Several DNA viruses evolved mechanisms to inhibit the cellular DNA damage response (DDR), which acts as an antiviral defense system. We present a novel mechanism by which the adenovirus (Ad) E4orf4 protein inhibits the DDR. E4orf4 interacts with the DNA damage sensor DNA-PK in a biphasic manner. Early during infection, E4orf4 requires DNA-PK activity to inhibit various branches of the DDR, whereas it later inhibits DNA-PK itself. Furthermore, although both E4orf4 and DNA-PK are recruited to virus replication centers (RCs), DNA-PK is later distanced from late-phase RCs. Delayed DNA-PK inhibition greatly contributes to Ad replication efficiency. When E4orf4 is expressed alone, it is recruited to DNA damage sites. Inhibition of DNA-PK prevents both recruitment and the previously reported ability of E4orf4 to kill cancer cells. Our results support an important role for the E4orf4–DNA-PK interaction in Ad replication and in facilitation of E4orf4-induced cancer-selective cell death.
KEYWORDS: DNA damage response, DNA-PK, E4orf4, adenoviruses
ABSTRACT
The adenovirus (Ad) E4orf4 protein contributes to virus-induced inhibition of the DNA damage response (DDR) by reducing ATM and ATR signaling. Consequently, E4orf4 inhibits DNA repair and sensitizes transformed cells to killing by DNA-damaging drugs. Inhibition of ATM and ATR signaling contributes to the efficiency of virus replication and may provide one explanation for the cancer selectivity of cell death induced by the expression of E4orf4 alone. In this report, we investigate a direct interaction of E4orf4 with the DDR. We show that E4orf4 physically associates with the DNA-dependent protein kinase (DNA-PK), and we demonstrate a biphasic functional interaction between these proteins, wherein DNA-PK is required for ATM and ATR inhibition by E4orf4 earlier during infection but is inhibited by E4orf4 as infection progresses. This biphasic process is accompanied by initial augmentation and a later inhibition of DNA-PK autophosphorylation as well as by colocalization of DNA-PK with early Ad replication centers and distancing of DNA-PK from late replication centers. Moreover, inhibition of DNA-PK improves Ad replication more effectively when a DNA-PK inhibitor is added later rather than earlier during infection. When expressed alone, E4orf4 is recruited to DNA damage sites in a DNA-PK-dependent manner. DNA-PK inhibition reduces the ability of E4orf4 to induce cancer cell death, likely because E4orf4 is prevented from arriving at the damage sites and from inhibiting the DDR. Our results support an important role for the E4orf4–DNA-PK interaction in Ad replication and in facilitation of E4orf4-induced cancer-selective cell death.
IMPORTANCE Several DNA viruses evolved mechanisms to inhibit the cellular DNA damage response (DDR), which acts as an antiviral defense system. We present a novel mechanism by which the adenovirus (Ad) E4orf4 protein inhibits the DDR. E4orf4 interacts with the DNA damage sensor DNA-PK in a biphasic manner. Early during infection, E4orf4 requires DNA-PK activity to inhibit various branches of the DDR, whereas it later inhibits DNA-PK itself. Furthermore, although both E4orf4 and DNA-PK are recruited to virus replication centers (RCs), DNA-PK is later distanced from late-phase RCs. Delayed DNA-PK inhibition greatly contributes to Ad replication efficiency. When E4orf4 is expressed alone, it is recruited to DNA damage sites. Inhibition of DNA-PK prevents both recruitment and the previously reported ability of E4orf4 to kill cancer cells. Our results support an important role for the E4orf4–DNA-PK interaction in Ad replication and in facilitation of E4orf4-induced cancer-selective cell death.
INTRODUCTION
The detection and repair of DNA damage in cells are accomplished by the DNA damage response (DDR) network. DNA damage is recognized by sensor proteins, including poly(ADP-ribose) polymerase 1 (PARP-1) (1, 2), KU proteins (3), or the MRN complex, consisting of the Mre11, Rad50, and Nbs1 proteins (4–7). The sensors recruit proteins that transduce the signal to chromatin, to cellular checkpoints, and to the repair machinery, facilitating DNA repair (8). Signal transducers include the phosphatidylinositol 3-kinase-like protein kinase (PIKK) family members ataxia-telangiectasia mutated (ATM), ATM and Rad3related (ATR), and the DNA-dependent protein kinase (DNA-PK) (reviewed in references 9 and 10).
When double-strand breaks (DSBs) occur, KU proteins rapidly sense the damage and bind at the damage site, recruiting the kinase catalytic subunit DNA-PKcs within seconds, to create the DNA-PK holoenzyme at the DSB. Subsequent translocations at the DSB allow DNA-PKcs to interact directly with the DSB end and to be activated. The DNA-PK kinase activity is required for DNA repair through nonhomologous end joining (NHEJ) and was also implicated in repair through homologous recombination (HR) (reviewed in references 3 and 11). At the DSB, DNA-PKcs itself is phosphorylated at more than 40 sites, some of which are clustered to specific regions in the protein. The phosphorylation events at these sites include autophosphorylation as well as phosphorylation by other DDR kinases (12–15). DNA-PK activation is manifested by autophosphorylation at S2056 and possibly also T2609 (12, 16), although some reports indicate that T2609 may be phosphorylated by ATM and ATR as well (13, 15, 17, 18). Phosphorylation at S2056 and T2609 has been suggested to cause conformation changes that are required for DNA-PK disassembly from DSB sites and facilitate the ligation of DNA ends (reviewed in references 19 and 20). DNA-PK has several functions besides its contribution to DNA repair, including roles in transcriptional regulation, metabolism, innate immunity, and mitosis, among others (11, 20, 21).
Viral DNA products accumulating during infection with DNA viruses, including linear double-stranded viral genomes and replication intermediates, may be recognized by the cell as DNA damage and may consequently activate the DDR. Unhindered DDR activation may lead to “repair” of viral genome ends by their ligation and could result in inhibition of virus replication (22). To increase their replication efficiency, many DNA viruses, including adenoviruses (Ads), acquired mechanisms to attenuate the DDR (23, 24). On the other hand, several viruses also recruit activated DDR enzymes to aid in processes required for virus replication (23, 25). Inhibition of the DDR during Ad infection requires several Ad E4 gene products as well as the E1B-55K protein (early region 1B, 55 kDa) (26–29).
DNA-PK was reported to functionally interact with three Ad proteins. The E4orf3 and E4orf6 proteins associate with DNA-PK to inhibit viral genome concatenation (30), whereas the L4-33K protein, an alternative splicing factor, is phosphorylated and inhibited by DNA-PK (31). In the absence of DNA-PK, the L4-33K protein is able to shift more efficiently Ad mRNA splicing from early to late mRNA patterns.
The Ad E4orf4 protein performs several functions in the course of infection, which contribute to efficient progression of virus replication. Its functions include regulation of early viral gene expression, induction of alternative splicing of viral RNAs, and modulation of protein translation (32–39). Furthermore, E4orf4 was recently shown to contribute to inhibition of the cellular DDR by Ad, through effects on ATM and ATR signaling, thus increasing the efficiency of Ad replication (26). When E4orf4 is expressed alone, outside the context of virus infection, it initiates a nonclassical cell death process that is both p53 and caspase independent. However, this process can lead eventually to classical caspase-dependent apoptosis in various types of cells (40–43). The known mechanisms that underlie E4orf4-induced cell death are well conserved in evolution and have been shown to contribute to cell killing by E4orf4 in several organisms, including Saccharomyces cerevisiae, Drosophila melanogaster, and mammalian cells (44–48). Interestingly, E4orf4 was reported to kill cancer cells more efficiently than normal cells (49), evoking the possibility that studies of the underlying mechanisms may provide new avenues for cancer therapy. The cancer selectivity of E4orf4-induced cell death could result from several E4orf4 activities and will be better understood once knowledge of the underlying mechanisms is improved (39, 50). Inhibition of the DDR by E4orf4 may contribute to this cancer-selective cell death because DDR modules are defective in many types of cancer, making the tumor cells more dependent on residual DNA damage signaling and, thus, more sensitive to DDR inhibitors, including E4orf4.
Protein phosphatase 2A (PP2A) is an important protein partner of E4orf4 which contributes to several E4orf4 functions, including attenuation of the DDR (26, 39, 49–53). PP2A is a heterotrimeric enzyme containing a catalytic subunit (C), a scaffolding subunit (A), and one of several regulatory B subunits that belong to at least four unrelated gene families and which determine the specificity of the PP2A holoenzyme toward various substrates (54). Studies of the mechanisms underlying E4orf4 action have revealed that E4orf4 associates with regulatory B subunits of PP2A (51, 53). All three types of PP2A subunits (A, B, and C) are present in the E4orf4-PP2A complex, and this complex possesses substantial phosphatase activity (47, 51, 53).
In this work, we investigated one of the mechanisms by which E4orf4 directly affects the DDR via a physical association with the DNA damage sensor DNA-PK. The characterization of this interaction and its consequences for virus infection and E4orf4-induced cell death is described.
RESULTS
E4orf4 associates with DNA-PKcs and KU70.
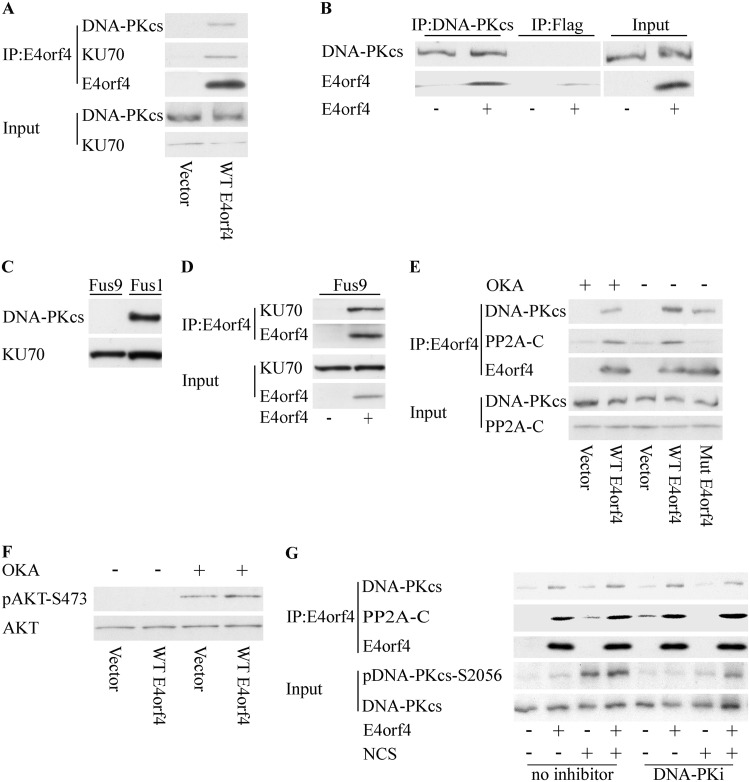
Mass spectrometry analysis of nuclear proteins that coimmunoprecipitated with the Ad E4orf4 protein suggested that E4orf4 associates with the DNA-PK catalytic subunit (DNA-PKcs) and with KU proteins. To validate this interaction, HCT116 cells expressing E4orf4 from a doxycycline (Dox)-inducible promoter were induced for 3 h by Dox to activate E4orf4 expression or were left untreated, and E4orf4 was immunoprecipitated from the cell extracts. As shown in Fig. 1A, DNA-PKcs was found in immune complexes precipitated from E4orf4-expressing cells but not in those from control cells. A reciprocal immunoprecipitation using a DNA-PKcs-specific antibody verified that E4orf4 associated with DNA-PKcs, and a parallel immunoprecipitation with a control isotype-matched antibody demonstrated the specificity of this assay (Fig. 1B). In addition, KU70, encoded by the XRCC5 gene, also associated with E4orf4 (Fig. 1A). To test whether the interaction with KU70 was DNA-PKcs dependent, a similar immunoprecipitation experiment was performed using extracts from MO59J-Fus9 cells lacking DNA-PK (Fig. 1C). Figure 1D demonstrates that E4orf4 could interact with KU70 in the absence of DNA-PKcs.
FIG 1.
E4orf4 associates with DNA-PKcs and KU70. (A) HCT116 cells were transfected with a plasmid expressing the tetracycline repressor together with a plasmid expressing doxycycline (Dox)-inducible E4orf4 or an empty vector. The cells were induced for 3 h by Dox to activate E4orf4 expression or were left untreated, and E4orf4 was immunoprecipitated (IP) from the cell extracts. Proteins from input lysates and immune complexes were separated on SDS gels and subjected to Western blot analysis with the indicated antibodies. A representative blot is shown. (B) Cell extracts as described above for panel A were subjected to immunoprecipitation with a DNA-PKcs-specific antibody or with a control, isotype-matched, Flag tag-specific antibody. Western blot analysis of input and immunoprecipitated proteins was performed, and a representative blot is shown. (C) MO59J-Fus9 and MO59J-Fus1 cell extracts were subjected to Western blot analysis with the indicated antibodies. (D) MO59J-Fus9 cells were transfected as described above for panel A, and cell extracts were subjected to immunoprecipitation with E4orf4-specific antibodies. A representative blot shows analysis with the indicated antibodies. (E) HCT116 cells were transfected with a plasmid expressing the tetracycline repressor together with plasmids expressing Dox-inducible WT-E4orf4 or the R81F84A mutant or an empty vector. The cells were induced for 3.5 h by Dox to activate E4orf4 expression or were left untreated. One set of cells was subjected to treatment with 0.4 μM okadaic acid for 1 h prior to harvest. A representative blot is shown. (F) HCT116 cells were treated as described above for panel E. Whole-cell extracts were prepared in SDS buffer, and a Western blot was stained with antibodies against pAkt-S473 and total Akt, which served as a loading control. (G) HCT116 cells were transfected with a plasmid expressing WT-E4orf4 from a Dox-inducible promoter or with an empty vector. The cells were induced with Dox for 3.5 h and treated with 0.5 ng/μl NCS or 0.01 mM the DNA-PK inhibitor NU7441 (DNA-PKi) for 1 h and 1.5 h prior to harvest, respectively. One set of cells was left untreated. Whole-cell extracts were prepared and subjected to immunoprecipitation with an E4orf4-specific antibody. The presence of DNA-PKcs and PP2A-C in the immune complexes or input lysates was determined by Western blot analysis using the specified antibodies, and a representative blot is shown.
Characterization of the interaction between E4orf4 and DNA-PKcs.
PP2A was reported to contribute to all E4orf4 functions known to date (reviewed in references 39 and 50), and we therefore examined whether PP2A contributed to the interaction observed between E4orf4 and DNA-PKcs. HCT116 cells inducibly expressing wild-type E4orf4 (WT-E4orf4) or the R81F84A E4orf4 mutant that does not bind PP2A (52) and cells containing the corresponding empty vectors were induced with Dox for 3 h, and protein extracts were subjected to immunoprecipitation with E4orf4-specific antibodies. Figure 1E demonstrates that DNA-PKcs was detected in immune complexes with both WT and mutant E4orf4 proteins, but the mutant protein interacted with DNA-PKcs slightly less efficiently than WT-E4orf4. To examine whether PP2A activity was required for the E4orf4–DNA-PKcs interaction, WT-E4orf4 was immunoprecipitated from cells that were treated with the phosphatase inhibitor okadaic acid for the last hour prior to harvest. As shown in Fig. 1E, treatment with okadaic acid reduced the levels of DNA-PKcs in the E4orf4 immune complexes but did not eliminate the interaction. To ascertain whether the okadaic acid concentrations used in the experiment were effective in reducing PP2A activity, phosphorylation of pAkt-S473, one of the known PP2A substrates, was examined. As shown in Fig. 1F, pAkt-S473 phosphorylation was strongly increased in the okadaic acid-treated cells, regardless of E4orf4 expression, indicating that the inhibitor worked well. The results suggest that PP2A activity improves the association of E4orf4 with DNA-PKcs, although it is not absolutely required to maintain it.
In addition to testing the contribution of PP2A to the E4orf4–DNA-PKcs interaction, we used two approaches to examine whether the activity of DNA-PKcs itself was required to facilitate this interaction. First, we examined whether DNA damage affected the E4orf4–DNA-PKcs association. E4orf4 expression was induced by Dox for 3 h in HCT116 cells, and the cells were subjected to treatment with the radiomimetic drug neocarzinostatin (NCS) for 1 h prior to harvest. NCS treatment resulted in activation of DNA-PK, manifested by increased phosphorylation of DNA-PKcs at the autophosphorylation site S2056, but the interaction with E4orf4 was not affected (Fig. 1G). In the second approach, we examined the interaction between E4orf4 and DNA-PKcs in the presence of a DNA-PK inhibitor. This inhibitor, NU7441, is a widely used, highly specific DNA-PK inhibitor, which is more than 1,000-fold less effective against ATM and ATR (55). As demonstrated in Fig. 1G, although the inhibitor efficiently reduced DNA-PKcs autophosphorylation at S2056, it did not affect the E4orf4–DNA-PKcs interaction. Therefore, DNA-PK activity is not required for the association of this enzyme with E4orf4.
Inhibition of DNA-PK activity obstructs the ability of E4orf4 to reduce virus- or DNA damage-induced phosphorylation of ATM and Chk1.
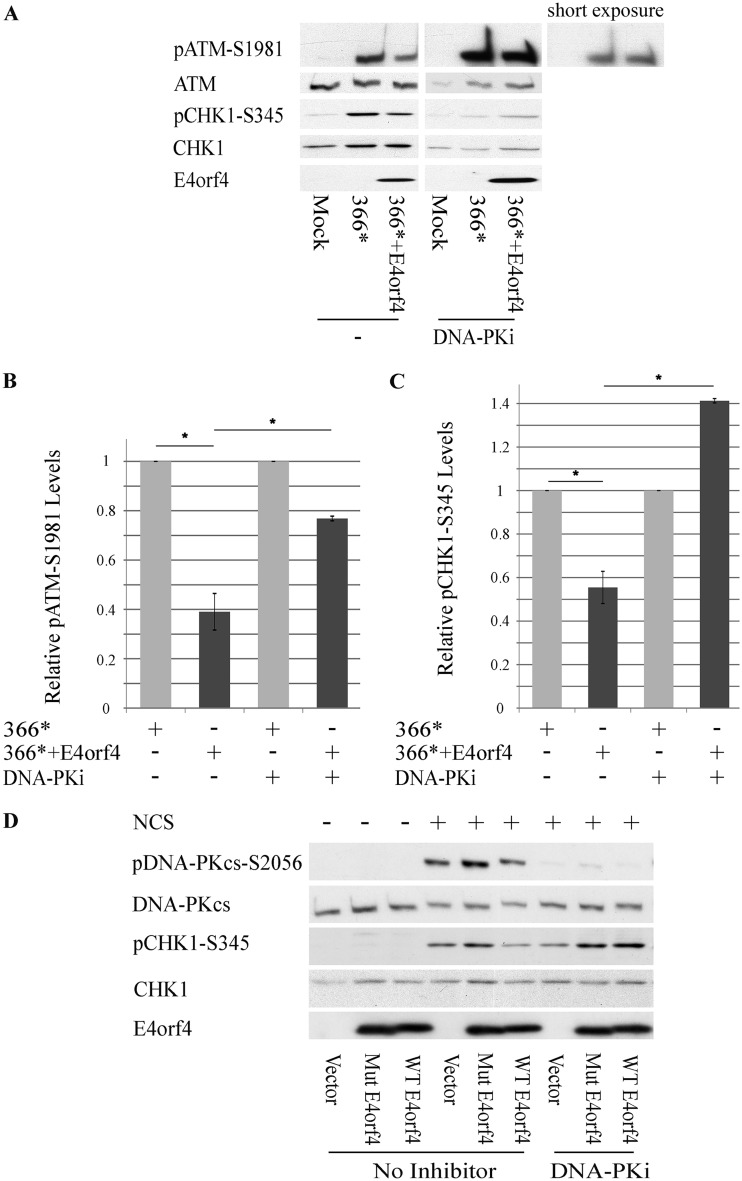
We have previously reported that E4orf4 inhibits DNA damage signaling through the ATM and ATR branches of the DDR, as demonstrated by its ability to reduce the phosphorylation of ATM and ATR substrates (26). However, we obtained no indication that E4orf4 interacts with ATM or ATR. Since E4orf4 associates with DNA-PK (Fig. 1), we examined whether DNA-PK activity was required for the ability of E4orf4 to reduce ATM and ATR signaling. HeLa cells were mock infected or infected with an Ad mutant lacking the E4 region (dl366*), which is unable to inhibit the DDR, or with the mutant virus dl366*+E4orf4, which lacks all E4 open reading frames (ORFs) except E4orf4. These mutant viruses were utilized to investigate the effect of E4orf4 on the virus-induced DDR without the additional complexity introduced by the presence of other E4 proteins that affect the DDR. As previously reported (26) and as shown in Fig. 2A to C, the dl366* mutant virus activated the DDR, as manifested by enhanced phosphorylation of ATM and the ATR substrate Chk1, whereas the presence of E4orf4 in the dl366*+E4orf4 virus resulted in significantly reduced ATM and Chk1 phosphorylation levels. In contrast, when the cells were infected with the same virus mutants in the presence of a DNA-PK inhibitor, phosphorylation of ATM and Chk1 was not reduced as efficiently by E4orf4. It should be noted that incubation of cells with the DNA-PK inhibitor for several hours consistently reduced total Chk1 protein levels, as shown in Fig. 2A. Overall, the results demonstrate that an active DNA-PK is required for inhibition of ATM and ATR signaling by E4orf4 during Ad infection.
FIG 2.
DNA-PK activity is required for inhibition of the ATM and ATR signaling pathways by E4orf4. (A) HeLa cells were either mock infected or infected with the Ad mutants dl366* lacking the whole E4 region and dl366*+E4orf4 expressing E4orf4 as the only E4 ORF. A DNA-PK inhibitor (DNA-PKi) (NU7441) was added to the infected cells for the duration of the infection starting at 2 h p.i., and another group of infected cells was left untreated. Proteins were harvested at 24 h p.i., and Western blot analysis was carried out with the indicated antibodies for phosphorylated and nonphosphorylated proteins. One representative blot is shown. The parts of this blot showing proteins in the presence or absence of a DNA-PK inhibitor are from the same exposed blot, but some lanes were removed from the middle. An additional short exposure of pATM in the presence of the DNA-PK inhibitor is shown to demonstrate more clearly the similarities in band intensities between the two infections. (B and C) Blots as described above for panel A from three independent experiments were subjected to densitometry. The levels of phosphorylated ATM and Chk1 as well as of the total proteins were calculated, and phosphoprotein levels were normalized to levels of the total corresponding protein. Normalized phosphoprotein levels in cells infected with dl366* (light gray bars) were defined as 1, and relative levels in dl366*+E4orf4-infected cells (dark gray bars) are shown (n = 3). Error bars represent standard errors. Statistical significance was determined by Student’s t test. *, P < 0.02. (D) HeLa cells were transfected with a plasmid expressing WT-E4orf4 from a Dox-inducible promoter or with an empty vector. The cells were induced with Dox for 4 h and treated with 0.5 ng/μl NCS or 0.01 mM the DNA-PK inhibitor NU7441 for 1 h and 1.5 h prior to harvest, respectively. One set of cells was left untreated. Whole-cell extracts were prepared and subjected to Western blot analysis with the specified antibodies, and a representative blot is shown.
Similar results were obtained when E4orf4 was expressed alone and DNA damage was induced by NCS treatment. Figure 2D demonstrates that WT-E4orf4 reduced NCS-induced Chk1 phosphorylation, whereas inhibition of DNA-PK prevented the reduction of Chk1 phosphorylation by E4orf4 and even led to an increase in pChk1 levels. As described above, the R81F84A E4orf4 mutant that is unable to bind PP2A did not decrease Chk1 phosphorylation. Furthermore, when expressed alone, WT-E4orf4 did not reduce DNA-PKcs autophosphorylation at S2056 to a significant degree (Fig. 1G and Fig. 2D).
Because the interaction between E4orf4 and PP2A was shown to be required for inhibition of ATM and ATR signaling by E4orf4 (26), we examined whether DNA-PK inhibition affected this interaction. However, as demonstrated in Fig. 1G, E4orf4 could bind the PP2A-C subunit equally well in the presence or absence of the inhibitor, indicating that inhibition of DNA-PK did not interfere with E4orf4 signaling by disrupting the E4orf4-PP2A interaction.
E4orf4 reduces DNA-PKcs autophosphorylation during Ad infection, but this is not an immediate early event.
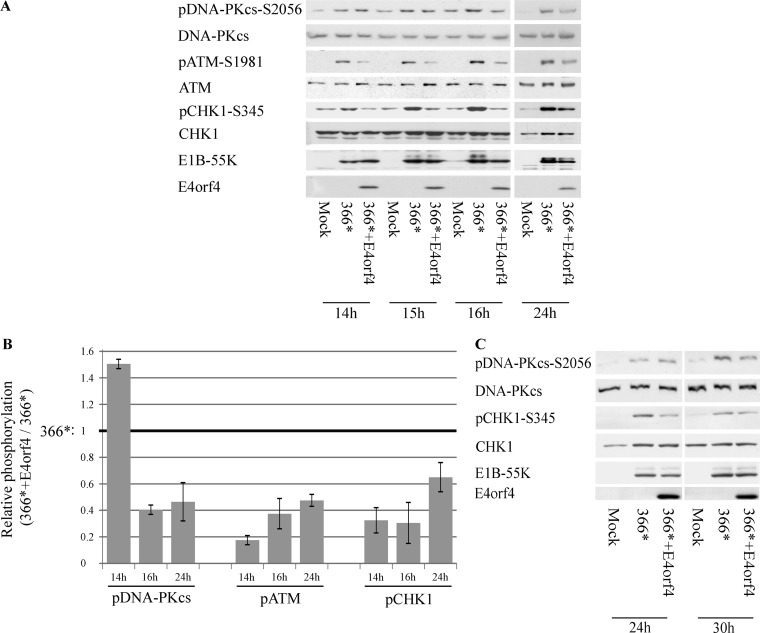
Because E4orf4 inhibited ATM- and ATR-regulated damage signaling in a DNA-PK-dependent manner, we examined whether DNA-PK activity was influenced by E4orf4 during Ad infection. HCT116 cells were infected with the mutant viruses dl366* and dl366*+E4orf4 or were left uninfected. Cell extracts were prepared at various times postinfection (p.i.), and DNA-PKcs autophosphorylation at S2056 was determined as an indication of DNA-PK activity. Figures 3A and B demonstrate that at 14 h postinfection, dl366* infection enhanced DNA-PKcs S2056 phosphorylation, and E4orf4 in the dl366*+E4orf4 virus did not reduce this phosphorylation and even augmented it. ATM and Chk1 phosphorylation levels were reduced in the presence of E4orf4 (dl366*+E4orf4) at the same time point, compared with their levels in cells infected with dl366*. However, as infection with the dl366*+E4orf4 virus progressed, DNA-PKcs autophosphorylation was decreased compared with its levels in dl366*-infected cells, and ATM and Chk1 phosphorylation continued to be reduced. The DNA-PKcs phosphorylation level remained low in the presence of E4orf4 at least up to 24 h p.i. The transition point from higher to lower DNA-PKcs phosphorylation levels observed in cells infected with the dl366*+E4orf4 virus was identified following examination of DNA-PKcs autophosphorylation at several time points during infection. Similarly, a transition from slightly increased pDNA-PKcs-S2056 phosphorylation to reduced phosphorylation at this site in the dl366*+E4orf4 virus infection was also observed in HeLa cells, although its timing was different (Fig. 3C). These E4orf4 effects did not result from reduced virus concentrations in the cells, as indicated by the similar levels of the early E1B-55K protein in various infections. Therefore, the results support the conclusion that E4orf4 inhibits DNA-PK activity, but this E4orf4 effect is delayed in comparison with inhibition of ATM and ATR signaling.
FIG 3.
E4orf4 effects on DNA-PKcs autophosphorylation. (A) HCT116 cells were either mock infected or infected with the Ad mutant dl366*, lacking the E4 region, or with dl366*+E4orf4, lacking all E4 ORFs except E4orf4. Proteins were harvested at the indicated times p.i., and Western blot analysis was carried out with the specified antibodies. Samples from the 14- to 16-h and from the 24-h time points of this experiment were run on separate gels. (B) Protein bands in blots from two independent experiments performed as described above for panel A were quantified by densitometry, the levels of phosphorylated proteins were normalized to the levels of the corresponding total proteins, and the ratios between normalized phosphorylation in dl366*-infected cells (defined as 1) and parallel values in dl366*+E4orf4-infected cells are shown in a graph (n = 2). Error bars represent the standard errors. (C) HeLa cells were infected as described above for panel A and harvested for analysis at the indicated time points. All lanes shown are from the same exposed blot, but some lanes were removed from the middle.
Inhibition of DNA-PK activity reduces E4orf4 recruitment to DNA damage sites but does not affect E4orf4 localization to viral replication centers.
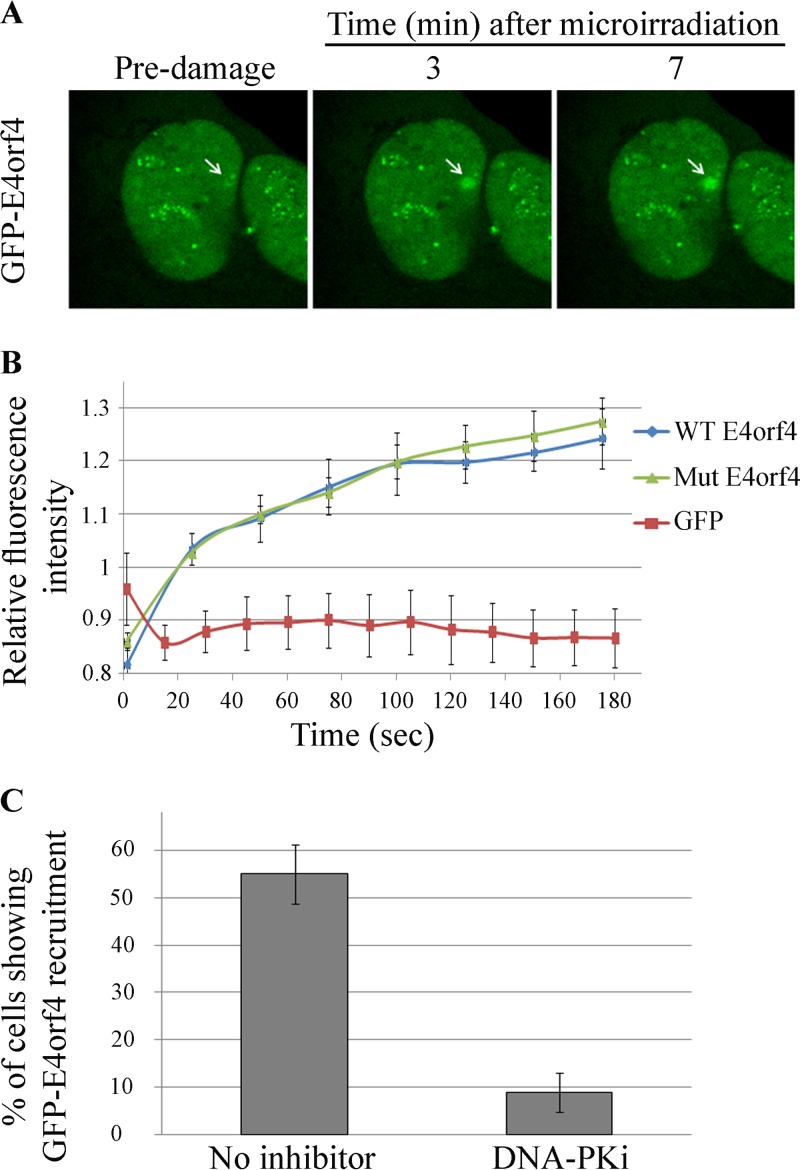
One mechanism that could account for the finding that inhibition of DNA-PK prevented E4orf4 from reducing ATM and ATR signaling is DNA-PK-dependent recruitment of E4orf4 to DNA damage sites or to viral replication centers (RCs). E4orf4 may not be able to influence DNA damage signaling if it is unable to reach the damage sites. To examine this possibility in the case of non-virus-induced DNA damage, we used laser microirradiation to induce localized DNA damage in the nuclei of living cells and determined the kinetics of recruitment of E4orf4 fused with green fluorescent protein (GFP) to the damage site by time-lapse microscopy. As shown in Fig. 4A and B, GFP-tagged E4orf4 was efficiently recruited to laser-microirradiated sites, whereas GFP alone was not. The E4orf4 mutant protein (R81F84A), which is unable to bind PP2A, was recruited to DNA damage sites as efficiently as the WT-E4orf4 protein (Fig. 4B) suggesting that an interaction with PP2A was dispensable for this recruitment. However, inhibition of DNA-PK activity by a highly specific inhibitor (NU7441 [55]) significantly reduced E4orf4 recruitment to laser-microirradiated regions (Fig. 4C). These results demonstrate that an active DNA-PK is required for E4orf4 recruitment to DNA damage sites.
FIG 4.
E4orf4 recruitment to DNA damage sites requires DNA-PK activity. (A) E4orf4-GFP was expressed for 3 h in U2OS cells. The cells were then treated with Hoechst stain and subjected to laser microirradiation. Images were taken by time-lapse microscopy. Recruitment of E4orf4-GFP to a laser-microirradiated site at various times after irradiation is shown (marked with a white arrow). (B) Recruitment of WT-E4orf4–GFP, the GFP-tagged R81F84A E4orf4 mutant, and GFP alone to laser-microirradiated sites was quantified at several time points postirradiation and is plotted in a graph (n > 10). Error bars represent standard deviations. (C) Recruitment of WT-E4orf4–GFP to DNA damage sites and the effect of a specific DNA-PK inhibitor (DNA-PKi) (NU7441) are shown as the percentage of cells in which E4orf4-GFP accumulated at the laser-microirradiated site (n > 30 from three independent experiments). Error bars represent standard deviations.
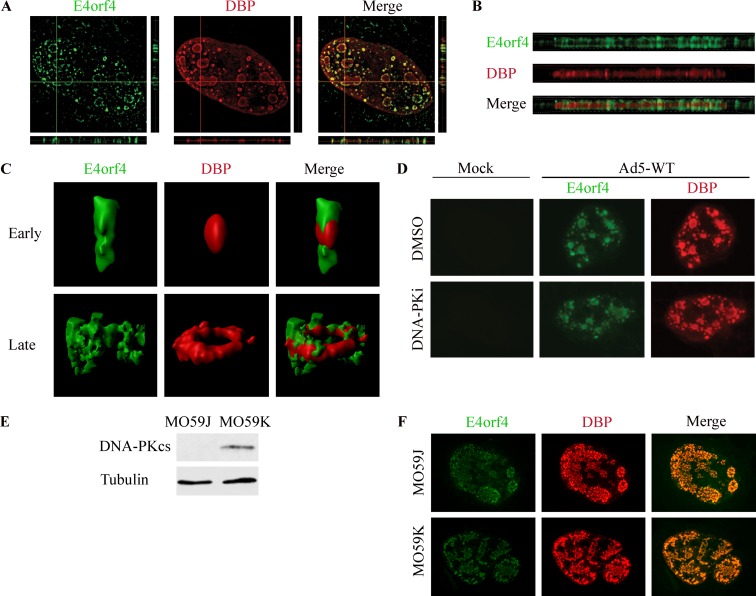
Next, we examined whether E4orf4 was recruited to virus RCs. HeLa cells were infected for 24 h with WT-Ad5, and the localization of the Ad DNA binding protein (DBP), marking viral RCs, and of E4orf4 was determined by immunofluorescence detected using high-resolution microscopy. Various views of a representative infected cell nucleus are shown in Fig. 5, including a topological view (Fig. 5A), orthogonal projections (Fig. 5A and B), and three-dimensional (3D) renderings of two types of RCs, an early RC in the shape of a small focus and a ring-shaped late RC structure (Fig. 5C) (56). These images demonstrate that E4orf4 surrounds and grasps DBP-marked RCs, with partial colocalization. To determine whether inhibition of DNA-PK influenced the recruitment of E4orf4 to viral RCs, infections were carried out in the presence or absence of a DNA-PK inhibitor and subjected to analysis by confocal microscopy. Figure 5D demonstrates that unlike the recruitment of E4orf4 to irradiation-induced damage sites, the recruitment of E4orf4 to viral RCs was not affected by inhibition of DNA-PK activity. Similarly, E4orf4 could be recruited equally well to Ad RCs in cells expressing DNA-PKcs (MO59K) and in cells lacking it (MO59J) (Fig. 5E and F).
FIG 5.
E4orf4 is recruited to viral RCs in a DNA-PK-independent manner. (A) HeLa cells were infected with WT-Ad5 and were fixed and stained at 24 h p.i. A high-resolution image of a representative infected nucleus is shown with horizontal and vertical orthogonal projections. (B) Enlarged image of a horizontal orthogonal projection from the nucleus presented in panel A. (C) Representative 3D renderings of early and late replication centers prepared with Imaris software. (D) HeLa cells were either mock infected or infected with WT-Ad5 in the presence of the solvent dimethyl sulfoxide (DMSO) or the DNA-PK inhibitor (DNA-PKi) NU7441. Representative confocal images of cells fixed at 24 h p.i. are shown. (E) MO59K and MO59J cell extracts were subjected to Western blot analysis with the indicated antibodies. (F) MO59K and MO59J cells were infected with WT-Ad5, fixed at 24 h p.i., and subjected to confocal microscopy. Representative images are shown.
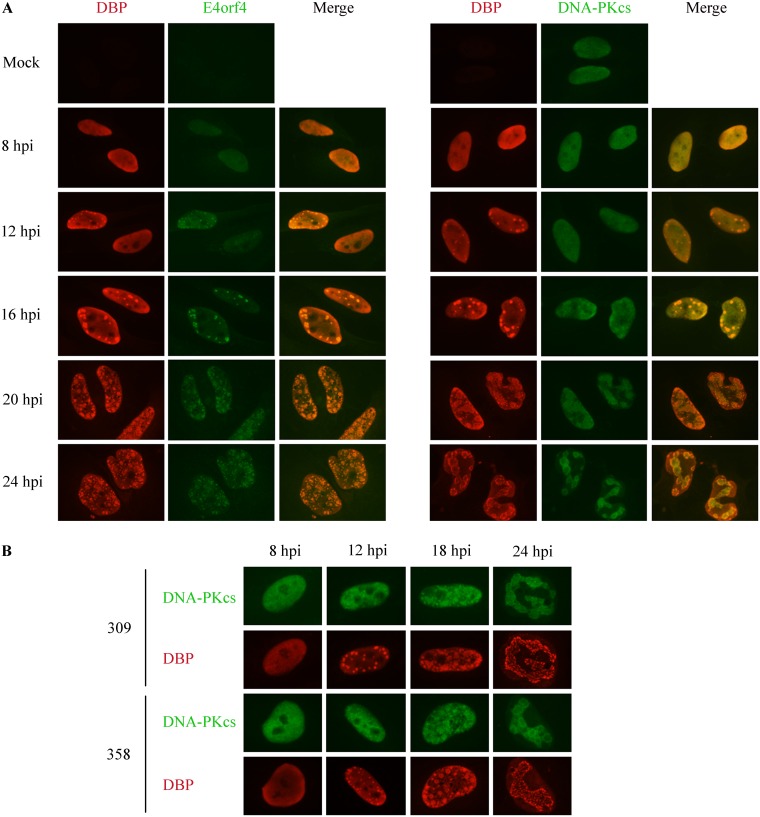
DNA-PKcs is recruited to viral replication centers, but colocalization with DBP is transient.
Because we found a biphasic functional interaction between E4orf4 and DNA-PKcs, wherein DNA-PK was first required for inhibition of ATM and ATR signaling by E4orf4 but was later inhibited by E4orf4 (Fig. 2 and 3), we investigated whether DNA-PKcs was recruited to viral RCs and examined the kinetics of recruitment of both proteins. As shown in Fig. 6A, DBP was found in a diffuse pattern in infected cell nuclei at 8 h p.i. but concentrated in small replication foci at 12 h p.i. Ringlike RC structures accumulated at 20 h p.i. and later. In most cells, E4orf4 localized to DBP-marked RCs synchronously and similarly to DBP. In contrast, DNA-PKcs staining was more heterogeneous at 12 h p.i., with clear RC colocalization seen in most cells only later. Furthermore, unlike the focused appearance of DBP and E4orf4 in RCs, DNA-PKcs staining in the vicinity of RCs appeared more diffuse. A comparison of DNA-PK recruitment to Ad RCs during infection with phenotypically WT-Ad5 (dl309) or a mutant Ad lacking E4orf4 (dl358) established that the recruitment of this enzyme to RCs was not dependent on E4orf4 (Fig. 6B).
FIG 6.
Comparison of the recruitment of DNA-PKcs and E4orf4 to virus replication centers. (A) HeLa cells were infected with WT-Ad5 and fixed at various times postinfection. One set of cells was stained with DBP and E4orf4 antibodies, while another set was stained with DBP and DNA-PKcs antibodies. Representative images are shown for each time point. (B) HeLa cells were infected with phenotypically WT-Ad5 (dl309) or a mutant Ad lacking E4orf4 (dl358) and processed as described above for panel A. hpi, hours postinfection.
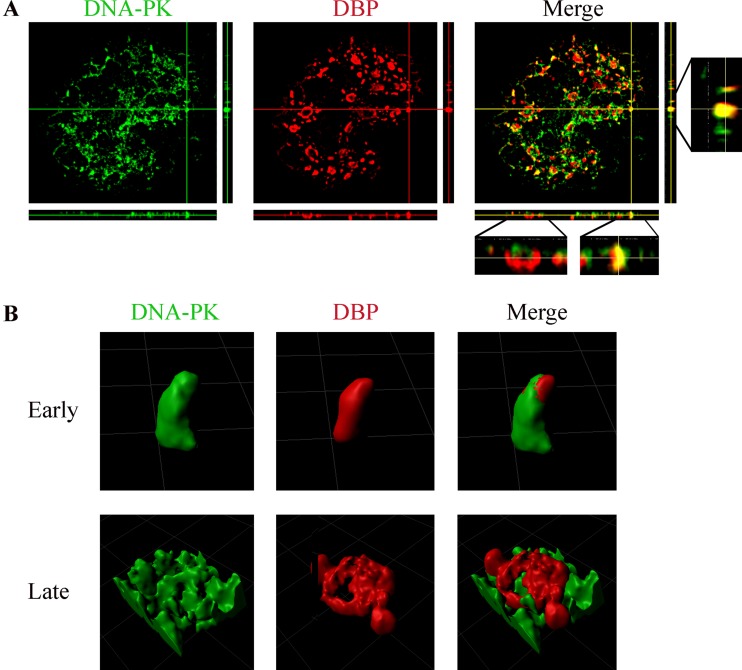
To examine more concisely the relative localization of DNA-PKcs and DBP, high-resolution microscopy was utilized to visualize the two proteins. Cells infected for 24 h were examined because they contained early as well as late RC structures, facilitating analysis of both. Orthogonal projections of images of the two proteins in a representative infected cell nucleus as well as 3D renderings of individual RCs indicate that DBP and DNA-PKcs significantly colocalized in early RCs, whereas DNA-PKcs only slightly colocalized with DBP in late ringlike RCs and appeared to have been distanced from these RCs to a neighboring location (Fig. 7).
FIG 7.
DNA-PKcs colocalizes with early replication centers but localizes at the vicinity of late, ringlike replication centers. (A) HeLa cells were infected with WT-Ad5, fixed at 24 h p.i., and stained with antibodies to DBP and DNA-PKcs. A high-resolution image of a representative infected nucleus is shown with horizontal and vertical orthogonal projections. (B) Representative 3D renderings of early and late replication centers prepared with Imaris software.
Inhibition of DNA-PK activity improves the efficiency of Ad replication.
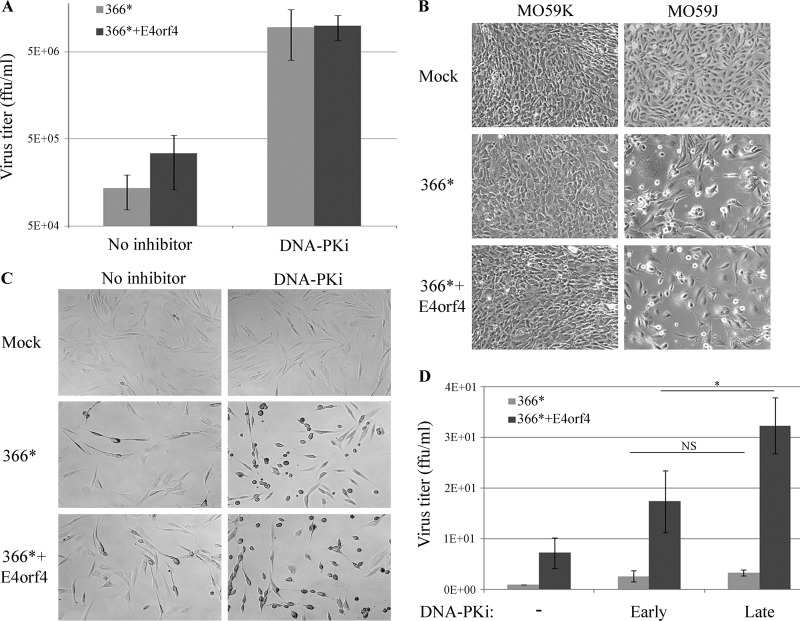
Because we found that during Ad infection, DNA-PK was eventually inhibited by E4orf4 (Fig. 3), and as it was reported that DNA-PK was the target of additional Ad proteins besides E4orf4 (30, 31), we investigated the effect of DNA-PK inhibition on the efficiency of Ad replication using three different cell lines. First, HeLa cells were infected with dl366* or dl366*+E4orf4 virus, in the presence or absence of a DNA-PK inhibitor added at a concentration that efficiently inhibited this enzyme (Fig. 2D), and the titer of progeny virus obtained at 24 h p.i. was determined. As shown in Fig. 8A, the DNA-PK inhibitor increased the dl366* virus titer significantly by an average of 71-fold, and the titer of dl366*+E4orf4 was increased 29-fold. To substantiate these results, we also compared the replication of the Ad mutants in MO59K cells containing WT DNA-PKcs and MO59J cells lacking it (Fig. 5E). Efficiencies of infection of MO59K and MO59J cells were found to be dissimilar, and flow cytometry analyses of infections with an Ad-GFP virus were used to assess relative infectivity. Infection of MO59J cells with Ad-GFP at 50 PFU/cell and infection of MO59K cells with the same virus at 250 PFU/cell resulted in >99.9% GFP-positive cells. The cells were then infected with the dl366* and dl366*+E4orf4 viruses at these multiplicities of infection, and the cytopathic effect of infection (CPE) was visualized at 3 days p.i. As shown in Fig. 8B, no CPE was observed in MO59K cells, while a strong CPE, manifested as rounding up and loss of cells, was seen in MO59J cells infected with 5-fold less virus. These results strongly support the conclusion that removal of DNA-PK enhanced Ad replication. Based on the findings that DNA-PK activity at early infection times was required for inhibition of ATM and ATR signaling by E4orf4 (Fig. 2 and 3) and that inhibition of ATM and ATR signaling also contributed to Ad infection (26, 57, 58), we explored the possibility that delayed inhibition of DNA-PK will enhance replication efficiency more effectively than inhibition starting at the onset of Ad infection. Immortalized human F-89 fibroblasts (59) were used in this experiment because Ad infection is much slower in these cells, providing enough time to track the production of progeny virus after the addition of a DNA-PK inhibitor at a later time point. First, we confirmed that the early addition of a DNA-PK inhibitor improved the efficiency of virus replication in these cells by monitoring the induction of CPE. CPE that accompanied Ad infection in F-89 cells included cell shrinking, rounding up, and detachment. F-89 cells infected with the mutant Ad viruses dl366* and dl366*+E4orf4 were subjected to the early addition of the DNA-PK inhibitor at the onset of virus infection. Figure 8C clearly demonstrates that inhibition of DNA-PK dramatically increased CPE in infected cells, establishing that virus infection proceeded more efficiently in the inhibitor-treated cells than in the nontreated cells. Next, the effects of early versus late addition of the DNA-PK inhibitor on virus titer were determined at 64 h p.i. Early addition started at 2 h p.i., and late addition started at 24 h p.i. Figure 8D demonstrates that a delayed inhibition of DNA-PK resulted in significantly increased replication efficiency of the dl366*+E4orf4 virus in comparison to an early addition of the inhibitor. However, during infection with the dl366* mutant virus, no significant difference in virus titers was observed between early and late additions of the inhibitor.
FIG 8.
DNA-PK inhibition improves the efficiency of Ad infection. (A) HeLa cells were infected with 30 FFU/cell of the Ad mutant viruses dl366* and dl366*+E4orf4. The DNA-PK inhibitor (DNA-PKi) NU7441 was added to the cells at 2 h p.i., and titers of viruses obtained at 24 h p.i. were determined. Average results of two independent experiments are shown in the graph. Error bars represent the standard errors. (B) MO59K and MO59J cells were infected with the Ad mutant viruses dl366* and dl366*+E4orf4 as described in the text. Representative images captured at a ×20 magnification at 72 h p.i. are shown. (C) F-89 cells were infected with 100 FFU/cell of the Ad mutant viruses dl366* and dl366*+E4orf4. The DNA-PK inhibitor NU7441 was added to the cells at 2 h p.i. Representative images captured at a ×20 magnification at 48 h p.i. are shown. (D) F-89 cells were infected as described above for panel C. The DNA-PK inhibitor prepared in DMSO was added to the cells every 12 h starting at either 2 h p.i. (“Early”) or 24 h p.i. (“Late”). One set of cells was treated with the solvent alone (−). Cells were harvested at 64 h p.i., and the virus titer was determined. Average results of three independent experiments are shown. Error bars represent standard errors. Statistical significance was determined by Student’s t test. *, P < 0.05; NS, nonsignificant.
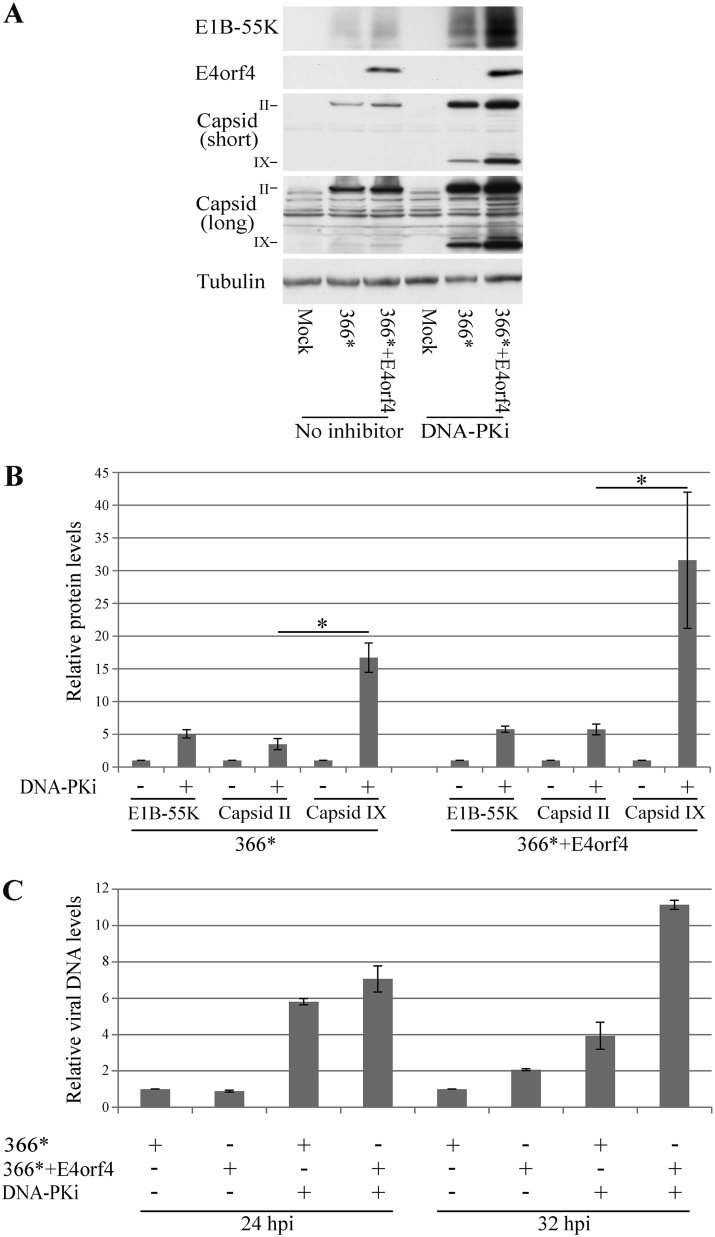
Since inhibition of DNA-PK increased the efficiency of virus propagation, we set out to determine which stages of viral infection were most affected by a decline in DNA-PK activity. Cell extracts were prepared from HeLa cells that were mock infected or infected with the mutant viruses in the presence or absence of the DNA-PK inhibitor, added at a concentration that efficiently inhibited this enzyme (Fig. 2D), and the levels of early and late Ad proteins were determined by Western blot analysis. As shown in Fig. 9A and B, inhibition of DNA-PK increased the expression of the early protein E1B-55K by an average of 5- to 6-fold in cells infected with the two mutant viruses compared to nontreated levels. Similarly, levels of a late protein, protein II (pII) (the hexon capsid protein), were increased 3.5- to 6-fold. In contrast, the levels of one of the virus cement proteins, protein IX, were increased more dramatically following DNA-PK inhibition, by an average of 16-fold in dl366* virus-infected cells and by an average of 31.6-fold in dl366*+E4orf4-infected cells. The effect of DNA-PK inhibition on Ad DNA levels was also determined and was found to be similar to its effect on E1B-55K and hexon protein levels (up to a 7-fold increase) (Fig. 9C).
FIG 9.
Effect of DNA-PK inhibition on various stages of Ad replication. HeLa cells were infected with the Ad mutants dl366* and dl366*+E4orf4 or were mock infected. A DNA-PK inhibitor (DNA-PKi) was added to one set of cells at 2 h p.i. as described in Materials and Methods. (A) Proteins were harvested 24 h later, and Western blot analysis was carried out with the indicated antibodies. A representative blot is shown. Two different exposures of the blot stained with a capsid-specific antibody are shown (short and long). (B) Graph summarizing data from three independent experiments performed as described above for panel A. Protein bands were quantified by densitometry. Protein levels in cells infected without the addition of an inhibitor were defined as 1, and relative protein levels in the presence of the inhibitor are shown for E1B-55K, capsid protein II (hexon), and capsid protein IX (n = 3). Error bars represent the standard errors. Statistical significance was determined by Student’s t test. *, P < 0.04. (C) Virus DNA levels at 24 and 32 h p.i. were quantified by quantitative PCR and normalized to the level of cellular DNA represented by the PRPH2 gene. Viral DNA levels in untreated dl366*-infected cells at each time point were defined as 1, and relative DNA levels in the other samples are shown in a graph. Two replicates were analyzed. Error bars represent standard errors.
Inhibition of DNA-PK activity impedes E4orf4-induced cell death.
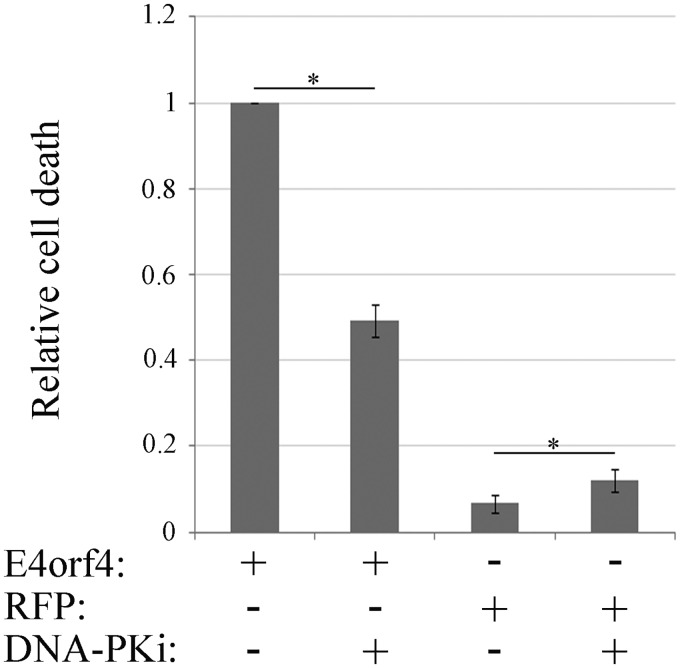
We have previously demonstrated that E4orf4 increased the accumulation of DNA damage following treatment of cells with DNA-damaging drugs, resulting in sensitization of cells to killing induced by sublethal concentrations of such drugs. We suggested that inhibition of the DDR could contribute to the ability of E4orf4 to induce cancer-selective cell death, as cancer cells already have deficiencies in DDR pathways and may be more sensitive to further DDR inhibition (26). Because DNA-PK inhibition prevented E4orf4 from being recruited to damage sites (Fig. 4) and from reducing DDR signaling (Fig. 2D), we hypothesized that it would consequently prevent E4orf4 from inducing cell death. To test this hypothesis, DAPI (4′,6-diamidino-2-phenylindole) assays for determination of E4orf4-induced cell death were performed in the presence or absence of a DNA-PK inhibitor, added at a concentration that efficiently inhibited this enzyme (Fig. 2D). It should be noted that many other apoptosis assays have been described as being unsuitable for measuring caspase-independent E4orf4-induced cell death in many cell lines (41, 60) and therefore were not employed here. As shown in Fig. 10, E4orf4 dramatically increased cell death levels compared with a control red fluorescent protein (RFP). Incubation of control cells with the DNA-PK inhibitor enhanced cell death, likely because of the accumulation of unrepaired DNA damage. However, when E4orf4-expressing cells were subjected to treatment with the DNA-PK inhibitor, E4orf4-induced cell death was significantly reduced, as expected.
FIG 10.
DNA-PK inhibition reduces E4orf4-induced cell death. HeLa cells were transfected with a plasmid expressing E4orf4 or with a control plasmid expressing RFP. The cells were treated with a DNA-PK inhibitor (DNA-PKi) at 3 h posttransfection, and another DNA-PK inhibitor aliquot was added after 12 h. The cells were fixed at 24 h posttransfection, immunostained with an antibody against E4orf4, and also stained with DAPI. Induction of cell death was measured by a DAPI assay. The percentage of E4orf4-expressing cells with apoptotic nuclei was determined by counting the total number of transfected cells and the number of transfected cells with abnormal, apoptotic nuclei. Average results of two independent experiments are shown. At least 150 transfected cells were analyzed in each experiment. Error bars represent standard errors. Statistical significance was determined by Student’s t test. *, P < 0.04.
DISCUSSION
In this report, we describe a novel interaction between the Ad E4orf4 protein and the DNA damage sensor DNA-PK, and we investigate the consequences of this interaction.
Contribution of DNA-PK to inhibition of ATM and ATR signaling by E4orf4.
Ad utilizes several mechanisms to inhibit the cellular DDR network, which acts as an antiviral defense mechanism. We have recently shown that the Ad E4orf4 protein contributes to DDR inhibition by reducing DNA damage- or virus-induced ATM and ATR signaling in a PP2A-dependent manner. It was also shown that ATM and ATR were not mutually required for inhibition of their signaling pathways by E4orf4 (26). One question arising from these results is whether E4orf4 inhibits ATM and ATR signaling individually or whether it affects a joint upstream regulator. We show here that E4orf4 associates with DNA-PK (Fig. 1) and that DNA-PK activity is essential for the ability of E4orf4 to reduce ATM and ATR signaling (Fig. 2). Consistent with these results, earlier during Ad infection, E4orf4 reduces ATM and ATR activation but does not decrease DNA-PK activation, represented by autophosphorylation at S2056, and even augments it (Fig. 3). However, at later time points, DNA-PK autophosphorylation is also reduced in the presence of E4orf4 (Fig. 3). The time scale of the transition from the utilization of DNA-PK activity to its inhibition by E4orf4 differs in diverse cell lines, but the principle remains the same. It should be noted that both E4orf4 and inhibition of DNA-PK do not decrease but rather increase the levels of Ad DNA (Fig. 9C), demonstrating that reduced ATM and ATR signaling does not result from reduced Ad DNA levels.
During Ad infection, E4orf4 is recruited to virus RCs concomitantly with DBP and appears to surround and grasp these centers (Fig. 5 and 6). This localization may allow E4orf4 to control events occurring at the RCs, including regulation of DDR processes. DNA-PKcs is also recruited to WT-Ad5 RCs (Fig. 6), although its staining pattern around them is more diffuse than DBP and E4orf4 staining. Interestingly, DNA-PKcs colocalizes with DBP more distinctly in early, focus-like RCs than in late, ring-shaped centers (Fig. 7). This finding suggests that DNA-PKcs is distanced from RCs during Ad infection, and this may contribute to the inhibition of DNA-PK, which occurs later in infection (Fig. 3). In contrast, the early colocalization of DNA-PKcs with viral RCs (Fig. 7) could facilitate the assistance that it provides to E4orf4 in inhibition of ATM and ATR signaling (Fig. 2).
When E4orf4 is expressed outside the context of virus infection, it does not consistently reduce damage-induced DNA-PK autophosphorylation to a large extent (Fig. 1 and 2). This suggests that a decrease of DNA-PK activation during Ad infection is induced by E4orf4 in collaboration with additional viral proteins encoded by transcription units other than E4. However, chemical inhibition of DNA-PK attenuated the ability of E4orf4 to decrease ATM and ATR signaling, even when expressed alone (Fig. 2D). In this regard, we have shown that E4orf4 is recruited to DNA damage sites induced by laser microirradiation and that this recruitment is significantly dependent on DNA-PK activity (Fig. 4). It is likely that E4orf4 must localize to DNA damage sites to affect damage-induced ATM and ATR signaling. In contrast, during virus infection, E4orf4 can localize to virus replication centers regardless of DNA-PK activity, but it still requires this activity to inhibit ATM and ATR signaling (Fig. 2 and 5). Therefore, it appears that inhibition of the ATM- and ATR-regulated DDR pathways by E4orf4 requires an additional DNA-PK function that is not associated with regulation of E4orf4 localization to damage sites.
PP2A is a major E4orf4 partner and was shown to be essential for the ability of E4orf4 to reduce ATM and ATR signaling (26). In this work, we find that an association of E4orf4 with PP2A is not required for E4orf4 recruitment to DNA damage sites (Fig. 4) and that PP2A activity only mildly affects the ability of E4orf4 to interact with DNA-PK (Fig. 1). Thus, PP2A must act after E4orf4 has been recruited to damage sites and has associated with DNA-PK. One possible mechanism by which DNA-PK inhibition could prevent attenuation of the DDR by E4orf4 is inhibition of the association of E4orf4 with PP2A. However, as shown in Fig. 1G, PP2A was efficiently coimmunoprecipitated with E4orf4 regardless of DNA-PK activity, revoking this possibility. It is also possible, but has yet to be examined, that DNA-PK phosphorylates PP2A subunits or E4orf4 and inhibits the enzymatic activity of the E4orf4-PP2A complex. It has been reported in the literature that phosphorylation of various PP2A subunits modulates the affinity of PP2A for diverse substrates and its activity toward them (61, 62). Sequence analysis of E4orf4 and PP2A subunits demonstrates that they contain PIKK phosphorylation consensus sites, and they may therefore act as DNA-PK substrates.
Contribution of DNA-PK inhibition to Ad replication.
As discussed above, a biphasic functional interaction exists between E4orf4 and DNA-PK during Ad infection, which is demonstrated by the finding that DNA-PK activity is first required for E4orf4 function but is eventually inhibited by E4orf4 at later times. The inhibition of DNA-PK is important for virus replication because the addition of a DNA-PK inhibitor from the onset of Ad infection until virus harvest or deletion of DNA-PKcs increased the virus titer significantly (Fig. 8). However, when DNA-PK was inhibited starting at a later time point during Ad infection, the virus titer was further increased, likely because ATM and ATR signaling could also be inhibited. The effect of delayed DNA-PK inhibition was more pronounced in infection with the virus expressing E4orf4, consistent with the conclusion that DNA-PK activity is required at the early stages of infection for E4orf4 effects on the DDR but should be inhibited at the later stages for maximal efficiency of virus replication.
Analysis of the effect of DNA-PK inhibition on Ad protein expression in HeLa cells revealed 3.5- to 6-fold increases in the levels of the early E1B-55K protein and of the major capsid protein hexon (pII). A similar, ≤7-fold increase in virus DNA levels was also observed. However, the increase in the level of the pIX cement protein was much more pronounced (16- to 31.6-fold in cells infected with dl366* and dl366*+E4orf4, respectively) (Fig. 9). Ad cement proteins, including proteins IIIa, VI, VIII, and IX, play crucial roles in virion assembly, disassembly, cell entry, and infection and exclusively stabilize the hexon shell (63). Therefore, it is possible that DNA-PK inhibition improves Ad replication by increasing the levels of the cement protein pIX, which results in capsid stabilization and increased infectivity of virions, rather than by causing a large increase in the number of virus particles represented by levels of the major capsid protein or the viral DNA.
Targeting DNA-PK by Ad appears to be very important for the virus, as this enzyme was reported to be the target of additional Ad proteins besides E4orf4. The Ad E4orf3 and E4orf6 proteins were reported to bind DNA-PK and to inhibit viral DNA concatenation and V(D)J recombination, which relies on NHEJ, a repair process regulated by DNA-PK (30). It was also shown that the Ad L4-33K protein associates with the catalytic subunit of DNA-PK (31). L4-33K functions as an alternative splicing factor involved in activating the shift from the earlier-appearing L1-52,55K mRNA to the later L1-IIIa transcript. L4-33K was shown to be highly phosphorylated by DNA-PK in vitro, and DNA-PKcs-deficient cells produced excessive L1-IIIa mRNA levels, suggesting an inhibitory role of DNA-PK in the temporal switch in L1 alternative RNA splicing. Thus, our results together with data from previous reports indicate that after DNA-PK activity is utilized to assist E4orf4 in inhibiting the ATM- and ATR-regulated DDR, inhibition of DNA-PK facilitates several additional important activities that increase the efficiency of virus replication. These include regulation of the progression of Ad infection from the early to the late phase by control of alternative splicing, increased expression of a viral cement protein required for increased infectivity of Ad virions, and any direct effects on DNA-PK-regulated repair processes that may occur.
Contribution of DNA-PK to E4orf4-induced cell death.
As reported previously, when E4orf4 is expressed alone, it induces cancer-selective cell death (49). We have previously suggested that one of the mechanisms that may account for the cancer selectivity of E4orf4-induced cell killing involves increased susceptibility of several types of cancer to DDR inhibition due to the inherent disruption of DDR pathways in cancer cells (26). In this report, we show that E4orf4 is rapidly recruited to DNA damage sites in a DNA-PK-dependent manner (Fig. 4) and that inhibition of this recruitment by a specific DNA-PK inhibitor attenuates the ability of E4orf4 to cause cell death (Fig. 10). These results support the notion that localization of E4orf4 to DNA damage sites and DDR inhibition at these sites contribute to E4orf4-induced cell death.
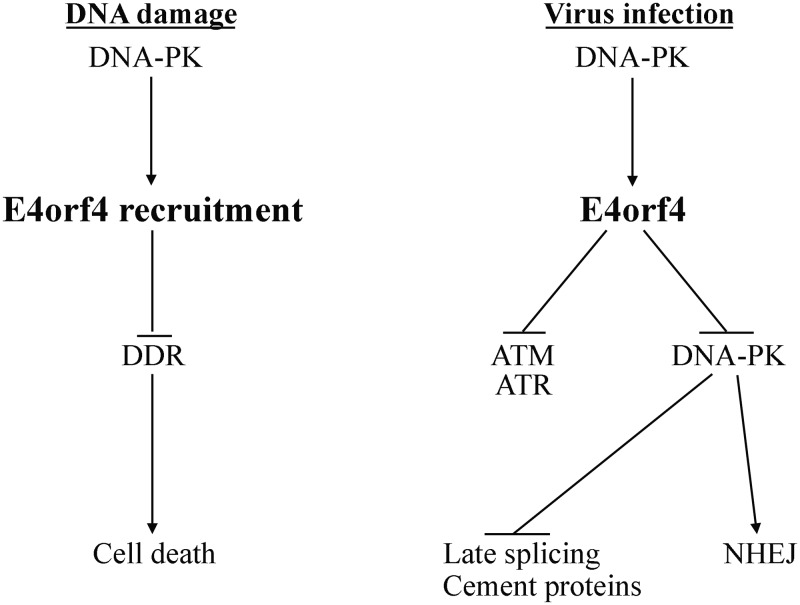
In summary, the findings that E4orf4 inhibits ATM and ATR signaling during virus infection and also inhibits DNA-PK suggest that it is involved in multipathway control of the DDR to assist other Ad proteins that neutralize various DDR branches. The biphasic functional interaction of E4orf4 with DNA-PK during infection allows this viral protein to take advantage of DNA-PK activity at early times of infection to inhibit DDR signaling by ATM and ATR, a beneficial event for Ad replication, and to inhibit DNA-PK itself at later times to achieve additional goals that further enhance Ad replication efficiency. DNA-PK also facilitates the induction of cancer cell death by E4orf4 via its effect on E4orf4 recruitment to DNA damage sites, where E4orf4 can manipulate the DDR network and cause cell death in cancer cells with inherent deficiencies in DDR pathways. These consequences of the E4orf4–DNA-PK functional interaction are summarized in Fig. 11.
FIG 11.
Contribution of the E4orf4–DNA-PK interaction to Ad replication and induction of cancer cell death. A model summarizing the consequences of the functional interaction between E4orf4 and DNA-PK during Ad infection and following induction of DNA damage is shown. The details are discussed in the text.
MATERIALS AND METHODS
Cells, plasmids, transfection, virus mutants, and infections.
HeLa, HCT116, U2OS, MO59J, and MO59K cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS). Immortalized human F-89 fibroblasts were a gift from Y. Shiloh’s laboratory at Tel Aviv University (59) and were cultured in the same medium as the other cell lines. MO59J-Fus9 and MO59J-Fus1 cells were a gift from M. Aladjem, NIH (64), and were grown in DMEM supplemented with 10% FCS and 250 μg/ml G418 (Invitrogen). Cells used for transfection with tetracycline-inducible plasmids were cultured for 3 days prior to the start of the experiment in medium containing 10% FCS guaranteed to be tetracycline free (BD Bioscience or PAN-Biotech GmbH).
WT-E4orf4 (32), WT-E4orf4–GFP, and the R81F84A E4orf4 mutant that does not bind PP2A (52) were cloned into the tetracycline-inducible pcDNA4/TO vector (Invitrogen). The plasmids were transfected together with a plasmid expressing the tetracycline repressor (pcDNA6/TR; Invitrogen), using TurboFect (Thermo Fisher Scientific) for HeLa cells and CalFectin (SignaGen) for HCT116 cells, according to the manufacturers’ instructions. One day later, the cells were induced with 1 μg/ml doxycycline (Dox) for 3 to 4 h. When required by the experiment, neocarzinostatin (NCS; Sigma) was added 1 h prior to harvest at a concentration of 0.5 μg/ml, the DNA-PK inhibitor (NU7441; Tocris Bioscience) was added 1.5 h prior to harvest at a final concentration of 10 μM, and okadaic acid (Santa Cruz Biotech) was added to a final concentration of 400 nM for the last hour of E4orf4 induction.
Adenoviral mutants dl366*, lacking the complete E4 region, and dl366*+E4orf4, lacking all E4 open reading frames (ORFs) except E4orf4, were previously described (65) and were propagated on W162 cells (from T. Shenk, Princeton University) (66). W162 cells were also used for determination of virus titers. Virus infections were performed at multiplicities of 30 fluorescence-forming units (FFU) per cell in HeLa and HCT116 cells and 100 FFU/cell in F-89 cells. MO59J and MO59K cells were infected with 50 and 250 PFU per cell, respectively, as described above. The DNA-PK inhibitor (NU7441; Tocris Bioscience) was added to infected cells at 2 h postinfection at a final concentration of 10 μM. This inhibitor was added again to HeLa cells at 13 h postinfection at a final concentration of 5 μM. For F-89 cells, the DNA-PK inhibitor was added at a concentration of 10 μM every 12 h during virus infection, up to 12 h before harvest. When late addition of the DNA-PK inhibitor was required, the first portion of this inhibitor was added at 24 h p.i. Untreated cells received equal quantities of the solvent. Infected cells were harvested for protein extraction, immunostaining, or virus collection at the indicated times.
Cell extracts, immunoprecipitation, and Western blot analysis.
Whole-cell extracts were prepared in sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 50 mM dithiothreitol [DTT], 10% glycerol), and viscosity was reduced by passing the extracts several times through a 27-gauge needle after a 3-min incubation at 65°C. Proteins were analyzed by Western blotting using the indicated antibodies. For immunoprecipitation assays, cell extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 100 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% IGEPAL CA-630, and 0.2% SDS, supplemented with 2× cOmplete protease inhibitor cocktail [Sigma], protease inhibitor cocktails for cells and yeast extracts [1:50 and 1:125, respectively; Sigma], and phosphatase inhibitor cocktails 2 and 3 [1:100; Sigma]).
Blot images were scanned with an Epson Photo 4990 scanner and processed using Adobe Photoshop 5.0 or 7.0. Band intensities were quantified using TotalLab software.
Antibodies specific for the following proteins were used in this work: E4orf4 (43); E1B-55K (clone 2A6) (67); Ad capsid proteins (a gift from T. Dobner); PP2A-C (BD Transduction Laboratories); hemagglutinin (HA) (Covance); pATM-S1981 (Epitomics); pChk1-S345, pAkt-S473, and Akt (Cell Signaling); ATM, Chk1, and DNA-PKcs (Santa Cruz); pDNA-PKcs-S2056 (Abcam); and alpha-tubulin (Sigma).
Relative quantitation of viral DNA.
Relative levels of viral DNA were determined by quantitative PCR, as previously described (26). Briefly, total infected cell extracts were prepared in RIPA buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS), sonicated, and treated with proteinase K. Quantitative real-time PCR was performed using the hexon-specific primers hexon-qPCR-fw (5′-CGCTGGACATGACTTTTGAG-3′) and hexon-qPCR-rev (5′-GAACGGTGTGCGCAGGTA-3′). Results were normalized to levels of the cellular PRPH2 gene determined by a parallel quantitative PCR with the primers TM684-fw (5′-CTGAAGCCGTACCTGGCTATC-3′) and TM685-rev (GTGTCCCGGTAGTACTTCATGC).
Laser microirradiation.
U2OS cells were presensitized with 10 μM Hoechst 3334 dye (Sigma) for 15 min at 37°C. Laser microirradiation was then performed using an inverted confocal microscope (LSM-700; Zeiss) equipped with a CO2 module and a 37°C heating chamber. A preselected spot within the nucleus was microirradiated with 10 iterations of a 405-nm laser with 100% power to generate localized DNA damage. Time-lapse images were then acquired using a 488-nm laser at 15-s time intervals. The fluorescence intensity of enhanced green fluorescent protein (EGFP) signals at laser-microirradiated sites was measured using Zen 2009 software (Carl Zeiss). Data obtained were corrected for the loss of total fluorescence and normalized to the initial intensity.
Cell death assay: the DAPI assay.
DAPI assays, which determine nuclear aberrations, have been described previously (68, 69). Briefly, cells were transfected with plasmids expressing E4orf4. At 24 h posttransfection, the cells were fixed with 4% paraformaldehyde and stained with antibodies to E4orf4 and with DAPI (Sigma). Fluorescent cells were visualized by using a Zeiss Axioskop microscope at a ×40 magnification. The fraction of E4orf4-expressing cells with condensed, abnormal, or fragmented nuclei was determined in each experiment by counting at least 150 transfected nuclei.
Immunofluorescence microscopy.
HeLa cells were seeded onto glass coverslips and infected as described above. At appropriate time points, the cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with PBS containing 0.25% Triton X-100 for 10 min at room temperature, and washed with PBS. The cells were blocked with PBS containing 10% goat serum and then incubated with primary antibodies diluted in blocking buffer. The antibodies used were anti-DBP rabbit polyclonal antibody (generously provided by Peter van der Vliet, University of Utrecht), anti-E4orf4 mouse monoclonal antibody (48), and anti-DNA-PK catalytic subunit mouse monoclonal antibody (Sigma-Aldrich). The cells were washed and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG (Invitrogen). The cells were washed a final time and mounted onto slides with Immu-Mount (Thermo Shandon). Images were captured using a 63× objective with a Zeiss Axiovert 200M digital deconvolution microscope and analyzed with AxioVision 4.8.2 SP3 software. For high-resolution microscopy, images were captured using a 100× objective with a Nikon N-SIM microscope. Image processing, reconstruction, and analysis were carried out using NIS-Elements Ar/NIS-Elements C software and NIS-A N-SIM analysis. 3D renderings were generated using Imaris software.
ACKNOWLEDGMENTS
We are grateful to Y. Shiloh and Y. Ziv (Tel Aviv University), T. Dobner and T. Speiseder (Leibniz Institute for Experimental Virology), and M. Aladjem (NIH) for cell lines, antibodies, and plasmids and to Shani Bendori (Technion) for help with the Imaris software.
This work was supported by a grant (no. 2013140) to T.K. and P.H. from the United States-Israel Binational Science Foundation and by a grant (no. 261/15) to T.K. from the Israel Science Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. 2010. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 3.Davis AJ, Chen BP, Chen DJ. 2014. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J 22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavin MF. 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 6.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 8.Sirbu BM, Cortez D. 2013. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 5:a012724. doi: 10.1101/cshperspect.a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin JF, Knudsen KE. 2014. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov 4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. 2007. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem 282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 14.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. 2003. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yajima H, Lee KJ, Chen BP. 2006. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol 26:7520–7528. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. 2005. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem 280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 17.Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, Lee BJ, Dubois RL, Liu C, Yu X, Lan L, Zha S. 2015. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol Cell 58:172–185. doi: 10.1016/j.molcel.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. 2007. trans autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol 27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackford AN, Jackson SP. 2017. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Jette N, Lees-Miller SP. 2015. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol 117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X, Shen Y, Jiang N, Fei X, Mi J. 2011. Emerging roles of DNA-PK besides DNA repair. Cell Signal 23:1273–1280. doi: 10.1016/j.cellsig.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Weitzman MD, Ornelles DA. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686–7696. doi: 10.1038/sj.onc.1209063. [DOI] [PubMed] [Google Scholar]
- 23.Weitzman MD, Lilley CE, Chaurushiya MS. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol 64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 24.Hollingworth R, Grand RJ. 2015. Modulation of DNA damage and repair pathways by human tumour viruses. Viruses 7:2542–2591. doi: 10.3390/v7052542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilley CE, Schwartz RA, Weitzman MD. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol 15:119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Brestovitsky A, Nebenzahl-Sharon K, Kechker P, Sharf R, Kleinberger T. 2016. The adenovirus E4orf4 protein provides a novel mechanism for inhibition of the DNA damage response. PLoS Pathog 12:e1005420. doi: 10.1371/journal.ppat.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnell AS, Grand RJ. 2012. DNA viruses and the cellular DNA-damage response. J Gen Virol 93:2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- 28.Carson CT, Orazio NI, Lee DV, Suh J, Bekker-Jensen S, Araujo FD, Lakdawala SS, Lilley CE, Bartek J, Lukas J, Weitzman MD. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J 28:652–662. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 30.Boyer J, Rohleder K, Ketner G. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307–312. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 31.Tormanen Persson H, Aksaas AK, Kvissel AK, Punga T, Engstrom A, Skalhegg BS, Akusjarvi G. 2012. Two cellular protein kinases, DNA-PK and PKA, phosphorylate the adenoviral L4-33K protein and have opposite effects on L1 alternative RNA splicing. PLoS One 7:e31871. doi: 10.1371/journal.pone.0031871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Israel H, Sharf R, Rechavi G, Kleinberger T. 2008. Adenovirus E4orf4 protein downregulates MYC expression through interaction with the PP2A-B55 subunit. J Virol 82:9381–9388. doi: 10.1128/JVI.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondesson M, Ohman K, Manervik M, Fan S, Akusjärvi G. 1996. Adenovirus E4 open reading 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol 70:3844–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mannervik M, Fan S, Strom AC, Helin K, Akusjarvi G. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256:313–321. doi: 10.1006/viro.1999.9663. [DOI] [PubMed] [Google Scholar]
- 35.Muller U, Kleinberger T, Shenk T. 1992. Adenovirus E4orf4 protein reduces phosphorylation of c-Fos and E1A proteins while simultaneously reducing the level of AP-1. J Virol 66:5867–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Estmer Nilsson C, Petersen-Mahrt S, Durot C, Shtrichman R, Krainer AR, Kleinberger T, Akusjärvi G. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J 20:864–871. doi: 10.1093/emboj/20.4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanopka A, Mühlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjärvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 38.O’Shea C, Klupsch K, Choi S, Bagus B, Soria C, Shen J, McCormick F, Stokoe D. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J 24:1211–1221. doi: 10.1038/sj.emboj.7600597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleinberger T. 2014. Induction of cancer-specific cell death by the adenovirus e4orf4 protein. Adv Exp Med Biol 818:61–97. doi: 10.1007/978-1-4471-6458-6_4. [DOI] [PubMed] [Google Scholar]
- 40.Lavoie JN, Nguyen M, Marcellus RC, Branton PE, Shore GC. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J Cell Biol 140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livne A, Shtrichman R, Kleinberger T. 2001. Caspase activation by adenovirus E4orf4 protein is cell line specific and is mediated by the death receptor pathway. J Virol 75:789–798. doi: 10.1128/JVI.75.2.789-798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcellus RC, Lavoie JN, Boivin D, Shore GC, Ketner G, Branton PE. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J Virol 72:7144–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shtrichman R, Kleinberger T. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol 72:2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afifi R, Sharf R, Shtrichman R, Kleinberger T. 2001. Selection of apoptosis-deficient adenovirus E4orf4 mutants in Saccharomyces cerevisiae. J Virol 75:4444–4447. doi: 10.1128/JVI.75.9.4444-4447.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornitzer D, Sharf R, Kleinberger T. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in S. cerevisiae and interacts with the anaphase promoting complex/cyclosome. J Cell Biol 154:331–344. doi: 10.1083/jcb.200104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maoz T, Koren R, Ben-Ari I, Kleinberger T. 2005. YND1 interacts with CDC55 and is a novel mediator of E4orf4-induced toxicity. J Biol Chem 280:41270–41277. doi: 10.1074/jbc.M507281200. [DOI] [PubMed] [Google Scholar]
- 47.Roopchand DE, Lee JM, Shahinian S, Paquette D, Bussey H, Branton PE. 2001. Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 20:5279–5290. doi: 10.1038/sj.onc.1204693. [DOI] [PubMed] [Google Scholar]
- 48.Pechkovsky A, Lahav M, Bitman E, Salzberg A, Kleinberger T. 2013. E4orf4 induces PP2A- and Src-dependent cell death in Drosophila melanogaster and at the same time inhibits classic apoptosis pathways. Proc Natl Acad Sci U S A 110:E1724–E1733. doi: 10.1073/pnas.1220282110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc Natl Acad Sci U S A 96:10080–10085. doi: 10.1073/pnas.96.18.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleinberger T. 2015. Mechanisms of cancer cell killing by the adenovirus E4orf4 protein. Viruses 7:2334–2357. doi: 10.3390/v7052334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleinberger T, Shenk T. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol 67:7556–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcellus RC, Chan H, Paquette D, Thirlwell S, Boivin D, Branton PE. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Balpha subunit of protein phosphatase 2A. J Virol 74:7869–7877. doi: 10.1128/JVI.74.17.7869-7877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shtrichman R, Sharf R, Kleinberger T. 2000. Adenovirus E4orf4 protein interacts with both Balpha and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Balpha. Oncogene 19:3757–3765. doi: 10.1038/sj.onc.1203705. [DOI] [PubMed] [Google Scholar]
- 54.Sents W, Ivanova E, Lambrecht C, Haesen D, Janssens V. 2013. The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J 280:644–661. doi: 10.1111/j.1742-4658.2012.08579.x. [DOI] [PubMed] [Google Scholar]
- 55.Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. 2004. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 56.Hidalgo P, Anzures L, Hernandez-Mendoza A, Guerrero A, Wood CD, Valdes M, Dobner T, Gonzalez RA. 2016. Morphological, biochemical, and functional study of viral replication compartments isolated from adenovirus-infected cells. J Virol 90:3411–3427. doi: 10.1128/JVI.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautam D, Bridge E. 2013. The kinase activity of ataxia-telangiectasia mutated interferes with adenovirus E4 mutant DNA replication. J Virol 87:8687–8696. doi: 10.1128/JVI.00376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah GA, O’Shea CC. 2015. Viral and cellular genomes activate distinct DNA damage responses. Cell 162:987–1002. doi: 10.1016/j.cell.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziv Y, Jaspers NG, Etkin S, Danieli T, Trakhtenbrot L, Amiel A, Ravia Y, Shiloh Y. 1989. Cellular and molecular characteristics of an immortalized ataxia-telangiectasia (group AB) cell line. Cancer Res 49:2495–2501. [PubMed] [Google Scholar]
- 60.Li S, Szymborski A, Miron MJ, Marcellus R, Binda O, Lavoie JN, Branton PE. 2009. The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene 28:390–400. doi: 10.1038/onc.2008.393. [DOI] [PubMed] [Google Scholar]
- 61.Ahn JH, McAvoy T, Rakhilin SV, Nishi A, Greengard P, Nairn AC. 2007. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci U S A 104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirchhefer U, Heinick A, Konig S, Kristensen T, Muller FU, Seidl MD, Boknik P. 2014. Protein phosphatase 2A is regulated by protein kinase Calpha (PKCalpha)-dependent phosphorylation of its targeting subunit B56alpha at Ser41. J Biol Chem 289:163–176. doi: 10.1074/jbc.M113.507996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci U S A 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimura T, Martin MM, Torres MJ, Gu C, Pluth JM, DeBernardi MA, McDonald JS, Aladjem MI. 2007. DNA-PK is involved in repairing a transient surge of DNA breaks induced by deceleration of DNA replication. J Mol Biol 367:665–680. doi: 10.1016/j.jmb.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang MM, Hearing P. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol 63:2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinberg DH, Ketner G. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J Virol 57:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarnow P, Sullivan CA, Levine AJ. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517. doi: 10.1016/0042-6822(82)90054-X. [DOI] [PubMed] [Google Scholar]
- 68.Avital-Shacham M, Sharf R, Kleinberger T. 2014. NTPDASE4 gene products cooperate with the adenovirus E4orf4 protein through PP2A-dependent and -independent mechanisms and contribute to induction of cell death. J Virol 88:6318–6328. doi: 10.1128/JVI.00381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brestovitsky A, Sharf R, Mittelman K, Kleinberger T. 2011. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res 39:6414–6427. doi: 10.1093/nar/gkr231. [DOI] [PMC free article] [PubMed] [Google Scholar]