The gut microbiota is a diverse and dynamic ecological community that is increasingly recognized to play important roles in host metabolic, immunological, and behavioral functioning. As such, identifying new routes for manipulating the microbiota may provide valuable additional methods for improving host health.
KEYWORDS: diet, fats, heavy metals, microbiota, physiology, polyphenols, protein
ABSTRACT
The gut microbiota is a diverse and dynamic ecological community that is increasingly recognized to play important roles in host metabolic, immunological, and behavioral functioning. As such, identifying new routes for manipulating the microbiota may provide valuable additional methods for improving host health. Dietary manipulations and prebiotic supplementation are active targets of research for altering the microbiota, but to date, this work has disproportionately focused on carbohydrates. However, many other resources can limit or shape microbial growth. Here, we provide a brief overview of the resource landscape in the mammalian gut and review relevant literature documenting associations between noncarbohydrate nutrients and the composition of the gut microbiota. To spur future work and accelerate translational applications, we propose that researchers take new approaches for studying the effects of diet on gut microbial communities, including more-careful consideration of media for in vitro experiments, measurement of absolute as well as relative abundances, concerted efforts to articulate how physiology may differ between humans and the animal models used in translational studies, and leveraging natural variation for additional insights. Finally, we close with a discussion of how to determine when or where to employ these potential dietary levers for manipulating the microbiota.
INTRODUCTION
The human microbiota make up a large and diverse community that is increasingly recognized as playing a critical role in human biology (1). Many aspects of physiology are now understood to be directly or indirectly modulated by the microbiota, particularly in the gut but also elsewhere in and on our bodies (2–4). Because of the relevance to biomedical science, there is growing interest in manipulating the gut microbiota to correct imbalances and promote healthy functioning (5–7). Such efforts are pursued in both academic and industry settings, with a large focus on diet, whether at the scale of single supplement prebiotics (8), foods rich in probiotic organisms (9), or cultural differences in nutrient intake (10). It is certainly logical to attempt to use diet manipulations to shape the gut microbiota, as diet directly affects nutritional niches in the gut, thereby altering the competitive landscape for gut microbes (11). Indeed, diet has been found to be a major driver of gut microbial composition (12, 13). However, the question remains of how to best use diet to manipulate the gut microbiota.
To date, dietary interventions targeting the gut microbiota have focused largely on carbohydrates, primarily fiber components, and their fermentation to short-chain fatty acids. This emphasis on carbohydrates, and in particular indigestible carbohydrates, derives from a simple calculus: the densest populations of microbes (for most animals) reside in the distal gut, these microbes require nutrients to grow and reproduce, and diet-derived indigestible carbohydrates reach the distal gut more reliably than do diet-derived lipids and proteins (14–16). The gut microbial production of short-chain fatty acids is important for the host, providing 60% to 70% of the energy used by gut epithelial cells and up to 30% of the host’s total energy (17). However, it remains unclear what fraction of the short-chain fatty acids benefiting the host is derived from carbohydrate fermentation, as short-chain fatty acids are also produced from microbial protein fermentation (18, 19). Moreover, it is unknown how much of the gut microbiota’s own energy pool is produced through carbohydrate fermentation.
Other factors beyond carbohydrate availability are known to shape overall microbial load and/or patterns of competition among specific members of the gut microbiota; these may therefore have direct consequences for human health. Aerobic and anaerobic respiration, protein fermentation, and chemosynthesis are continually taking place (17, 20), and all microbial metabolism requires more than carbon substrates. The microbiota have been shown to be responsive to other dietary components (Table 1), for example, fat and protein (21), as is discussed more extensively later in this piece. Indeed, there is evidence that other elements may be more important than carbon for host management of the microbiota (e.g., nitrogen [22–24]). As such, provisioning or depleting noncarbohydrate microbial resources, limiting or otherwise, would provide additional, potentially even more efficacious, routes for manipulating the gut microbiota.
TABLE 1.
Select experimental findings on microbial responses to dietary interventions showing that dietary elements beyond fiber often contribute to microbial variation
| Resource(s) | Finding(s) | Host | Reference |
|---|---|---|---|
| Fat | Consumption of high-saturated-fat diet increased relative abundance of Bilophila wadsworthia and promoted inflammation in genetically susceptible mice via bile acid alterations | Mouse | 83 |
| Fat | High-fat diets altered the gut microbial community, but these responses were idiosyncratic based on fat source | Mouse | 112 |
| Fat | High-fat-diet-associated small intestinal microbial community altered lipid digestion even when mice were fed a low-fat diet | Mouse | 109 |
| Fat | Switch to Westernized diet produced relative increases in Firmicutes and decreases in Bacteroidetes, a decrease in microbial diversity, and a greater increase in body fat than in controls | Mouse | 110 |
| Fat, fiber | Bacterial communities in mice fed low-fat/high-fiber diets or high-fat/high-sugar diets differed in composition but were mostly resilient to diet changes; in contrast, viral communities responded rapidly to switch between diets | Mouse | 111 |
| Fat, fiber | Microbial responses to introduction of high-fat/low-fiber or low-fat/high-fiber diets were documented within 24 h but were insufficient to overcome interindividual variability | Human | 13 |
| Fat, sugar | High-fat/high-sugar and low-fat/high-fiber diets shaped the gut microbiota consistently across mice with different genotypes and metabolic/immune phenotypes; blending the diets led to proportional changes in the gut microbiota | Mouse | 12 |
| Iron | Infant iron supplementation increased enterobacterial and Clostridium abundances, including many pathogens | Human | 38 |
| Iron | Child iron supplementation altered the gut microbiota, with a relative increase in enterobacteria and decrease in lactobacilli, even without changing human iron status | Human | 146 |
| Protein | Higher dietary casein levels increased total microbial DNA; some taxa, including members of the Clostridia and the sulfate reducer Desulfovibrio, decreased | Mouse | 120 |
| Protein | Gut microbial community was responsive to dietary fat content but not protein/sucrose ratio; host adiposity and survival were shaped by protein/sucrose ratio | Mouse | 121 |
| Protein | Increasing protein levels led to higher total microbial loads and changes in composition, including Bacteroidaceae absolute abundance | Mouse | 24 |
| Protein | High-protein diets changed fecal short-chain-fatty-acid concentrations, most notably reducing butyrate levels while also reducing the proportion of some Firmicutes and members of the Bacteroides | Human | 122 |
| Protein | Changes in dietary protein or fiber amt did not alter the microbial community at the phylum level, but high-protein diets were associated with an increase in Oscillibacter and a decrease in Collinsella aerofaciens | Human | 123 |
| Protein, fiber | Microbial relative-abundance and diversity responses to altered protein and fiber levels were more significant than responses to changes in fat or energy density across a range of diets | Mouse | 23 |
| Protein, fat | Short-term human diet interventions involving high-protein/high-fat diets resulted in rapid changes in the microbiota, including increases in bile-tolerant bacteria like Bilophila wadsworthia and members of the Bacteroides, with concurrent reductions in some Firmicutes | Human | 21 |
| Protein, fiber, fat, sugars | Microbiota changes and associated inflammation were consistently recorded in response to various levels of multiple fiber and protein sources but not digestible carbohydrates or most fats | Mouse | 189 |
Here, we propose that the field of microbiome research should expand thinking about dietary interventions. We intend not to undermine the value of carbohydrate-focused work but instead to extend the toolkits of researchers and clinicians by highlighting additional targets for research. We begin by briefly outlining the nutrient environment experienced by the microbiota and the mechanisms shaping that landscape. Next, we discuss noncarbohydrate resources in the gut that are likely contenders to impact the microbiota and new approaches for studying the effects of these resources. Finally, to emphasize the value of expanded approaches to dietary interventions, we also address what the outcome of such interventions could be, including alterations to both the metabolic and nonmetabolic contributions of microbiota to the host.
HOST DIGESTIVE PHYSIOLOGY AND THE RESOURCE ENVIRONMENT OF THE GUT MICROBIOTA
The mammalian gut microbiota as a whole is dominated by obligate anaerobes from the Firmicutes and Bacteroidetes phyla. However, gut microbial abundance, composition, and function differ radially and longitudinally across regions in response to fine-scale differences in the resource environment (1, 25).
Radial variation in the microbiome is driven by host immune defense, mucus production, epithelial sloughing, and environmental gradients in nutrients and oxygen (26, 27). Lateral communities, adhering to the epithelium or mucosal layers, are generally less diverse and less densely colonized than luminal bacterial communities (27–29). The microbes that are present are more likely to be oxygen tolerant and specialize in the metabolism of protein than luminal microbes (26). Due to their relatively high dependence on host secretions rather than diet-derived nutrients, epithelial and mucosal adherent microbial communities are generally more stable over time than luminal microbial communities (27) and likely more resistant to manipulation via diet. Because of their stability, and the fact that surveying these communities is invasive and therefore impractical for temporal tracking of the effects of a dietary intervention on the gut microbiome, we focus here on the luminal microbial community.
Across mammals, longitudinal variation in the gut microbiota is driven largely by host gut ecophysiology. Diet directly impacts the presence of macronutrients, micronutrients, and phytochemicals in the lumen and, over evolutionary time, has also shaped the basic structure of the gut and related host-derived environmental factors like pH, passage rate, and enzyme production. At the broadest level, this is evident in the differences between foregut and hindgut fermenters. Ruminant and pseudoruminant animals, such as cows, sloths, and colobus monkeys, have independently evolved specialized multichambered stomachs that allow microbial fermentation to occur prior to the passage of chyme into the small intestine, where the majority of host digestion and absorption occurs. In these gastric fermentation chambers, conditions are maintained to encourage efficient microbial metabolism of cellulose, including relatively high pH and relatively low passage rates for individual particles. In contrast, hindgut fermenters, like horses and rhinoceroses, have physiological adaptations to promote microbial residence and fermentation distal to the stomach and small intestine in an enlarged hindgut and a well-developed cecum. In both cases, these animals, which forage primarily on plant materials but lack the enzymes required to break down cellulose and hemicellulose themselves, actively promote microbial growth and fermentation and in turn gain a substantial fraction of their total energy budget from the absorption and metabolism of microbiota-derived volatile fatty acids. In contrast, animals with simple guts synthesize the enzymes required to digest the majority of their diet and exhibit a range of adaptations that serve to restrict the largest populations of microbes to the colon, ensuring that the host retains first access to nutrients.
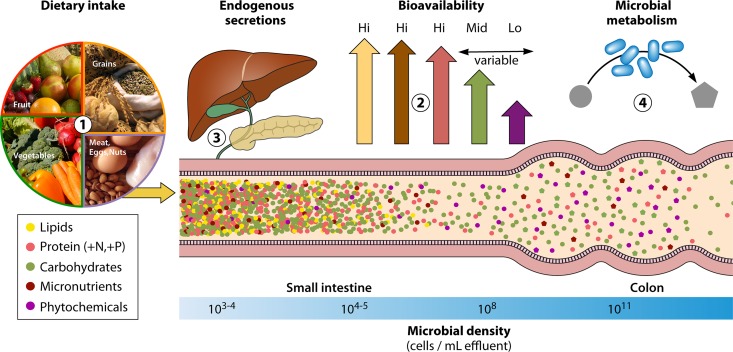
Humans exemplify this first-pass resource access strategy. Gut microbial density increases exponentially along the human gastrointestinal tract, with 102 to 103 microbes per ml of effluent in the stomach, 103 to 105 in the proximal small intestine, 108 in the ileum, and 1011 in the colon (1, 30, 31). Concomitant with this increase in microbial density are gradients in oxygen, pH, antimicrobial peptides (AMPs), and immunoglobulins (1). These gradients are the outcome of coevolution between humans and the human gut microbiota, a shared history marked by cooperation but also competition, in which humans have maximized the fraction of ingested nutrients serving our own metabolism by isolating primary digestion from the bulk of the microbial community.
Ultimately, the ecological niches available for microbial colonization are determined by the interaction of host physiology and dietary ecology, with diet being more readily modifiable than physiology. Below, we review how diet interacts with host physiology to shape gut microbial communities, with the goal of highlighting some properties of diet that serve as promising targets for therapeutic manipulation.
HOST DIET AND NUTRIENT AVAILABILITY IN THE HUMAN GUT
Nutrient availability in the gut is largely a function of four distinct processes: (i) dietary intake, (ii) host uptake of diet-derived nutrients, (iii) host endogenous secretions, and (iv) microbial metabolism of diet-derived, endogenous, and microbiota-derived compounds (Fig. 1). The broad relevance of dietary intake is clear, as most resources in the gut, as in the rest of the body, are ultimately acquired via diet. However, downstream processing of diet-derived compounds, by both host and microbiota, contributes importantly to the resource environment of the lumen. We focus here on these other processes shaping nutrient availability.
FIG 1.
Nutrient landscape of the gut as shaped by host and microbial processes. Gut microbial absolute and relative abundances are expected to be sensitive to the nutrients available within the gut lumen. Nutrient composition in the gut lumen is, in turn, dependent on (i) dietary intake of macronutrients, micronutrients, and phytochemicals; (ii) the bioavailability of those nutrients within the small intestine, which alters the fraction of ingested nutrients reaching the densest microbial communities in the colon; (iii) endogenous secretions such as bile acids and digestive enzymes that alter competitive dynamics among gut microbial taxa and/or modulate bioavailability; and (iv) microbial metabolism of nutrients, producing metabolites that may have downstream effects on competitive dynamics within the gut microbiota.
Host uptake of diet-derived nutrients.
Given that the densest and most diverse gut microbial populations in humans reside in the distal gut, in considering the effect of diet on the nutrient environment, it is critical to consider the fraction of diet-derived nutrients that actually reaches the colon.
Distinct pathways exist for the digestion of carbohydrates, proteins, and fats, but their absorption is not equally efficient. Free dietary lipids are almost completely absorbed by the terminal ileum, although fats contained within plant cell walls may remain inaccessible to digestion unless cell walls are ruptured by processing (32, 33). In contrast, dietary proteins and dietary carbohydrates show high degrees of variability in bioavailability based on their chemical form (16). Denaturation of proteins via heat or acid causes the protein structure to unfold, promoting bioavailability; for instance, there is a 2-fold increase in the ileal digestibility of egg protein served raw (51% to 65%) versus cooked (91% to 94%) (34, 35). In humans, most protein-rich foods are routinely cooked, and approximately 80 to 90% of proteins are absorbed in the small intestine (36). The bioavailability of carbohydrate also depends on its structure, with simple sugars being readily absorbed in the small intestine, complex polysaccharides (e.g., cellulose, lignin, pectin, and oligosaccharides) resisting digestion in the small intestine, and starch being either highly digestible or resistant, depending on its form (15, 37). Starch, the most common carbohydrate in the human diet, exists in a native state that is resistant to digestion by amylases, but here too, cooking produces consistent and significant increases in ileal digestibility ranging from 28% to 109% for substrates tested in humans (16).
The small intestine is also the site of vitamin, mineral, and metal absorption. Most water-soluble vitamins (e.g., vitamin C) are readily absorbed via active transport in the jejunum, while protein-bound B vitamins and fat-soluble vitamins (e.g., vitamins A, D, E, and K) are primarily absorbed further along the small intestine. Minerals are absorbed in the small intestine via either active transport in the proximal small intestine or passive diffusion around tight junctions in the distal small intestine. Divalent metals such as iron can be transported from the duodenal lumen via the divalent metal transporter or enter enterocytes in the form of heme via endocytosis, although not all dietary iron is taken up (38). Other diet-derived compounds, such as phytochemicals and chemical by-products of food processing, exhibit various degrees of absorption in the small intestine. Many common compounds, such as plant-derived polyphenols (39), are known to pass through the mammalian small intestine largely intact. Such poorly absorbed compounds may be especially likely to affect the microbiome because they concentrate in the colon as digestible nutrients and water are removed.
Ultimately, dietary material depleted of macronutrients and micronutrients passes into the colon, where water and electrolytes are recovered directly and residual nutrients become substrates for microbial metabolism. The mammalian colon has a limited ability to absorb diet-derived nutrients directly, but volatile short-chain fatty acids produced during microbial fermentation of carbohydrates and proteins diffuse into enterocytes, where they serve as important metabolic fuel for both local and systemic energy metabolism. Nutrients may be salvaged after processing in the large intestine through coprophagy, but this process is common only in some nonhuman species (40).
Host endogenous secretions.
In addition to diet-derived nutrients, the intestinal lumen is also rich in products produced by the host. For instance, epithelial cells are sloughed constantly, having the highest turnover rate of any fixed-cell population in the body. Upon entry into the intestinal lumen, these shed cells become substrates for both host and microbial metabolism (41). Epithelial cell life cycles interact with nutritional factors, with luminal nutrients directly modulating the rate of intestinal cell proliferation (42, 43).
Beyond the luminal nutrients introduced by epithelial cell sloughing, the host gut actively secretes compounds that modulate the gut microbiota via nutritional or antimicrobial effects. Goblet cells located along the entire intestinal tract secrete a continuous layer of mucus, composed of mucin glycoproteins (primarily MUC2) and trefoil peptides. Mucus serves as a critical barrier between the epithelium and the gut lumen, whose production is modulated by both host genetics and diet (44–46). However, mucus is also both a habitat and a nutrient source that differentially promotes the growth of specific microbial taxa. Although commensal bacterial genomes are generally rich in glycan-degrading enzymes (47, 48), some bacterial taxa (e.g., Akkermansia muciniphila [49] and Ruminococcus gnavus [50]) produce specialized glycosidases that confer an enhanced ability to metabolize MUC2 oligosaccharides. Gut microbiota composition can further alter patterns of oligosaccharide glycosylation (51), and the activity of mucolytic bacteria like A. muciniphila may actually stimulate mucus production in the host (52).
The host gut also produces a range of antimicrobial compounds to restrict microbial growth and prevent invasion into the mucosa. For instance, intestinal epithelial cells secrete IgA into the lumen via the polymeric immunoglobulin receptor (53), releasing an estimated 3 to 5 g daily (54). Paneth cells secrete a wide range of antimicrobial peptides (AMPs) that disrupt gut microbial cell membrane integrity and/or cellular function, including α-defensins and lysozymes (55). Jointly, IgA and AMPs regulate the growth of commensal gut bacteria, exerting selective effects that shape gut microbial community structure (56, 57). However, the extent to which diet shapes IgA and AMP production in the lumen remains to be determined.
Animals also secrete bile acids with robust direct (58, 59) and indirect (60) bactericidal properties. Bile is released into the duodenal lumen in response to ingestion of fats and serves to emulsify dietary lipids. Roughly 95% of bile acids released into the small intestine are reabsorbed in the ileum for recycling, but approximately 400 to 800 mg of bile acids daily escapes enterohepatic circulation (61). Residual bile acids can reach high-millimolar concentrations in the colon, where differential resistance to the antimicrobial effects of bile acids contributes to shaping gut microbial structure and function (62–65). Given any differential sensitivity of gut microbial taxa, dietary-fat-induced variation in bile acid production can be expected to influence gut microbial community structure and function.
Microbial metabolism of luminal compounds.
The gut microbiome harbors a metabolic capacity that far exceeds our own (66). Microbes can transform carbohydrates in ways that extend beyond fermentation. For instance, some microbes can degrade host-produced glycans such as intestinal mucus and human milk oligosaccharides, releasing nutrients into the lumen and exposing the residual glycan structure to further degradation by other members of the commensal community (67–69). The colonic gut microbiota can metabolize a wide variety of noncarbohydrate dietary compounds that resist digestion in the small intestine, including proteins (18, 70), polyphenols such as lignans (71, 72), and xenobiotic compounds like polycyclic aromatic hydrocarbons (73). Gut bacteria can also synthesize numerous macronutrients and micronutrients, including essential amino acids like lysine. Indeed, isotopic labeling studies in humans have found microbiota-derived lysine to contribute substantially to the plasma lysine and body protein pools, even on nitrogen-adequate diets (74, 75). Such activities can modify the resource environment of the gut, independent of and in conjunction with host action, with further consequences for gut microbial structure and function.
Bile acids offer a prime example of the ecological consequences of noncarbohydrate microbial metabolism. As mentioned above, bile acids that escape enterohepatic circulation and pass into the colon can possess strong antimicrobial properties. A key line of gut bacterial defense is to manipulate the antimicrobial properties of the primary bile acids produced by the host via deconjugation or transformation into secondary bile acids with altered chemical properties. Deconjugation involves the action of bile salt hydrolases (BSHs) that hydrolyze the bond linking the bile acid to the amino acid conjugate, which in humans is either taurine or glycine. BSH activity appears unique to the gut environment (76) and can show high redundancy within carrier taxa (e.g., Lactobacillus plantarum carries 4 functional BSH genes [77]), in which it may confer a survival advantage by reducing the stronger detergent properties of the conjugated forms. BSH activity is encoded in multiple Gram-positive and archaeal taxa but appears less widely distributed among Gram-negative bacteria, with functional BSH genes detected to date only among Bacteroides strains (62). Interestingly, taurine versus glycine conjugation of bile acids appears to depend on habitual diet, with animal-rich diets increasing the rate of taurine conjugation and plant-based diets enriching for glycine conjugation (78, 79). The higher rate of taurine conjugation on animal-rich diets is presumably due to an increased availability of taurine, which is derived exclusively from animal tissue and which humans have lost the ability to synthesize efficiently (80, 81). Thus, diet might shape gut microbial community structure via BSH both by changing the abundance of microbes possessing hydrolase activity and by modulating luminal concentrations of free taurine and glycine, which can be used as sources of nitrogen and carbon to enhance growth (82).
Notably, gut microbial responses to taurine versus glycine conjugation can also impact human health directly. For instance, because taurine catabolism releases sulfite in addition to ammonia and carbon dioxide, taurine deconjugation may select for sulfidogenic bacteria such as Bilophila wadsworthia, with downstream consequences for intestinal inflammation (83). In addition, germination of the nosocomial enteric pathogen Clostridium difficile from spores is maximized in the presence of taurocholic acid and glycine (84); indeed, most clinical isolates of C. difficile appear to require the taurine conjugate to germinate (85). Together, such data suggest a microbiota-mediated link between taurine-rich diets and the risk of gastrointestinal pathology.
Gut bacteria also transform host-produced primary bile acids into secondary forms with altered chemical properties. Cholic acid (CA) and chenodeoxycholic acid (CDCA) together represent 80% of the primary bile acid pool in humans (86) and are transformed by microbial metabolism into the two most abundant secondary bile acids in the human gut, deoxycholic acid (DCA) and lithocholic acid (LCA), respectively (87). Conversion of primary to secondary bile acids typically involves 7α-dehydroxylation, a multistep biochemical pathway found exclusively in anaerobic gut bacteria, most notably among members of Clostridia cluster XIVa (87–90). Correspondingly, the antibiotic-induced reduction in bacterial taxa capable of 7α-dehydroxylation shifts the fecal bile acid pool from secondary to primary bile acids (84, 91). Gut bacteria are also capable of oxidizing and epimerizing other hydroxy groups in the primary bile acid structure, altogether yielding more than 20 secondary structures (62). As a result of this microbial activity, the fecal bile acid pool consists almost exclusively of unconjugated and secondary bile acids (87).
Bacteria capable of transforming primary to secondary bile acids might benefit by outcompeting or inhibiting taxa more sensitive to secondary bile acids (62), with downstream effects on host health. For instance, Clostridium scindens and Clostridium hiranonis show 10-fold-higher 7α-dehydroxylating activity than other Clostridia, a phenotype linked to the production of an NAD(H)-dependent 3-dehydro-4-bile acid oxidoreductase encoded by the baiCD gene cluster (92, 93). This 7α-dehydroxylating activity is thought to underlie the protection conferred by these taxa against C. difficile colonization (94, 95), an effect potentially mediated by the inhibitory effect of secondary bile acids on C. difficile germination (84, 96). Notably, symptomatic C. difficile infection in humans was recently associated with negative or low baiCD gene cluster abundance, and a case study reported that successful treatment of recurrent C. difficile infection via fecal microbiota transplant (FMT) was associated with a shift from a baiCD-negative status pre-FMT to a baiCD-positive status post-FMT (91). Similarly, experimental administration of C. scindens was recently shown to confer resistance to C. difficile infection, with effects dependent on the synthesis of secondary bile acids (95).
Microbial processing of mucus is another example of the complex metabolic capacity of the gut microbiota. Although the capacity to degrade mucin is phylogenetically widespread (97), in vivo stable isotope experiments have found that there is significant interspecific and intraspecific variation in mucin foraging (98). There is also differential uptake of various components of mucin, with most cells incorporating mucin-sourced nitrogen, likely via its circulation in the free ammonium pool and incorporation during amino acid synthesis, while only a small proportion of the community takes up mucin-sourced carbon through direct metabolism of mucin oligosaccharides (98). Mucus serves as an important resource for the gut microbiota, especially when dietary resources are reduced. For example, mucin metabolism increases under dietary fiber deprivation (99), leading to a thinner mucosal layer and increased pathogen susceptibility. Correspondingly, mucus-degrading bacteria are relatively more abundant in hibernating or fasting animals (100, 101).
Ultimately, as these examples attest, the resource landscape of the distal gut lumen is a complex function of host physiology, nutritional ecology, and microbial activities. However, the system reacts to dietary levers in ways that, with additional research encompassing a range of substrates, we may ultimately be able to control precisely.
NONCARBOHYDRATE DIETARY COMPONENTS AND THE GUT MICROBIOTA
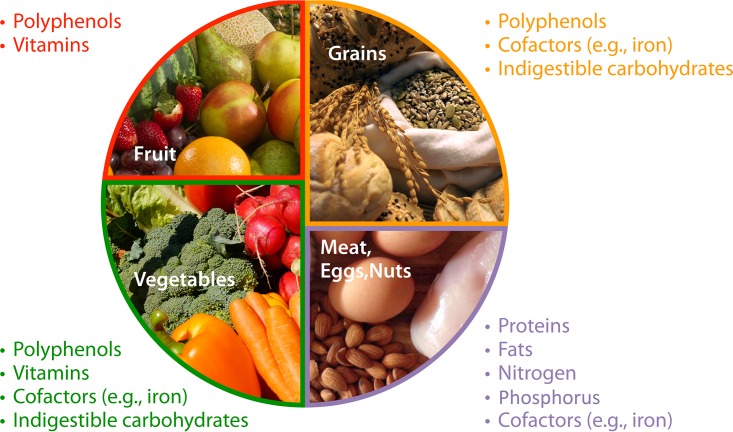
Much research effort has focused on relating variation in the gut microbiota to differences in diet. Diet dissimilarities between countries (102, 103), seasons (104, 105), and lifestyles (106, 107) are all associated with variation in the gut microbiota. More broadly, contrasts, and similarities, between animal species in their gut microbiota are also ascribed to diet (108). However, isolating what specific aspects of diet drive gut microbial composition and function is a complex and multifaceted problem. Gut microbial taxa may respond to differences in macronutrients, micronutrients, and elemental availability, all in various intersecting manners. Does a change following altered protein intake reflect responsiveness to protein availability? The macronutrient(s) replacing or replaced by protein? Nitrogen availability? Iron or other minerals found in protein-rich food? Most experiments unfortunately do not allow for ascribing causality precisely. Here, we briefly review some recent experimental work exploring the relationship between the noncarbohydrate fraction of diet and the gut microbiota and highlight some of the dietary constituents that we expect contribute to gut microbial responses (Fig. 2).
FIG 2.
Many resources relevant to microbial growth and function are naturally sourced from the typical human diet. Identifying what specific resources drive microbial differences in humans whose diets vary is a complicated endeavor, however. Here, we show how the USDA MyPlate-recommended diet components include various microbially relevant resources such that changing diet broadly may not allow researchers to ascribe causality to microbial changes. Further theory on limiting resources and controlled experiments isolating individual aspects of diet are necessary to develop a dynamic range of manipulations and to understand the degree to which host physiology modulates microbial responses to diet.
As many studies have found differences in the gut microbiota and health outcomes of Western and non-Western populations, much research has been performed in humans and mice using Western-style high-fat diets (12, 13, 83, 109, 110) or other altered-fat diets (111, 112). The gut microbiota is typically found to respond to changes in dietary fat, perhaps unsurprisingly since the gut microbiota is known to contribute to host metabolism of fats (21, 109, 113, 114). The effect of increased fat consumption can often be specifically tied to changes in bile acid production and metabolism (83, 109, 115, 116). Interruption of microbial bile acid metabolism by antibiotics or dietary shifts has been associated with negative health outcomes, including inflammation (117) and C. difficile infection (94, 95). As such, alterations to dietary fat consumption and thus bile acid production are expected to serve as meaningful tools for manipulating the gut microbiota, particularly in the context of disease. In most humans consuming Western-style diets, this may mean focusing on reducing fat intake, altering the proportional intake of fats from different sources (118, 119), and/or combatting downstream microbial effects through alternative diet changes.
Less research has been conducted on dietary protein levels than on fats or carbohydrates. However, work that has been carried out has shown significant responses in the gut microbiota following increases in protein intake (21, 23, 120–123). Indeed, changes in protein levels have been found to be one of the most significant predictors of gut microbial responses in studies that compare populations (13) or multiple dietary manipulations (23, 120). It has been proposed that this is because nitrogen is a limiting nutrient in the gut (24), and therefore, nitrogen availability would determine overall biomass in the system (24) and also set the terms for competition over other resources like carbohydrates (23). Nitrogen is commonly limiting in environmental microbial communities (124–127) as well as for animal hosts (128). Nevertheless, how particular forms of nitrogen (e.g., protein, amino acid, and free circulating ammonium [129]) contribute to microbial dynamics in the gut requires further study. Microbes both utilize and produce amino acids, for example, modulating the availability of the essential amino acid tryptophan, with downstream implications for inflammation, disease, and nervous system signaling (18, 130–132). Dietary sources will need to be considered alongside host secretions like mucus (98) and urea (133) as well as microbial biosynthesis of nitrogen-rich products (75) that can also play a prominent role in gut nitrogen dynamics. Overall, due to the theoretical and empirical support for nitrogen limitation, it seems likely that changing dietary protein will be a powerful tool for manipulating the gut microbiota.
Beyond the macronutrients, there are many other components of diet that matter for host nutrition and likely also gut microbial growth and functioning. For example, phosphorus is another element that is commonly limiting for microbes in the environment (126, 134–136) and has been shown to limit gut bacterial growth in vitro (137), but its role in vivo has not yet been studied. Better understood are trace metals, which are essential micronutrients for animals and can also contribute to gut microbial dynamics (138). Most research has focused on the importance of metal-based molecules for pathogen colonization and growth (38, 139–141) or their antimicrobial functions (142), but the competition for metals observed in pathogens (143) can also affect commensals (129, 137, 144, 145). Iron supplementation can alter gut microbial composition, most notably with increases in potential pathogens, even without changing host iron levels (38, 146). Zinc deficiency or excess has also been shown to produce altered gut microbial profiles and metabolism (147, 148). Similarly, polyphenols from plant sources, including tea, wine, and berries, can also modulate the gut microbiota by depleting sensitive taxa, promoting the growth of resistant taxa, and/or manipulating host-microbe interactions in a manner linked to positive health outcomes (149–152), including attenuated development of obesity and glucose intolerance in mice fed a high-fat/high-sugar diet (150, 153). Characterizing differences in nonmacronutrient dietary components accurately is challenging in observational studies, particularly ones that rely on dietary recall or population-wide trends. Therefore, relationships between these aspects of diet and gut microbial communities likely have gone undetected in many studies. Nevertheless, the underlying biology implies that they may prove to be rich untapped drivers of the microbiota.
Diet also impacts substrates for microbial respiration. While these molecules are not typically sourced directly from the diet, their availability as well as their usage are in part determined by host metabolism of diet. Various electron acceptors have been found to play an important role in pathogenesis, providing a unique metabolic niche that few strains can utilize to colonize (154–158). More generally, changes in electron acceptor availability underlie competition between respiring bacteria and fermenters and contribute to community dynamics following antibiotic disturbance (159). Similarly, competition between acetogens, methanogens, and sulfate-reducing bacteria for hydrogen gas, an end product of carbohydrate fermentation (160), likely dictates the levels of these organisms in the human gut (161), as has been shown in ruminants (162, 163). The availability of sulfate will also determine the abundance of sulfate reducers like Desulfovibrio piger (164, 165), although the potentially toxic effects of their metabolism, mediated by the sulfide end product, will be shaped by the abundance of methanogens which can utilize the hydrogen in H2S and thereby detoxify it (166).
Various other molecules and substrates likely play a role in microbial community dynamics in the gut. Numerous therapeutic and diet-derived xenobiotics are known to change gut microbial growth in vivo (167) and in vitro (168). For instance, a recent in vitro screen of more than 1,000 nonantibiotic drugs found that 24% of drugs with human targets inhibited the growth of at least 1 of the 40 human gut bacterial strains tested (168). Resource availability may also determine the ability of gut bacteria to make signaling molecules or other molecules mediating microbial interactions. Diet and microbial metabolism can affect gut pH (169) or redox state (159) and have further downstream effects on microbial composition. Although an in-depth review of all relevant conditions is not possible here, we hope that these examples highlight the diversity of diet-associated properties beyond fermentable carbohydrates that impact the gut microbiota.
RECOMMENDATIONS FOR FUTURE RESEARCH
As outlined in the section above, various noncarbohydrate diet interventions can be expected to manipulate the gut microbiota. We would argue, however, that adjustments to the way that we analyze such manipulations will help identify and refine appropriate substrates of focus above and beyond just testing more resource types. Although there are numerous potential pathways to enhance the detection and mechanistic characterization of dietary levers, we offer four initial recommendations for new tacks to studying dietary properties that shape and can potentially manipulate the gut microbiota.
First, we propose that in vitro studies should utilize more-diverse media to ensure that all aspects of nutrient limitation might be captured. Minimal media are typically carbon limited and thus are most likely to identify interactions based on carbon. Studies with complex media most often use modified Gifu anaerobic medium (MGAM), a rich medium with high success for isolating gut strains (170) but one mismatched to the gut with regard to the abundance of various substrates. For example, at a very simple level, MGAM has a much lower carbon-to-nitrogen ratio (i.e., much higher nitrogen availability) than the mammalian gut (24). Varying nutrient availability even among rich media can result in highly differential growth (171), and more-thorough assays, including variation in the concentrations as well as kinds of nutrients available, should be done to identify nutrient requirements of different taxa. However, if a single medium is to be used, it should be designed to more closely hew to conditions found in the gut.
Second, we propose that research more often include measurements of changes in absolute abundance. Amplicon sequencing produces compositional data, which have many statistical limitations (172, 173). Recent work by Vandeputte and colleagues (174) demonstrated that variation in absolute abundance underpins changes in host phenotype more so than variation in relative abundance. However, only a small fraction of research on gut microbial responses to diet incorporates absolute-abundance data. It has been shown that changes in dietary protein altered overall concentrations of fecal bacteria in mice (24), supporting the inference that nitrogen is limiting in the gut. In contrast, while adding an indigestible carbohydrate (porphyran) to mouse diets allowed colonization by an exogenous strain of Bacteroides, there was not an overall increase in the number of bacteria (175); that is, the new strain ousted an equal number of other bacteria, a distinction not evident from relative-abundance data alone. Between these two studies, we can observe that some dietary manipulations (like changes in protein content) change the total biomass of the ecosystem, whereas others (like the addition of porphyran) change only community composition. Each may be a worthwhile goal, but they are nevertheless distinct. Appreciating and tailoring experiments to capture these differences will be necessary for interventions targeting specific health outcomes.
Third, we recommend more prudence when translating data from mouse research to humans. The mouse gastrointestinal tract differs in important ways from that of humans from mouth to anus and as such likely harbors a unique resource environment. For example, mice have lower ratios of small intestine to large intestine length (176) and area (177), which may decrease nutrient uptake proximal to the bulk of the microbial community, thereby dampening resource limitations present in humans. Mouse studies frequently report microbial community structural and functional profiles based on endpoint cecal samples; these profiles are unlikely to be recapitulated in humans, who lack an enlarged, functionally separate cecum (176). More generally, the mouse microbiota is not identical to that of humans (178); thus, humanized mice (i.e., germfree mice colonized by a human gut microbial community) are considered a closer alternative (179). However, humanized mice retain the anatomical differences from humans and also have numerous physiological differences from conventional mice, including a deficient immune system (180), which may make them less appropriate for use in studies of disease. It is also important to note that the environment experienced by mice in experiments differs from typical human conditions. Most notably, mouse diets do not vary under standard husbandry, so the introduction of a new diet or diet component may represent a more significant disturbance than when a human introduces a new food atop their normally variable diet. As omnivorous mammals, mice are ecologically similar to humans in many ways and have a long history of coresidence with humans. Moreover, given that laboratory mice have been reared for hundreds of generations in direct contact with humans and consuming highly processed (milled and cooked) diets, it is possible that mice and humans exhibit a degree of convergent adaptation in digestion that remains unexplored. Certainly, given the exceptional genetic, environmental, and microbiological control possible with mice, and the depth of systemic understanding arising from their frequent use in biological studies (181), mice will remain critical experimental models for studies of the microbiota. However, we would advocate that researchers should keep the differences outlined above in mind and seek to treat them more transparently.
Finally, we encourage leveraging variability in responses to more clearly understand the nuances inherent in dietary interventions. It is likely not the case that the same nutrients will be limiting in every gut due to individual host variation in diet or physiology. Focusing on individual responses will provide a clearer picture of what nutrients can shape the gut microbial community and in what contexts. For example, studies targeting resistant starch as a possible prebiotic have observed variable responses, with nearly half of individuals showing limited responses in gut microbial structure or microbial function, as measured by short-chain-fatty-acid concentrations (182). Similarly, some individuals have been found to pass most added resistant starch intact, whereas others have microbial communities that ferment >95% of it (123). With more-extensive studies of what defines a responsive community, it may be possible to design personalized interventions tailored to an individual’s baseline microbial community composition and functional potential.
MANIPULATIONS TO WHAT END?
While many resources can shape the composition and function of the gut microbiota, different resource manipulations will be appropriate depending on the context. The goal of intervention to either promote or eliminate particular microbially mediated host phenotypes should determine the area of focus. In general, such phenotypes can be considered to belong to either of two categories: nutritional and nonnutritional contributions of the gut microbiota.
Unsurprisingly, nutritional contributions of the gut microbiota can be manipulated by altering the nutrients available in the lumen. Microbes break down diet components to grow and in so doing may produce metabolites taken up by the host, such as short-chain fatty acids but also amino acids, lactate, and ammonia (11, 17). Microbial symbionts can also produce nutrients that are missing from or insufficient in the diet, including essential vitamins (183) and amino acids (75). Changing diets can produce alterations to these metabolic pathways, resulting in changes in host provisioning (e.g., see reference 110). Focusing diet interventions on substrates that are necessary in the metabolic process of interest, and in particular those substrates that are limiting to the process, will allow for greater dynamic control over microbial metabolic provisioning. However, it is important to note that the gut microbiota can also compete with the host for resources (184) or hurt the host when overproducing certain metabolites (185), so increasing populations through provisioning will not always be beneficial.
Beyond nutritional contributions, the gut microbiota shapes the physiology of many other host organ systems. The gut microbiota plays a crucial role in colonization resistance (186), it modulates the immune system (4), and it produces or alters host production of signaling molecules, with effects on organs as distant as the brain (187, 188). Manipulating the gut microbiota through diet to promote these functions is less straightforward, at least when it comes to determining what manipulations to use. Substrates necessary to promote the growth of species of interest or to make the signaling molecules involved will need to be identified first either through in vitro screens or through more-expansive measurements of functioning in diet intervention studies. The effects of dietary changes on immune functioning or signaling may be more complicated than those on metabolism. For instance, increasing mucosal secretions to support a physical barrier and colonization resistance may involve any or all of the following: promoting growth of bacterial taxa (e.g., Firmicutes) that produce the short-chain fatty acid butyrate, whose uptake promotes mucus secretion (45); limiting the growth of taxa that catabolize mucus (e.g., Bacteroidetes) (98); promoting the growth of mucin degraders (e.g., Verrucomicrobia) that nevertheless stimulate robust barrier function (52); altering cross-feeding interactions among gut bacteria at the mucus barrier (69); or fostering a complex gut microbiota that interacts with various aspects of host biology to improve overall immunity.
More broadly, to support host health, it will be necessary to determine what a healthy gut microbial community looks like. Of course, the answer will not necessarily be the same for all individuals, but in the absence of a particular functional target, general structural guidelines will be necessary before beginning interventions. Some diets, for example, those including protein supplementation, could increase the overall bacterial load (24, 189), but it remains unknown what constitutes an ideal bacterial load for a human gut. Similarly, while low diversity is often considered a marker for community imbalance, there is little observational or experimental evidence documenting ideal diversity levels or their functional implications (190). Better defining of the goals in microbial manipulation will require answering these questions and others, with particular attention paid to both the evolutionary context in which our relationship to the microbiota arose as well as how contemporary contexts may differ. Gut microbial profiles that were healthy historically and those most improving health among traditional subsistence, developing, and industrialized populations can all be expected to differ. Rational attempts to engineer the gut microbiota via diet will need to appreciate these differences and to capitalize on existing variation to accelerate the pace of discovery.
REFERENCES
- 1.Walter J, Ley R. 2011. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol 65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Sommer F, Bäckhed F. 2013. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen JJ, Sartor RB. 2015. Therapeutic manipulation of the microbiome in IBD: current results and future approaches. Curr Treat Options Gastroenterol 13:105–120. doi: 10.1007/s11938-014-0042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kootte RS, Vrieze A, Holleman F, Dallinga-Thie GM, Zoetendal EG, de Vos WM, Groen AK, Hoekstra JB, Stroes ES, Nieuwdorp M. 2012. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes Metab 14:112–120. doi: 10.1111/j.1463-1326.2011.01483.x. [DOI] [PubMed] [Google Scholar]
- 7.Kinross JM, Darzi AW, Nicholson JK. 2011. Gut microbiome-host interactions in health and disease. Genome Med 3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 9.Marco ML, Heeney D, Binda S, Cifelli CJ, Cotter PD, Foligne B, Ganzle M, Kort R, Pasin G, Pihlanto A, Smid EJ, Hutkins R. 2017. Health benefits of fermented foods: microbiota and beyond. Curr Opin Biotechnol 44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 10.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O’Toole PW, Ercolini D. 2016. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 11.Hooper LV, Midtvedt T, Gordon JI. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 12.Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. 2015. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmody RN, Turnbaugh PJ. 2012. Gut microbes make for fattier fish. Cell Host Microbe 12:259–261. doi: 10.1016/j.chom.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englyst KN, Englyst HN. 2005. Carbohydrate bioavailability. Br J Nutr 94:1–11. doi: 10.1079/BJN20051457. [DOI] [PubMed] [Google Scholar]
- 16.Carmody RN, Wrangham RW. 2009. The energetic significance of cooking. J Hum Evol 57:379–391. doi: 10.1016/j.jhevol.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Cummings JH, Macfarlane GT. 1991. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 18.Neis EP, Dejong CH, Rensen SS. 2015. The role of microbial amino acid metabolism in host metabolism. Nutrients 7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordgaard I, Mortensen PB, Langkilde AM. 1995. Small intestinal malabsorption and colonic fermentation of resistant starch and resistant peptides to short-chain fatty acids. Nutrition 11:129–137. [PubMed] [Google Scholar]
- 20.Yadav M, Verma MK, Chauhan NS. 2018. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol 200:203–217. doi: 10.1007/s00203-017-1459-x. [DOI] [PubMed] [Google Scholar]
- 21.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schluter J, Foster KR. 2012. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol 10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes AJ, Chew YV, Colakoglu F, Cliff JB, Klaassens E, Read MN, Solon-Biet SM, McMahon AC, Cogger VC, Ruohonen K, Raubenheimer D, Le Couteur DG, Simpson SJ. 2017. Diet-microbiome interactions in health are controlled by intestinal nitrogen source constraints. Cell Metab 25:140–151. doi: 10.1016/j.cmet.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Reese AT, Pereira FC, Schintlmeister A, Berry D, Wagner M, Hale LP, Wu A, Jiang S, Durand HK, Zhou X, Premont R, Diehl AM, O’Connell TM, Alberts SC, Kartzinel TR, Pringle RM, Dunn RR, Wright JP, David LA. 2018. Microbial nitrogen limitation in the mammalian large intestine. Nat Microbiol 3:1441–1450. doi: 10.1038/s41564-018-0267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierer N, Ferrenberg S, Flores GE, Gonzalez A, Kueneman J, Legg T, Lynch RC, McDonald D, Mihaljevic JR, O’Neill SP, Rhodes ME, Song SJ, Walters WA. 2012. From animalcules to an ecosystem: application of ecological concepts to the human microbiome. Annu Rev Ecol Evol Syst 43:137–155. doi: 10.1146/annurev-ecolsys-110411-160307. [DOI] [Google Scholar]
- 26.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones RB, Zhu X, Moan E, Murff HJ, Ness RM, Seidner DL, Sun S, Yu C, Dai Q, Fodor AA, Azcarate-Peril MA, Shrubsole MJ. 2018. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep 8:4139. doi: 10.1038/s41598-018-22408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stearns JC, Lynch MDJ, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. 2011. Bacterial biogeography of the human digestive tract. Sci Rep 1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg RD. 1996. The indigenous gastrointestinal microflora. Trends Microbiol 4:430–435. doi: 10.1016/0966-842X(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 32.Novotny JA, Gebauer SK, Baer DJ. 2012. Discrepancy between the Atwater factor predicted and empirically measured energy values of almonds in human diets. Am J Clin Nutr 96:296–301. doi: 10.3945/ajcn.112.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groopman EE, Carmody RN, Wrangham RW. 2015. Cooking increases net energy gain from a lipid-rich food. Am J Phys Anthropol 156:11–18. doi: 10.1002/ajpa.22622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evenepoel P, Claus D, Geypens B, Hiele M, Geboes K, Rutgeerts P, Ghoos Y. 1999. Amount and fate of egg protein escaping assimilation in the small intestine of humans. Am J Physiol Gastrointest Liver Physiol 277:935–943. [DOI] [PubMed] [Google Scholar]
- 35.Evenepoel P, Geypens B, Luypaerts A, Hiele M, Ghoos Y, Rutgeerts P. 1998. Digestibility of cooked and raw egg protein in humans as assessed by stable isotope techniques. J Nutr 128:1716–1722. doi: 10.1093/jn/128.10.1716. [DOI] [PubMed] [Google Scholar]
- 36.Borgstrom B, Dahlqvist A, Lundh G, Sjovall J. 1957. Studies of intestinal digestion and absorption in the human. J Clin Invest 36:1521–1536. doi: 10.1172/JCI103549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvester KR, Englyst HN, Cummings JH. 1995. Ileal recovery of starch from whole diets containing resistant starch measured in vitro and fermentation of ileal effluent. Am J Clin Nutr 62:403–411. doi: 10.1093/ajcn/62.2.403. [DOI] [PubMed] [Google Scholar]
- 38.Jaeggi T, Kortman GA, Moretti D, Chassard C, Holding P, Dostal A, Boekhorst J, Timmerman HM, Swinkels DW, Tjalsma H, Njenga J, Mwangi A, Kvalsvig J, Lacroix C, Zimmermann MB. 2015. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 64:731–742. doi: 10.1136/gutjnl-2014-307720. [DOI] [PubMed] [Google Scholar]
- 39.Selma MV, Espin JC, Tomas-Barberan FA. 2009. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem 57:6485–6501. doi: 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- 40.Hörnicke H, Björnhag G. 1980. Coprophagy and related strategies for digesta utilization, p 707–730. In Ruckebusch Y, Thivend P (ed), Digestive physiology and metabolism in ruminants. Springer, Dordrecht, Netherlands. [Google Scholar]
- 41.Blaut M. 2002. Relationship of prebiotics and food to intestinal microflora. Eur J Nutr 41:i11–i16. doi: 10.1007/s00394-002-1102-7. [DOI] [PubMed] [Google Scholar]
- 42.Johnson LR. 1988. Regulation of gastrointestinal mucosal growth. Physiol Rev 68:456–502. doi: 10.1152/physrev.1988.68.2.456. [DOI] [PubMed] [Google Scholar]
- 43.Dahly E, Gillingham M, Guo Z, Murali S, Nelson D, Holst J, Ney D. 2003. Role of luminal nutrients and endogenous GLP-2 in intestinal adaption to mid-small bowel resection. Am J Physiol Gastrointest Liver Physiol 284:G670–G682. doi: 10.1152/ajpgi.00293.2002. [DOI] [PubMed] [Google Scholar]
- 44.Barcelo A, Claustre J, Moro F, Chayvialle JA, Cuber JC, Plaisancie P. 2000. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut 46:218–224. doi: 10.1136/gut.46.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. 2009. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J 420:211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 46.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. 2015. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. 2011. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 50.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. 2013. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arike L, Holmen-Larsson J, Hansson GC. 2017. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 27:318–328. doi: 10.1093/glycob/cww134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mostov KE. 1994. Transepithelial transport of immunoglobulins. Annu Rev Immunol 12:63–84. doi: 10.1146/annurev.iy.12.040194.000431. [DOI] [PubMed] [Google Scholar]
- 54.Brandtzaeg P, Pabst R. 2004. Let’s go mucosal: communication on slippery ground. Trends Immunol 25:570–577. doi: 10.1016/j.it.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Maldonado-Contreras AL, McCormick BA. 2011. Intestinal epithelial cells and their role in innate mucosal immunity. Cell Tissue Res 343:5–12. doi: 10.1007/s00441-010-1082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato LM, Kawamoto S, Maruya M, Fagarasan S. 2014. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev 260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 58.Coleman R, Lowe PJ, Billington D. 1980. Membrane lipid composition and susceptibility to bile salt damage. Biochim Biophys Acta 599:294–300. doi: 10.1016/0005-2736(80)90075-9. [DOI] [PubMed] [Google Scholar]
- 59.Heuman DM, Bajaj RS, Lin Q. 1996. Adsorption of mixtures of bile salt taurine conjugates to lecithin-cholesterol membranes: implications for bile salt toxicity and cytoprotection. J Lipid Res 37:562–573. [PubMed] [Google Scholar]
- 60.D’Aldebert E, Biyeyeme Bi Mve MJ, Mergey M, Wendum D, Firrincieli D, Coilly A, Fouassier L, Corpechot C, Poupon R, Housset C, Chignard N. 2009. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology 136:1435–1443. doi: 10.1053/j.gastro.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 61.Vlahcevic ZR, Heuman DM, Hylemon PB. 1996. Physiology and pathophysiology of enterohepatic circulation of bile acids, p 376–417. In Zakim D, Boyer T (ed), Hepatology: a textbook of liver disease, 3rd ed, vol 1 Saunders, Philadelphia, PA. [Google Scholar]
- 62.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. 2014. Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urdaneta V, Casadesus J. 2017. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med (Lausanne) 4:163. doi: 10.3389/fmed.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wahlstrom A, Kovatcheva-Datchary P, Stahlman M, Backhed F, Marschall HU. 2017. Crosstalk between bile acids and gut microbiota and its impact on farnesoid X receptor signalling. Dig Dis 35:246–250. doi: 10.1159/000450982. [DOI] [PubMed] [Google Scholar]
- 66.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. 2018. Glycan utilization and cross-feeding activities by Bifidobacteria. Trends Microbiol 26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, de Vos WM. 2017. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 8:e00770-17. doi: 10.1128/mBio.00770-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Portune KJ, Beaumont M, Davila AM, Tome D, Blachier F, Sanz Y. 2016. Gut microbiota role in dietary protein metabolism and health-related outcomes: the two sides of the coin. Trends Food Sci Technol 57:213–232. doi: 10.1016/j.tifs.2016.08.011. [DOI] [Google Scholar]
- 71.Bess EN, Bisanz JE, Spanogiannopoulos P, Ang QY, Bustion A, Kitamura S, Alba DL, Wolan DW, Koliwad SK, Turnbaugh PJ. 2018. The genetic basis for the cooperative bioactivation of plant lignans by a human gut bacterial consortium. bioRxiv doi: 10.1101/357640. [DOI] [PMC free article] [PubMed]
- 72.Duda-Chodak A, Tarko T, Satora P, Sroka P. 2015. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review. Eur J Nutr 54:325–341. doi: 10.1007/s00394-015-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuohy KM, Hinton DJ, Davies SJ, Crabbe MJ, Gibson GR, Ames JM. 2006. Metabolism of Maillard reaction products by the human gut microbiota: implications for health. Mol Nutr Food Res 50:847–857. doi: 10.1002/mnfr.200500126. [DOI] [PubMed] [Google Scholar]
- 74.Metges CC, El-Khoury AE, Henneman L, Petzke KJ, Grant I, Bedri S, Pereira PP, Ajami AM, Fuller MF, Young VR. 1999. Availability of intestinal microbial lysine for whole body lysine homeostasis in human subjects. Am J Physiol Endocrinol Metab 277:597–607. [DOI] [PubMed] [Google Scholar]
- 75.Metges CC. 2000. Contribution of microbial amino acids to amino acid homeostasis of the host. J Nutr 130:1857S–1864S. doi: 10.1093/jn/130.7.1857S. [DOI] [PubMed] [Google Scholar]
- 76.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lambert JM, Bongers RS, de Vos WM, Kleerebezem M. 2008. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol 74:4719–4726. doi: 10.1128/AEM.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hardison WG. 1978. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology 75:71–75. [PubMed] [Google Scholar]
- 79.Sjövall J. 1959. Dietary glycine and taurine on bile acid conjugation in man; bile acids and steroids. Proc Soc Exp Biol Med 100:676–678. doi: 10.3181/00379727-100-24741. [DOI] [PubMed] [Google Scholar]
- 80.Chesney RW, Helms RA, Christensen M, Budreau AM, Han XB, Sturman JA. 1998. The role of taurine in infant nutrition. Adv Exp Med Biol 442:463–476. doi: 10.1007/978-1-4899-0117-0_56. [DOI] [PubMed] [Google Scholar]
- 81.Hayes KC, Sturman JA. 1981. Taurine in metabolism. Annu Rev Nutr 1:401–425. doi: 10.1146/annurev.nu.01.070181.002153. [DOI] [PubMed] [Google Scholar]
- 82.Huijghebaert SM, Eyssen HJ. 1982. Specificity of bile salt sulfatase activity from Clostridium sp. strains S1. Appl Environ Microbiol 44:1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Devkota S, Wang YW, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10(−/−) mice. Nature 487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carlson PE, Kaiser AM, McColm SA, Bauer JM, Young VB, Aronoff DM, Hanna PC. 2015. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. 2013. Pleiotropic roles of bile acids in metabolism. Cell Metab 17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 88.Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. 2003. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7alpha-dehydroxylating bacteria in human feces. Clin Chim Acta 331:127–134. doi: 10.1016/S0009-8981(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 89.Kitahara M, Takamine F, Imamura T, Benno Y. 2000. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 50:971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 90.Kitahara M, Takamine F, Imamura T, Benno Y. 2001. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int J Syst Evol Microbiol 51:39–44. doi: 10.1099/00207713-51-1-39. [DOI] [PubMed] [Google Scholar]
- 91.Solbach P, Chhatwal P, Woltemate S, Tacconelli E, Buhl M, Gerhard M, Thoeringer CK, Vehreschild MJGT, Jazmati N, Rupp J, Manns MP, Bachmann O, Suerbaum S. 2018. BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PLoS One 13:e0196977. doi: 10.1371/journal.pone.0196977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wells JE, Hylemon PB. 2000. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 66:1107–1113. doi: 10.1128/AEM.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kang DJ, Ridlon JM, Moore DR, Barnes S, Hylemon PB. 2008. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim Biophys Acta 1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McGuckin MA, Lindén SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 98.Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. 2013. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A 110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carey HV, Walters WA, Knight R. 2013. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am J Physiol Regul Integr Comp Physiol 304:R33–R42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Costello EK, Gordon JI, Secor SM, Knight R. 2010. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J 4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y. 2014. Seasonal variation in human gut microbiome composition. PLoS One 9:e90731. doi: 10.1371/journal.pone.0090731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zimmer J, Lange B, Frick J-S, Sauer H, Zimmermann K, Schwiertz A, Rusch K, Klosterhalfen S, Enck P. 2012. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr 66:53–60. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- 107.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, De Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, Crittenden AN. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martinez-Guryn K, Hubert N, Frazier K, Urlass S, Musch MW, Ojeda P, Pierre JF, Miyoshi J, Sontag TJ, Cham CM, Reardon CA, Leone V, Chang EB. 2018. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 23:458–469. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Howe A, Ringus DL, Williams RJ, Choo ZN, Greenwald SM, Owens SM, Coleman ML, Meyer F, Chang EB. 2016. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J 10:1217–1227. doi: 10.1038/ismej.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. 2013. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. JPEN J Parenter Enteral Nutr 37:746–754. doi: 10.1177/0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, Rawls JF. 2012. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Costantini L, Molinari R, Farinon B, Merendino N. 2017. Impact of omega-3 fatty acids on the gut microbiota. Int J Mol Sci 18:E2645. doi: 10.3390/ijms18122645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. 2011. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 116.Yokota A, Fukiya S, Islam KBMS, Ooka T, Ogura Y, Hayashi T, Hagio M, Ishizuka S. 2012. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 3:455–459. doi: 10.4161/gmic.21216. [DOI] [PubMed] [Google Scholar]
- 117.Devkota S, Chang EB. 2015. Interactions between diet, bile acid metabolism, gut microbiota, and inflammatory bowel diseases. Dig Dis 33:351–356. doi: 10.1159/000371687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. 2015. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab 22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coelho OGL, Cândido FG, Alfenas RDCG. 11 July 2018. Dietary fat and gut microbiota: mechanisms involved in obesity control. Crit Rev Food Sci Nutr doi: 10.1080/10408398.2018.1481821. [DOI] [PubMed] [Google Scholar]
- 120.Faith JJ, McNulty NP, Rey FE, Gordon JI. 2011. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kiilerich P, Myrmel LS, Fjære E, Hao Q, Hugenholtz F, Sonne SB, Derrien M, Pedersen LM, Petersen RK, Mortensen A, Licht TR, Rømer MU, Vogel UB, Waagbø LJ, Giallourou N, Feng Q, Xiao L, Liu C, Liaset B, Kleerebezem M, Wang J, Madsen L, Kristiansen K. 2016. Effect of a long-term high-protein diet on survival, obesity development, and gut microbiota in mice. Am J Physiol Endocrinol Metab 310:E886–E899. doi: 10.1152/ajpendo.00363.2015. [DOI] [PubMed] [Google Scholar]
- 122.Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, Duncan G, Johnstone AM, Lobley GE, Wallace RJ, Duthie GG, Flint HJ. 2011. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr 93:1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- 123.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Craine JM, Morrow C, Fierer N. 2007. Microbial nitrogen limitation increases decomposition. Ecology 88:2105–2113. doi: 10.1890/06-1847.1. [DOI] [PubMed] [Google Scholar]
- 126.Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, Galbraith ED, Geider RJ, Guieu C, Jaccard SL, Jickells TD, La Roche J, Lenton TM, Mahowald NM, Maranon E, Marinov I, Moore JK, Nakatsuka T, Oschlies A, Saito MA, Thingstad TF, Tsuda A, Ulloa O. 2013. Processes and patterns of oceanic nutrient limitation. Nat Geosci 6:701–710. doi: 10.1038/ngeo1765. [DOI] [Google Scholar]
- 127.Sistla SA, Asao S, Schimel JP. 2012. Detecting microbial N-limitation in tussock tundra soil: implications for Arctic soil organic carbon cycling. Soil Biol Biochem 55:78–84. doi: 10.1016/j.soilbio.2012.06.010. [DOI] [Google Scholar]
- 128.Sterner RW, Elser JJ. 2002. Ecological stoichiometry: the biology of elements from molecules to the biosphere. Princeton University Press, Princeton, NJ. [Google Scholar]
- 129.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay J-M, Langella P, Xavier RJ, Sokol H. 2016. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, Alvarez JI, Kébir H, Anandasabapathy N, Izquierdo G, Jung S, Obholzer N, Pochet N, Clish CB, Prinz M, Prat A, Antel J, Quintana FJ. 2016. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Agus A, Planchais J, Sokol H. 2018. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 133.Ni J, Shen TD, Chen EZ, Bittinger K, Bailey A, Roggiani M, Sirota-Madi A, Friedman ES, Chau L, Lin A, Nissim I, Scott J, Lauder A, Hoffmann C, Rivas G, Albenberg L, Baldassano RN, Braun J, Xavier RJ, Clish CB, Yudkoff M, Li H, Goulian M, Bushman FD, Lewis JD, Wu GD. 2017. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med 9:eaah6888. doi: 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cleveland CC, Townsend AR, Schmidt SK. 2002. Phosphorus limitation of microbial processes in moist tropical forests: evidence from short-term laboratory incubations and field studies. Ecosystems 5:680–691. doi: 10.1007/s10021-002-0202-9. [DOI] [Google Scholar]
- 135.Souza V, Eguiarte LE, Siefert J, Elser JJ. 2008. Microbial endemism: does phosphorus limitation enhance speciation? Nat Rev Microbiol 6:559–564. doi: 10.1038/nrmicro1917. [DOI] [PubMed] [Google Scholar]
- 136.Sundareshwar PV, Morris JT, Koepfler EK, Fornwalt B. 2003. Phosphorus limitation of coastal ecosystem processes. Science 299:563–565. doi: 10.1126/science.1079100. [DOI] [PubMed] [Google Scholar]
- 137.Dabrock B, Bahl H, Gottschalk G. 1992. Parameters affecting solvent production by Clostridium pasteurianum. Appl Environ Microbiol 58:1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lopez CA, Skaar EP. 2018. The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe 23:737–748. doi: 10.1016/j.chom.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Becker KW, Skaar EP. 2014. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev 38:1235–1249. doi: 10.1111/1574-6976.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Payne SM. 1993. Iron acquisition in microbial pathogenesis. Trends Microbiol 1:66–69. doi: 10.1016/0966-842X(93)90036-Q. [DOI] [PubMed] [Google Scholar]
- 141.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bäumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kortman GA, Raffatellu M, Swinkels DW, Tjalsma H. 2014. Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol Rev 38:1202–1234. doi: 10.1111/1574-6976.12086. [DOI] [PubMed] [Google Scholar]
- 146.Zimmermann MB, Chassard C, Rohner F, N’Goran EK, Nindjin C, Dostal A, Utzinger J, Ghattas H, Lacroix C, Hurrell RF. 2010. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d’Ivoire. Am J Clin Nutr 92:1406–1415. doi: 10.3945/ajcn.110.004564. [DOI] [PubMed] [Google Scholar]
- 147.Reed S, Neuman H, Moscovich S, Glahn RP, Koren O, Tako E. 2015. Chronic zinc deficiency alters chick gut microbiota composition and function. Nutrients 7:9768–9784. doi: 10.3390/nu7125497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Starke IC, Pieper R, Neumann K, Zentek J, Vahjen W. 2014. The impact of high dietary zinc oxide on the development of the intestinal microbiota in weaned piglets. FEMS Microbiol Ecol 87:416–427. doi: 10.1111/1574-6941.12233. [DOI] [PubMed] [Google Scholar]
- 149.Smith AH, Mackie RI. 2004. Effect of condensed tannins on bacterial diversity and metabolic activity in the rat gastrointestinal tract. Appl Environ Microbiol 70:1104–1115. doi: 10.1128/AEM.70.2.1104-1115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roopchand DE, Carmody RN, Kuhn P, Moskal K, Rojas-Silva P, Turnbaugh PJ, Raskin I. 2015. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 64:2847–2858. doi: 10.2337/db14-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dolara P, Luceri C, De Filippo C, Femia AP, Giovannelli L, Caderni G, Cecchini C, Silvi S, Orpianesi C, Cresci A. 2005. Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat Res 591:237–246. doi: 10.1016/j.mrfmmm.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 152.Lee HC, Jenner AM, Low CS, Lee YK. 2006. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol 157:876–884. doi: 10.1016/j.resmic.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 153.Anhe FF, Roy D, Pilon G, Dudonne S, Matamoros S, Varin TV, Garofalo C, Moine Q, Desjardins Y, Levy E, Marette A. 2015. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64:872–883. doi: 10.1136/gutjnl-2014-307142. [DOI] [PubMed] [Google Scholar]
- 154.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. 2017. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, Baumler AJ. 2016. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Baumler AJ. 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, Sifuentes-Dominguez L, Huff-Hardy K, Wilson RP, Gillis CC, Tükel Ç, Koh AY, Burstein E, Hooper LV, Bäumler AJ, Winter SE. 2018. Precision editing of the gut microbiota ameliorates colitis. Nature 553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Nimmagadda S, O’Connell TM, Wright JP, Deshusses MA, David LA. 2018. Antibiotic induced changes in microbial respiration disrupt redox dynamics in the mouse gut. Elife 7:e35987. doi: 10.7554/eLife.35987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chassard C, Gaillard-Martinie B, Bernalier-Donadille A. 2005. Interaction between H-2-producing and non-H-2-producing cellulolytic bacteria from the human colon. FEMS Microbiol Lett 242:339–344. doi: 10.1016/j.femsle.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 161.Gibson GR, Macfarlane GT, Cummings JH. 1988. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J Appl Bacteriol 65:103–111. doi: 10.1111/j.1365-2672.1988.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 162.Attwood G, McSweeney C. 2008. Methanogen genomics to discover targets for methane mitigation technologies and options for alternative H2 utilisation in the rumen. Aust J Exp Agric 48:28–37. doi: 10.1071/EA07203. [DOI] [Google Scholar]
- 163.Fonty G, Joblin K, Chavarot M, Roux R, Naylor G, Michallon F. 2007. Establishment and development of ruminal hydrogenotrophs in methanogen-free lambs. Appl Environ Microbiol 73:6391–6403. doi: 10.1128/AEM.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marquet P, Duncan SH, Chassard C, Bernalier-Donadille A, Flint HJ. 2009. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol Lett 299:128–134. doi: 10.1111/j.1574-6968.2009.01750.x. [DOI] [PubMed] [Google Scholar]
- 165.Gibson GR, Cummings JH, Macfarlane GT. 1991. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Ecol 86:103–111. doi: 10.1111/j.1574-6941.1991.tb01742.x. [DOI] [Google Scholar]
- 166.Ozuolmez D, Na H, Lever MA, Kjeldsen KU, Jørgensen BB, Plugge CM. 2015. Methanogenic archaea and sulfate reducing bacteria co-cultured on acetate: teamwork or coexistence? Front Microbiol 6:492. doi: 10.3389/fmicb.2015.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Carmody RN, Turnbaugh PJ. 2014. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest 124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Duncan SH, Louis P, Thomson JM, Flint HJ. 2009. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 170.Rettedal EA, Gumpert H, Sommer MO. 2014. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun 5:4714. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 171.Tramontano M, Andrejev S, Pruteanu M, Klunemann M, Kuhn M, Galardini M, Jouhten P, Zelezniak A, Zeller G, Bork P, Typas A, Patil KR. 2018. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat Microbiol 3:514–522. doi: 10.1038/s41564-018-0123-9. [DOI] [PubMed] [Google Scholar]
- 172.Lovell D, Taylor J, Zwart A, Helliwell C. 2010. Caution! Compositions! Can constraints on omics data lead analyses astray? CSIRO, Canberra, Australia. [Google Scholar]
- 173.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLoS Comput Biol 8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 175.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. 2018. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557:434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Treuting PM, Dintzis SM. 2012. Lower gastrointestinal tract, p 177–192. In Dintzis SM, Frevert CW, Liggitt HD, Montine KS, Treuting PM (ed), Comparative anatomy and histology—a mouse and human atlas. Elsevier, Inc, Amsterdam, Netherlands. [Google Scholar]
- 177.Casteleyn C, Rekecki A, Van der Aa A, Simoens P, Van den Broeck W. 2010. Surface area assessment of the murine intestinal tract as a prerequisite for oral dose translation from mouse to man. Lab Anim 44:176–183. doi: 10.1258/la.2009.009112. [DOI] [PubMed] [Google Scholar]
- 178.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nguyen TL, Vieira-Silva S, Liston A, Raes J. 2015. How informative is the mouse for human gut microbiota research? Dis Model Mech 8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Venkataraman A, Sieber JR, Schmidt AW, Waldron C, Theis KR, Schmidt TM. 2016. Variable responses of human microbiomes to dietary supplementation with resistant starch. Microbiome 4:33. doi: 10.1186/s40168-016-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. 2013. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 184.Foster KR, Chluter JS, Oyte KZC, Rakoff-Nahoum S. 2017. The evolution of the host microbiome as an ecosystem on a leash. Nature 548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. 2013. The influence of diet on the gut microbiota. Pharmacol Res 69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 186.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rhee SH, Pothoulakis C, Mayer EA. 2009. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol 6:306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sampson TR, Mazmanian SK. 2015. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Llewellyn SR, Britton GJ, Contijoch EJ, Vennaro OH, Mortha A, Colombel JF, Grinspan A, Clemente JC, Merad M, Faith JJ. 2018. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology 154:1037–1046. doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reese AT, Dunn RR. 2018. Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. mBio 9:e01294-18. doi: 10.1128/mBio.01294-18. [DOI] [PMC free article] [PubMed] [Google Scholar]