The strigolactone composition of sorghum root exudates greatly influences germination and infection by the root parasitic weed Striga hermonthica, with orobanchol correlating with resistance and 5-deoxystrigol with susceptibility.
Keywords: Germination stimulant, parasitic weed, resistance, sorghum, Striga, strigolactones
Abstract
Sorghum is an important food, feed, and industrial crop worldwide. Parasitic weeds of the genus Striga constitute a major constraint to sorghum production, particularly in the drier parts of the world. In this study we analysed the Striga germination stimulants, strigolactones, in the root exudates of 36 sorghum genotypes and assessed Striga germination and infection. Low germination-stimulating activity and low Striga infection correlated with the exudation of low amounts of 5-deoxystrigol and high amounts of orobanchol, whereas susceptibility to Striga and high germination-stimulating activity correlated with high concentrations of 5-deoxystrigol and low concentrations of orobanchol. Marker analysis suggested that similar genetics to those previously described for the resistant sorghum variety SRN39 and the susceptible variety Shanqui Red underlie these differences. This study shows that the strigolactone profile in the root exudate of sorghum has a large impact on the level of Striga infection. High concentrations of 5-deoxystrigol result in high infection, while high concentrations of orobanchol result in low infection. This knowledge should help to optimize the use of low germination stimulant-based resistance to Striga by the selection of sorghum genotypes with strigolactone profiles that favour normal growth and development, but reduce the risk of Striga infection.
Introduction
The root-parasitic weed Striga (Striga hermonthica [Del.] Benth.) is a serious threat to food security in sub-Saharan Africa. The parasite is one of the most serious pests limiting yields of the major cereal crops, maize (Zea mays L.), pearl millet [Pennisetum glaucum (L.) R. Br.], and sorghum (Sorghum bicolor [L.] Moench) (Taylor, 2003; Gressel et al., 2004; Ejeta, 2007; Scholes and Press, 2008; Badu-Apraku et al., 2013). Methods used to control Striga range from cheap traditional means such as hand pulling, crop rotation, fallow, and variety choice, employed by resource-limited farmers, to modern, more expensive interventions such as the use of herbicides and fertilizers, practised by resource-rich farmers (Oswald, 2005; Parker, 2009). For decades, the development of satisfactory, low-cost, and efficient control technologies has been a major and difficult challenge, owing to the complexity of the parasite’s life cycle, its production of large amounts of tiny seeds with prolonged viability, and the serious damage inflicted on the host by the parasite while it is still hidden underground (Scholes and Press, 2008; Spallek et al., 2013). The increase in frequency and magnitude of crop yield losses and the risk of potential future spread have led to an intensification of studies aiming to improve Striga control through cultural and chemical measures and the generation of Striga-resistant crop varieties. Striga-resistant crop genotypes have been proposed to provide the simplest, cheapest, and most durable solution to the problem (Ejeta, 2005; Tesso and Ejeta, 2011).
In general, Striga resistance refers to the ability to reduce or prevent infection with and reproduction of the parasite, while tolerance refers to the ability to support equally severe levels of infection as other varieties of the same host species without the associated impairment of growth or loss in grain yield (Rodenburg et al., 2005; Rodenburg and Bastinaans, 2011). One of the most studied and best documented mechanisms of resistance against Striga is based on the fact that a host-derived signal is required for Striga seeds to germinate. Low production of the germination signal could potentially be a mechanism of resistance (Ejeta et al., 1992; Ejeta et al., 2000; Ejeta et al., 2007; Yoder and Scholes, 2010; Cardoso et al., 2011). The sorghum cultivar Framida, originating from Uganda, was reported as early as 1958 to be a low germination-stimulant producer (Williams, 1958). In Sudan, the sorghum cultivars Tetron and SRN39 were also reported to be low stimulant producers (Kambal and Musa, 1979; Bebawi, 1981). In addition, positive correlations between the amount of germination stimulant produced and Striga infection levels in the field were reported by several authors (Vasudeva Rao, 1984; Rich et al., 2004; Mohemed et al., 2016).
Parker et al. (1977) used the double-pot technique to screen a large collection of sorghum genotypes for low germination stimulant-based resistance to Striga asiatica. In this technique, 7-day-old sorghum seedlings were grown in sterile quartz sand in a pot with a perforated base, which was placed in another pot without perforations to collect root exudates. An aliquot of the root exudate was applied to preconditioned Striga seeds to assess its germination-inducing activity. Use of the double pot and other similar techniques resulted in the identification of several low-stimulant genotypes. Field screening for resistance to Striga in Sudan revealed that field resistance is more frequent among low stimulant producers than among high stimulant producers (Babiker, 2002). The agar gel assay was developed to screen sorghum genotypes for resistance to Striga based on the low capacity of the root exudates to stimulate the germination of Striga seeds under controlled conditions (Hess et al., 1992; Haussmann et al., 2000). The agar gel assay was also used for mapping of a low-germination-stimulant quantitative trait locus (QTL), lgs, which was subsequently used as a marker to transfer the low-stimulant trait into other sorghum genotypes (Hess et al., 1992; Scholes and Press, 2008; Satish et al., 2012). In all the screening approaches described above, sorghum genotypes were identified as low or high stimulant producers on the basis of the germination-inducing activity of their root exudate rather than on the composition and nature of the signalling molecules. Identification of the sorghum-derived germination stimulant has been an important research target for decades. Sorgoleone and dihydrosorgoleone were the first identified sorghum-derived Striga germination stimulants in cultivar IS 4225 (Chang et al., 1986). However, sorghum cultivars with resistance to Striga based on low stimulant production were found to produce the same amounts of these compounds as the susceptible cultivars, undermining an in planta role for sorgoleone in the induction of Striga seed germination (Siame et al., 1993). Indeed, sorghum also produces strigolactones, and these were found to be more active in inducing Striga germination at extremely low concentrations, and to correlate better with resistance to Striga, than dihydrosorgoleone (Bouwmeester et al., 2003; Ejeta et al., 2007; Xie et al., 2010). Strigolactones including strigol, sorgolactone, sorgomol, and 5-deoxystrigol were identified in a number of sorghum cultivars (Hauck et al., 1992; Siame et al., 1993; Awad et al., 2006; Xie et al., 2008) (Fig. 1). However, variation in the composition and quantity of strigolactones produced by different sorghum varieties have been discussed in only a few reports. For example, Awad et al. (2006) characterized 5-deoxystrigol as the major stimulant in three sorghum cultivars (M 800, Hybrid, and Swarna), sorgomol as a second major stimulant in two of them (M 800 and Hybrid), and strigol, but not sorgomol, as the second major stimulant in the root exudate of the third cultivar (Swarna). In addition, sorgolactone—which was previously isolated by Hauck et al. (1992) from the sorghum cultivar Haygrazer—was not detected in any of the cultivars investigated by Awad et al. (2006). Structural variation among the strigolactones present in root exudates of different sorghum genotypes has been suggested to influence susceptibility to Striga in the field (Yoneyama et al., 2015). For example, the Striga-susceptible sorghum cultivar Tabat exuded 5-deoxystrigol as the major stimulant (Yoneyama et al., 2010), while the Striga-resistant cultivar SRN39 exuded only small amounts of 5-deoxystrigol and had sorgomol as its major stimulant, in addition to a number of minor putative unidentified strigolactones (Yoneyama et al., 2010). The authors suggested that susceptibility to Striga of some sorghum genotypes is associated with the production and/or exudation of more stable non-hydroxy strigolactones, that is, 5-deoxystrigol and sorgolactone, rather than the more unstable strigolactone alcohols such as sorgomol (Yoneyama et al., 2010). Although strigolactones have been analysed in sorghum root exudates and genotypic variation for strigolactone exudation in sorghum has been demonstrated, more studies on the direct association between host genotype, strigolactone exudation, and Striga germination, host specificity, and parasitism are still needed.
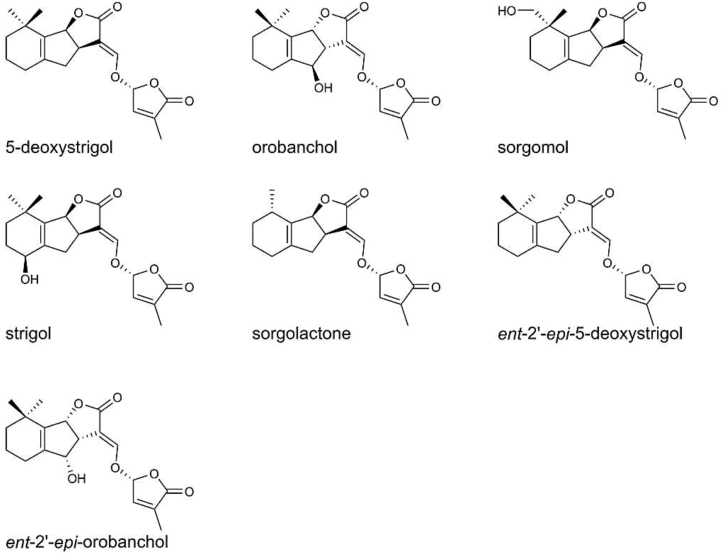
Fig. 1.
Chemical structures of the strigolactones detected in root exudates of sorghum genotypes.
The present investigation, which included 36 sorghum genotypes comprising landraces and improved varieties collected from Sudan, was carried out to study the influence of genotypic variation in the quality and quantity of strigolactones in the sorghum root exudates and their role in Striga infection.
Materials and methods
Seeds of 36 sorghum genotypes were obtained from the gene bank of the Agricultural Research Co-operation (ARC) Wad Medani, Sudan (see Supplementary Table S1 at JXB online). Data on field performance, agronomic traits, and susceptibility to Striga were obtained from the sorghum breeding program of the ARC. Seeds of a sorghum ecotype of Striga hermonthica (Del.) Benth. were collected in 2009/2010 in a sorghum field at the Abu Naama Research Station, Sudan, and were supplied by Dr A. Hamid, Sinnar University, Sudan.
Analysis of root exudates
For the collection of root exudates, germinated sorghum seeds of the 36 genotypes were planted in 3 litre plastic pots filled with 1.5 litres of sand. One week after planting, the seedlings were thinned to five plants per pot. Half-strength modified Hoagland’s nutrient solution was applied to each pot (500 ml at 48 h intervals). The plants were allowed to grow under controlled conditions in a climate room with artificial light at 450 µmol m−2 s−1 and at a temperature of 28 °C (day; 10 h)/25 °C (night; 14 h) and 70% relative humidity for 4 weeks. In the fifth week, phosphorus (P) deficiency was created in each pot to increase strigolactone production (López-Ráez et al., 2008). To achieve this, 3 litres of P-deficient nutrient solution (half-strength modified Hoagland’s nutrient solution minus P) were added to each pot and allowed to drain freely from the pot. The plants were kept under P deficiency for 1 week prior to flushing each pot with 3 litres of P-deficient nutrient solution to remove accumulated strigolactones. The plants were then allowed to grow for an additional 48 h, after which root exudates were collected in a 1 litre plastic bottle by passing 3 litres of nutrient solution without P through each pot. The collected exudate was passed over a solid phase extraction (SPE) C18 column (500 mg 3 ml–1) and strigolactones were eluted with 6 ml acetone. For further purification, the acetone was evaporated under a vacuum at 25 °C using a rotary evaporator. The residue was dissolved in 4 ml hexane and loaded on to a pre-equilibrated Silica gel Grace Pure SPE (200 mg 3 ml–1) column; strigolactones were eluted with 2 ml hexane:ethyl acetate (1:9). The solvent was evaporated and the residue dissolved in 200 µl of 25% aqueous acetonitrile. The sample was filtered through a Minisart SRP4 0.45 µm filter (Sartorius, Germany) prior to LC-MS/MS analysis.
Strigolactone analysis
The strigolactones 5-deoxystrigol, sorgomol, and orobanchol were identified and quantified using ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) as previously described by López-Ráez et al. (2008). The samples were analysed by a Waters Xevo triple quadrupole tandem mass spectrometer (Waters, Milford, MA, USA) equipped with electrospray ionization source and coupled to an Acquity UPLC system (Waters). Multiple reactions monitoring (MRM) was used for quantification of strigolactones in sorghum root exudates. 5-Deoxystrigol was detected at a retention time of 7.8 min in MRM channels m/z 331>97, 331>216, and 331>234; sorgomol at 4.8 min in MRM channels m/z 347>317, 317>97, and 317>133; orobanchol at 4.5 min in MRM channels m/z 347>97, 347>205, and 347>233; sorgolactone at 4.7–4.8 min in MRM channels m/z 317>97 and 317>133; strigol at 4.50 min in MRM channels m/z 347>97, 329> 97, and 329>215; ent-2ʹ-epi-orobanchol at 4.5 min in MRM channels m/z 347>97, 347>205, and 347>233; and ent-2ʹ-epi-5-deoxystrigol at 8.0 min in MRM channels m/z 331>97, 331>216, and 331>234. Data acquisition and analysis were performed using Mass Lynx 4.1 (TargetLynx) software (Waters).
Assessment of germination-stimulating activity
Striga seeds placed on 8 mm glass fibre filter paper discs (~50 seeds each) were surface sterilized and preconditioned as previously described by Matusova et al. (2004). The discs containing preconditioned seeds were treated with aliquots (50 µl) of authentic strigolactones (5-deoxystrigol, sorgomol, orobanchol, strigol, sorgolactone, ent-2ʹ-epi-5-deoxystrigol, and ent-2ʹ-epi-orobanchol) (0.02 µM in water) or a 100-fold diluted, C18-purified root exudate, and then incubated at 30 °C for 48 h and subsequently examined for germination (radicle protrusion). Distilled water and the synthetic strigolactone GR24 (0.02 µM) were included as negative and positive controls, respectively. All treatments were replicated six times.
Assessment of Striga infection
Based on differences in strigolactone production and profile, 22 sorghum genotypes were selected for a pot experiment using soil infested with Striga seeds to examine whether differences in strigolactone content in the exudate resulted in differences in Striga infection. Plants were grown in 18 litre pots (0.25 m length × 0.25 m width × 0.30 m height) containing a mixture of sand and soil, collected from the top layer (0–0.25 m) of an arable field near Wageningen. Striga seeds (8 mg) were added and mixed thoroughly with the soil in each pot. The pots were watered and kept for 10 days in the greenhouse to allow for conditioning of Striga seeds. Subsequently, five seeds of each of the 22 sorghum genotypes were sown in the middle of the pot. Seedlings were thinned to one plant per pot 4 days after emergence. Plants were grown in a temperature-controlled greenhouse at a temperature of 28 °C (day; 10 h)/25 °C (night; 14 h), with natural sunlight supplemented with artificial light and at 70% relative humidity. Half-strength modified Hoagland’s nutrient solution was applied in the first week (250 ml at 48 h intervals). For the remainder of the experimental period, a nutrient solution with 20% P (250 ml per pot at 48 h intervals) was applied to stimulate strigolactone exudation.
Counting of Striga seedlings started when Striga emergence was first observed on the susceptible genotype Tabat, ~2 weeks after sowing. Subsequently, the number of emerged Striga seedlings per pot per genotype was assessed every 3 days until Striga had emerged on all of the 22 genotypes. Further Striga counts were made at weekly intervals for 10 weeks. The total number of Striga plants per host plant was determined after harvesting each genotype at maturity by root washing and counting the attached Striga tubercles and plants.
Gene expression analysis
To investigate the role of strigolactone biosynthetic genes as a possible explanation for genotypic differences in strigolactone production among sorghum genotypes, we selected three resistant genotypes, SRN39, IS9830, and Tetron, which produce low amounts of 5-deoxystrigol and sorgomol but higher amounts of orobanchol, and which induce low Striga germination and exhibit low levels of Striga infection; and one susceptible genotype, Fakimustahi, which produces high amounts of 5-deoxystrigol and sorgomol but low amounts of orobanchol, and which elicits high Striga seed germination and displays high levels of Striga infection. Differences in the expression of the sorghum orthologs of the rice strigolactone biosynthetic genes DWARF27 (Sb05g022855.1), DWARF17 (CAROTENOID CLEAVAGE DIOXYGENASE7; CCD7) (Sb06g024560), DWARF10 (CAROTENOID CLEAVAGE DIOXYGENASE 8; CCD8) (Sb03g034400.1), and MORE AXILLARY BRANCHING 1 (MAX1) (Sb03g032220) were analysed for these genotypes (Supplementary Tables S2–S5).
Primers were designed on the basis of the predicted mRNA sequences (Supplementary Table S6) in the National Center for Biotechnology Information (NCBI) database. The D27 (DWARF27) gene sequence was obtained from the published sequence in the NCBI database (http://www.ncbi.nlm.nih.gov; clone ID-FJ641055) and the corresponding sorghum orthologous gene was identified by BLASTn analysis (Supplementary Table S2). For SbCCD8 and SbCCD7, the corresponding rice gene sequence was used to identify the orthologs in sorghum (http://www.gramene.org) as described previously (Vallabhaneni et al., 2010; Guan et al., 2012; Priya and Siva, 2014) (Supplementary Tables S3 and S4). For SbMAX1, Arabidopsis thaliana MAX1 (AT2G26170) was obtained from the published sequence (http://www.arabidopsis.org) and the corresponding sorghum orthologous gene was identified by BLASTn analysis in PlantGDB (http://www.plantgdb.org/) (Supplementary Table S5). Primer pairs for individual genes were designed with Primer Express 2.0 software (Applied Biosystems, Foster City, CA, USA). Primer sequences were confirmed using the BLASTn program to ensure amplification of unique and appropriate cDNA fragments (Supplementary Table S6).
Total RNA was extracted from roots of the genotypes that were grown under conditions of P deprivation for 2 weeks. The RNA was extracted from 150 mg of homogenized ground roots using 500 ml Trizol (Invitrogen) and subsequently purified with chloroform and precipitated with isopropanol. Pellets were washed with ethanol (70% v/v) and then resuspended in 20 µl Milli-Q water, and DNA was removed using a DNAase I Kit (Qiagen) according to the manufacturer’s instructions. The RNA was cleaned by using a DNase treatment prior to quantitative (q) PCR (Promega).
Gene expression was assessed by using cDNA from each sorghum genotype. cDNA was synthesized by using the iScript cDNA Synthesis Kit (BioRad) using 1 μg of total RNA per sample, following the manufacturer’s instructions. The qPCR reactions were performed using iQ SYBR Green Supermix (BioRad). The qPCR reaction consisted of 5 µl of SYBR Green Supermix, 0.5 µl of forward primer, 0.5 µl of reverse primer (with each primer at a concentration of 0.1 μM), 1 µl of 10-fold diluted template cDNA, and 2 µl double-deionized water. The qRT–PCR program was one cycle of 95 °C for 10 s, followed by 45 cycles of 94 °C for 10 s, 60 °C for 10 s, and 72 °C for 15 s. The amplifications were detected by using a BioRad CFX Connect. The relative levels of RNA for each gene were calculated from cycle threshold values according to the ΔΔCt method (Schmittgen and Livak, 2008). The specificity of the reactions was verified by melting curve analysis. The expression data presented are the average of three control and three biological replicates. The sorghum actin gene SbACTIN (Sb01g010030.1) (Supplementary Table S6) was used as a reference.
Marker analysis
PCR was performed in a total reaction volume of 25 µl. The reaction was set up as follows: sterile distilled water 14.76 µl, buffer (5×) 5 µl, forward primer (10 mM) 0.5 µl, reverse primer (10 mM) 0.5 µl, dNTPs (10 mM) 0.5 µl, MgCl2 (25 mM) 2.5 µl, Taq DNA polymerase (5U) 0.24 µl, and template DNA (50 ng µl–1) 1 µl. PCR amplification conditions were as follows: initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, and elongation at 72 °C for 2 min., followed by a final elongation step at 72 °C for 5 min. The reaction products were loaded on to 3% agarose gels, stained with ethidium bromide, and run in 1× Tris–acetate–EDTA buffer at 100 V for 1 h, with a 50 bp DNA (0.5 μg per lane) ladder (Thermo Scientific). Gels were visualized using an EpiChemi II Darkroom (UVP Ltd, Cambridge, UK) gel documentation system. The sequences of the forward and reverse primers for the SB3344 markers are provided in Supplementary Table S7.
Statistical analysis
The statistical package SAS (version 15) was used for analysis of variance (ANOVA) and Pearson’s correlation analyses. Duncan’s honest significant difference test was subsequently performed to establish the significance of differences. The relationship between various strigolactones and the number of emerged Striga seedlings, Striga biomass, and in vitro Striga germination was analysed by correlation analysis and stepwise regressions using the IBM SPSS Statistics 20 package. The Origin Pro 9 (64-bit) statistical software package was used for principal component analysis (PCA). To meet the assumption of normality, the strigolactone peak area and number of emerged Striga seedlings were subjected to logarithmic transformation prior to ANOVA.
Results
Strigolactone analysis
The major strigolactones that were detected in the root exudates of all sorghum genotypes under investigation were 5-deoxystrigol, sorgomol, and orobanchol (Fig. 1; Supplementary Table S8). Other strigolactones, including sorgolactone, strigol, ent-2ʹ-epi-orobanchol and ent-2ʹ-epi-5-deoxystrigol, were detected in low concentrations in the root exudates of some genotypes (data not shown). The amount of strigolactones secreted and the ratio between the individual strigolactones differed considerably among sorghum genotypes (Supplementary Table S8;Fig. 2). Among all the genotypes studied, Debeikri, Feterita Geshaish, Hemisi, Kolom, Najad, Tabat, and N13 were the highest 5-deoxystrigol producers (Supplementary Table S8). Bari, Dari, Fakimustahi, Wad Fahel, and Wad Elmardi were the highest sorgomol producers, and SRN39, Tetron, IS9830, Framida, and Hakika were the highest orobanchol producers (Supplementary Table S8). The latter genotypes were also among the lowest 5-deoxystrigol producers. Arfa Gadamak, Korokollow, Mogud, Tafsagabeid, Tokarawe, Wad Ahmed, and Wad Baco secreted intermediate amounts of 5-deoxystrigol and low amounts of sorgomol and orobanchol (Supplementary Table S8).
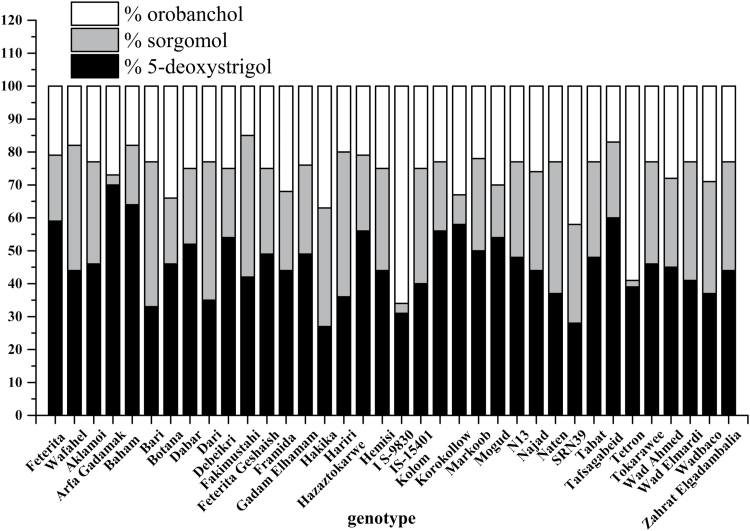
Fig. 2.
Genotypic variation in the proportions of the strigolactones 5-deoxystrigol, sorgomol, and orobanchol in root exudates of sorghum.
Proportions of 5-deoxystrigol, sorgomol, and orobanchol in the root exudates
The relative proportion of 5-deoxystrigol in the composition of the strigolactone blend in root exudate was very high (59–70%) in Arfa Gadamak, Baham, Tafsagabeid, and Feterita; high (50–58%) in Korokollow, Hazaztokarwe, Kolom, Mogud, Debeikri, Dabar, and Markoob; moderately high (40–49%) in Gadam Elhamam, Feterita Geshaish, Tabat, N13, Botana, Aklamoi, Tokarawe, Wad Ahmed, Najad, Zahrat Elgadambalia, Hemisi, Framida, Wad Fahel, Fakimustahi, Wad Elmardi and IS15401; moderate (30–39%) in Tetron, Wad Baco, Naten, Hariri, Dari, Bari, and IS9830; and low (27–28%) in SRN39 and Hakika (Fig. 2). The relative proportion of orobanchol in the composition of the strigolactone blend was high (42–66%) in IS9830, Tetron, and SRN39; moderate (29–37%) in Hakika, Botana, Korokollow, Framida, Mogud, Wad Baco, Wad Ahmed, Arfa Gadamak, Najad, Dabar, Feterita Geshaish, IS15401, Hemisi, and Debeikri; low (20–24%) in Gadam Elhamam, Tabat, N13, Naten, Zahrat Elgadambalia, Aklamoi, Bari, Dari, Tokarawe, Wad Elmardi, Kolom, Markoob, Feterita, Hazaztokarwe, and Hariri; and very low (2–9%) in Wad Fahel, Baham, Tafsagabeid, and Fakimustahi (Fig. 2). The relative proportion of sorgomol in the strigolactone blend was high (40–44%) in Hariri, Bari, Fakimustahi, Dari, and Naten; moderate (29–38%) in Wad Fahel, Hakika, Wad Elmardi, IS15401, Wad Baco, Zahrat Elgadambalia, Aklamoi, Hemisi, Tokarawe, Najad, SRN39, Tabat, and N13; moderately low (24–28%) in Markoob, Gadam Elhamam, Wad Ahmed, Feterita Geshaish, and Framida; low (16–23%) in Dabar, Hazaztokarwe, Tafsagabeid, Debeikri, Kolom, Botana, Feterita, Baham, and Mogud; and very low (2–9%) in Korokollow, IS9830, Arfa Gadamak, and Tetron (Fig. 2).
Germination-inducing activity of sorghum root exudates
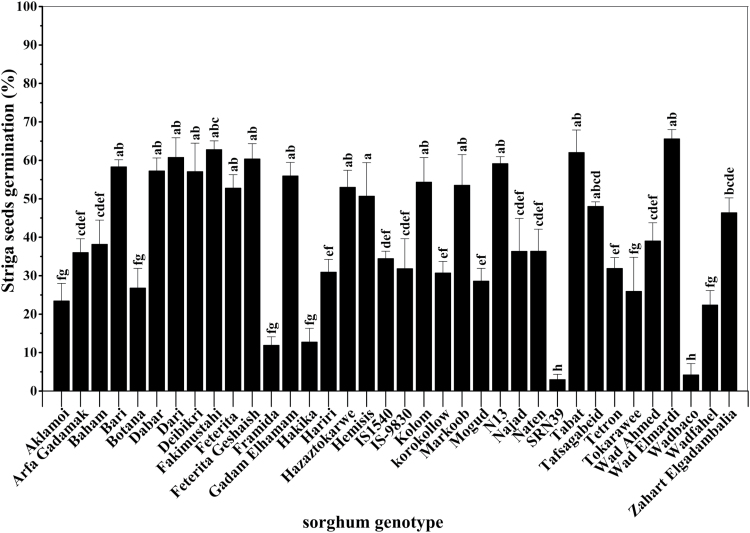
The C18-purified, 100-fold-diluted root exudates displayed significant variation in germination-inducing activity (P<0.01) (Fig. 3). The germination-inducing activity of root exudates was very high (60–67%) for Bari, Dabar, Dari, Fakimustahi, Feterita Geshaish, Kolom, Markoob, N13, Tabat, and Wad Elmardi; high (50–59%) for Debeikri, Feterita, Hemisi, Gadam Elhamam and Hazaztokarwe; moderate (30–49%) for Baham, Tafsagabeid, Zahrat Elgadambalia, Wad Ahmed, Naten, Arfa Gadamak, Najad, IS15104, IS9830, Tetron, Hariri, Mogud, and Korokollow; and low (0–29%) for Aklamoi, Botana, Framida, Hakika, SRN39, Wad Baco, Wad Fahel and Tokarawe. Among the genotypes, the germination-inducing activity of the root exudates was highest for Wad Elmardi, Fakimustahi, and Tabat, and lowest for SRN39, Wad Baco, and Framida (Fig. 3). Distilled water and the synthetic strigolactone GR24 (0.02 µM) induced negligible (0%) and 50% germination, respectively.
Fig. 3.
Genotypic variation in the germination-inducing activity of sorghum root exudates. Bars represent means±SE (n=5). Least significant differences of means at P=0.05 by ANOVA. Letters above the bars indicate different significance groups after Duncan’s pairwise comparisons (P<0.05).
Germination-stimulating activity of standard strigolactones
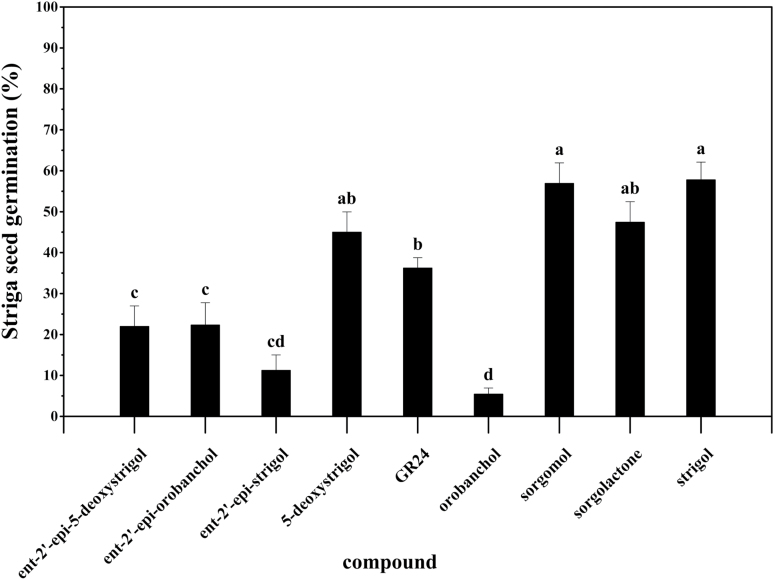
The natural strigolactones 5-deoxystrigol, sorgomol, orobanchol, strigol, sorgolactone, ent-2ʹ-epi-5-deoxystrigol, and ent-2ʹ-epi-orobanchol, together with the synthetic strigolactone GR24, each at 0.02 µM, displayed significant differences in germination-inducing activity (Fig. 4). GR24 induced 36% germination. Sorgomol, strigol, sorgolactone and 5-deoxystrigol induced 57%, 58%, 47%, and 45% germination, respectively. Ent-2ʹ-epi-orobanchol, ent-2ʹ-epi-5-deoxystrigol, and ent-2ʹ-epi-strigol induced 22%, 22%, and 11% germination, respectively. Orobanchol, by contrast, induced only 5% germination.
Fig. 4.
Germination response of Striga seeds to natural and synthetic strigolactones at 0.02 μM. Results represent the mean of five replicates. Error bars indicate SE. Different letters above the bars indicate significant differences between mean values (P<0.05; ANOVA).
Striga infection
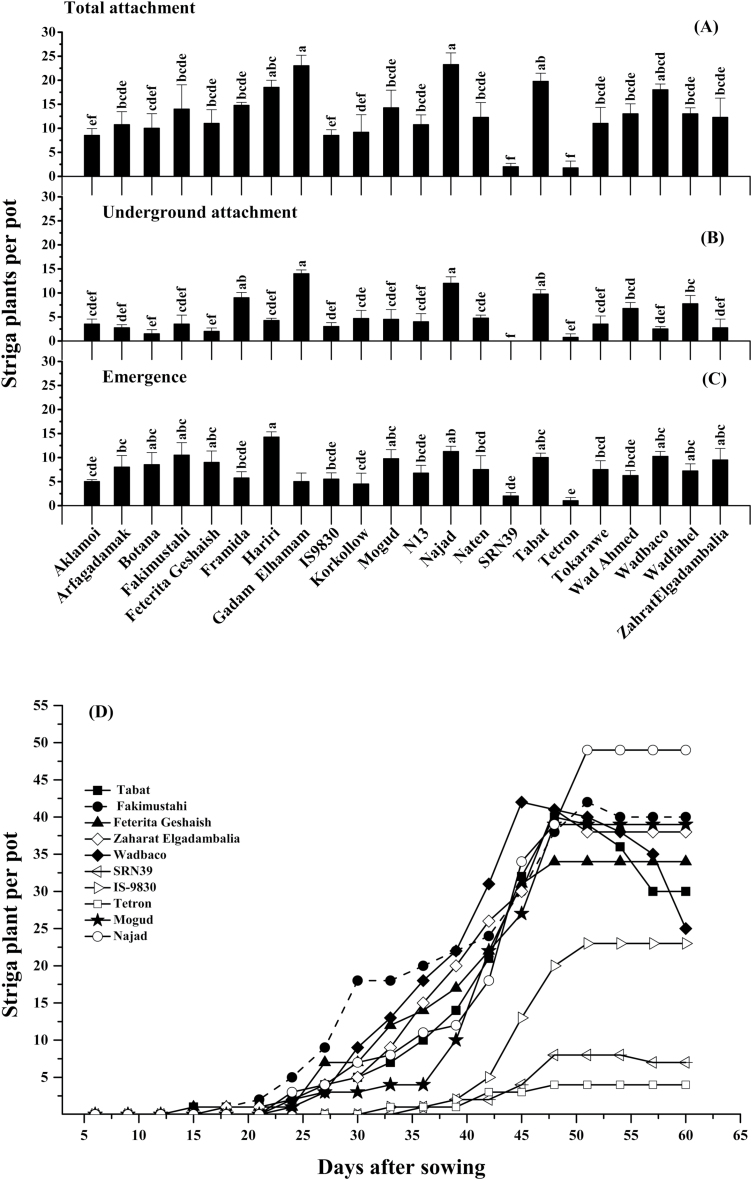
To investigate the effect of the observed differences in germination-stimulating activity on Striga infection, a selection of 22 sorghum genotypes was grown in pots with soil infected with Striga seeds. The mean total number of attached Striga plants per pot was highest (18–24) on Najad, Gadam Elhamam, Tabat, Hariri, and Wad Baco; moderate (11–15) on Framida, Mogud, Fakimustahi, Wad Ahmed, Wad Fahel, Naten, Zahrat Elgadambalia, Feterita Geshaish, Tokarawe, Arfa Gadamak, and N13; low (9–10) on Botana, Korokollow, and Aklamoi; and IS9830; and negligible (2) on SRN39 and Tetron (Fig. 5A).
Fig. 5.
Emergence of Striga plants in a greenhouse pot experiment. (A–C) Mean±SE total attached (A), below-ground attached, (B) and emerged (C) Striga plants per pot at harvest (n=4). The significance of a treatment effect was determined by one-way ANOVA for all genotypes; different letters above the bars indicate significant differences after Duncan’s pairwise comparison (P<0.05). (D) Time course of Striga emergence in the greenhouse pot experiment on 10 selected sorghum genotypes. Data are the mean total emergence of Striga plants per pot during a period of 76 days (n=4 replicates).
The number of non-emerged attached Striga plants per pot was highest (9–15) on Gadam Elhamam, Najad, Tabat, and Framida; moderate (7–8) on Wad Ahmed and Wad Fahel; low (3–5) on Naten, Korokollow, Mogud, Hariri, N13, Aklamoi, Fakimustahi, Tokarawe, IS9830, Arfa Gadamak, Zahrat Elgadambalia, Wad Baco, and Feterita Geshaish; and negligible (0–2) on Botana, Tetron, and SRN39 (Fig. 5B).
The number of emerged Striga plants per pot was highest (9–15) on Hariri, Najad, Fakimustahi, Wad Baco, Tabat, Mogud, Zahrat Elgadambalia, Feterita Geshaish, Botana, and Arfa Gadamak; moderate (6–8) on Naten, Tokarawe, Wad Fahel, N13, Wad Ahmed, Framida, and IS9830; low (5) on Aklamoi, Korokollow, and Gadam Elhamam; and negligible (1–2) on SRN39 and Tetron (Fig. 5C).
Emergence time
The number of days to first emergence of Striga showed significant (P<0.01) dependence on the sorghum genotype (Supplementary Table S9). Emergence was very early [15–18 days after sowing (DAS)] on Aklamoi, Fakimustahi, Feterita Geshaish, Tabat, Gadam Elhamam, Wad Fahel, and Zahrat Elgadambalia; moderately early (23–27 DAS) on Arfa Gadamak, Botana, Framida, Korokollow, Mogud, Naten, N13, Najad, Tokarawe, and Wad Baco; and late (31–36 DAS) on Hariri, IS9830, SRN39, Tetron, and Wad Ahmed. Fig. 5D shows the emergence of Striga for a selection of 10 genotypes.
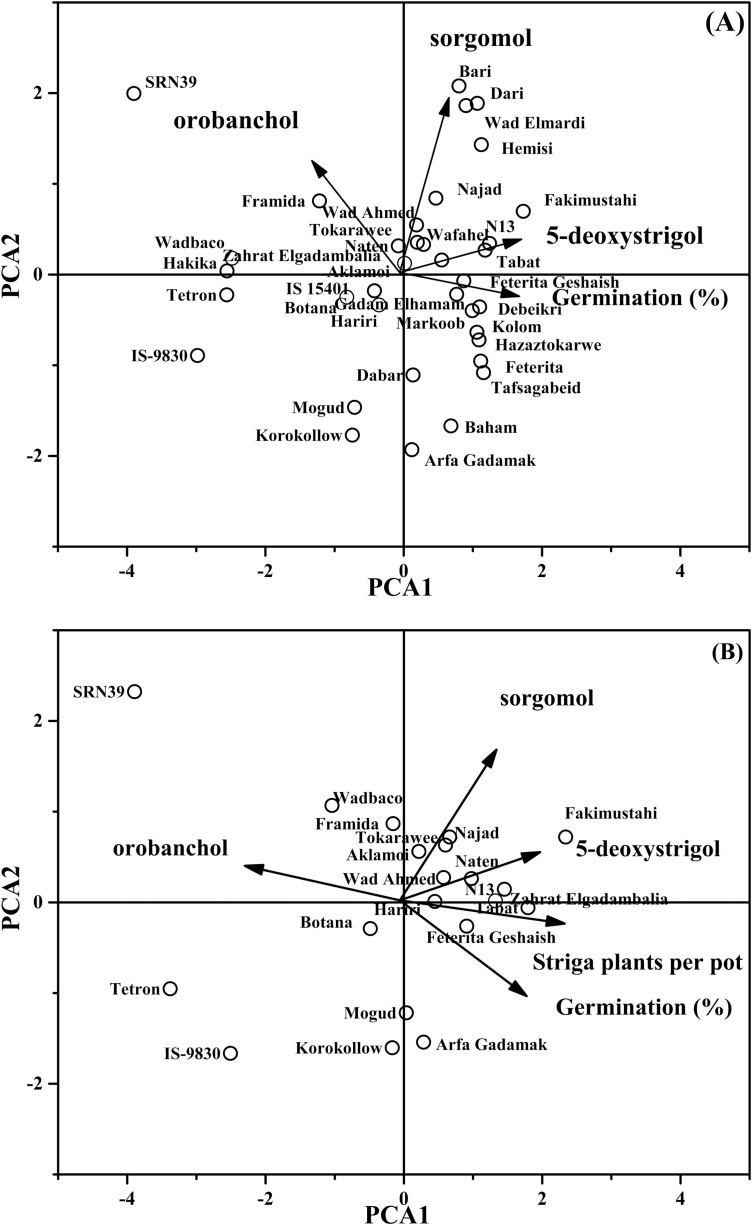
Principal component analysis
The peak areas of 5-deoxystrigol, sorgomol, and orobanchol were used in PCA to visualize the relationship between the different sorghum genotypes, their strigolactone profile, and the germination-inducing activity of their root exudates (Fig. 6A) and, for a subset of the genotypes, the number of Striga per pot (Fig. 6B). The first two principal components in Fig. 6A had an eigenvalue higher than 1, and explained 74% of the variation in strigolactone content (Supplementary Table S10). The first principal component (PC1) explained 47% of the variation, with positive loadings for both 5-deoxystrigol and germination and a negative loading for orobanchol (Supplementary Table S10). The second principal component (PC2) explained 27% of the total variation, with a high positive loading for sorgomol (Supplementary Table S10). Along PC1, genotypes are separated based on 5-deoxystrigol and orobanchol concentration, with the highest 5-deoxystrigol-producing genotypes, such as Debeikri, Fakimustahi, Hazaztokarwe, N13, Tabat, and Wad Fahel, clustering on the right side of the plot, and the highest orobanchol-producing genotypes, such as Hakika, Framida, SRN39, and Wad Baco, clustering in the upper left quadrant (Fig. 6A). The PC2 differentiated genotypes with high and low concentrations of sorgomol. The high sorgomol producers (Bari, Dari, and Wad Elmardi) clustered in the upper part of the PCA, while low-sorgomol-producing genotypes, for example, Arfa Gadamak, Korokollow, Mogud, Baham, and Dabar, clustered in the lower part.
Fig. 6.
(A) Bi-plot of the first two components of a PCA based on strigolactone peak area, displaying the genotypic differences in strigolactone production and composition and the correlation between the different strigolactones and germination-stimulating activity. (B) PCA showing the relationship between strigolactone production and composition in selected sorghum genotypes grown in pots, germination of Striga seeds as induced by root exudates, and the number of emergent Striga plants per pot on these genotypes.
A PCA plot of the selection of genotypes that was assessed in a pot experiment for Striga infection showed a similar clustering of genotypes, as it was based on the same strigolactone data (Fig. 6B). There was a striking strong correlation between the vectors for germination (in root exudate) and number of Striga plants per pot. According to the directions and angles of the vectors, 5-deoxystrigol exhibited a positive correlation with the germination-stimulating activity of the root exudates, while orobanchol exhibited a negative correlation with the germination-stimulating activity (Fig. 6A, B). The perpendicular angle between sorgomol and germination-stimulating activity suggests no correlation (Fig. 6A, B). The angle between 5-deoxystrigol and Striga plants per pot suggests a positive correlation, while that between orobanchol and Striga plants per pot suggests a negative correlation (Fig. 6B).
Correlation analysis
In addition to multivariate PCA, correlation analysis was used to investigate the relationship between the concentrations of 5-deoxystrigol, sorgomol, and orobanchol in root exudates, and Striga germination and infection. The amount of 5-deoxystrigol in the root exudates displayed a significant positive correlation with Striga seed germination (r=0.38; P<0.01), while orobanchol showed a significant negative correlation (r=–0.25; P<0.01) and sorgomol showed a non-significant negative correlation (Table 1).
Table 1.
Correlation coefficients between strigolactone levels in the root exudates, in vitro Striga germination, and Striga emergence in a pot experiment using 22 sorghum genotypes
| 5-Deoxystrigol | Sorgomol | Orobanchol | |
|---|---|---|---|
| Germination | 0.38** | –0.17 NS | –0.25** |
| Striga plants per pot | 0.36** | 0.23* | –0.41** |
| 5-Deoxystrigol | 1 | 0.19* | –0.32** |
| Sorgomol | 0.19* | 1 | –0.12 NS |
| Orobanchol | –0.32** | –0.12 NS | 1 |
**Correlation is significant at the P<0.01 level (one-tailed). NS, not significant.
Correlation analysis across all 22 genotypes between strigolactone peak areas in the root exudate and number of emerged Striga plants in the pot experiment showed that 5-deoxystrigol significantly and positively correlated with the number of emerged Striga (r=0.36; P<0.01), while orobanchol displayed a significant negative correlation (r=–0.41; P≤0.05) and sorgomol a significant positive correlation (r=0.23; P<0.05) (Table 1).
Stepwise regression with the peak areas of the strigolactones as predictors for Striga emergence resulted in a model (R2=0.27; P≤0.001) (Table 2) containing 5-deoxystrigol, sorgomol, and orobanchol as significant predictors for Striga emergence. Of these, 5-deoxystrigol showed the highest positive contribution, with a regression coefficient of 0.29 (P≤0.006) (Table 2). The regression coefficients for sorgomol and orobanchol were 0.25 (P≤0.01) and –0.26 (P≤0.01), respectively (Table 2).
Table 2.
Summary for the stepwise best regression model (R2=0.27) predicting the contribution of the strigolactone level in the root exudates of sorghum genotypes to Striga emergence
| Variables | Coefficient of regression | P |
|---|---|---|
| 5-Deoxystrigol | 0.29 | 0.006 |
| Sorgomol | 0.25 | 0.01 |
| Orobanchol | –0.26 | 0.01 |
Variation in strigolactone biosynthetic genes
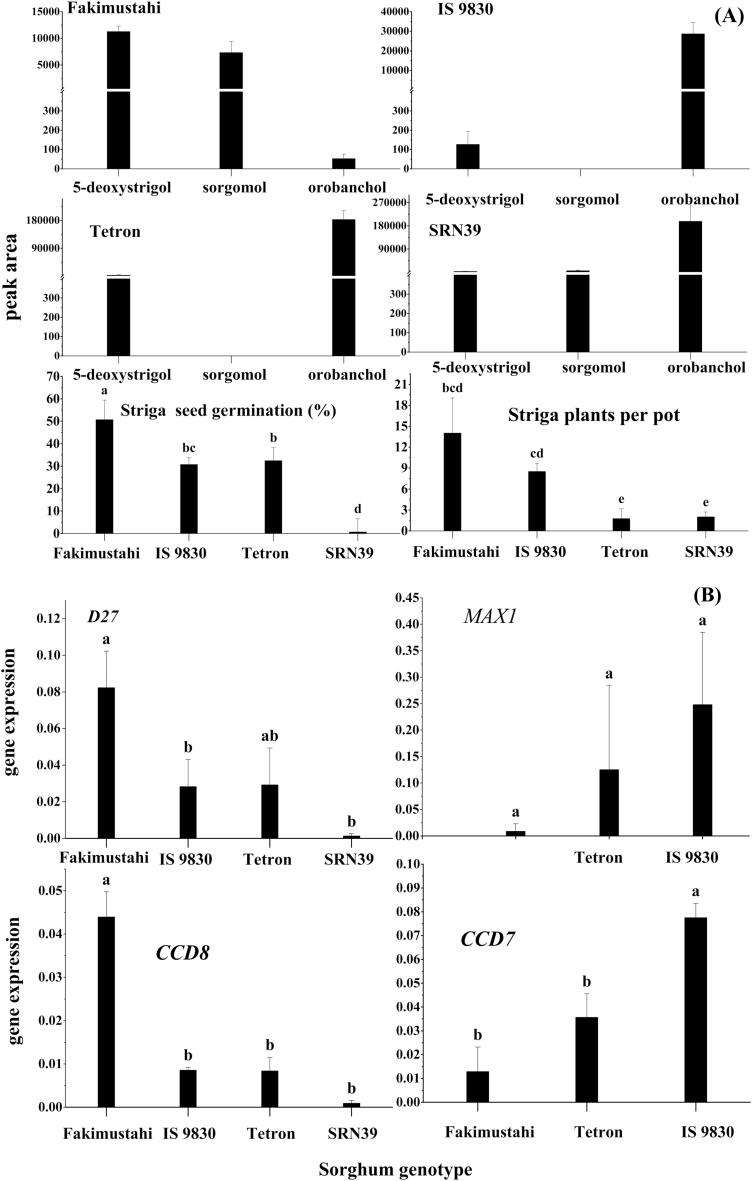
To assess the relationship between expression of strigolactone biosynthetic genes and differences in the strigolactone profile, induction of Striga seed germination, and Striga infection, we analysed four sorghum genotypes differing in Striga resistance (Fig. 7A). RT–qPCR showed that the expression of D27, CCD7, and CCD8 differed significantly (P<0.025) among the genotypes (Fig. 7B). The expression of both D27 and CCD8 was higher in the Striga-susceptible genotype Fakimustahi than in the Striga-resistant genotypes IS9830, Tetron, and SRN39 (Fig. 7B). In contrast, the expression of CCD7 was higher in genotypes IS9830 and Tetron than in Fakimustahi (Fig. 7B). Although there was no significant difference in the expression of MAX1 among the genotypes, it showed a tendency towards the same pattern of expression as CCD7, that is, higher in the more resistant genotypes. The amount of 5-deoxystrigol and sorgomol in the exudate correlated positively with the expression of D27 (r=0.70; P<0.05, and r=0.73; P<0.05) and negatively with that of CCD8 (significant only for 5-deoxystrigol: r=–0.80; P<0.01) (Supplementary Table S11, Fig. 7B).
Fig. 7.
(A) Amounts of the strigolactones 5-deoxystrigol, sorgomol, and orobanchol in the root exudates of one Striga-susceptible (Fakimustahi) and three Striga-resistant (IS9830, Tetron, and SRN39) sorghum genotypes used to analyse gene expression, as well as the germination of preconditioned Striga seeds induced by the root exudates of these genotypes, and Striga emergence (plants per pot) in the pot experiment. Data are the means±SE of peak area, germination (%), and total number of emerged Striga plants at harvest (n=4). (B) Relative expression of D27, CCD8, CCD7, and MAX1 in the roots of the same four genotypes. Data are means±SE (n=3). The significance of a treatment effect was determined by one-way ANOVA. different letters above the bars indicate significant differences after Duncan’s pairwise comparison (P<0.05).
Markers for germination-stimulating activity in sorghum
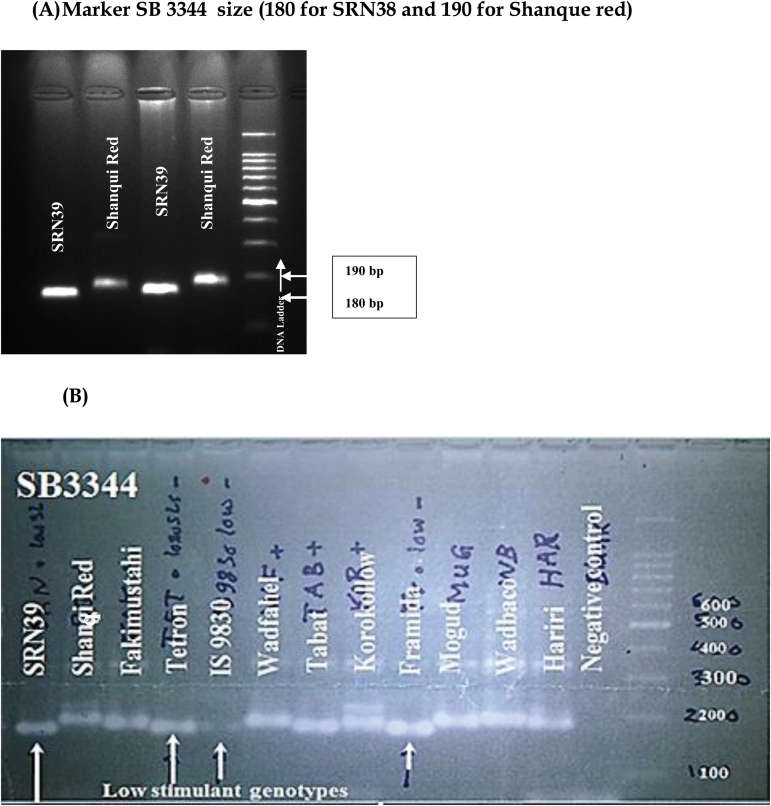
In an attempt to understand the genetics underlying the differences in strigolactone profiles and Striga resistance found in the present study, 12 sorghum genotypes were genotyped. Primers were used corresponding to the marker SB3344, recently reported for the lgs QTL for the low germination stimulant-based resistance of genotype SRN39 (Satish et al., 2012). PCR using these primers generated a polymorphic band pattern discriminating the resistant low-stimulant genotype SRN39 and the susceptible high-stimulant genotype Shanqui Red, which lacks the resistance allele (Satish et al., 2012) (Fig. 8A). The size of the PCR-amplified bands ranged from 170 to 190 bp. For the genotypes Tetron, IS9830, and Framida, a similar haplotype pattern to that of SRN39 was observed (Fig. 8B); the same was observed for Hakika (data not shown). For the genotypes Wad Fahel, Mogud, Wad Baco, and Hariri, a haplotype pattern similar to that of Shanqui Red was observed (Fig. 8B). Two bands were amplified for the genotype Korokollow, while for the genotypes Fakimustahi and Tabat an intermediate-sized band was amplified (Fig. 8B).
Fig. 8.
(A) PCR products obtained with a primer pair for the marker SB3344 and two reference genotypes of sorghum resistant (SRN39) and susceptible (Shanqui Red) to Striga. The fragment sizes for the reference genotypes are ~170 bp for SRN39 and ~190 bp for Shanqui Red. (B) PCR products obtained with the marker SB3344 primer pair for 10 further sorghum genotypes. Arrows indicate the amplified fragment of the low-stimulant genotypes.
Discussion
Our results reveal that, irrespective of morphological group, geographical location, climatic adaptation, and field reaction to Striga, all sorghum genotypes release 5-deoxystri- gol, sorgomol, and/or orobanchol as major strigolactones (Supplementary Table S8). In addition, low concentrations of strigol, sorgolactone, ent-2ʹ-epi-orobanchol, and ent-2ʹ-epi-5-deoxystrigol were detected in the root exudates of some genotypes. The results also reveal significant genotypic variation in the total concentrations of strigolactones and their relative amounts (Supplementary Table S8; Fig. 2). The production of mixtures of strigolactones and the variation in their amount and composition in sorghum root exudates are in line with several previous reports (Awad et al., 2006; Yoneyama et al., 2010; Jamil et al., 2013). Despite the reports on the identification and characterization of several strigolactones, ent-2ʹ-epi-5-deoxystrigol and ent-2ʹ-epi-orobanchol have never been reported before in root exudates or extracts of sorghum. In rice, high levels of ent-2ʹ-epi-5-deoxystrigol were reported in the root exudates of some cultivars (IAC 165, IAC 1246, and Gangweondo; Jamil et al., 2012); whereas ent-2ʹ-epi-orobanchol was reported and characterized in tobacco root exudates as a potent germination stimulant for Phelipanche ramosa L. seeds (Xie et al., 2007). Furthermore, the present study, in line with a previous report (Mohemed et al., 2016), unequivocally confirmed the production of orobanchol by a range of sorghum genotypes, including IS15401 and the Striga-resistant genotypes Framida, Hakika, SRN39, Tetron, and IS9830. Orobanchol was first characterized in red clover by Yokota et al. (1998) and subsequently reported by Jamil et al. (2012) in rice.
In the germination bioassay, 5-deoxystrigol, sorgomol, sorgolactone, and strigol exhibited relatively high germination-inducing activity in Striga seeds compared with the other strigolactones, consistent with previous reports (Hauck et al., 1992; Yasuda et al., 2003; Xie et al., 2008; Cardoso et al., 2014a). Likewise, we showed that at a concentration of 0.02 µM ent-2′-epi-5-deoxystrigol, ent-2′-epi-strigol, or GR24 displayed a moderate yet potent activity (>30% germination), while orobanchol showed negligible activity (<10% germination). The low germination-inducing activity of orobanchol in Striga is consistent with previous reports on the low sensitivity of Striga (sorghum strain) to the orobanchol-type strigolactones (Nomura et al., 2013; Cardoso et al., 2014a). The low sensitivity of Striga to orobanchol suggests that selection for high orobanchol producers may be an effective strategy to obtain sorghum genotypes with good arbuscular mycorrhizal fungi recruitment, but lower Striga germination-inducing activity. It is noteworthy that orobanchol is an effective inducer of mycorrhizal hyphal branching (Akiyama et al., 2010). Indeed, high-orobanchol-producing genotypes were less infected by Striga in a pot experiment (Fig. 6B). Interestingly, we also found a correlation between the level of Striga infection and the Striga emergence rate (Supplementary Table S12). Early emergence of Striga may reflect a highly compatible host–parasite interaction, while late emergence suggests the presence of physical and/or physiological barriers delaying early infection and/or hindering subsequent development (Arnaud et al., 1999; Gurney et al., 1999; Haussmann et al., 2000; Van Ast et al., 2000). Delaying the time of first infection not only influences Striga parasitism and reproduction, but also strongly reduces its damaging effects on host plants (Van Ast and Bastiaans, 2006) (Frost et al., 1997). Whether orobanchol plays a role in the creation of this physical or physiological barrier, or is simply causing lower germination and therefore delayed attachment, remains unclear.
The genotypes used in the present study can be classified into three groups with respect to the timing of Striga emergence (Fig. 5D; Supplementary Table S9). The first group includes Fakimustahi, Feterita Geshaish, Zahrat Elgadambalia, and Tabat, in which Striga emergence was early (15–18 DAS) (Fig. 5A). The second group consists of Arfa Gadamak, Botana, Framida, Korokollow, Mogud, Naten, N13, Najad, Tokarawe, and Wad Baco, in which Striga emergence was moderately early (23–27 DAS), and a third group comprises Hariri, IS9830, SRN39, Tetron, and Wad Ahmed, in which Striga emergence was late (31–36 DAS) (Fig. 5A). These results, which are consistent with a report by Timko and Scholes (2013), suggest that the first group has the least mechanical and/or physiological barriers that delay ingress of the parasite and/or reduce its growth rate, while in the latter two groups constitutive and/or induced barriers may delay the onset of parasitism and/or the growth rate of the parasite (Fig. 5A).
It is noteworthy that all genotypes that showed late Striga emergence possess a strigolactone blend low in 5-deoxystrgol or sorgomol and rich in the less active stimulant orobanchol. Furthermore, the results revealed that differences in the proportions of 5-deoxystrigol, sorgomol, and orobanchol in root exudates of different sorghum genotypes influenced their germination-inducing activity and Striga infection (Figs. 3, 4, 6B). The genotypes Feterita Geshaish, Gadam Elhamam and Tabat, whose root exudates contain a high proportion of 5-deoxystrigol and a low proportion of orobanchol, were associated with high Striga germination and displayed high infection levels in pots (Fig. 5). Similarly, the genotypes Fakimustahi and Hariri, the root exudates of which contain a high proportion of sorgomol and a low proportion of orobanchol, were associated with high in vitro germination and high Striga infection. Conversely, the genotypes SRN39, IS9830, Hakika, and Wad Baco, whose the root exudates contain a high proportion of orobanchol and a low proportion of 5-deoxystrigol, were associated with low Striga germination and displayed low levels of infection by the parasite. These findings, which are in agreement with previous reports that showed a significant negative relationship between orobanchol, in vitro germination, and field infection by the parasite (Vasudeva Rao, 1984; Mohemed et al., 2016; Gobena et al., 2017), are further substantiated by correlation analysis, which showed the existence of a significant negative association between orobanchol and in vitro Striga germination and Striga emergence in pots (Table 1). The notable negative correlation between orobanchol and 5-deoxystrigol (Fig. 6B) suggests a negative biosynthetic correlation between the two strigolactones, resulting in low production of the strong Striga germination stimulant 5-deoxystrigol when there is a high level of orobanchol. However, a direct inhibitory effect of orobanchol on Striga germination cannot be excluded without further investigation.
The present study further revealed that high stimulant production and in vitro germination are not solely responsible for high infection by the parasite in pot experiments. The genotypes N13 and Tabat have high proportions of 5-deoxystrigol (Supplementary Table S8; Fig. 2) and induced high in vitro germination of Striga (Fig. 3), but N13 displayed far less infection by the parasite than Tabat in the pot experiment (Fig. 5). The differential response in N13 could possibly be attributed to mechanical resistance, as reported by Grenier et al. (2007) and Mbuvi et al. (2017). In addition, high proportions of sorgomol in some genotypes, such as Wad Fahel, did not necessarily result in higher germination-stimulating activity, although it was associated with a higher Striga infection level in the pot experiment (Fig. 5B). The discrepancy between the results of in vitro germination assays and performance in the pot experiment of some genotypes may be attributable to inherent differences between the two assay methods. Root exudates may vary in composition with the species, cultivar, growth stage, and conditions of growth. Hence, in soils the seeds of the parasite may be exposed to root exudates with a constantly changing composition of strigolactones and other signalling chemicals which may be synergistic or antagonistic to the action of strigolactones (Yoneyama et al., 2009). Nevertheless, there is a good correlation between in vitro germination induced by root exudates and Striga infection in the pot experiment (Fig. 6B). Our findings suggest that, in the absence of other Striga resistance mechanisms, high strigolactone production with a high proportion of 5-deoxystrigol and sorgomol together with low orobanchol in the root exudates results in high susceptibility to Striga. These findings confirm the positive relationship between the level of 5-deoxystrigol in the root exudates of genotypes grown in a greenhouse with Striga infection of the same genotypes in the field previously reported by Mohemed et al. (2016). On the other hand, high proportions of orobanchol in the root exudates enhanced resistance to the parasite. The PCA bi-plot (Fig. 6B) clearly shows that low Striga infection in the pot experiment (in genotypes SRN39, Tetron, IS9830, Framida, Mogud, and Wad Baco) is associated with high proportions of orobanchol, and conversely, susceptibility to the parasite is clearly associated with high proportions of 5-deoxystrigol.
The fact that most of the genotypes we investigated contained mostly the same strigolactones suggests that strigolactone biosynthesis in sorghum is highly conserved. Nevertheless, selection pressure or selection by breeders for Striga resistance seems to have resulted in preferential selection of genotypes that produce strigolactones with low Striga germination-inducing activity, that is, orobanchol (Gobena et al., 2017). The tendency of Striga-resistant sorghum genotypes—previously described as low-germination-stimulant producers— to produce higher levels of orobanchol shows that it is not the flux through the strigolactone pathway that was affected by selection for Striga resistance in these genotypes, but the flux towards orobanchol, a strigolactone with opposite stereochemistry to 5-deoxystrigol.
It is therefore also not unexpected that there were no large differences in the expression of the strigolactone biosynthetic genes, all of which catalyse the core strigolactone biosynthetic pathway. Nevertheless, the expression of D27 and CCD8 in root tissues of the susceptible sorghum genotype Fakimustahi was 3- and 4-fold higher, respectively, than in the resistant genotypes IS9830 and Tetron (Fig. 7B). In contrast, expression of CCD7 was higher in the resistant genotype IS9830 than in the susceptible genotype Fakimustahi (Fig. 7B). In rice, Zhang et al. (2014) showed that the MAX1 homologs Os900 and Os1400 catalyse the conversion of carlactone to ent-2ʹ-epi-5-deoxystrigol (deoxyorobanchol) and of ent-2ʹ-epi-5-deoxystrigol (deoxyorobanchol) to orobanchol, respectively. The catalytic function of the four sorghum MAX1s has not been elucidated yet, and there were no significant differences in the expression of one of these MAX1 homologs in the sorghum lines investigated in this study. Accordingly, it is difficult to find a direct correlation between gene expression at this one time point and the levels of the strigolactones in the root exudates; more genotypes need to be studied to pinpoint the roles of these and other genes in the changes in strigolactone biosynthesis in the resistant genotypes.
Genotypes IS9830, Framida, Hakika, and Tetron showed the same amplification of the marker SB3344 as that of the resistant genotype SRN39, confirming the results of Satish et al. (2012). Hakika (also known as P9405) is in fact derived from SRN39 (Mbwaga et al., 2007). The presence of the SB3344 marker coincided with high relative proportions of orobanchol in the root exudates of Tetron (59%) and IS9830 (66%), but the root exudates of Framida and Hakika showed less high relative proportions of orobanchol (32% and 37%, respectively; Fig. 2). It is noteworthy that, based on peak area, IS9830 produced far less 5-deoxystrigol than Tetron and SRN39, and Tetron and IS9830 produced far less sorgomol than SRN39 (Fig. 7).
In essence, the results show that in sorghum genotypes SRN39, IS9830, Framida, Tetron, and Hakika the low-stimulant trait is associated with the same marker (SB3344), and the low-stimulant allele for these genotypes might therefore be identical. However, in the genotypes Wad Fahel, Mogud, and Wad Baco, which are susceptible to the parasite despite the relatively low germination-inducing activity of their root exudates (Figs. 3, 7, 6B), the SRN39 low germination stimulant (lgs) linked marker was not amplified. Instead, amplification resulted in a fragment pattern similar to that of the high-stimulant, Striga-susceptible Chinese sorghum genotype Shanqui Red, suggesting that these genotypes lack the resistance allele at the lgs locus. Genotypes Tabat and Fakimustahi, which produce high levels of 5-deoxystrigol, do not show the expected Shanqui Red-type fragment pattern (Fig. 8B). This may be caused by the possession of different alleles at the lgs locus in these genotypes or recombination between the marker amplified by the primers and the LGS allele. Korokollow also does not show the typical fragment pattern of either SRN39 or Shanqui Red (Fig. 8B), but this genotype has a quite atypical strigolactone profile (Fig. 2), suggesting the absence of the standard lgs locus.
Our results confirm previous reports that low 5-deoxystrigol and high orobanchol production are associated with low germination traits and low infection by the Striga parasite, and that delayed emergence of the parasite reduces its damaging effects on the host (Gobena et al., 2017; Mohemed et al., 2016). Moreover, the results suggest the involvement of genes other than lgs in controlling the low germination traits in some of the genotypes investigated. These genes could provide additional sources for the introgression of low germination traits from donors other than SRN39 into new preferred sorghum varieties, such as Feteritas, using marker-assisted selection and backcrossing.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Local names, status and collection site of sorghum landraces, improved cultivars, and exotic material used.
Table S2. D27 (DWARF 27) sorghum orthologous putative gene sequence.
Table S3. CCD7 (CAROTENOID CLEAVAGE DIOXYGENASE 7) orthologous putative gene sequence.
Table S4. CCD8 (CAROTENOID CLEAVAGE DIOXYGENASE 8) orthologous putative gene sequence.
Table S5. SbMAX1 orthologous putative gene sequence.
Table S6. Primers used for quantitative RT–PCR.
Table S7. Primers used for quantitative PCR for marker analysis.
Table S8. Genotypic variation in strigolactone production in sorghum.
Table S9. Emergence rate, expressed as days to the first Striga emergence.
Table S10. Principal components, eigenvalues, loadings, and percentage of total variance explained in principal component analysis.
Table S11. Correlation coefficients between strigolactone peak area in the root exudates and expression of biosynthetic genes D27 and CCD8, Striga germination, and Striga emergence.
Table S12. Pearson correlation between number of emerged Striga per pot, total Striga biomass per pot, and emergence rate.
Acknowledgements
The authors acknowledge funding by the Nuffic Fellowship Program, the Netherlands and the Wageningen sandwich fellowship (to NEM), the Netherlands Organization for Scientific Research (to HJB, VICI grant 865.06.002 and equipment grant 834.08.001), the European Research Council (ERC) for the ERC Advanced grant CHEMCOMRHIZO (670211 to HJB) and the Bill and Melinda Gates Foundation (Promise to HJB). The authors would like to thank Dr Altahir Ibrahim Mohamed, Plant Genetic Resource Unit, Agricultural Research Corporation, Wad Medani, Sudan, for providing seeds and characterizations of the sorghum genotypes. We would like to thank Prof. Binne Zwanenburg, Prof. Koichi Yoneyama, and Prof. Tadao Asami for providing strigolactone standards, and Dr Patrick Rich for helpful comments on the manuscript.
References
- Akiyama K, Ogasawara S, Ito S, Hayashi H. 2010. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant & Cell Physiology 51, 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud MC, Véronési C, Thalouarn P. 1999. Physiology and histology of resistance to Striga hermonthica in Sorghum bicolor var. Framida. Australian Journal of Plant Physiology 26, 63–70. [Google Scholar]
- Awad AA, Sato D, Kusumoto D, Kamioka H, Takeuchi Y, Yoneyama K. 2006. Characterization of strigolactones, germination stimulants for the root parasitic plants Striga and Orobanche, produced by maize, millet and sorghum. Plant Growth Regulation,48, 221–227. [Google Scholar]
- Babiker AGT. 2002. Striga control in Sudan: an integrated approach. In: Leslie JF, ed. Sorghum and millets diseases. Ames: Iowa State Press, 159–163. [Google Scholar]
- Badu-Apraku B, Yalloub CG, Oyekunle M. 2013. Genetic gains from selection for high grain yield and Striga resistance in early maturing maize cultivars of three breeding periods under Striga-infested and Striga-free environments. Field Crop Research 147, 54–67. [Google Scholar]
- Bebawi F. 1981. Intraspecific physiological variants of Striga hermonthica. Experimental Agriculture 17, 419–423. [Google Scholar]
- Bouwmeester HJ, Matusova R, Zhongkui S, Beale MH. 2003. Secondary metabolite signalling in host–parasitic plant interactions. Current Opinion in Plant Biology 6, 358–364. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Charnikhova T, Jamil M, Delaux PM, Verstappen F, Amini M, Lauressergues D, Ruyter-Spira C, Bouwmeester H. 2014. Differential activity of Striga hermonthica seed germination stimulants and Gigaspora rosea hyphal branching factors in rice and their contribution to underground communication. Plos One 9, e104201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Ruyter-Spira C, Bouwmeester HJ. 2011. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Science 180, 414–420. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Zhang Y, Jamil M et al. 2014b. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proceedings of the National Academy of Sciences, USA 111, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Netzly DH, Butler LG, Lynn DG. 1986. Chemical regulation of distance. Characterization of the first natural host germination stimulant for Striga asiatica. Journal of the American Chemical Society 108, 7858–7860. [DOI] [PubMed] [Google Scholar]
- Ejeta G. 2005. Integrating biotechnology, breeding, and agronomy in the control of the parasitic weed Striga spp in sorghum. In: Tuberosa R, Phillips RL, Gale M, eds. In the wake of the double helix: from the green revolution to the gene revolution. Bologna: Avenue Media, 239–251. [Google Scholar]
- Ejeta G. 2007. The Striga scourge in Africa: a growing pandemic. In: Ejeta G, Gressel J, eds. Integrating new technologies for Striga control: towards ending the witch-hunt. Singapore: World Scientific, 3–16. [Google Scholar]
- Ejeta G, Butler LG, Babiker AG. 1992. New approaches to the control of Striga. Striga Research at Purdue University, Research Bulletin 991.West Lafayette: Agricultural Experiment Station, Purdue University. [Google Scholar]
- Ejeta G, Mohammed A, Rich P, Melake-berhan A, Housley TL, Hess DE. 2000. Selection for mechanisms of resistance to Striga in sorghum. In: Haussmann BIG, Hess DE, Koyama ML, Grivet L, Rattunde HFW, Geiger HH, eds. Breeding for Striga resistance in cereals. Proceedings of a workshop held at IITA, Ibadan, Nigeria Weilkersheim: Margraf Verlag, 29–37. [Google Scholar]
- Ejeta G, Rich PJ, Mohamed A. 2007. Dissecting a complex trait to simpler components for effective breeding of sorghum with a high level of Striga resistance. In: Ejeta G, Gressel J, eds. Integrating new technologies for Striga control: towards ending the witch-hunt. Singapore: World Scientific, 87–98. [Google Scholar]
- Gobena D, Shimels M, Rich PJ, Ruyter-Spira C, Bouwmeester HJ, Satish Kanuganti S, Mengiste T, Ejeta G. 2017. Mutation in sorghum LOW GERMINATION STIMULANT1 alters strigolactones and causes Striga resistance. Proceedings of the National Academy of Sciences, USA 114, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier C, Ibrahim Y, Haussmann BI, Kiambi D, Ejeta G. 2007. Marker-assisted selection for Striga resistance in sorghum. In Ejeta G, Gressel J, eds. Integrating New technologies for Striga control: towards ending the witch-hunt. Singapore: World Scientific, 159–172. [Google Scholar]
- Gressel J, Hanafi A, Head G, Marasas W, Obilana AB, Ochanda J, Souissi T, Tzotzos G. 2004. Major heretofore intractable biotic constraints to African food security that may be amenable to novel biotechnological solutions. Crop Protection 23, 661–689. [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR. 2012. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiology 160, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney AL, Press MC, Scholes JD. 1999. Infection time and density influence the response of sorghum to the parasitic angiosperm Striga hermonthica. New Phytologist 143, 573–580. [DOI] [PubMed] [Google Scholar]
- Hauck C, Müller S, Schildknecht H. 1992. A germination stimulant for parasitic flowering plants from Sorghum bicolor, a genuine host plant. Journal of Plant Physiology 139, 474–478. [Google Scholar]
- Haussmann BIG, Hess DE, Welz HG, Geiger HH. 2000. Improved methodologies for breeding striga-resistant sorghums. Field Crops Research 66, 195–211. [Google Scholar]
- Hess DE, Ejeta G, Butler LG. 1992. Selecting sorghum genotypes expressing a quantitative biosynthetic trait that confers resistance to Striga. Phytochemistry 31, 493–497. [Google Scholar]
- Jamil M, Charnikhova T, Houshyani B, van Ast A, Bouwmeester HJ. 2012. Genetic variation in strigolactone production and tillering in rice and its effect on Striga hermonthica infection. Planta 235, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil M, Van Mourik TA, Charnikhova T, Bouwmeester HJ. 2013. Effect of diammonium phosphate application on strigolactone production and Striga hermonthica infection in three sorghum cultivars. Weed Research 53, 121–130. [Google Scholar]
- Kambal AEL, Musa BM. 1979. Breeding Sorghum cultivars for resistance to Striga. In Technical Progress Report No I. Striga research in the Sudan. Khartoum: Agricultural Research Corporation, Wad Medani, and Faculty of Agriculture, University of Khartoum. [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V et al. 2008. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytologist 178, 863–874. [DOI] [PubMed] [Google Scholar]
- Matusova R, van Mourik T, Bouwmeester HJ. 2004. Changes in the sensitivity of parasitic weed seeds to germination stimulants. Seed Science Research 14, 335–344. [Google Scholar]
- Mbuvi DA, Masiga CW, Kuria EK, Masanga J, Wamalwa M, Mohamed A, Odeny DA, Hamza N, Timko MP, Runo S. 2017. Novel sources of witchweed (Striga) resistance from wild sorghum accessions. Frontiers in Plant Science 8, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbwaga AM, Riches C, Ejeta G. 2007. Integrated Striga management to meet sorghum demand in Tanzania. In: G. Ejeta and J. Gressel, eds. Integrating new technologies for Striga control: towards ending the witch-hunt. Singapore: World Scientific, 253–264. [Google Scholar]
- Mohemed N, Charnikhova T, Bakker EJ, van Ast A, Babiker AG, Bouwmeester HJ. 2016. Evaluation of field resistance to Striga hermonthica (Del.) Benth. in Sorghum bicolor (L.) Moench. The relationship with strigolactones. Pest Management Science 72, 2082–2090. [DOI] [PubMed] [Google Scholar]
- Nomura S, Nakashima H, Mizutani M, Takikawa H, Sugimoto Y. 2013. Structural requirements of strigolactones for germination induction and inhibition of Striga gesnerioides seeds. Plant Cell Reports 32, 829–838. [DOI] [PubMed] [Google Scholar]
- Oswald A. 2005. Striga control—technologies and their dissemination. Crop Protection 24, 333–342. [Google Scholar]
- Parker C. 2009. Observations on the current status of Orobanche and Striga problems worldwide. Pest Management Science 65, 453–459. [DOI] [PubMed] [Google Scholar]
- Parker CH, Hitchcock AM, Ramaiah KV. 1977. The germination of Striga species by crop root exudates; techniques for selecting resistant crop cultivars. In Proceedings of the Asian-Pacific Weed Science Society 6th Conference 67–74. [Google Scholar]
- Priya R, Siva R. 2014. Phylogenetic analysis and evolutionary studies of plant carotenoid cleavage dioxygenase gene. Gene 548, 223–233. [DOI] [PubMed] [Google Scholar]
- Rich PJ, Grenier C, Ejeta G. 2004. Striga resistance in the wild relatives of sorghum. Crop Science 44, 2221–2229. [Google Scholar]
- Rodenburg J, Bastinaans L. 2011. Host-plant defence against Striga spp.: reconsidering the role of tolerance. Weed Research 51, 438–441. [Google Scholar]
- Rodenburg J, Bastinaans L, Weltzien E, Hess DE. 2005. How can field selection for Striga resistance and tolerance in sorghum be improved?Field Crops Research 93, 34–50. [Google Scholar]
- Satish K, Gutema Z, Grenier C, Rich PJ, Ejeta G. 2012. Molecular tagging and validation of microsatellite markers linked to the low germination stimulant gene (lgs) for Striga resistance in sorghum [Sorghum bicolor (L.) Moench]. Theoretical and Applied Genetics 124, 989–1003. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Scholes JD, Press MC. 2008. Striga infestation of cereal crops – an unsolved problem in resource limited agriculture. Current Opinion in Plant Biology 11, 180–186. [DOI] [PubMed] [Google Scholar]
- Siame BA, Weerasuriya Y, Wood K, Ejeta G, Butler LG. 1993. Isolation of strigol, a germination stimulant for Striga asiatica, from host plants. Journal of Agricultural and Food Chemistry 41, 1486–1491. [Google Scholar]
- Spallek T, Mutuku M, Shirasu K. 2013. The genus Striga: a witch profile. Molecular Plant Pathology 14, 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JRN. 2003. Overview: importance of sorghum in Africa. In Enhancing Nutritional and Functional Properties for Africa. Proceedings of a Workshop on the Proteins of Sorghum and Millet, April 2–4 2003, Pretoria, South Africa Available from: http://www.afripro.org.uk. 1–21. [Google Scholar]
- Tesso TT, Ejeta G. 2011. Integrating multiple control options enhances Striga management and sorghum yield on heavily infested soils. Agronomy Journal 103, 1464. [Google Scholar]
- Timko MP, Scholes JD. 2013. Host reaction to attack by root parasitic plants. In Parasitic Orobanchaceae. Berlin: Springer, 115–141. [Google Scholar]
- Vallabhaneni R, Bradbury LM, Wurtzel ET. 2010. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Archives of Biochemistry and Biophysics 504, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ast A, Bastiaans L. 2006. The role of infection time in the differential response of sorghum cultivars to Striga hermonthica infection. Weed Research 46, 264–274. [Google Scholar]
- Van Ast A, Bastiaans L, Kropff MJ. 2000. A comparative study on Striga hermonthica interaction with a sensitive and a tolerant sorghum cultivar. Weed Research 40, 479–493. [Google Scholar]
- Vasudeva Rao MJ. 1984. Patterns of resistance to Striga asiatica in sorghum and millets, with special reference to Asia. In: Ayensu ES, Doggett H, Keynes RD, Marton Lefevre J, Musselman LJ, Parker C, Pickering A, eds. Proceedings of the International Workshop on the Biology and Control of Striga, Dakar. Paris: ICSU Press, 71–92. [Google Scholar]
- Williams C. 1958. The parasitism of witchweed—a review. West African Journal of Biological Chemistry 2, 57–73. [Google Scholar]
- Xie X, Kusumoto D, Takeuchi Y, Yoneyama K, Yamada Y, Yoneyama K. 2007. 2ʹ-epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. Journal of Agricultural and Food Chemistry 55, 8067–8072. [DOI] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Kusumoto D, Yamada Y, Takeuchi Y, Sugimoto Y, Yoneyama K. 2008. Sorgomol, germination stimulant for root parasitic plants, produced by Sorghum bicolor. Tetrahedron Letters 49, 2066–2068. [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. 2010. The strigolactone story. Annual Review of Phytopathology 48, 93–117. [DOI] [PubMed] [Google Scholar]
- Yasuda N, Sugimoto Y, Kato M, Inanaga S, Yoneyama K. 2003. (+)-Strigol, a witchweed seed germination stimulant, from Menispermum dauricum root culture. Phytochemistry 62, 1115–1119. [DOI] [PubMed] [Google Scholar]
- Yoder JI, Scholes JD. 2010. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Current Opinion in Plant Biology 13, 478–484. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sakai H, Okuno K, Yoneyama K, Takeuchi Y. 1998. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry 49, 1967–1973. [Google Scholar]
- Yoneyama K, Arakawa R, Ishimoto K et al. 2015. Difference in Striga-susceptibility is reflected in strigolactone secretion profile, but not in compatibility and host preference in arbuscular mycorrhizal symbiosis in two maize cultivars. New Phytologist 206, 983–989. [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Awad AA, Xie X, Yoneyama K, Takeuchi Y. 2010. Strigolactones as germination stimulants for root parasitic plants. Plant & Cell Physiology 51, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Yoneyama K, Takeuchi Y. 2009. Strigolactones: structures and biological activities. Pest Management Science 65, 467–470. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Van Dijk AD, Scaffidi A et al. 2014. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nature Chemical Biology 10, 1028–1033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.