulldown with glutathione Sepharose beads and immunoblotted with GST and FLAG antibodies.
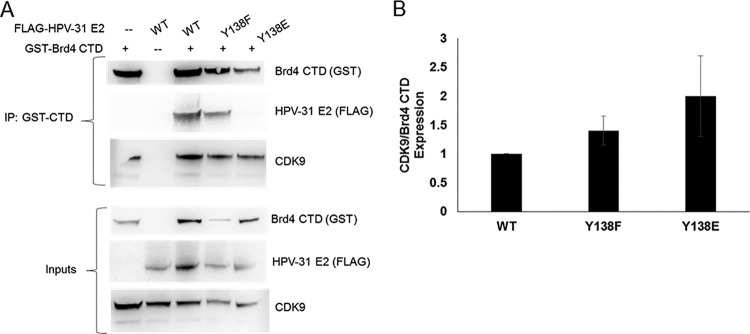
These experiments implied that the interaction of E2 with the CTD was necessary for induction of viral replication. The Brd4-CTD is known to interact with P-TEFb (30). P-TEFb is a transcriptional complex composed of CDK9 and cyclin T1 that stimulates RNA polymerase (pol) II processivity (31). As E2 and CDK9 are known to compete for Brd4-CTD binding (32), we hypothesized that since Y138E is unable to bind the Brd4-CTD, more CDK9 might remain in complex with Brd4-CTD in comparison to WT or Y138F. To test this, HEK293TT cells were transfected with E2 and the Y138 mutants along with the Brd4-CTD. The Brd4-CTD pulldown showed association with WT and Y138F proteins but not with Y138E (Fig. 8A). CDK9 association with the CTD was quantified and normalized to CTD immunoprecipitation levels, since E2-CTD binding increases the stability of these proteins (26, 29). Consistent with our hypothesis, CDK9 binding to the Brd4-CTD increased about 2-fold following Y138E overexpression and compared to that of WT E2 (Fig. 8B).
FIG 8.
The Brd4 CTD increased binding to CDK9 in the presence of Y138E. (A) HEK293TT cells were transfected with GST-CTD and FLAG-HPV-31 E2 Y138 mutants. GST pull down with glutathione Sepharose beads and immunoblotted with GST, FLAG, and CDK9 antibodies. (B) Western blot images were quantified for CDK9 and Brd4 CTD expression. Values are expressed as means ± SEM, n = 3. CDK9 expression was normalized to Brd4 CTD expression for coimmunoprecipitation, WT = 1.
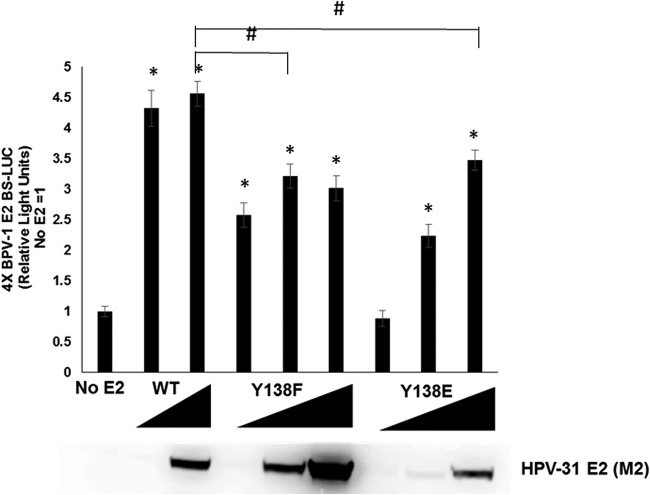
HPV E2 protein can act as a transcriptional activator or repressor, which is in part dependent on the context of the positions of E2 binding sites relative to the transcriptional initiation sites of the promoter. We therefore questioned whether loss of CTD association affected the transcriptional properties of E2 Y138E. Displacement of P-TEFb from the CTD is required for the processivity of RNA pol II during transcription (33). To test the transcriptional activity of the Y138 mutants, C33A cells were transfected with a reporter construct that contains four canonical E2 binding sites upstream of the luciferase gene (pGL2-E2BS-Luc) with various concentrations of the Y138 mutants. Both Y138F and Y138E activated transcription severalfold but not quite to levels comparable with those of WT E2 (Fig. 9). This implies that association with Brd4, but not the CTD itself, is necessary for partial E2 transcriptional activity.
FIG 9.
Y138E has impaired transcriptional activity. C33A cells were transfected with WT (10 and 100 ng), Y138F (10, 100, and 200 ng), or Y138E (10, 100, and 400 ng) and the pGL2-E2BS-Luc reporter plasmid. After 48 h, cells were lysed and firefly luciferase levels measured using Dual-Glo luciferase reagent. Values are expressed as means ± SEM (n = 8). *, P ≤ 0.01 by two-tailed t test when compared to the no-E2 group. #, P ≤ 0.01 by two-tailed t test compared to E2 WT.
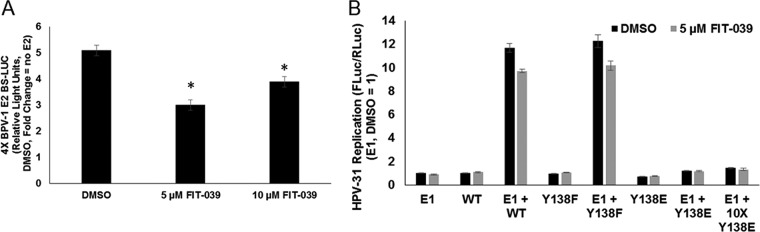
Since we observed that the Brd4-CTD bound more CDK9 in the presence of Y138E and that E2 Y138E is defective for induction of E1-dependent DNA replication, we wondered whether CDK9 release from Brd4 CTD prevents origin firing. In this model, E2 would bind the CTD and release P-TEFb, allowing the CDK9 kinase to not only activate RNA polymerase but inhibit DNA polymerase, thereby preventing collision of the transcription and DNA replication complexes (34). We therefore questioned whether a CDK9 inhibitor might restore the replication function of E2 Y138E. FIT-039 binds to the ATP-binding pocket of CDK9 and inhibits the kinase activity of P-TEFb (35). FIT-039 was reported to repress HPV replication and transcription in human neonatal foreskin keratinocytes (HFK) raft cultures maintaining HPV-18 genomes (36). C33A cells were treated with 5 μM or 10 μM FIT-039 for 48 h, and transcriptional activity of HPV-31 E2 was measured using the four-E2-binding-site luciferase reporter. As expected, 5 and 10 μM FIT-039 inhibited HPV-31 transcriptional activity without any notable cell toxicity, confirming that the CDK9 inhibitor was active at these concentrations (Fig. 10A). C33A cells were transfected with HPV-31 E1, and the E2 Y138 mutants and transient replication was measured in the presence of 5 μM FIT-039 (Fig. 10B). E1-dependent replication was not rescued with expression of E2 Y138E in the presence of FIT-039, even at higher concentrations of Y138E, suggesting that CDK9/P-TEFb is not a repressor of HPV replication.
FIG 10.
FIT-039 impairs HPV-31 transcription but does not rescue Y138E replication. (A) C33A cells were transfected with control plasmid or HPV-31 E2 and the pGL2-E2BS-Luc reporter plasmid. After 6 h, 5 or 10 μM FIT-039 was added to the cells. After 42 h, cells were lysed and firefly luciferase levels were measured. Values are expressed as fold change compared to no E2. Values are expressed as means ± SEM (n = 8). P < 0.0001 by 1-way ANOVA. *, P < 0.0001 by two-tailed t test compared to dimethyl sulfoxide (DMSO). (B) C33A cells were transfected with HPV-31 E1 and HPV-31 Y138 mutants, along with pFLORI31 and pRL constructs. After 6 h, 5 μM FIT-039 was added, and after an additional 42 h, cells were harvested and firefly and Renilla luciferase levels measured. Firefly luciferase levels were normalized to Renilla luciferase levels. Values are expressed as means ± SEM (n = 8).
DISCUSSION
The papillomavirus E2 protein is posttranslationally modified by the host cell to coordinate viral replication and transcription with the epithelial growth and differentiation program. We previously reported that phosphorylation of Y102 impaired viral replication and transcription in bovine papillomavirus (BBPV) E2 (7). Our subsequent publication revealed that FGFR3 induced phosphorylation of E2 and inhibited viral DNA replication; however, this outcome also occurred with Y102F, inferring that this tyrosine is not the target of this kinase (12). We therefore sought to identify the tyrosine residue(s) that is responsible for the inhibitory effects of FGFR3. The phosphodefective E2 mutant Y138F complexed with and showed decreased levels of tyrosine phosphorylation with kinase-activated FGFR3 and was not susceptible to its inhibitory effects on E1-dependent transient DNA replication. E2 Y138F coimmunoprecipitated with E1 and was observed to form nuclear foci.
In contrast, the phosphomimetic Y138E was defective in transient DNA replication assays. While the previously described BPV-1 Y102E mutant was impaired, HPV-31 E2 Y138E coimmunoprecipitated with the E1 DNA helicase about half as efficiently as the WT; however, recruitment of E1 to the viral ori, as observed by ChIP assays, was not impaired. Overexpression of E2 in HeLa cells represses transcription through occupancy of E2 binding sites within the p97 promoter of integrated HPV genomes. Y138E repressed E6 transcription in HeLa cells and, together with the ChIP data, confirmed that the Y138E mutant protein accesses chromatin and binds to its cognate DNA site. GPS2/AMF1 exerts its transcriptional control on E2 through recruitment of the p300 acetyltransferase (37). It has been previously shown that amino acids 134 to 216 of the BPV1 E2 TAD bind to GPS2. While BPV-1 E2 Y138H was impaired in GPS2 binding and transcriptional activation, the phenylalanine mutant was not, and the HPV-31 Y138F and Y138E mutants bound GPS2 to a similar extent as that of the WT (22). Therefore, the inability of E2 Y138E to stimulate DNA replication is not due to defective E1, GPS2, or DNA interactions.
Brd4 was reported to regulate HPV DNA replication, and can both activate and repress viral transcription (27). Brd4 colocalizes with E2 to tether the viral genome to chromosomes during mitotic segregation (24), which also involves the ChlR1 and TopBP1 proteins (38–40). Because of these multiple activities, the precise role of the Brd4-E2 interaction in the viral replicative program remains uncertain. Brd4 and TopBP1 colocalize with E1 and E2 in nuclear foci where viral genome replication occurs (39, 41). Immunofluorescence studies reveal that Brd4 dissociates from these foci after initiation (42). There are conflicting replication data using HPV-16 and HPV-31 E2 mutants that do not bind Brd4; however, these mutants have not been fully characterized and could be partially defective for Brd4 association or other interacting partners. During viral amplification, Brd4 is not detectable in these foci and may not be necessary for E1- and E2-dependent replication (42). Therefore, one prevailing concept is that the interaction between E2 and Brd4 is only required to initiate DNA replication (39, 42). Our experiments support the hypothesis that E2 interaction with the CTD is necessary for DNA replication and is regulated by Y138 phosphorylation.
Brd4 functions as a chromatin adaptor through its two bromodomains that bind preferentially to acetylated histones (43). The bromodomains also interact with the RFC-140 subunit of replication factor C and with signal-induced proliferation-associated protein 1 (SPA1) (44). The extra terminal (ET) domain interacts with NSD3, JMJD6, and GLTSCR1 to impart the transcriptional activation function (45). The CTD interacts with the cyclin T1 and CDK9 subunits of the P-TEFb complex, and this activated P-TEFb phosphorylates RNA pol II-CTD to pause-release transcription (33). Recently, Brd4 was found to interact with CDC6 in the DNA prereplication complex (46) and with treslin, a DNA replication initiation regulator (47). The DBD of high- and low-risk HPV E2 proteins bind the BID domain of Brd4, while the PDID domain of Brd4 associates only with the DBD of high-risk E2 (18). The interactions between high-risk E2 proteins and Brd4 BID are mediated by CK2 phosphorylation (26). In contrast, the TADs of HPV E2 proteins bind to the Brd4 CTD with variable efficiencies (48). In our coimmunoprecipitation experiments, full-length Brd4 and the BID region of Brd4 bound WT HPV-31 E2 and the Y138F and Y138E mutant proteins. This was expected, as mutational changes were not made to the DBD of E2. The Y138E mutant displayed a reduced association with the CTD, suggesting that phosphorylation at Y138 would abrogate Brd4 CTD interaction with the TAD. These results imply that E2 complex formation with Brd4 is needed for transient viral replication. In the available cocrystal structure, a 20-amino acid Brd4 C-terminal peptide forms a helix that binds across the three α-helices on the outer surface of the BPV E2 TAD, while E1 binds to the inner surface of E2 (49, 50). E2 Y138 is present in a β-strand adjacent to the α3 helix. A detailed structure of an extended Brd4 peptide in complex with E2 that depicts all the contacts between these proteins is necessary to fully understand their interaction.
Our findings are consistent with previous observations for Y138. In an earlier study that analyzed HPV-16 E2 TAD mutants, Y138 was replaced by alanine and was found to be moderately impaired for transcriptional activation (40% to 80% of WT), DNA replication (10% to 40% of WT), and E1 binding (40% to 80% of WT), and exhibited reduced Brd4-CTD association (51, 52). BPV Y138F bound E1, and although it could stimulate transcription and transient replication, it was not as active as the WT (22).
It was previously reported that papillomavirus E2 transcription functions require Brd4 binding to the TAD (52, 53). The presence of Brd4 at a promoter and its recruitment of P-TEFb is required for transcriptional elongation by RNA polymerase II. Several viral proteins regulate P-TEFb (31, 54). Recently, it was reported that the CDK9 inhibitor FIT-39 repressed HPV early promoter activity, resulting in reduction of E6 and E7 transcripts (36). From our experiments, we presume that both WT E2 and Y138F can compete with CDK9 for Brd4-CTD binding more favorably than Y138E, and we subsequently tested Y138E and Y138F for transcriptional activation. Interestingly, Y138E, although not completely defective, was significantly impaired and required higher levels of protein expression to reach transcriptional activity comparable to that of the WT. Unexpectedly, Y138F was also impaired, consistent with the notion that the replication and transcriptional activities of phosphorylated E2 are independent. Impaired transcriptional activity of Y138E may be explained by its inability to bind the CTD and release CDK9 to stimulate RNA polymerase processivity. Nonetheless, Y138E activated transcription, with high protein expression but not transient replication. Chemical inhibition of CDK9 kinase activity did not restore the replication function of Y138E; hence, it is unlikely that CDK9 is involved in phospho-E2-mediated restriction of the HPV replicative process.
There is recent evidence of a liquid-phase separation within the nucleus that contains transcriptional regulators and replication factors in high-concentration clusters using low-complexity disorder regions (LCDRs) in these proteins (55). Brd4 was reported to be present in such subnuclear foci (56). Interactions between tyrosine residues in LCDRs promote phase separation (57). We speculate that the HPV E2 protein may be present in these dynamic condensates and that the charge changes subsequent to tyrosine phosphorylation decrease its hydrophobicity and release E2 from such domains. The inability of Y138E to form nuclear foci may be attributable to impaired phase separation. This could explain a regulatory mechanism used by the virus to prevent overreplication.
MATERIALS AND METHODS
Plasmids and antibodies.
The following plasmids were used: pCDNA3-HA-HPV-31 E1 (HA, hemagglutinin), codon-optimized triple-FLAG-HPV-31 E1 (19), pSG5-HA-HPV-31 E2 (58), codon-optimized pCDNA3-FLAG-HPV-31 E2, pCDNA-FGFR3 K650E (D. Donoghue, UC San Diego), and pCDNA3-HA-AMF-1 (37). pcDNA3-F:hBrd4 FL, pCN-GST:hBrd4 CTD (amino acids 1224 to 1362), and pCN-GST:hBrd4 BID (amino acids 524 to 579) were previously described (48). FGFR2 and FLAG-FGFR3 K650E plasmids were provided by L. Thompson (UC Irvine). Reporter constructs pFLORI31 (ori firefly luciferase [FLuc]) and pRL (Renilla luciferase [RLuc]) were previously described (19). The following antibodies were used: rabbit-PY-1000 (Cell Signaling), rat anti-HPV-16 E1 (12), mouse anti-FLAG M2, mouse anti-β-actin (Sigma), rabbit anti-AMF-1 (37), rabbit anti-FGFR3 (Sigma), SD12 (rabbit anti-glutathione S-transferase [GST]), rabbit anti-CDK9 (Cell Signaling), and rabbit anti-BRD4 (Cell Signaling). Tyr-to-Phe (Y138F) and Tyr-to-Glu (Y138E) mutations were made in pSG5-HA-HPV-31 E2 and the pDCDNA3-FLAG-HPV-31 E2 constructs using the Quick-Change XL Lightning kit (Agilent Technologies). FLAG-HPV-31 E2 fragments (aa 107 to 372, aa 176 to 372, and aa 205 to 372) were amplified by PCR from codon-optimized HPV-31 E2, described above, with a FLAG tag and inserted into pCDNA3 with BamH1 and HindIII. For FLAG-HPV-31 E2 fragments 1 to 106 and 176 to 372, amino acids 1 to 106 were cloned into pCDNA3 using the BamH1 and HindIII restriction sites. The 176-to-372 fragment was cloned into this construct using the EcoR1 and ApaI restriction sites.
Cell culture.
HEK293TT (from J. Schiller and C. Buck), C33A (from D. Lowy), HeLa, and CV-1 were cultured in Dulbecco’s Modified Eagle Medium (Life Technologies) with 10% fetal bovine serum (Peak Serum) and penicillin-streptomycin (100 U/ml; Life Technologies). All cells were grown at 37̊°C and 5% CO2. FIT-039 was purchased from Aobious and diluted to 10 mM stock dimethyl sulfoxide (DMSO).
Coimmunoprecipitations and immunoblotting.
Cells were transfected with polyethylenimine (PEI; 2 mg/ml) at a ratio of 2 mg/μl PEI to 1 μg DNA. After 48 h, cells were lysed in 50 mM HEPES (pH 7.4), 300 mM NaCl, 1 mM Na3VO4, 0.5% NP-40, 1 mM dithiothreitol (DTT), and 1× protease inhibitor cocktail. M2 affinity gel (Sigma) or Glutathione Sepharose High Performance (GE Healthcare) was added to the lysates and rotated overnight at 4°C. Beads were washed three times with lysis buffer and once with high-salt buffer (50 mM HEPES [pH 7.4], 500 mM NaCl, 1 mM Na3VO4, 0.5% NP-40, 1 mM DTT, and 1× protease inhibitor cocktail). Proteins were removed from beads with 2× SDS lysis buffer, separated by 4 to 12% SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore), and detected by Western blotting using indicated primary antibodies.
Transient DNA replication assays.
C33A cells were seeded into a 96-well plate, and each well was transfected with 100 ng pCDNA3-FGFR3 K650E, 0.5 ng pRL (Rluc), 2.5 ng pFLORI31, 10 ng codon-optimized triple FLAG-HPV-31-E1 (19), and 10 ng pSG5-HA-HPV-31 E2 or 1 ng pCDNA3-HPV-31 E2 using lipofectamine 2000. After 72 h, cells were lysed and luciferase activity was measured using Dual Glo (Promega). Firefly luciferase levels were normalized to Renilla luciferase levels.
Chromatin immunoprecipitation.
ChIP assays were performed using a ChIP-IT Express chromatin immunoprecipitation kit (Active Motif). Real-time PCR was performed using SsoFast EvaGreen mastermix (Bio-Rad). Primers for the HPV-18 long control region (LCR) were forward (F), 5′-GGGACCGAAAACGGTGTAT-3′, and reverse (R), 5′-TTCCGTGCACAGTACAGGTA-3′. Results were analyzed using Bio-Rad CFX manager software.
Immunofluorescence.
CV-1 cells were transfected onto coverslips with 1 μg pCDNA3-FLAG-HPV-31 E2 constructs using lipofectamine (Life Technologies). After 24 h, coverslips were washed twice in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 15 min, and washed three times in PBS. Coverslips were placed in 10% goat serum in PBS-Triton X (PBS-T; 0.25%) for 30 min, followed by M2 antibody in PBS-T overnight; they were then washed three times with PBS, incubated in a 1:5,000 dilution of Alexa Fluor goat anti-mouse 594 (Invitrogen) for 1 h, washed three times in PBS, and mounted onto slides with Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI). Cells were analyzed with a Nikon microscope.
HeLa transcriptional repression assay.
HeLa cells were grown to 50% confluence and transfected with 10 μg FLAG-HPV 31 E2 (WT or mutants) with lipofectamine 2000. At 48 h posttransfection, cells were lysed in TRIzol, and RNA was isolated. RNA (1 μg) was used for cDNA synthesis using a SuperScript III first-strand synthesis system (Thermo Fisher Scientific). cDNA was analyzed by quantitative PCR for HPV18 E6 and actin using previously published primers (59).
Statistical analysis.
One-way analysis of variance (ANOVA) and two-tailed t tests were used for analysis. Means are expressed ± standard error of the mean (SEM). P values are indicated in figure legends. All experiments were performed at least 3 independent times.
ACKNOWLEDGMENTS
We appreciate the generosity of the following for plasmids: Alison McBride (NIAID/NIH) for the codon-optimized HPV-31 E2, Jacques Archambault (McGill University) for HPV-31 ori luciferase plasmids, Daniel Donoghue (University of California, San Diego) for pCDNA-FGFR3, Leslie Thompson (UC Irvine) for the FGFR2 and FLAG-FGFR3 constructs, and Cheng-Ming Chiang (University of Texas Southwestern Medical Center) for the Brd4 constructs.
Research discussed in this publication was supported by the National Cancer Institute (grant R01CA058376 to E.J.A.) and by the National Institute of Allergy and Infectious Diseases (grant T32AI060519 to M.D.).
The content is solely the responsibility of the authors and does not represent the official views of the NIH.
REFERENCES
- 1.Kajitani N, Satsuka A, Kawate A, Sakai H. 2012. Productive lifecycle of human papillomaviruses that depends upon squamous epithelial differentiation. Front Microbiol 3:152. doi: 10.3389/fmicb.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham SV. 2010. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol 5:1493–1506. doi: 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 4.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. 2015. Human papillomavirus molecular biology and disease association. Rev Med Virol 25:2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride AA. 2013. The papillomavirus E2 proteins. Virology 445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller M, Demeret C. 2012. The HPV E2-host protein-protein interactions: a complex hijacking of the cellular network. Tovj 6:173–189. doi: 10.2174/1874357901206010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culleton SP, Kanginakudru S, DeSmet M, Gilson T, Xie F, Wu S-Y, Chiang C-M, Qi G, Wang M, Androphy EJ. 2017. Phosphorylation of the bovine papillomavirus E2 protein on tyrosine regulates its transcription and replication functions. J Virology 91:e01854-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekhar V, McBride AA. 2012. Phosphorylation regulates binding of the human papillomavirus type 8 E2 protein to host chromosomes. J Virol 86:10047–10058. doi: 10.1128/JVI.01140-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y-C, Roark AA, Bian X-L, Wilson VG. 2008. Modification of papillomavirus E2 proteins by the small ubiquitin-like modifier family members (SUMOs). Virology 378:329–338. doi: 10.1016/j.virol.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinlan EJ, Culleton SP, Wu S-Y, Chiang C-M, Androphy EJ. 2013. Acetylation of conserved lysines in bovine papillomavirus E2 by p300. J Virol 87:1497–1507. doi: 10.1128/JVI.02771-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Poel S, Dreer M, Velic A, Macek B, Baskaran P, Iftner T, Stubenrauch F. 2018. Identification and functional characterization of phosphorylation sites of the human papillomavirus 31 E8̂E2 Protein. J Virology 92:e01743-17. doi: 10.1128/JVI.01743-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie F, DeSmet M, Kanginakudru S, Jose L, Culleton SP, Gilson T, Li C, Androphy EJ. 2017. Kinase activity of fibroblast growth factor receptor-3 regulates activity of the papillomavirus E2 protein. J Virology doi: 10.1128/jvi.01066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart KC, Robertson SC, Donoghue DJ. 2001. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, stat activation, and phosphatidylinositol 3-kinase activation. Mol Biol Cell 12:931–942. doi: 10.1091/mbc.12.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L'Hôte CGM, Knowles MA. 2005. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Experimental Cell Res 304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Choi CH, Chung J-Y, Kim J-H, Kim B-G, Hewitt SM. 2016. Expression of fibroblast growth factor receptor family members is associated with prognosis in early stage cervical cancer patients. J Transl Med 14:124. doi: 10.1186/s12967-016-0874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koole K, van Kempen PMW, Swartz JE, Peeters T, van Diest PJ, Koole R, van Es RJJ, Willems SM. 2016. Fibroblast growth factor receptor 3 protein is overexpressed in oral and oropharyngeal squamous cell carcinoma. Cancer Med 5:275–284. doi: 10.1002/cam4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F. 2010. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 18.Wu S-Y, Nin DS, Lee AY, Simanski S, Kodadek T, Chiang C-M. 2016. BRD4 phosphorylation regulates HPV E2-mediated viral transcription, origin replication, and cellular MMP-9 expression. Cell Rep 16:1733–1748. doi: 10.1016/j.celrep.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fradet-Turcotte A, Morin G, Lehoux M, Bullock PA, Archambault J. 2010. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 399:65–76. doi: 10.1016/j.virol.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSmet M, Kanginakudru S, Jose L, Xie F, Gilson T, Androphy EJ. 2018. Papillomavirus E2 protein is regulated by specific fibroblast growth factor receptors. Virology 521:62–68. doi: 10.1016/j.virol.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popescu NC, DiPaolo JA, Amsbaugh SC. 1987. Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet Cell Genet 44:58–62. doi: 10.1159/000132342. [DOI] [PubMed] [Google Scholar]
- 22.Breiding DE, Sverdrup F, Grossel MJ, Moscufo N, Boonchai W, Androphy EJ. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol Cell Biol 17:7208–7219. doi: 10.1128/MCB.17.12.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee A-Y, Chiang C-M. 2009. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J Biol Chem 284:2778–2786. doi: 10.1074/jbc.M805835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349–360. doi: 10.1016/S0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 25.You J, Schweiger M-R, Howley PM. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J Virol 79:14956–14961. doi: 10.1128/JVI.79.23.14956-14961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng G, Schweiger M-R, Martinez-Noel G, Zheng L, Smith JA, Harper JW, Howley PM. 2009. Brd4 regulation of papillomavirus protein E2 stability. J Virol 83:8683–8692. doi: 10.1128/JVI.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iftner T, Haedicke-Jarboui J, Wu S-Y, Chiang C-M. 2017. Involvement of Brd4 in different steps of the papillomavirus life cycle. Virus Res 231:76–82. doi: 10.1016/j.virusres.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinney CC, Kim MJ, Chen D, McBride AA. 2016. Brd4 activates early viral transcription upon human papillomavirus 18 infection of primary keratinocytes. mBio 7:e01644-16. doi: 10.1128/mBio.01644-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McPhillips MG, Ozato K, McBride AA. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J Virol 79:8920–8932. doi: 10.1128/JVI.79.14.8920-8932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A 104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Justyna Z, F IN, Shona M. 2016. P‐TEFb goes viral. Bioessays 38:S75–S85. doi: 10.1002/bies.201670912. [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Li Q, Lievens S, Tavernier J, You J. 2010. Abrogation of the Brd4-positive transcription elongation factor b complex by papillomavirus E2 protein contributes to viral oncogene repression. J Virol 84:76–87. doi: 10.1128/JVI.01647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen FX, Smith ER, Shilatifard A. 2018. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19:464–478. doi: 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y-L, Pasero P. 2017. Transcription-replication conflicts: orientation matters. Cell 170:603–604. doi: 10.1016/j.cell.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M, Onogi H, Kii I, Yoshida S, Iida K, Sakai H, Abe M, Tsubota T, Ito N, Hosoya T, Hagiwara M. 2014. CDK9 inhibitor FIT-039 prevents replication of multiple DNA viruses. J Clin Invest 124:3479–3488. doi: 10.1172/JCI73805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ajiro M, Sakai H, Onogi H, Yamamoto M, Sumi E, Sawada T, Nomura T, Kabashima K, Hosoya T, Hagiwara M. 2018. CDK9 inhibitor FIT-039 suppresses viral oncogenes E6 and E7 and has a therapeutic effect on HPV-induced neoplasia. Clinical Cancer Res doi: 10.1158/1078-0432.Ccr-17-3119. [DOI] [PubMed] [Google Scholar]
- 37.Peng Y-C, Kuo F, Breiding DE, Wang Y-F, Mansur CP, Androphy EJ. 2001. AMF1 (GPS2) modulates p53 transactivation. Mol Cell Biol 21:5913–5924. doi: 10.1128/MCB.21.17.5913-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris L, McFarlane-Majeed L, Campos-León K, Roberts S, Parish JL. 2017. The cellular DNA helicase ChlR1 regulates chromatin and nuclear matrix attachment of the human papillomavirus 16 E2 protein and high-copy-number viral genome establishment. J Virology 91:e01853-16. doi: 10.1128/JVI.01853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gauson EJ, Donaldson MM, Dornan ES, Wang X, Bristol M, Bodily JM, Morgan IM. 2015. Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2-mediated DNA replication. J Virol 89:4980–4991. doi: 10.1128/JVI.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parish JL, Bean AM, Park RB, Androphy EJ. 2006. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol Cell 24:867–876. doi: 10.1016/j.molcel.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Helfer CM, Pancholi N, Bradner JE, You J. 2013. Recruitment of Brd4 to the human papillomavirus type 16 DNA replication complex is essential for replication of viral DNA. J Virol 87:3871–3884. doi: 10.1128/JVI.03068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakakibara N, Chen D, Jang MK, Kang DW, Luecke HF, Wu S-Y, Chiang C-M, McBride AA. 2013. Brd4 is displaced from HPV replication factories as they expand and amplify viral DNA. PLoS Pathog 9:e1003777. doi: 10.1371/journal.ppat.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S-Y, Chiang C-M. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 45.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31:2641. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Dulak AM, Hattersley MM, Willis BS, Nikkilä J, Wang A, Lau A, Reimer C, Zinda M, Fawell SE, Mills GB, Chen H. 2018. BRD4 facilitates replication stress-induced DNA damage response. Oncogene 37:3763–3777. doi: 10.1038/s41388-018-0194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sansam CG, Pietrzak K, Majchrzycka B, Kerlin MA, Chen J, Rankin S, Sansam CL. 2018. A mechanism for epigenetic control of DNA replication. Genes Dev 32:224–229. doi: 10.1101/gad.306464.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S-Y, Lee AY, Lai H-T, Zhang H, Chiang C-M. 2013. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell 49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbate EA, Voitenleitner C, Botchan MR. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol Cell 24:877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Abbate EA, Berger JM, Botchan MR. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev 18:1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakai H, Yasugi T, Benson JD, Dowhanick JJ, Howley PM. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J Virol 70:1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schweiger M-R, You J, Howley PM. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J Virol 80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helfer C, Yan J, You J. 2014. The cellular bromodomain protein Brd4 has multiple functions in E2-mediated papillomavirus transcription activation. Viruses 6:3228. doi: 10.3390/v6083228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schröder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnölzer M, Verdin E, Zhou M-M, Ott M. 2012. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem 287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plys AJ, Kingston RE. 2018. Dynamic condensates activate transcription. Science 361:329. doi: 10.1126/science.aau4795. [DOI] [PubMed] [Google Scholar]
- 56.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA. 2018. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Powell MLC, Smith JA, Sowa ME, Harper JW, Iftner T, Stubenrauch F, Howley PM. 2010. NCoR1 mediates papillomavirus E8̂E2C transcriptional repression. J Virology 84:4451–4460. doi: 10.1128/JVI.02390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas Y, Androphy EJ. 2018. Human papillomavirus replication regulation by acetylation of a conserved lysine in the E2 protein. J Virology 92:e01912-17. doi: 10.1128/JVI.01912-17. [DOI] [PMC free article] [PubMed] [Google Scholar]