Temporary changes in the composition of the microbiota, for example, by oral administration of probiotics, can modulate the host immune system. However, the underlying mechanisms by which probiotics interact with the host are often unknown. Here, we show that Lactobacillus reuteri R2lc and 2010 harbor an orthologous PKS gene cluster that activates the aryl hydrocarbon receptor (AhR). AhR is a ligand-activated transcription factor that plays a key role in a variety of diseases, including amelioration of intestinal inflammation. Understanding the mechanism by which a bacterium modulates the immune system is critical for applying rational selection strategies for probiotic supplementation. Finally, heterologous and/or optimized expression of PKS is a logical next step toward the development of next-generation probiotics to prevent and treat disease.
KEYWORDS: Lactobacillus reuteri, probiotic, aryl hydrocarbon receptor, biosynthetic gene cluster, gut symbiont, pigment, polyketides, secondary metabolism
ABSTRACT
A mechanistic understanding of microbe-host interactions is critical to developing therapeutic strategies for targeted modulation of the host immune system. Different members of the gut symbiont species Lactobacillus reuteri modulate host health by, for example, reduction of intestinal inflammation. Previously, it was shown that L. reuteri activates the aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor that plays an important role in the mucosal immune system, by the production of tryptophan catabolites. Here, we identified a novel pathway by which L. reuteri activates AhR, which is independent of tryptophan metabolism. We screened a library of 36 L. reuteri strains and determined that R2lc and 2010, strains with a pigmented phenotype, are potent AhR activators. By whole-genome sequencing and comparative genomics, we identified genes unique to R2lc and 2010. Our analyses demonstrated that R2lc harbors two genetically distinct polyketide synthase (PKS) clusters, functionally unknown (fun) and pks, each carried by a multicopy plasmid. Inactivation of pks, but not fun, abolished the ability of R2lc to activate AhR. L. reuteri 2010 has a gene cluster homologous to the pks cluster in R2lc with an identical gene organization, which is also responsible for AhR activation. In conclusion, we identified a novel PKS pathway in L. reuteri R2lc and 2010 that is responsible for AhR activation.
IMPORTANCE Temporary changes in the composition of the microbiota, for example, by oral administration of probiotics, can modulate the host immune system. However, the underlying mechanisms by which probiotics interact with the host are often unknown. Here, we show that Lactobacillus reuteri R2lc and 2010 harbor an orthologous PKS gene cluster that activates the aryl hydrocarbon receptor (AhR). AhR is a ligand-activated transcription factor that plays a key role in a variety of diseases, including amelioration of intestinal inflammation. Understanding the mechanism by which a bacterium modulates the immune system is critical for applying rational selection strategies for probiotic supplementation. Finally, heterologous and/or optimized expression of PKS is a logical next step toward the development of next-generation probiotics to prevent and treat disease.
INTRODUCTION
The mammalian intestinal tract contains trillions of bacteria, which collectively contribute to our well-being (1). Metagenomic studies suggest that microbial metabolites play a central role in microbe-host interactions, including regulation of the host immune system (2). Therefore, a mechanistic understanding of metabolite-mediated microbe-host interactions is critical for developing therapeutic strategies for targeted modulation of the host immune system.
Polyketide synthases (PKS) and nonribosomal peptide synthases (NRPS) are secondary metabolites produced by biosynthetic gene clusters (BCGs) that assemble simple molecules, such as acetyl-CoA, into complex metabolites, some of which (i.e., erythromycin) are important to the pharmaceutical industries (3). Many medically important biosynthetic gene clusters were identified from soil and marine microbes, including the antitumor polyketide onnamide (4) and the immunosuppressant rapamycin (5). Although PKS gene clusters have previously been identified in the human gut microbiome (6), our understanding of their role in immunomodulation is limited. One of the few examples describing the role of PKS clusters in immunomodulation is the pks island cluster in Escherichia coli Nissle 1917. Inactivation of the PKS island in Nissle 1917 results in a loss of anti-inflammatory function in the adoptive T-cell transfer model of inflammation (7).
PKS and NRPS clusters have been identified in other lactic acid bacteria (LAB). For example, the oral pathogen Streptococcus mutans encodes an NRPS/PKS system that provides the organism with an advantage in multispecies biofilms in dental caries (8). LAB that have been associated with food, like Lactococcus lactis KF147 (mung bean sprouts) and the sourdough isolates Lactobacillus reuteri LTH2584, TMW1.106, TMW1.112, and TMW1.656, also encode NRPS/PKS systems (9, 10). It was suggested that the NRPS/PKS cluster in L. reuteri contributes to persistence in sourdough (10). Thus, the identified NRPS/PKS clusters in those LAB appear to be advantageous to the organisms’ ability to thrive in their niches.
The host can recognize microbial metabolites via different pathways, including via pattern recognition receptors and the aryl hydrocarbon receptor (AhR) (11, 12). Pigmented virulence factors like phenazines and naphthoquinone phthiocol are detected by the host immune system and degraded through AhR-mediated cytokine and chemokine production (12). AhR is a ligand-activated transcription factor that recognizes environmental pollutants, dietary compounds (i.e., glucobrassicin and flavonoids), and microbial-derived secondary metabolites (i.e., indole-3-carbinol). Upon ligand binding, AhR translocates into the nucleus to induce target gene expressions. The role of AhR has been extensively studied in relation to metabolism of environmental toxins, but the focus has recently shifted to its role in modulation of the adaptive and innate immune system. The activation of AhR, for example, is important for the production of interleukin-22 (IL-22), which enhances the innate immune response by inducing the production of antimicrobial peptides (Reg3 lectins) to fight off intestinal pathogens and to protect intestinal tissues from inflammation damage by inducing tight junction proteins (13, 14).
Recently, different strains of L. reuteri have been shown to activate AhR and modulate the immune system through tryptophan (Trp) metabolism (15–17). For example, L. reuteri 100-23 metabolizes dietary tryptophan into bioactive indole derivatives, which subsequently activates AhR. AhR activation by L. reuteri 100-23 increases Il22 expression in the intestine, which in turn provides protection from Candida infection in mice that could not take up tryptophan (13). In mammals, 99% of dietary tryptophan is taken up by the host (18), thereby limiting tryptophan availability to the microbiota to produce AhR ligands. Therefore, identification of specific gut microbes, including L. reuteri strains, that activate AhR independent of tryptophan metabolism is important for developing AhR-mediated biotherapeutic strategies to target intestinal diseases.
Here, we screened an L. reuteri library of 36 strains with different host origins for their ability to activate AhR. We employed an in vitro screening method and identified that L. reuteri R2lc and 2010 are potent AhR activator strains. By whole-genome sequencing, comparative genomics, and targeted gene inactivation, we discovered that an orthologous PKS cluster is responsible for the AhR activation phenotype in L. reuteri R2lc and 2010.
RESULTS
L. reuteri R2lc and 2010 are potent AhR activators.
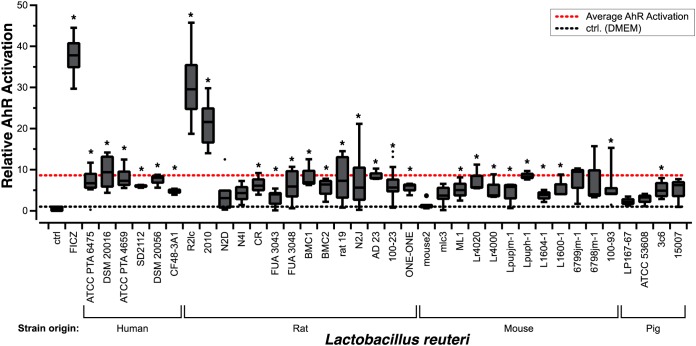
To determine the capacity of L. reuteri to activate AhR, we exploited an in vitro reporter cell line, H1L6.1c3, in which luminescence levels are positively correlated with AhR activation (19). We screened cell-free supernatants of 36 L. reuteri strains of human, pig, and rodent origins (Table 1). Compared to the medium control, 27 out of 36 strains activated AhR (P < 0.05) (Fig. 1). Importantly, the cell-free bacterial supernatants had no effect on cell viability (see Fig. S1 in the supplemental material). We found that two pigmented strains, R2lc and 2010, were considerably more potent in their ability to activate AhR (30.2-fold ± 8.3-fold and 20.9-fold ± 5.6-fold, respectively) than other L. reuteri strains (mean activation, 8.6-fold). To understand the underlying mechanism by which R2lc and 2010 activate AhR, we focused first on L. reuteri R2lc for several reasons. First, R2lc has been safely consumed by humans (20, 21), which, to some extent, paves the way to apply this strain in future human trials. Second, several research groups demonstrated that R2lc reduces intestinal inflammation in different inflammation disease models (22–26), reduces acute liver injury (27), and reduces bacteremia (28).
TABLE 1.
Bacterial strains used in this studya
| Species | Strain | VPL identifier | Origin or description | Source and/or reference |
|---|---|---|---|---|
| Lactobacillus reuteri | CF48-3A1 | VPL1086 | Human | BioGaia AB, JGI 2502171173 |
| Lactobacillus reuteri | SD2112 | VPL1013 | Human | BioGaia AB |
| Lactobacillus reuteri | ATCC PTA 6475 | VPL1014 | Human | BioGaia AB |
| Lactobacillus reuteri | DSM 20016 | VPL1046 | Human | JGI 640427118 |
| Lactobacillus reuteri | ATCC PTA 4659 | VPL1031 | Human | BioGaia AB |
| Lactobacillus reuteri | DSM20056 | VPL1061 | Human | JGI 642555135 |
| Lactobacillus reuteri | R2lc | VPL1053 | Rat | Siv Ahrné, JGI 2716884882 |
| Lactobacillus reuteri | 2010 | VPL1054 | Rat | BioGaia AB, JGI 2710724192 |
| Lactobacillus reuteri | N2D | VPL1067 | Rat | Siv Ahrné |
| Lactobacillus reuteri | FUA3043 | VPL1062 | Rat | 49 |
| Lactobacillus reuteri | FUA3048 | VPL1063 | Rat | 49 |
| Lactobacillus reuteri | BMC1 | VPL1093 | Rat | Stafan Roos |
| Lactobacillus reuteri | BMC2 | VPL1057 | Rat | Stefan Roos |
| Lactobacillus reuteri | 100-23 | VPL1049 | Rat | JGI 2500069000 |
| Lactobacillus reuteri | CR | VPL1059 | Rat | 49 |
| Lactobacillus reuteri | Rat 19 | VPL1069 | Rat | 49 |
| Lactobacillus reuteri | AD 23 | VPL1048 | Rat | 49 |
| Lactobacillus reuteri | N2J | VPL1052 | Rat | Siv Ahrné |
| Lactobacillus reuteri | N4I | VPL1063 | Rat | 49 |
| Lactobacillus reuteri | mouse 2 | VPL1070 | Mouse | 49 |
| Lactobacillus reuteri | one-one | VPL1060 | Mouse | 49 |
| Lactobacillus reuteri | 6799jm-1 | VPL1051 | Mouse | 49 |
| Lactobacillus reuteri | L1600-1 | VPL1064 | Mouse | 49 |
| Lactobacillus reuteri | 100-93 | VPL1047 | Mouse | 49 |
| Lactobacillus reuteri | Lr4020 | VPL1072 | Mouse | 49 |
| Lactobacillus reuteri | 6798jm-1 | VPL1055 | Mouse | 49 |
| Lactobacillus reuteri | mlc3 | VPL1050 | Mouse | JGI 2506381016 |
| Lactobacillus reuteri | Lpuph-1 | VPL1056 | Mouse | JGI 2506381017 |
| Lactobacillus reuteri | Lr4000 | VPL1071 | Mouse | BioGaia AB |
| Lactobacillus reuteri | ML1 | VPL1058 | Mouse | 49 |
| Lactobacillus reuteri | L1604-1 | VPL1066 | Mouse | 49 |
| Lactobacillus reuteri | lpupjm1 | VPL1065 | Mouse | 49 |
| Lactobacillus reuteri | 3c6 | VPL1083 | Pig | JGI 2599185333 |
| Lactobacillus reuteri | Lp167-67 | VPL1085 | Pig | BioGaia AB, JGI 2599185361 |
| Lactobacillus reuteri | I5007 | VPL1082 | Pig | JGI 2554235423 |
| Lactobacillus reuteri | ATCC 53608 | VPL1090 | Pig | BioGaia AB, EMBL accession no. LN906634 |
| Lactobacillus reuteri | TMW1.112 | VPL1089 | Sourdough | JGI 2534682347 |
| Lactobacillus reuteri | TMW1.656 | VPL1088 | Sourdough | JGI 2534682350 |
| Escherichia coli | EC1000 | VPL1009 | In trans RepA provider; Kanr | 50 |
| Escherichia coli | VPL3002 | VPL3002 | EC1000 harboring pVPL3002; Emr | 29 |
| Lactobacillus reuteri | ATCC PTA 6475 | VPL3025 | Harboring plasmid pJP042 | 44 |
| Lactobacillus reuteri | R2lc ΔaraT | VPL4192 | Deletion of araT gene in R2lc | 29 |
| Lactobacillus reuteri | R2lc Δfun | VPL4208 | Deletion of genes funE, funF, and funG in R2lc | This study |
| Lactobacillus reuteri | R2lc Δpks::Cm | VPL4209 | Replacement of genes pksG, pksH, and pksI with the gene encoding chloramphenicol resistance | This study |
| Lactobacillus reuteri | 2010 Δpks | VPL4183 | Deletion of genes pksG, pksH, and pksI in 2010 | This study |
VPL, van Pijkeren Laboratory strain identification number. Further information on the listed Joint Genome Institute (JGI) source sequences can be found at the JGI genome portal (http://genome.jgi.doe.gov).
FIG 1.
Assessment of Lactobacillus reuteri AhR activation potential. L. reuteri AhR activation is strain specific. Data are represented as fold changes relative to the DMEM control (ctrl). The positive control is DMEM supplemented with 500 nM the AhR ligand 6-formylindolo[3,2-b]carbazole (FICZ). Data are presented as box-and-whisker plots. The whiskers represent the maximum and minimal values, and the lower, middle, and upper lines of the box represent first quartile, median, and third quartile, respectively. Open circles represent suspected outliers, which are data points that are 1.5 times below the first quartile or 1.5 times above the third quartile. Closed circles represent outliers, which are 3 times the value of the first or third quartile. Data represent averages of results from 3 independent experiments. Statistical significance between strains and the DMEM control was determined by the one-sample t test. Asterisks show statistical significance (P < 0.05).
L. reuteri R2lc and 2010 produce bright orange pigment.
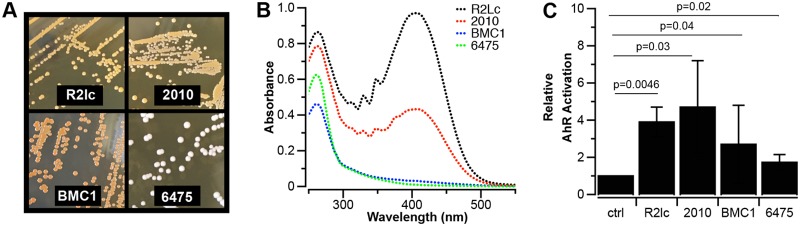
Three out of 36 strains, R2lc, 2010, and BMC1, are pigmented (Fig. 2A). Although BMC1 did not show a strong AhR activation phenotype, we reasoned that this might be due to differences in the chemical structures of the pigments. To analyze this, we first performed methanol extraction, followed by UV-visible spectroscopy. We identified that both R2lc and 2010 have an absorption wavelength maximum (λmax) of 405 to 409 nm, while this peak was absent in BMC1 and 6475 (Fig. 2B). These data show that the pigments of L. reuteri R2lc and 2010 have similar properties, while the pigment of BMC1 is distinctly different. Second, we tested all extracts for their ability to activate AhR. We found that all extracts, including that of a nonpigmented strain (6475), increased AhR activation (Fig. 2C); however, each of the extracts was much less potent than the cell-free supernatants of R2lc and 2010. Together, these data suggested that the orange pigments produced by R2lc and 2010 are not the main drivers of the strong AhR activation phenotype.
FIG 2.
Orange pigments extracted from R2lc, 2010, and BMC1 are not strong AhR ligands. (A) R2lc, 2010, and BMC1 produce an orange pigment, while most strains, like 6475, are opaque. (B) R2lc and 2010 have λmax values of 405 to 409 nm. BMC1 and 6475 did not absorb at this region. (C) Methanol extracts from R2lc, 2010, and BMC1 do not show strong AhR activation. Data shown represent averages from three biological replicates, and error bars represent standard deviations. Statistical significance between extracts and the negative control was determined by the one-sample t test. AhR activation was not different between strains (P > 0.05). Statistical significance was determined by Student’s t test (a P value of <0.05 was considered significant).
L. reuteri R2lc AraT-mediated tryptophan metabolism is not the main driver of AhR activation.
Previously, several groups demonstrated that L. reuteri activates AhR via indole molecules acquired by aromatic amino acid amino transferase (AraT)-mediated tryptophan metabolism (15, 17). To determine to what extent L. reuteri R2lc AraT plays a role in AhR activation, we used a derivative of R2lc in which araT was deleted (R2lc ΔaraT) (29). In our experimental setup, R2lc ΔaraT did not reduce the AhR-activating potential of L. reuteri R2lc (Fig. 3A), suggesting that another mechanism is driving AhR activation in R2lc. To test if AraT, which is conserved in L. reuteri, contributes to the basal level of AhR activation, we inactivated araT in L. reuteri 6475, a strain that activates AhR at a reduced level compared to R2lc. L. reuteri 6475 ΔaraT displayed a nonsignificant 20% reduction in AhR activation compared to the wild type (Fig. 3B). Thus, at least in our model system, AraT marginally impacts AhR activation of L. reuteri 6475 and is not a major driver by which L. reuteri R2lc activates AhR.
FIG 3.
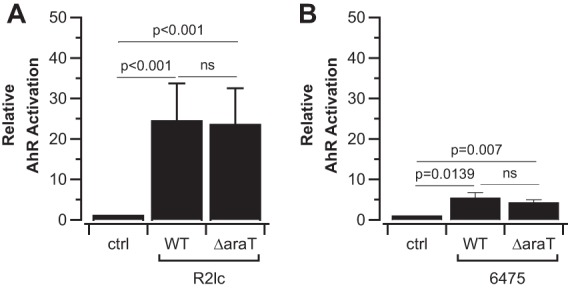
R2lc activates AhR independent of AraT. (A) AraT is not the main driver of AhR activation by R2lc. (B) Deletion of the araT gene in 6475 does not significantly change the AhR activation level in L. reuteri 6475. Data shown represent averages of results from three biological replicates, and error bars represent standard deviations. Statistical significance between strains and the negative control was determined by the one-sample t test. Statistical significance between strains was determined with Student’s t test. ns, not significant; WT, wild type.
L. reuteri R2lc harbors two multicopy plasmids.
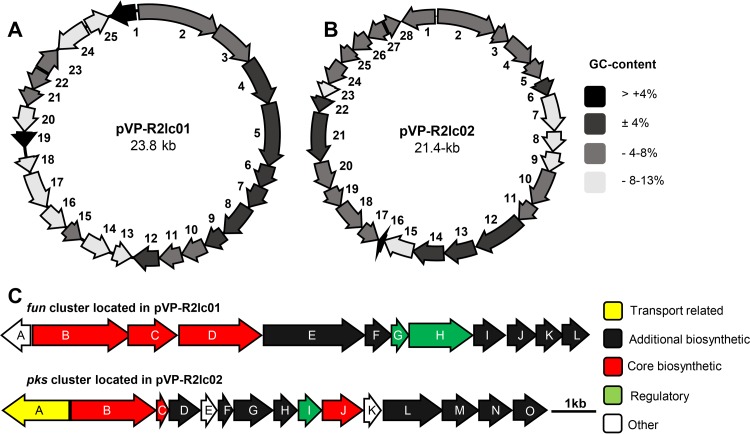
To identify the AhR activation pathway in L. reuteri R2lc, our strategy was to first identify genes unique to R2lc, followed by gene inactivation to assess the phenotype. To this end, we compared the draft genome of L. reuteri R2lc with the genomes of 10 L. reuteri strains that we determined have a relatively low AhR activation potential. We identified 79 putative genes unique to L. reuteri R2lc (Table S2). Interestingly, within this pool of unique genes, several genes putatively encode proteins involved in polyketide production and plasmid replication, which were distributed over five contigs. By primer walking and gap closure, we identified a 23.8-kb plasmid and a 21.4-kb plasmid, here referred to as pVP-R2lc01 and pVP-R2lc02, respectively. Plasmid pVP-R2lc01 contains 25 open reading frames (ORFs) (Fig. 4A and Table 2), while pVP-R2lc02 contains 28 ORFs (Fig. 4B and Table 3). The pVP-R2lc01 and pVP-R2lc02 plasmids have similar GC contents (32.4% and 32.1%, respectively), which are lower than the predicted chromosomal GC content (38.5%) (Table 4). By quantitative real-time PCR, we determined that, relative to the chromosome, pVP-R2lc01 and pVP-R2lc-02 are present at 3.75 ± 1.14 and 3.82 ± 0.62 copies, respectively.
FIG 4.
Overview of the pVP-R2lc01 and pVP-R2lc02 plasmids and their predicted PKS clusters. (A) The 23.8-kb pVP-R2lc01 plasmid contains 25 ORFs (indicated by arrows), including the fun cluster (ORFs 1 to 12). (B) The 21.4-kb pVP-R2lc02 plasmid contains 28 ORFs and carries the pks cluster (ORFs 1 to 15). Differences in GC contents of each gene (compared to the chromosomal GC content) are represented with different colors, as shown on the right. (C) The fun cluster (top) spans 13.4 kb containing 12 ORFs, and the pks cluster (bottom) spans 11.3 kb containing 15 ORFs. Transport-related, additional biosynthetic, core biosynthetic, regulatory, and other genes are represented (for gene annotations, see Tables 2 and 3).
TABLE 2.
Predicted annotations of genes in pVP-R2lc01a
| ORF (gene) | Locus tag | Predicted function | Length (amino acids) |
|---|---|---|---|
| 1 (funA) | C5O77_00105 | Biotin-(acetyl-CoA-carboxylase) ligase | 250 |
| 2 (funB) | C5O77_00110 | Beta-ketoacyl synthase domain | 798 |
| 3 (funC) | C5O77_00115 | 8-Amino-7-oxononanoate synthase | 399 |
| 4 (funD) | C5O77_00120 | Hypothetical protein (acyl carrier protein) | 663 |
| 5 (funE) | C5O77_00125 | Hypothetical protein [NAD(P)-dependent dehydrogenase, short-chain alcohol dehydrogenase family] | 847 |
| 6 (funF) | C5O77_00130 | Hypothetical protein (phosphopantetheinyl transferase) | 228 |
| 7 (funG) | C5O77_00135 | Hypothetical protein (transcriptional regulator, TetR family) | 203 |
| 8 (funH) | C5O77_00140 | Acetyl-CoA carboxylase biotin carboxyl carrier protein subunit | 135 |
| 9 (funI) | C5O77_00145 | Acetyl-CoA carboxylase biotin carboxylase subunit | 447 |
| 10 (funJ) | C5O77_00150 | Acetyl-CoA carboxylase carboxyl transferase subunit beta | 270 |
| 11 (funK) | C5O77_00155 | Acetyl-CoA carboxylase carboxyl transferase subunit alpha | 249 |
| 12 (funL) | C5O77_00160 | NAD(P)H dehydrogenase | 194 |
| 13 | C5O77_00165 | Replication-associated protein RepC | 126 |
| 14 | C5O77_00170 | ATPase | 292 |
| 15 | C5O77_00050 | Hypothetical protein | 106 |
| 16 | C5O77_00055 | Hypothetical protein | 219 |
| 17 | C5O77_00060 | Hypothetical protein (relaxase/mobilization nuclease domain-containing protein) | 461 |
| 18 | C5O77_00065 | Mobilization protein (MobC) | 124 |
| 19 | C5O77_00070 | Hypothetical protein | 78 |
| 20 | C5O77_00075 | Hypothetical protein | 96 |
| 21 | C5O77_00080 | XRE family transcriptional regulator | 100 |
| 22 | C5O77_00085 | Hypothetical protein (RelE toxin of the RelE/RelB toxin-antitoxin system) | 129 |
| 23 | C5O77_00090 | Hypothetical protein | 122 |
| 24 | C5O77_00095 | Site-specific integrase | 195 |
| 25 | C5O77_00100 | Hypothetical protein | 95 |
Annotations were obtained from the JGI-IMG automated annotation pipeline.
TABLE 3.
Predicted annotations of genes in pVP-R2lc02a
| ORF (gene) | NCBI locus tag | Predicted functionb | Length (amino acids) |
|---|---|---|---|
| 1 (pksA) | C5O77_01185 | MFS transporter | 549 |
| 2 (pksB) | C5O77_01050 | 3-Oxoacyl-ACP synthase | 688 |
| 3 (pksC) | C5O77_01055 | Acyl carrier protein | 90 |
| 4 (pksD) | C5O77_01060 | Hypothetical protein (acetyl-CoA reductase 3-oxoacyl-[acyl carrier protein] reductase) | 239 |
| 5 (pksE) | C5O77_01065 | Hypothetical protein | 109 |
| 6 (pksF) | C5O77_01070 | Hypothetical protein (3-hydroxyacyl-[acyl carrier protein] dehydratase) | 111 |
| 7 (pksG) | C5O77_01075 | Hypothetical protein (glyoxylase, beta-lactamase superfamily II) | 298 |
| 8 (pksH) | C5O77_01080 | Hypothetical protein (4′-phosphopantetheinyl transferase superfamily protein) | 178 |
| 9 (pksI) | C5O77_01085 | PadR family transcriptional regulator | 177 |
| 10 (pksJ) | C5O77_01090 | (Acyl carrier protein) S-malonyl transferase | 317 |
| 11 (pksK) | C5O77_01095 | Acetyl-CoA carboxylase, biotin carboxyl carrier protein | 142 |
| 12 (pksL) | C5O77_01100 | Acetyl-CoA carboxylase, biotin carboxylase subunit | 459 |
| 13 (pksM) | C5O77_01105 | Acetyl-CoA carboxylase carboxyl transferase subunit beta | 279 |
| 14 (pksN) | C5O77_01110 | Acetyl-CoA carboxylase carboxyl transferase subunit alpha | 257 |
| 15 (pksO) | C5O77_01115 | Biotin-(acetyl-CoA carboxylase) ligase | 262 |
| 16 | C5O77_01120 | tRNA_Met_CAT | |
| 17 | C5O77_01125 | Hypothetical protein | 104 |
| 18 | C5O77_01130 | ATPase | 272 |
| 19 | C5O77_01135 | Hypothetical protein | 95 |
| 20 | C5O77_01140 | Hypothetical protein | 230 |
| 21 | C5O77_01145 | Hypothetical protein (relaxase/mobilization nuclease domain-containing protein) | 470 |
| 22 | C5O77_01150 | Hypothetical protein (mobilization protein C) | 124 |
| 23 | C5O77_01155 | Hypothetical protein | 78 |
| 24 | C5O77_01160 | Hypothetical protein | 152 |
| 25 | C5O77_01165 | XRE family transcriptional regulator | 91 |
| 26 | C5O77_01170 | Type II toxin-antitoxin system RelE/ParE family toxin | 120 |
| 27 | C5O77_01175 | Site-specific integrase | 195 |
| 28 | C5O77_01180 | Hypothetical protein | 92 |
Annotations were obtained from the JGI-IMG automated annotation pipeline. MFS, major facilitator superfamily.
Gene names were obtained from JGI-IMG annotation pipeline results. If there is another name for a specific gene, it is provided in parentheses or brackets.
TABLE 4.
Lengths, percent G+C contents, and accession numbers for draft genomes, plasmids, and PKS clusters
| Strain | Draft genome |
Plasmid or contig |
PKS cluster |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Predicted length (bp) | G+C content (%) | NCBI accession no. | No. of contigs | Name | Length (bp) | G+C content (%) | Name | Length (bp) | G+C content (%) | |
| L. reuteri R2lc | 2,084,790 | 38.45 | PTLS00000000 | 58 | pVP-R2lc01 | 23,739 | 32.42 | fun | 13,356 | 30.12 |
| pVP-R2lc02 | 21,446 | 32.12 | pks | 12,091 | 31.85 | |||||
| L. reuteri 2010 | 2,214,494 | 38.52 | PUXG00000000 | 38 | Contig 27 | 69,047 | 35.57 | pks | 12,247 | 30.97 |
In silico analyses of fun and pks clusters.
Since we identified polyketide synthase (PKS) genes on both pVP-R2lc01 and pVP-R2lc02, we used the secondary metabolite prediction software antiSMASH to identify potential clusters. This analysis revealed that each pVP-R2lc plasmid was predicted to carry a single PKS cluster, here referred to as fun and pks clusters, located on pVP-R2lc01 and pVP-R2lc02, respectively (Fig. 4C).
Further analysis by MultiGeneBlast (30) did not identify a biosynthetic gene cluster (BGC) similar to those found on pVP-R2lc02. However, when we used single protein sequences to search the NCBI Swiss-Prot database, we identified homologs (27 to 63% amino acid identity) that have been biochemically characterized (Table S3). Nonredundant searches revealed putative homologs in Lactococcus lactis subsp. cremoris KW10. Although this gene cluster in KW10 is not intact and separated in different contigs, we identified that all 15 genes in the pks cluster have a homolog in the KW10 genome (Table S4).
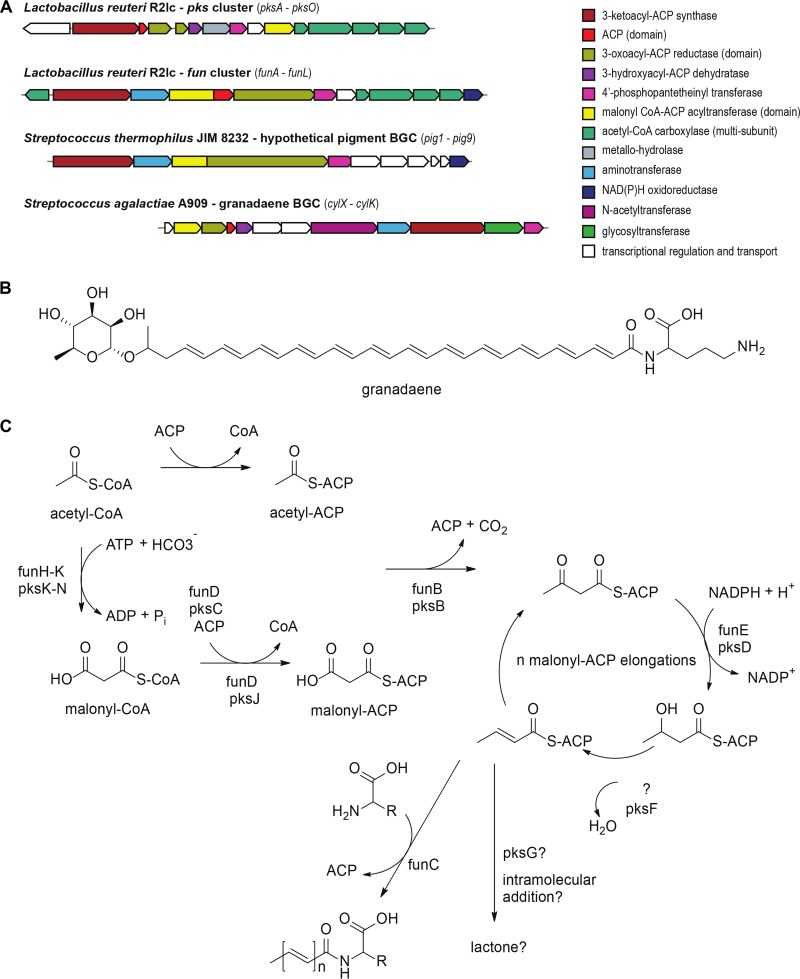
MultiGeneBlast analyses revealed two gene clusters homologous to the fun cluster one in Leuconostoc gasicomitatum LMG 18811 (LEGAS_1823 to LEGAS_1830) and another in Streptococcus thermophilus JIM 8232 (pig-1 to pig-9) (Fig. 5A). The latter BGC is predicted to be involved in group B Streptococcus pigment biosynthesis (31). These types of pigments, known as granadaene, have been structurally characterized for Streptococcus agalactiae A909 (Fig. 5B) (32) and Propionibacterium (33, 34).
FIG 5.
In silico analyses of the fun and pks clusters. (A) Schematic representation of Lactobacillus and Streptococcus type II polyketide-like fatty acid BGCs. Genes are color-coded by predicted function. (B) Chemical structure of granadaene, produced by group B Streptococcus. The polyene chain is formed by 13 elongation rounds, conjugated to the nonproteinogenic amino acid ornithine, and terminally glycosylated with rhamnose. (C) Comparison of the proposed biosynthetic pathways and predicted compounds for fun and pks clusters.
The final product chain length (number of extensions) cannot be predicted from the L. reuteri R2lc-derived PKS clusters, although BLAST searches of the fun and pks clusters identified enzymes related to long-chain fatty acid machinery and short-chain fatty acid machinery, respectively. Both BGCs have a dedicated multisubunit acetyl-CoA carboxylase (funH to funK [funH-K] and pksI-N) for the biotin-dependent carboxylation of acetyl-CoA to form malonyl-CoA. A malonyl CoA:ACP (acyl carrier protein) acyltransferase (pksJ and funD) is predicted to transfer the malonate to a dedicated ACP domain in the same polypeptide (funD) in the fun cluster and is encoded separately (by pksC) in the pks cluster. A predicted ketosynthase (funB and pksB) catalyzes the decarboxylative condensation of an acetyl-ACP primer and malonyl-ACP extender units. A 3-oxoacyl-ACP reductase (funE and pksD) reduces the 3-keto to a 3-hydroxyl group, which is subsequently reduced to an alkene by a 3-hydroxyacyl-ACP dehydratase (pksF). Since the fun cluster is not predicted to encode a dehydratase, it is plausible that this function is complemented by pksF or the regular fatty acid dehydratase. Both BGCs lack an enoyl-ACP reductase for further reduction of the carbon chain, resulting in the conjugated double-bond pattern of the predicted products. The growing unsaturated chain is further extended by iterative cycles incorporating malonyl-ACP, after which both biosynthetic pathways are predicted to diverge. The polyene fatty acid produced by the fun cluster is predicted to be condensed to a free l-amino acid by the aminotransferase funC, similar to the ornithine conjugation in granadaene. The final product of pks is hypothesized to be cyclized to form a lactone. pksG encodes a predicted metallo-hydrolase, which is potentially involved in intramolecular addition or lactone cleavage (Fig. 5C). Based on our results following the analyses of the PKS clusters with antiSMASH and MultiGeneBlast software, we conclude that the fun and pks clusters are predicted to encode type II polyketide/fatty-acid-like biosynthesis machineries.
The pks cluster is responsible for AhR activation in R2lc.
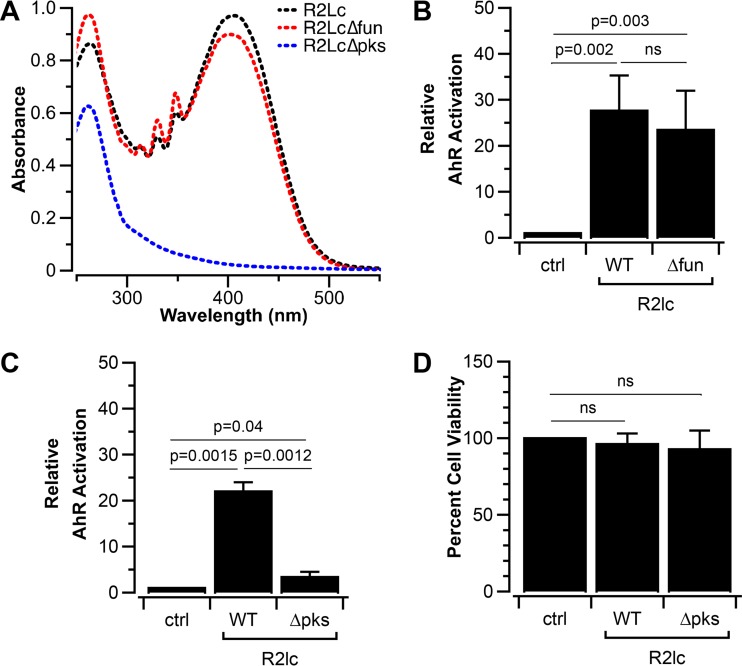
To investigate if the PKS clusters play a role in AhR activation, we generated R2lc derivatives in which the fun or pks cluster was inactivated. For R2lc Δfun, we deleted approximately 1.4 kb that corresponds to funE-G. This region included the gene encoding a 228-amino-acid phosphopantetheinyl transferase (PPTase) protein, which catalyzes the posttranslational modification of acyl carrier proteins in PKS pathways. R2lc Δfun, but not R2lc Δpks, retained its pigmented color, and R2lc Δfun exhibited absorption characteristics similar to those of wild-type R2lc (λmax = 405 to 409 nm) (Fig. 6A). Also, R2lc Δfun activated AhR similarly to wild-type R2lc (Fig. 6B).
FIG 6.
PKS is responsible for AhR activation. (A) R2lc∆pks does not absorb at 395 to 410 nm, in the region where R2lc and R2lc∆fun exhibited maximum absorbance. (B) The fun cluster does not drive AhR activation of L. reuteri R2lc. (C) Inactivation of the pks cluster significantly reduces the ability to activate AhR. (D) Extracts used for AhR activation assays do not impact cell viability. Data shown in bar graphs are averages of results from at least 3 biological replicates, and error bars represent standard deviations. A one-sample t test was used to compare strains and the negative control, and Student’s t test was used to compare wild-type and mutant strains (a P value of <0.05 was considered significant). ns, not significant; WT, wild type.
To investigate the role of the pks cluster in AhR activation, we replaced the pksG-I genes with a single promoter-gene fusion encoding chloramphenicol (Cm) resistance to yield R2lc Δpks::Cm. When we assessed the AhR activation potential of R2lc Δpks::Cm, we observed a significantly reduced ability to activate AhR (Fig. 6C), while cell viability upon exposure of the different bacterial supernatants was unaffected (Fig. 6D). In conclusion, the pks cluster from pVP-R2lc02 is responsible for the AhR-activating phenotype of L. reuteri R2lc.
L. reuteri 2010 activates AhR via a predicted gene cluster orthologous to pks.
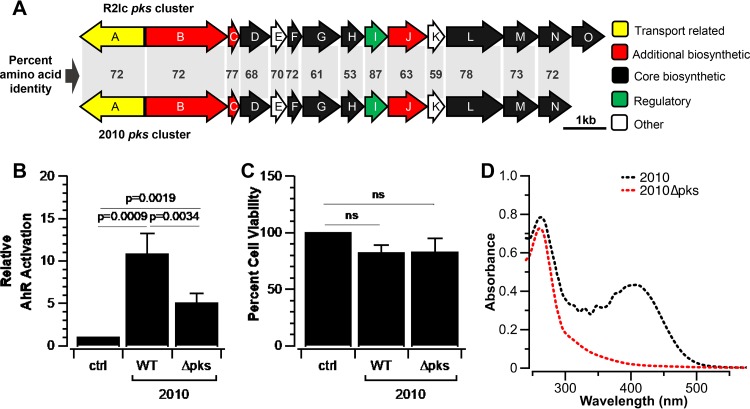
To identify the mechanism by which L. reuteri 2010 activates AhR, we first determined the genome sequence. Since we have identified the pathway by which R2lc activates AhR, we searched the 2010 genome for R2lc pks homologs. We identified a homologous gene cluster in 2010 with an identical gene organization (Fig. 7A), which was located on a single 69-kb contig. By inverse PCR, we were not able to find the adjacent contigs, and we did not identify open reading frames that putatively encode a replication-associated protein on the 69-kb contig. Also, unlike the plasmids in R2lc, we did not detect differences in the depth of sequencing coverage, which led us to suggest that the L. reuteri 2010 pks cluster could be encoded from the chromosome. Compared to R2lc pks, the 2010 pks cluster shares considerable identity on the amino acid level, ranging from 53% to 87%. Not surprisingly, the antiSMASH analysis predicted that this cluster encodes an aryl polyene (APE) biosynthetic synthase cluster. To test if the pks cluster is responsible for AhR activation of L. reuteri 2010, we deleted the pksG-I region, followed by an AhR activation assay. Indeed, 2010 Δpks had a significantly reduced AhR activation capacity compared to the wild type (Fig. 7B). Also, here, the reduced AhR activation capacity was not linked to differences in cell viability, since supernatants derived from L. reuteri 2010 wild-type and 2010 Δpks strains yielded comparable cell survival patterns (Fig. 7C). Similarly, 2010 Δpks is not pigmented and did not show an absorption band at 405 to 409 nm (Fig. 7D).
FIG 7.
2010 carries a PKS cluster similar to pks in R2lc. (A) Percent amino acid identities of homologous genes in pks clusters in R2lc and 2010. Transport-related, additional biosynthetic, core biosynthetic, regulatory, and other genes are represented. (B) Deletion of a 1.5-kb region (ORFs pksG-I) in 2010 (2010 Δpks) reduces the strain’s ability to activate AhR. (C) Extracts used for AhR activation assays derived from L. reuteri 2010 and L. reuteri 2010 Δpks do not impact cell viability. (D) 2010 shows a maximum absorption band at 405 to 409 nm, which was not observed in 2010 Δpks. Data shown represent averages of results from three biological replicates, and error bars represent standard deviations. A one-sample t test was used to compare strains and the negative control, and Student’s t test was used to compare wild-type and mutant strains (a P value of <0.05 was considered significant). ns, not significant; WT, wild type.
DISCUSSION
In this study, our aim was to gain an understanding of the potential of Lactobacillus reuteri to activate AhR at the species level. We screened a library of L. reuteri strains, derived from different hosts, and identified two orange-pigmented strains of rodent origin that are potent activators of AhR. By genome sequencing, comparative genomics, and targeted gene deletion, we identified that an orthologous PKS cluster is responsible for the AhR-activating phenotype.
In L. reuteri 100-23, the aromatic amino acid aminotransferase (AraT) has been shown to play a key role in AhR activation by metabolizing dietary tryptophan (Trp) into AhR ligands (15). In our model, inactivation of the R2lc strain’s araT gene, whose gene product has 96% amino acid homology to AraT of 100-23, did not alter the AhR-activating phenotype compared to the wild-type strain. One possibility is that the effect of R2lc AraT was masked by the robust PKS-mediated AhR activation. However, the double mutant (R2lc ΔaraT Δpks) revealed an AhR activation phenotype similar to that of R2lc Δpks (data not shown). Thus, AraT does not seem to contribute to AhR activation in our experimental model. Although Trp is present in Dulbecco’s modified Eagle’s medium (DMEM), the rate of metabolism, for example, driven by reduced araT gene expression compared to in vivo conditions, may not yield enough ligand to robustly activate AhR.
The fact that nearly all L. reuteri isolates show basal AhR activation leads us to hypothesize that select metabolic end products may result in AhR activation. This is somewhat substantiated by our finding that other lactic acid bacteria, including Lactobacillus plantarum and Lactobacillus casei, also yielded a basal level of AhR activation (data not shown). A link between bacterial metabolic end products and AhR activation would open new avenues of research to exploit a combination of probiotics and prebiotics to enhance the basal AhR activation potential, an intriguing idea, especially given the role of AhR in ameliorating intestinal inflammation (35).
We identified PKS clusters as the main driver of AhR activation in L. reuteri R2lc and 2010. PKS clusters are not commonly identified in L. reuteri; the only example described in the literature so far is reutericyclin, a chromosomally encoded secondary metabolite from sourdough isolates (10). Reutericyclin has bacteriostatic and bactericidal activities (36) and is proposed to provide these strains with an ecological advantage (37). The reutericyclin producer L. reuteri strains have been used in animal trials without any adverse effects (38). However, in our AhR assay, supernatants of reutericyclin producer strains L. reuteri TMW1.112 and TMW1.656 were toxic to hepatoma cells; thus, we were not able to assess the potential of these PKS-encoding sourdough isolates to activate AhR, and it was outside the scope of this work to test if the toxicity was driven by reutericyclin.
We identified that the pks cluster in R2lc has no homology with any BGCs in the NCBI database. The fun cluster has homology with the granadaene pigment pathway in Streptococcus thermophilus JIM 8232. Granadaenes and aryl polyenes (APEs) are similar from a biosynthetic point of view (related to fatty acid biosynthesis). Since there is some functional redundancy between both PKS clusters, they could cross talk with each other. For example, the fun cluster does not seem to have a 3-hydroxyacyl-ACP dehydratase to reduce the 3-hydroxyl groups in the growing chain to alkenes. This could be done by pksE or potentially by borrowing the equivalent enzyme from the fatty acid biosynthesis machinery. Therefore, at this point, we cannot rule out the possibility that R2lc Δfun retained activity due to cross talk between the fun and pks clusters, and this will be analyzed by future liquid chromatography-mass spectrometry (LC-MS) analyses. However, the fact that the inactivation of pks in L. reuteri 2010, a strain that does not carry a fun cluster homolog, yielded a phenotype similar to what we observed in R2lc Δpks supports the conclusion that the orthologous pks clusters in R2lc and 2010 are responsible for AhR activation.
In conclusion, we identified novel PKS gene clusters in L. reuteri that activate the aryl hydrocarbon receptor. Future studies will focus on the role of L. reuteri PKS in immunomodulation and ecology. Recent work has shown that AhR plays an important role in microbiota establishment in mice. AhR contributes to a selective pressure on the gut microbiota subphylum taxonomic groups, potentially by inducing IL-22-mediated antimicrobial production (39). This, combined with the fact that R2lc is a competitive strain in cocolonization studies with other L. reuteri isolates (21), could indicate a role for AhR in providing R2lc with a competitive advantage in host colonization; however, a direct antimicrobial effect, like reutericyclin, cannot be excluded. Aside from a potential ecological role, the secondary metabolite could contribute to amelioration of intestinal inflammation (40). While R2lc has been demonstrated to ameliorate colitis, the probiotic mechanism has not been fully uncovered. Therefore, we plan to determine the role of the R2lc pks cluster in amelioration of colitis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1, Table 5, and Table 6, respectively. Lactobacillus reuteri strains were cultured in De Man-Rogosa-Sharpe (MRS) medium (Difco, BD Biosciences). Unless stated otherwise, we prepared bacterial cultures as follows. Lactobacilli were incubated at 37°C under hypoxic conditions (5% CO2, 2% O2). Escherichia coli EC1000 was used as a general cloning host and cultured at 37°C in lysogeny broth (LB; Teknova). Electrocompetent E. coli EC1000 cells were prepared as described previously (41). Electrocompetent L. reuteri cells were prepared as described previously (42), with slight modifications. Briefly, L. reuteri cells were grown to an optical density at 600 nm (OD600) of 0.6 and harvested by centrifugation (4°C at 3,200 × g for 5 min). Cell pellets were washed twice with wash butter (0.5 M sucrose, 10% [vol/vol] glycerol). If applicable, erythromycin was supplemented at 5 μg/ml for Lactobacillus strains and at 300 μg/ml for E. coli EC1000 strains.
TABLE 5.
Plasmids used in this studya
| Plasmid | Characteristic(s) | Reference |
|---|---|---|
| pVPL3002 | pORI19 harboring L. reuteri-derived ddlF258Y | 29 |
| pVPL3669 | Emr; derivative of vector pVPL3002 in which the L. reuteri R2lc Δfun deletion cassette was cloned into the MCS | This study |
| pVPL3805 | Emr; derivative of vector pVPL3002 in which the L. reuteri R2lc Δpks deletion cassette was cloned into the MCS | This study |
| pVPL31010 | Emr; derivative of vector pVPL3002 in which the L. reuteri 2010 Δpks deletion cassette was cloned into the MCS | This study |
| pVPL31041 | Emr; derivative of vector pVPL3002 in which the L. reuteri R2lc Δpks::Cm insertion cassette was cloned into the MCS | This study |
pVPL, van Pijkeren Laboratory plasmid identification number; MCS, multiple-cloning site; ddlF258Y, a derivative of ddl in which mutations are made yielding the amino acid change phenylalanine to tyrosine at position 258 in the d-Ala-d-Ala (Ddl) protein.
TABLE 6.
Oligonucleotides used in this studya
| Oligonucleotide | Sequence (5′–3′) | Description |
|---|---|---|
| oVPL49 | ACAATTTCACACAGGAAACAGC | F; insert screening of pVPL3002 |
| oVPL97 | CCCCCATTAAGTGCCGAGTGC | R; insert screening of pVPL3002 |
| oVPL187 | TACCGAGCTCGAATTCACTGG | R; amplifies the pVPL3002 backbone |
| oVPL188 | ATCCTCTAGAGTCGACCTGC | F; amplifies the pVPL3002 backbone |
| oVPL1730 | TGAACCTCAATGTGCCTAGC | F; amplifies the u/s flanking region of the R2lc Δfun deletion cassette |
| oVPL1731 | AATTTAGTTGGGTTATGCTA | R; amplifies the u/s flanking region of the R2lc Δfun deletion cassette |
| oVPL1732 | TTAAAGGTACTGATAATTTCTATC | F; amplifies the d/s flanking region of the R2lc Δfun deletion cassette |
| oVPL1733 | TAGCGGACGTCCTGTAAAGT | R; amplifies the d/s flanking region of the R2lc Δfun deletion cassette |
| oVPL1734 | AAACGACGGCCAGTGAATTCGAGCTCGGTATGAACCTCAATGTGCCTAGCTGGCTTTATA | LCR bridging oligonucleotide to ligate the plasmid backbone + the u/s flanking region of the R2lc Δfun deletion cassette |
| oVPL1735 | TTTTCCCAAATAGCATAACCCAACTAAATTAAGGTACTGATAATTTCTATCAGTAAGTCT | LCR bridging oligonucleotide to ligate u/s and d/s flanking regions of the R2lc Δfun deletion cassette |
| oVPL1736 | AATATTCTTAACTTTACAGGACGTCCGCTAATCCTCTAGAGTCGACCTGCAGGCATGCAA | LCR bridging oligonucleotide to ligate the d/s flanking region of the R2lc Δfun deletion cassette and plasmid backbone |
| oVPL1737 | CGCTATTACGCCAGCTGGCG | F; sequencing of the R2lc Δfun deletion region |
| oVPL1738 | TCTGCTGATGGGCCTATAAAT | R; sequencing of the R2lc Δfun deletion region |
| oVPL1739 | TCGCTGCAAAGAGCAATCT | F; DCO PCR screening for R2lc Δfun |
| oVPL1740 | GGTGATAAAGTCTTGGCTGGAG | R; DCO PCR screening for R2lc Δfun |
| oVPL2334 | AAATATCTCCATGTCCTGGCAATAC | F; amplifies the u/s flanking region of the R2lc Δpks deletion cassette |
| oVPL2335 | TATCCCGACGAGCAAGTAAAG | R; amplifies the u/s flanking region of the R2lc Δpks deletion cassette |
| oVPL2336 | AATGGGGCTGTTATCGTTTTCC | F; amplifies the d/s flanking region of the R2lc Δpks deletion cassette |
| oVPL2337 | AAGCTGTATGGCAGGGCTTTC | R; amplifies the d/s flanking region of the R2lc Δpks deletion cassette |
| oVPL2338 | ACATTTAACCTTTACTTGCTCGTCGGGATAATCCTCTAGAGTCGACCTGCAGGCATGCAA | LCR bridging oligonucleotide to ligate the plasmid backbone and u/s flanking region of the R2lc Δpks deletion cassette |
| oVPL2339 | AAACGACGGCCAGTGAATTCGAGCTCGGTAAATGGGGCTGTTATCGTTTTCCTGTTTTCT | LCR bridging oligonucleotide to ligate the d/s flanking region of the R2lc Δpks deletion cassette and the plasmid backbone |
| oVPL2340 | TCTCCTAAAGAAAGCCCTGCCATACAGCTTAAATATCTCCATGTCCTGGCAATACTAGGT | LCR bridging oligonucleotide to ligate u/s and d/s flanking regions of the R2lc Δpks deletion cassette |
| oVPL2341 | TGTCCTAGCTGATGCTGCAAC | F; DCO PCR screening for R2lc Δpks |
| oVPL2342 | AATAGTTCCAGGGGTGCTTC | R; DCO PCR screening for R2lc Δpks |
| oVPL2518 | TGAAAGTGAGTTGTATGGGTGG | F; amplifies the u/s flanking region of the 2010 Δpks deletion cassette |
| oVPL2519 | TCTAGTTCTCTATAATAATTTACGCGC | R; amplifies the u/s flanking region of the 2010 Δpks deletion cassette |
| oVPL2520 | AACTGTTGGATTTCTTGAAAGTCC | F; amplifies the d/s flanking region of the 2010 Δpks deletion cassette |
| oVPL2521 | AGTCGGGTATTTAGCGCAAATTG | R; amplifies the d/s flanking region of the 2010 Δpks deletion cassette |
| oVPL2522 | AAAACGACGGCCAGTGAATTCGAGCTCGGTAAACTGTTGGATTTCTTGAAAGTCCATAAA | LCR bridging oligonucleotide to ligate the plasmid backbone and u/s flanking region of the 2010 Δpks deletion cassette |
| oVPL2523 | AAGAAAGGCCACCCATACAACTCACTTTCATCTAGTTCTCTATAATAATTTACGCGCTGA | LCR bridging oligonucleotide to ligate u/s + d/s flanking regions of the 2010 Δpks deletion cassette |
| oVPL2524 | GCTTTTTCAATTTGCGCTAAATACCCGACTATCCTCTAGAGTCGACCTGCAGGCATGCAA | LCR bridging oligonucleotide to ligate the d/s flanking region of the 2010 Δpks deletion cassette with the plasmid backbone |
| oVPL2525 | TCTGAAGTAGGTGACGGTGC | F; sequencing of the 2010 Δpks deletion region |
| oVPL2526 | AATCCAATTGTCCCAGGAGTC | R; sequencing of the 2010 Δpks deletion region |
| oVPL2527 | GCTTTTTGTGCTCCTTGACC | F; DCO PCR screening for 2010 Δpks |
| oVPL2528 | TGCCGTTTTCTGAGGTGTCG | R; DCO PCR screening for 2010 Δpks |
| oVPL2856 | AGTGTCATGGCGCATTAACG | F; amplifies the Cm gene of the R2lc Δpks::Cm insertion cassette |
| oVPL2857 | TTATAAAAGCCAGTCATTAGGCC | R; amplifies the Cm gene of the R2lc Δpks::Cm insertion cassette |
| oVPL2858 | TCTCCTAAAGAAAGCCCTGCCATACAGCTTTTATAAAAGCCAGTCATTAGGCCTATCTGA | LCR bridging oligonucleotide to ligate the d/s flanking region of the R2lc Δpks::Cm deletion cassette with the Cm gene |
| oVPL2859 | CCCTTTATTCCGTTAATGCGCCATGACACTAAATATCTCCATGTCCTGGCAATACTAGGT | LCR bridging oligonucleotide to ligate the u/s flanking region of the R2lc Δpks::Cm deletion cassette with the Cm gene |
| oVPL2860 | TGGGAAACAATTTCCCCGAAC | Internal PCR screening for the Cm gene |
| oVPL665 | TCCTCACTCAAGTGGTGCTG | F; amplifies the GAPDH gene in R2lc and its mutants; used for qPCR analyses |
| oVPL666 | ACCGAATGCTGGGTTAGTAG | R; amplifies the GAPDH gene in R2lc and its mutants; used for qPCR analyses |
| oVPL3095 | TGGCAAACCTTTTTGTTGTTCTGG | F; amplifies the funB gene in R2lc and its mutants; used for qPCR analyses |
| oVPL3096 | TCGCATTAATACCTCCAAATCCG | R; amplifies the funB gene in R2lc and its mutants; used for qPCR analyses |
| oVPL3097 | ATGTCAGAATGGGTTTTTGCTGG | F; amplifies the pksB gene in R2lc and its mutants; used for qPCR analyses |
| oVPL3098 | TGATAAGCCGTGCCCTAAAATTTC | R; amplifies the pksB gene in R2lc and its mutants; used for qPCR analyses |
| oVPL395 | ATGCCAGCTACTAAAAAAGAAATCCTTAG | F; amplifies araT in 6475; used for MAMA-PCR |
| oVPL396 | TTAATCCTCCTTATTAATGAAGGCCG | MAMA-PCR oligonucleotide; used for screening L. reuteri 6475 ΔaraT |
| oVPL401 | ACAAAGATTCTTGGTGGGATTCCGATTGAAGTTGATACTTAAGGCGATGATTTTGTTCTCACACCCGCAAGACTCCAAAG | Lagging-strand oligonucleotide that mutates S150X; when incorporated, yields a silent mutation and in-frame stop codon |
| oVPL402 | GGATTCCGATTGAAGTTGATACTTAA | R; amplifies araT in 6475; used for MAMA-PCR |
oVPL, van Pijkeren Laboratory oligonucleotide identification number; F, forward; R, reverse; u/s, upstream; d/s, downstream; qPCR, quantitative PCR. The sequence in boldface type indicates a stop codon.
Reagents and enzymes.
To amplify DNA fragments for cloning and screening, we used Phusion Hot Start DNA polymerase II (Thermo Scientific) and Taq DNA polymerase (Denville Scientific), respectively. We used T4 DNA ligase (Thermo Scientific) for blunt-end ligations. If applicable, we treated purified PCR products with DpnI (Thermo Scientific) to remove the plasmid template DNA. Phosphorylation of double-stranded DNA (dsDNA) fragments was performed with T4 polynucleotide kinase (Thermo Scientific). Ligase cycling reactions (LCRs) were performed as described previously (43).
Construction of suicide shuttle vectors.
To generate mutant strains in lactobacilli, we used our recently developed counterselection plasmid (pVPL3002) (29). To generate R2lc Δfun, R2lc Δpks, and 2010 Δpks deletion cassettes, 500 to 1,000 bp of upstream and downstream flanking regions of target genes were amplified by colony PCR. To amplify the flanking sequences, we used oVPL1730-oVPL1731 (upstream; R2lc Δfun), oVPL1732-1733 (downstream; R2lc Δfun), oVPL2334-2335 (upstream; R2lc Δpks), oVPL2336-2337 (downstream; R2lc Δpks), oVPL2518-2519 (upstream; 2010 Δpks), and oVPL2520-2521 (downstream; 2010 Δpks), followed by column purification (GeneJET PCR purification kit; Thermo Scientific).
The pVPL3002 backbone was amplified with oVPL187-oVPL188, followed by column purification (GeneJET PCR purification kit; Thermo Scientific) and DpnI treatment. Column-purified amplicons were quantified (Qubit; Life Technologies). The amplicons were mixed at equimolar quantities (0.25 pmol), followed by phosphorylation, ethanol precipitation, and LCR. Following LCR, DNA was precipitated with pellet paint (Novagen), resuspended in 5 μl sterile water, and transformed (at 2.5 kV, 25 μF, and 200 Ω) (Gene Pulser Xcell; Bio-Rad) into electrocompetent E. coli EC1000 cells. By PCR, we screened for insertion of our target sequences using oligonucleotides that flank the multiple-cloning site (oVPL49-oVPL97). Finally, the integrity of deletion cassettes was determined by Sanger sequencing.
To replace the pksG-I genes in the R2lc pks cluster with the gene encoding chloramphenicol (Cm) resistance, we used our laboratory stock of L. reuteri 6475::Cm to amplify the Cm gene with the PHELP promoter (oVPL2856-2857). The amplicon was cloned into pVPL3805 (R2lc Δpks deletion cassette) by LCR to yield R2lc Δpks∷Cm, which we named VPL31041.
Generation of L. reuteri mutant strains by homologous recombination.
Three micrograms of plasmid DNA was electroporated into electrocompetent L. reuteri cells. To identify plasmid integration, colonies were screened by PCR with oligonucleotide mixtures oVPL1739-oVPL1740-oVPL97 (upstream single crossover [SCO]; R2lc Δfun), oVPL1739-oVPL1740-oVPL97 (downstream SCO; R2lc Δfun), oVPL2341-oVPL2342-oVPL97 (upstream SCO; R2lc Δpks::Cm), oVPL2341-oVPL2342-oVPL49 (downstream SCO; R2lc Δpks::Cm), oVPL2525-oVPL2526-oVPL97 (upstream SCO; 2010 Δpks), and oVPL2525-oVPL2526-oVPL49 (downstream SCO; 2010 Δpks). Following confirmation of SCO, bacterial cells were cultured for two passages in MRS broth without antibiotic selection, and cells were plated on MRS agar plates containing 400 μg/ml vancomycin. Vancomycin-resistant colonies represent cells in which a second homologous recombination took place. To identify cells in which a double-crossover (DCO) event took place, we performed PCR using oligonucleotides oVPL1739-oVPL1740 (R2lc Δfun) and oVPL2341-oVPL2342 (R2lc Δpks::Cm). We used Sanger sequencing to verify the integrity of the recombinant strains.
Construction of L. reuteri 6475 ΔaraT by recombineering.
The gene encoding AraT in L. reuteri 6475 was inactivated by single-stranded DNA recombineering as described previously (44). Briefly, at OD600 values of >0.55 and <0.65, L. reuteri 6475 harboring the recombineering plasmid pJP042 was supplemented with 10 ng/ml induction peptide (MAGNSSNFIHKIKQIFTHR; Peptide2.0 Inc.) for 20 min to induce the expression of RecT. Electrocompetent cells were prepared as described above, and 100 μg of the recombineering oligonucleotide (oVPL401) was transformed accordingly. We used a mismatch amplification mutation assay-PCR (MAMA-PCR) (45) with oligonucleotides oVPL395, oVPL396, and oVPL402 to identify recombinant genotypes, which were confirmed by Sanger sequencing.
Preparation of cell-free bacterial supernatants for AhR activation assays.
Lactobacillus cultures were grown in MRS broth for approximately 16 h and subsequently subcultured in prewarmed MRS broth (0.1%, vol/vol) until the OD600 reached 2. Cell pellets were harvested by centrifugation (3,200 × g for 5 min) and washed with an equal volume of Dulbecco’s modified Eagle’s medium (DMEM). Cells were resuspended in an equal volume of DMEM harboring 5% (vol/vol) newborn calf serum (NBCS; Life Technologies) and 1% (vol/vol) nonessential amino acids (NEAA; Gibco) and incubated for 18 h under hypoxic conditions (5% CO2 and 2% O2). To prevent settling of the cells, and to maintain hypoxic conditions throughout the culture, we gently mixed the cells on an orbital shaker (standard orbital shaker; VWR) (orbital speed, 165 rpm). After incubation, the supernatants were collected following centrifugation (3,200 × g for 5 min), the pH was adjusted to 7.6 with 500 nM NaOH, and the supernatants were subsequently filter sterilized (0.22 μm, polyvinylidene difluoride [PVDF]; Millipore).
In vitro AhR activation assays.
The murine hepatoma cell line H1L6.1c3 (a gift from Gregory Kennedy) was used as a reporter cell line to determine AhR activation (19). Expression of the firefly luciferase gene is driven by a dioxin response element (DRE). The level of luminescence is a direct indicator of the level of AhR activation. Briefly, H1L6.1c3 cells were resuspended to a concentration of 1 × 105 cells/ml in DMEM containing 10% (vol/vol) NBCS, 1% (vol/vol) NEAA, and a 1% (vol/vol) penicillin-streptomycin mixture (Lonza). Per biological replicate, we seeded two 96-well plates (clear-bottom white polystyrene; Corning) with 2 × 104 cells/well; one plate was used to determine the activation of AhR, while the other plate was used to determine cell viability (see below). After a 24-h incubation (37°C with 5% CO2), cells were gently washed with 200 μl phosphate-buffered saline (PBS), after which 200 μl of the cell-free bacterial supernatant was added in quadruplicate. As controls, we included DMEM containing 5% NBCS and 1% NEAA or the same medium supplemented with the AhR ligand 6-formylindolo[3,2-b]carbazole (FICZ) (500 nM). Cells were incubated for 18 h, followed by assessment of AhR activation and cell viability. For AhR activation, cells were gently washed with 200 μl PBS, followed by the addition of 50 μl/well luciferase assay reagent (Bright-Glo luciferase assay; Promega). After a 5-min incubation at room temperature (RT), total luminescence was measured with a luminometer (BD moonlight 3010; BD Biosciences). Fold AhR activation was calculated as the fold increase in luminescence relative to the DMEM control.
Cell viability.
To determine cell viability, we used the CellTiter-Glo 2.0 assay kit (Promega) that quantifies the amount of ATP present, which indicates the percentage of metabolically active cells. Briefly, a cell culture plate was incubated at room temperature for 30 min. Next, 100 μl/well of the supernatant was removed, followed by the addition of 100 μl cell viability assay reagent. Samples were incubated for 10 min at 22°C. The luminescence level per well was determined as described above. Percent cell viability for each sample was calculated based on the percent luminescence relative to the DMEM control that contained 5% NBCS and 1% NEAA.
Pigment extraction and UV-visible spectrophotometry.
L. reuteri cell pellets (2 ml; OD600 = 2) were washed twice with PBS and resuspended in 2 ml of 90% methanol. The cell suspension was vortexed for 30 s, followed by centrifugation (14,000 × g for 2 min) and filter sterilization (0.22 μm, PVDF; Millipore). The filtrate was diluted in methanol and transferred into a 1-cm quartz cell. This solution was used to investigate the absorption profile, with scanning at a wavelength range of 240 to 600 nm on a UV-visible spectrophotometer (Shimadzu UV-1601 PC; Shimadzu Corp., Kyoto, Japan) at RT (20°C to 22°C). For AhR assays, the methanol extract was evaporated at room temperature overnight. The precipitate was resuspended in 2 ml complete DMEM containing 5% NBCS and 1% NEAA, which was subsequently used to determine the AhR activation potential of orange pigment with the AhR reporter cell line as described above.
Whole-genome sequencing.
Genomic DNAs were prepared with a genomic DNA purification kit (Wizard; Promega), and DNA concentrations were determined using the Qubit dsDNA high-sensitivity assay kit (Life Technologies). Sequencing was performed at the University of Wisconsin—Madison Biotechnology Center. Samples were prepared according to the TruSeq Nano DNA LT library prep kit (Illumina Inc.). Briefly, samples were sheared using a Covaris M220 ultrasonicator (Covaris Inc.) and size selected for an average insert size of 550 bp using solid-phase reversible-immobilization bead-based size exclusion. The quality and quantity of the finished libraries were analyzed using an Agilent DNA1000 chip and a Qubit dsDNA high-sensitivity assay kit. Libraries were standardized to 2 nM. Paired-end, 250-bp sequencing was performed using the Illumina MiSeq sequencer and a MiSeq 500-bp (v2) sequencing cartridge. Images were analyzed using the standard Illumina pipeline (version 1.8.2).
Comparative genome and bioinformatics analyses.
De novo assemblies of sequence reads were performed using the SeqMan NGen software package (DNASTAR version 12.3.1.4) with standard settings. We closed the gaps between contigs that consist of unique genes in R2lc by Sanger sequencing and primer walking. Briefly, we determined adjacent contigs by PCR using oligonucleotides located at the end of contigs. Next, we amplified gaps between contigs by PCR, and these DNA fragments were sequenced. For primer walking, new oligonucleotides were designed, and an approximately 500- to 900-bp gap was closed after each round of Sanger sequencing. Oligonucleotides used for gap closure are listed in Table S1 in the supplemental material. Assembled draft genomes were uploaded to the Joint Genome Institute Integrated Microbial Genomes and Microbes (JGI-IMG) database to perform annotation and genome comparison. We determined unique genes in L. reuteri R2lc and 2010 genomes by comparison against the following L. reuteri genomes: 100-23, 3c6, CF48-3A1, DSM20016, I5007, JCI1112, LP167-67, Lpuph1, mlc3, and SD2112. We next uploaded draft genomes to the antiSMASH tool, a Web-based secondary metabolite prediction software (46), to identify PKS clusters. We determined the percent amino acid identity of PKS gene products in R2lc and 2010 using the basic local alignment search tool for proteins (BLASTP) at the National Center for Biotechnology Information.
Plasmid copy number determination.
Relative copy numbers of the pVP-R2lc01 and pVP-R2lc02 plasmids were determined by quantitative PCR as described previously (47), with slight modifications. We used the chromosomal gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference, which is present as a single copy. Bacterial cultures were grown until the OD600 reached 2. Five-hundred-microliter bacterial cultures were pelleted by centrifugation (21,130 × g for 2 min). Cell pellets were washed twice with 500 μl sterile water (21,130 × g for 2 min) and resuspended in 400 μl sterile water, followed by microwave treatment (1,100 W for 2 min). Suspensions diluted 100-fold were used as the template for PCR analyses. PCR mixtures contained 8 μl of the cell suspension (100-fold diluted), 1 μl (250 nM each) of the primer pair, and 10 μl of SYBR green master mix (Bio-Rad). Primer pairs were designed for the single-copy housekeeping gene GAPDH (oVPL665-666), pVP-R2lc01 (oVPL3095-2096), and pVP-R2lc02 (oVPL3097-2098). The PCR efficiency for each primer pair was determined. The threshold cycle (CT) values were determined using the CFX96 real-time system (Bio-Rad). Relative copy number analyses were performed using the Pfaffl method (48). A total of three biological replicates were performed.
Accession number(s).
The DNA sequences corresponding to Lactobacillus reuteri R2lc and 2010 have been deposited in GenBank with accession numbers PTLS00000000 and PUXG00000000, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Siv Ahrné (Lund University, Sweden) for providing L. reuteri R2lc, N2J, N2D, and N4I; Jens Walter (University of Alberta) for sharing part of his L. reuteri strain collection; and BioGaia AB (Stockholm, Sweden) for providing L. reuteri strains ATCC PTA 6475, ATCC PTA 4659, and 2010.
This work was supported by startup funds from the University of Wisconsin—Madison to J.-P.V.P., the UW-Madison Food Research Institute, and the United States Department of Agriculture, National Institute of Food and Agriculture (USDA NIFA) Hatch award MSN185615 and grant no. 2018-6717-27523. M.Ö. received financial support from the Turkish Ministry of National Education, from the Department of Food Science, and is the recipient of the Robert H. and Carol L. Deibel Distinguished Graduate Fellowship in Probiotic Research, which is awarded by the Food Research Institute (UW-Madison).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01661-18.
REFERENCES
- 1.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisch KM. 2013. Biosynthesis of natural products by microbial iterative hybrid PKS-NRPS. RSC Adv 3:18228. doi: 10.1039/c3ra42661k. [DOI] [Google Scholar]
- 4.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci U S A 101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwecke T, Aparicio JF, Molnár I, König A, Khaw LE, Haydock SF, Oliynyk M, Caffrey P, Cortés J, Lester JB. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci U S A 92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, Bacquié V, Pénary M, Gaultier E, Nougayrède J-P, Fioramonti J, Oswald E. 2012. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3:501–509. doi: 10.4161/gmic.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, Cichewicz R, Li Y, Liu J, Roe B, Ferretti J, Merritt J, Qi F. 2010. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl Environ Microbiol 76:5815–5826. doi: 10.1128/AEM.03079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golomb BL, Yu AO, Coates LC, Marco ML. 2018. The Lactococcus lactis KF147 nonribosomal peptide synthetase/polyketide synthase system confers resistance to oxidative stress during growth on plant leaf tissue lysate. Microbiologyopen 7:e00531. doi: 10.1002/mbo3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin XB, Lohans CT, Duar R, Zheng J, Vederas JC, Walter J, Gänzle M. 2015. Genetic determinants of reutericyclin biosynthesis in Lactobacillus reuteri. Appl Environ Microbiol 81:2032–2041. doi: 10.1128/AEM.03691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Moura-Alves P, Faé K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, Barison N, Diehl A, Munder A, Constant P, Skrahina T, Guhlich-Bornhof U, Klemm M, Koehler A-B, Bandermann S, Goosmann C, Mollenkopf H-J, Hurwitz R, Brinkmann V, Fillatreau S, Daffe M, Tümmler B, Kolbe M, Oschkinat H, Krause G, Kaufmann SHE. 2014. AhR sensing of bacterial pigments regulates antibacterial defence. Nature 512:387–392. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- 13.Lamas B, Natividad JM, Sokol H. 2018. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol 11:1024–1038. doi: 10.1038/s41385-018-0019-2. [DOI] [PubMed] [Google Scholar]
- 14.Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, Macdonald TT, Pallone F, Monteleone G. 2011. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141:237.e1–248.e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay J-M, Langella P, Xavier RJ, Sokol H. 2016. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cervantes-Barragan L, Chai JN, Tianero MD, DiLuccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh C-S, Colonna M. 2017. Lactobacillus reuteri induces gut intraepithelial CD4+ CD8αα+ T cells. Science 357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbongue JC, Nicholas DA, Torrez TW, Kim N-S, Firek AF, Langridge WHR. 2015. The role of indoleamine 2,3-dioxygenase in immune suppression and autoimmunity. Vaccines 3:703–729. doi: 10.3390/vaccines3030703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Langenhove K, Croes K, Denison MS, Elskens M, Baeyens W. 2011. The CALUX bio-assay: analytical comparison between mouse hepatoma cell lines with a low (H1L6.1c3) and high (H1L7.5c1) number of dioxin response elements. Talanta 85:2039–2046. doi: 10.1016/j.talanta.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 20.Johansson M-L, Molin G, Jeppsson B, Nobaek S, Ahrne S, Bengmark AS. 1993. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol 59:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, Peterson DA, Blom J, Wenzel CQ, Szymanski CM, Walter J. 2017. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol 83:e00132-17. doi: 10.1128/AEM.00132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holma R, Salmenperä P, Lohi J, Vapaatalo H, Korpela R. 2001. Effects of Lactobacillus rhamnosus GG and Lactobacillus reuteri R2LC on acetic acid-induced colitis in rats. Scand J Gastroenterol 36:630–635. doi: 10.1080/003655201750163114. [DOI] [PubMed] [Google Scholar]
- 23.Fabia R, Ar’rajab A, Johansson ML, Willén R, Andersson R, Molin G, Bengmark S. 1993. The effect of exogenous administration of Lactobacillus reuteri R2LC and oat fiber on acetic acid-induced colitis in the rat. Scand J Gastroenterol 28:155–162. doi: 10.3109/00365529309096063. [DOI] [PubMed] [Google Scholar]
- 24.Mao Y, Yu J-L, Ljungh Å, Molin G, Jeppsson B. 1996. Intestinal immune response to oral administration of Lactobacillus reuteri R2LC, Lactobacillus plantarum DSM 9843, pectin and oatbase on methotrexate-induced enterocolitis in rats. Microb Ecol Health Dis 9:261–269. doi: 10.3109/08910609609166466. [DOI] [Google Scholar]
- 25.Mao Y, Nobaek S, Kasravi B, Adawi D, Stenram U, Molin G, Jeppsson B. 1996. The effects of Lactobacillus strains and oat fiber on methotrexate-induced enterocolitis in rats. Gastroenterology 111:334–344. doi: 10.1053/gast.1996.v111.pm8690198. [DOI] [PubMed] [Google Scholar]
- 26.Ahl D, Liu H, Schreiber O, Roos S, Phillipson M, Holm L. 2016. Lactobacillus reuteri increases mucus thickness and ameliorates dextran sulphate sodium-induced colitis in mice. Acta Physiol (Oxf) 217:300–310. doi: 10.1111/apha.12695. [DOI] [PubMed] [Google Scholar]
- 27.Adawi D, Kasravi FB, Molin G, Jeppsson B. 1997. Effect of Lactobacillus supplementation with and without arginine on liver damage and bacterial translocation in an acute liver injury model in the rat. Hepatology 25:642–647. doi: 10.1002/hep.510250325. [DOI] [PubMed] [Google Scholar]
- 28.Thorlacius H, Nobaek S, Wang XD, Andersson R, Molin G, Bengmark S, Jeppsson B. 2003. Lactobacilli attenuate bacteremia and endotoxemia associated with severe intra-abdominal infection. Surgery 134:467–473. doi: 10.1067/S0039-6060(03)00246-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Oh J-H, Alexander LM, Özçam M, Van Pijkeren J-P. 2018. d-Ala-d-Ala ligase as a broad-host-range counterselection marker in vancomycin-resistant lactic acid bacteria. J Bacteriol 200:e00607-17. doi: 10.1128/JB.00607-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medema MH, Takano E, Breitling R. 2013. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol 30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delorme C, Bartholini C, Luraschi M, Pons N, Loux V, Almeida M, Guédon E, Gibrat J-F, Renault P. 2011. Complete genome sequence of the pigmented Streptococcus thermophilus strain JIM8232. J Bacteriol 193:5581–5582. doi: 10.1128/JB.05404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa-Fraile M, Dramsi S, Spellerberg B. 2014. Group B streptococcal haemolysin and pigment, a tale of twins. FEMS Microbiol Rev 38:932–946. doi: 10.1111/1574-6976.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosa-Fraile M, Rodríguez-Granger J, Haidour-Benamin A, Cuerva JM, Sampedro A. 2006. Granadaene: proposed structure of the group B Streptococcus polyenic pigment. Appl Environ Microbiol 72:6367–6370. doi: 10.1128/AEM.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanberg C, Lutnaes BF, Langsrud T, Nes IF, Holo H. 2007. Propionibacterium jensenii produces the polyene pigment granadaene and has hemolytic properties similar to those of Streptococcus agalactiae. Appl Environ Microbiol 73:5501–5506. doi: 10.1128/AEM.00545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Díaz-Díaz CJ, Ronnekleiv-Kelly SM, Nukaya M, Geiger PG, Balbo S, Dator R, Megna BW, Carney PR, Bradfield CA, Kennedy GD. 2016. The aryl hydrocarbon receptor is a repressor of inflammation-associated colorectal tumorigenesis in mouse. Ann Surg 264:429–436. doi: 10.1097/SLA.0000000000001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Höltzel A, Gänzle MG, Nicholson GJ, Hammes WP, Jung G. 2000. The first low molecular weight antibiotic from lactic acid bacteria: reutericyclin, a new tetramic acid. Angew Chem Int Ed Engl 39:2766–2768. doi:. [DOI] [PubMed] [Google Scholar]
- 37.Gänzle MG, Vogel RF. 2003. Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int J Food Microbiol 80:31–45. doi: 10.1016/S0168-1605(02)00146-0. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Zhao X, Le MHA, Zijlstra RT, Gänzle MG. 2015. Reutericyclin producing Lactobacillus reuteri modulates development of fecal microbiota in weanling pigs. Front Microbiol 6:762. doi: 10.3389/fmicb.2015.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray IA, Nichols RG, Zhang L, Patterson AD, Perdew GH. 2016. Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice. Sci Rep 6:33969. doi: 10.1038/srep33969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao C, Major A, Rendon D, Lugo M, Jackson V, Shi Z, Mori-Akiyama Y, Versalovic J. 2015. Histamine H2 receptor-mediated suppression of intestinal inflammation by probiotic Lactobacillus reuteri. mBio 6:e01358-15. doi: 10.1128/mBio.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Russell DW. 2006. Transformation of E. coli by electroporation. CSH Protoc 2006:pdb.prot3933. doi: 10.1101/pdb.prot3933. [DOI] [PubMed] [Google Scholar]
- 42.Oh J-H, van Pijkeren J-P. 2014. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kok S, De Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, Newman JD, Chandran SS. 2014. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol 3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- 44.van Pijkeren JP, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha RS, Zarbl H, Keohavong P, Thilly WG. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl 2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 46.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Kranenburg R, Golic N, Bongers R, Leer RJ, de Vos WM, Siezen RJ, Kleerebezem M. 2005. Functional analysis of three plasmids from Lactobacillus plantarum. Appl Environ Microbiol 71:1223–1230. doi: 10.1128/AEM.71.3.1223-1230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NCK, Patil PB, Juge N, MacKenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J. 2011. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet 7:e1001314. doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.